The direct monitoring of enzyme activities is broadly useful in many fields, ranging from biochemistry to medicinal chemistry and biology.[1] A biological process often involves multiple enzymes, which can work independently or cooperatively to control a specific biological event. Monitoring their activities can provide markers of the progress of such a process (such as the cascade of caspase activity that occurs in apoptosis), and yield information about the mechanism and timing of the interacting species. The ability to track multiple targets in a single event or different processes simultaneously could greatly improve our basic understanding, and facilitate biological and clinical studies as well.
However, the real-time fluorescent multi-color sensing of enzymes faces some technical limitations due to the properties of available fluorophores. Traditional organic fluorophores in distinct colors have disparate absorption wavelengths, requiring different filter sets for monitoring each species. Moreover, the use of different fluorophores can require the development of different conjugation strategies, and complicated fluorophore–quencher pair selection. One potential approach to addressing these problems is the use of inorganic quantum dots (QDs), which allow single-wavelength excitation, and can generate size- and composition-tunable emissions.[2] However, difficulties in uniform chemical modification and cellular delivery, along with their relatively large size (15–35 nm) and cytotoxicity[3] present some limitations of their own for application in sensing, especially in the cellular context.
We have adopted a different approach for multispectral labeling which makes use of short sequences of fluorophores assembled on a DNA backbone, with the fluorophores replacing DNA bases. In this oligodeoxyfluoroside (ODF) design, the phosphodiester backbone confers aqueous solubility and acts as a scaffold to hold the fluorophores close, promoting electronic interactions.[4, 5] Multiple forms of energy and excitation transfer, such as excimer and exciplex formation, and highly efficient FRET, have been observed.[6, 7] A broad spectrum of ODFs can be excited at one wavelength,[8] which offers the possibility of real-time multi-color application in biological systems. In a recent initial test of applying ODF labels to enzyme sensing, a cyan-colored tetrapyrene ODF was conjugated to a dabcyl quencher via ester linkages, and used as a fluorescent reporter for esterases and lipases.[9] The ODF-based probe showed aqueous solubility, high quenching efficiency, and large fluorescent signal turn-on in vitro and in intact human cells.
A promising strategy for monitoring multiple enzyme pathways would be the development of sets of dark (quenched) chemosensors that can be monitored simultaneously by use of single-excitation multispectral labeling. However, for ODF labels it is unknown whether they can be applied to enzymes other than esterases. For example, in the case of proteases, the active sites are relatively large, requiring longer substrates and presenting a more difficult challenge for quenching strategies and for synthesis and conjugation. Thus we undertook a study to address these issues, and we chose the well-studied caspases as a set of biomedically relevant and selective protease enzymes as the targets.
Caspases are a class of cysteine proteases that play central roles in the cellular processes of apoptosis, necrosis, and inflammation.[10] They are under investigation as drug targets since they play key roles in these medically important processes.[11] Caspase-controlled apoptosis has a characteristic enzyme cascade, which involves multiple caspases at different stages and pathways.[12] The family of enzymes has varied sequence selectivity but all are known to require an aspartate (Asp) residue at the substrate cleavage site.[13] Noteworthy from the sensing standpoint is the fact that caspases are usually in inactive forms before apoptosis, and can be activated by external or internal chemical and physical signals.[14]
Because of the broad biomedical importance of caspases, development of sensors for these enzymes has received a good deal of attention. Fluorescent caspase sensors can be used to report on the apoptosis process in living cells, and multiple types of such sensors have been reported.[15–18] Most are based on traditional organic fluorophores conjugated to caspase substrate peptides, and in some cases they have limited solubility in water due to the poor solubility of both the peptides and the organic fluorophores. Many probes are not inherently dark, and so cannot be used in homogeneous assays, and require washing procedures to be used in cells.[16, 17] Inorganic nanoparticle-based sensors of caspases have also been reported,[19] but their practical application in living systems remains to be demonstrated. Finally, engineered dimers of fluorescent proteins bridged by caspase substrate peptides have also been described, but as complex macromolecules they are not used in vitro, and require gene delivery or genetically modified cells to be active.[20, 21] To our knowledge there are no examples to date of multicolor quenched probe sets selective for different caspases.
Here we describe the synthesis, fluorescence and enzyme substrate properties of a set of three multicolor caspase chemosensors built with distinct ODF labels. This novel DNA–peptide conjugate sensor design utilizes different ODF sequences as fluorophores, dabcyl as a general quencher and contains short caspase substrate peptides in between. We demonstrate selective reporting of three different caspases by distinct color responses. Further, an ODF caspase probe is shown to function in cell culture to report on apoptosis.
Sensor design and synthesis
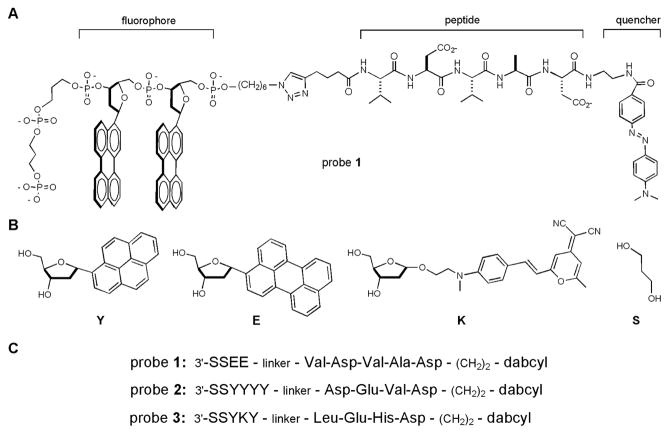
Each caspase chemosensor contains three parts: an ODF sequence as the fluorophore, a quencher, and a short peptide as the caspase substrate in the middle (see Fig. 1). We employed a convergent strategy for their synthesis (Scheme S1, Supporting Information). First, three ODF sequences with different emission colors were prepared on a DNA synthesizer: EE (yellow), YYYY (cyan), and YKY (orange). Two C3 spacers (S, with one phosphate group each) were placed at the 3′end of each ODF sequence to improve the aqueous solubility, and an azide group was placed at the 5′-end for bioorthogonal conjugation by “click” cycloaddition.[9]
Figure 1.
Structures of ODF-based fluorescent caspase chemosensors and their components. (A) Complete structure of an ODF sensor (probe 1 is shown as an example); (B) Structures of monomers incorporated into ODF labels; (C) Sequences of the three probes designed for caspases 2, 7, and 9, respectively.
Three short caspase substrate peptides were synthesized: Val-Asp-Val-Ala-Asp for caspase-2, Asp-Glu-Val-Asp for caspase-3/7, and Leu-Glu-His-Asp for caspase-9.[22–25] 5-Hexynoic acid was coupled to the N-terminus of each peptide to provide an alkyne for conjugation, and Dabcyl was coupled to the C-termini as a prospective general quencher.
Three caspase chemosensors with peptide-DNA conjugate structure were synthesized (see sequences in Fig. 1c): Probe 1 for caspase-2, Probe 2 for caspase-7, and Probe 3 for caspase-9. The compounds were purified by HPLC, and identities were confirmed by MALDI-MS and by spectral characterization. The absorption spectra indicate that probes 2 and 3 can be excited at 340 nm. Because perylene (E) does not have strong absorbance at 340 nm, the optimum excitation wavelength for probe 1 is 380 nm. All three sensors can be excited using one excitation filter (330–380 nm) under an epifluorescence microscope (see below).
ODF chemosensor spectral properties
Absorption spectra were obtained both for the sensors and for the ODF components. The spectra of the ODF sensors show the expected bands that are characteristic of the dye components (Fig. S1). Bands are visible for pyrene, perylene, and monomer K; in addition, the long-wavelength absorption band of the dabcyl (methyl red) quencher is also present. To test the inherent quenching efficiency of dabcyl for the three ODF sequences, we separately prepared dabcyl-quenched conjugates with a previously reported ester linkage rather than amide linkages.[9] These model compounds allowed all three fluorophore-quencher pairs to be completely released by one enzyme under identical conditions. Experiments showed that the YYYY sequence has the highest fluorescence turn-on ratio, about 60-fold at 480 nm.[9] EE and YKY also showed strong fluorescence turn-on, ca. 20-fold at the long-wavelength emission bands (Fig. S2). The liberated ODFs appeared cyan, yellow-white, and red under a UV lamp after the cleavage reactions were complete (Fig. S2). These results indicate that a simple dabcyl group can be used to quench all tested multiply-colored ODFs with good quenching efficiency, thus significantly reducing the complexity of fluorophore–quencher pair selection in chemosensor design.
For the three peptide-linked caspase probes (1–3) in this study, spectral comparison before and after cleavage can be made indirectly, by comparing residual emission spectra of the unreacted (dark) probes with the spectra of the corresponding ODF fluorophores alone. Results (Fig. S3) are similar to the ester-linked model systems, showing (for the unreacted probes) strong quenching at the long-wavelength bands.
Enzymatic reporting in vitro
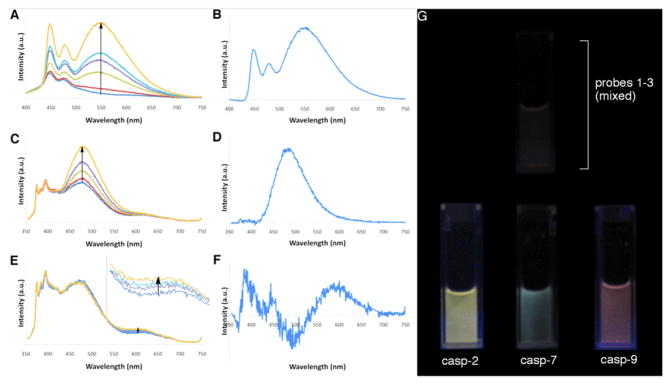
Next we turned to the enzymatic reporting potential of caspase substrate probe compounds 1–3. Homogeneous enzymatic assays for the three candidate sensors were performed with recombinant human caspases 2, 7, and 9, respectively, in 10 mM HEPES buffer (pH 7.3). Results showed that significant fluorescence enhancement was observed in each case (Fig. 2 and S4), indicating the occurrence of a measurable degree of peptide cleavage over the 4 hour time course. The probes 1–3 yielded fluorescence enhancements at ca. 550, 480, and 620 nm respectively, consistent with the results of the above model studies. The reaction rates were slower than the esterase references, particularly for probe 3 which yielded a relatively small 76% enhancement after 4 h. We expect that the rates of caspase cleavage can be affected by multiple factors, including inherent enzyme activity, instability of the enzymes over the assay period, and possibly unfavorable interactions between ODFs and the caspase active sites.
Figure 2.
In vitro enzymatic assay of caspase probes 1, 2 and 3. Each vial contains 0.5 μM probe and 625 U/mL of corresponding caspase enzyme. Fluorescence spectra were recorded in HEPES buffer (10 mM, with 2 mM DTT and 0.1% Prionex®, pH 7.33) at 30 °C in a time course of 4 h. Left: full range fluorescence spectra; right: fluorescence change after 4 h (spectra were obtained by subtracting the spectra at 0 min from the spectra at 4 h). (A, B) Probe 1 with caspase-2; (C, D) Probe 2 with caspase-7; (E, F) Probe 3 with caspase-9. EE was excited at 380 nm, while YYYY and YKY were excited at 340 nm.
To test whether the probes exhibited selectivity for one caspase over another, assays were performed using an equimolar mixture of the three probes. One caspase was added into a cocktail containing equal amounts of all three caspase chemosensors, and the reaction mixture was incubated at 30 °C for 4 h. Fluorescence changes were analyzed by emission spectra (Fig. 3) and visual inspection (Fig. 3G and S5). Results showed fluorescence changes that were similar to the changes with the same enzyme/sensor pair alone (Figs. 2 and 3). The selectivity is readily observed by eye (Figs. 3G and S5). Caspase-2 is highly specific to short peptide Val-Asp-Val-Ala-Asp,[22] and strong yellow fluorescence was observed when it was added into the sensor mixture, characteristic of the response of probe 1. Caspase-9 is selective for the peptide Leu-Glu-His-Asp,[23] and orange-red fluorescence was observed after enzyme addition, similar to the observation with probe 3 alone. Finally, caspase-7 is known to recognize the peptide Asp-Glu-Val-Asp (the peptide in probe 2), while it also shows weak activity with Val-Asp-Val-Ala-Asp.[24] Green fluorescence was observed with caspase-7, matching our expectation of sensor response for this enzyme.
Figure 3.
In vitro selectivity assay with different caspase enzymes. Each vial contained 0.5 μM of all three caspase probes and 625 U/mL of one caspase enzyme. Conditions were as in Fig. 2 with a time course of 4 h. Left: full range fluorescence spectra; right: spectral change after 4 h (spectra obtained by subtracting the spectra at 0 min from the spectra at 4 h). (A, B) With caspase-2, excited at 380 nm; (C, D) with caspase-7, excited at 340 nm; (E, F) with caspase-9, excited at 340 nm (inset shows magnified scale). (G) Images of caspase selectivity assays taken under a UV lamp with 2.5 μM caspase probes. Top: mixed probes only; bottom: same probe mixture with each caspase added separately (image after 4 h).
Cellular monitoring of apoptosis
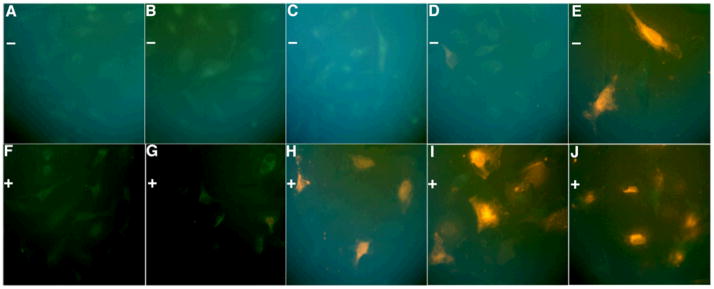
Next we tested whether an ODF probe could detect caspase activity in intact HeLa cells. Initial tests of the probes in cultured cells revealed that signals from probes 1 and 2 were difficult to observe above background (see Fig. S6), which could be due to low activity of corresponding caspases, and/or from obscuring of the relatively low signal by background fluorescence. For continuing experiments we chose probe 3, which showed strong signals (Fig. 4), likely due to high caspase-9 activity, which was activated by anisomycin through the chemically-induced apoptosis pathway,[26] and possibly also to the red color, which is expected to suffer from less cellular background interference. The commercial cationic lipid Exgen 500 was used as the delivery reagent to enhance uptake of the polyanionic sensor into the cells.
Figure 4.
True-color images of cellular apoptosis experiments. Experiments were performed with 2.5 μM probe 3 in HeLa cells under 100X microscope using Exgen 500 as delivery agent, and time was recorded after adding anisomycin. Top: control experiments without anisomycin (−); bottom line: experiments with 125 μg/mL anisomycin (+). (A, F) 1 h; (B, G) 2 h; (C, H) 4 h; (D, I) 12 h; (E, J) 24 h.
As expected, the addition of anisomycin resulted in strong changes in cell morphology after 3–4 h, characteristic of apoptosis. Experiments with the probe added showed strong orange fluorescence after two hours post-addition of the apoptosis-inducing agent, while no significant fluorescence was observed in the controls lacking anisomycin (Figs. 4, S6). Clear differences in fluorescence between the anisomycin-treated cells and untreated controls could still be observed 12 hours after treatment. After 24 hours, the controls also showed orange fluorescence, likely due to non-specific cleavage by other protease(s) in the cells (Fig. S7). It is noteworthy that the signals remain associated with cells for at least 24 hours; we attribute this to the polyanionic charge of the probe, which is expected to prevent passage through the cell membrane. This favorable property allows a timecourse of activity to be observed without loss of the sensor to the medium. This obviates the need for protein crosslinking (a common strategy in caspase probes), which inactivates the caspases in the act of detecting them.[16, 17] Moreover, the presence of the DNA backbone here allows for convenient use of commercial DNA uptake reagents for intracellular delivery, thus eliminating the need for complex conjugation strategies.
The probes could also function together in cells as a cocktail, where the orange signal of probe 3 dominated at later time points but green signals (indicating caspase-3 activity) were visible at 2 h (Fig. S8). The signals observed with probe 3 are consistent with known caspase activities in apoptosis. Anisomycin activates c-Jun N-terminal kinase (JNK), which then activates caspase-8. The apoptosis signal is amplified by caspase-9 (the probe’s intended target), which then facilitates the activation of the effector enzyme caspase-3 to start apoptosis.[26] Anisomycin induces apoptosis in a concentration-dependent fashion,[27] and treatment of HeLa cells with 10 μg/mL anisomycin was reported to cause 38% cell death in 24 h.[28] We used a higher concentration (125 μg/mL) to induce a more rapid apoptosis signal, and the observed orange fluorescence increase reached the maximum after ca. 6 h (Fig. 4 and S6). Compared to commercially available caspase probes,[15, 16, 21] our quenched probes do not require cell lysis or additional additives, such as DNA. Because the ODF probes are dark in their unreacted state, they can be used both in homogeneous assay and in live cells without washes. The probes make cellular monitoring much easier, enabling the observation of enzyme activity in real time.
We expect that this chemosensor design could be readily modified for other enzymes by simply changing the substrate portion of the structure, and could be easily changed to other emission colors by using different ODF sequences.[8] Future studies will be directed to evaluating cocktails of mixed probes in cellular pathways.
Experimental Section
Probe synthesis
ODF monomers with 5′-DMT and 3′-phosphoramidite were prepared by published methods,[5, 8, 29] and the ODF sequences were synthesized with standard DMT/phos-phoramidite chemistry. The 5′-azide was added via a commercial 5′-bromo-modifier. Substrate peptides were prepared with standard solid phase peptide synthesis on a 2-chlorotrityl chloride resin, with 5-hexynoic acid coupled to the N-terminus. Dabcyl was coupled to the C-terminus and the peptide-quencher compounds were purified by reverse-phase HPLC. They were then coupled to the corresponding ODF sequences via Cu(I)-mediated Huisgen-Sharpless cycloaddition.[9] The final caspase chemosensors were purified by HPLC on a reverse phase (C5) column, and the identities were confirmed by MALDI-MS. See details in the Supporting Information.
Optical Measurements
UV and fluorescence spectra were obtained in pure water at room temperature. See SI for details.
In vitro assays
Fluorescence spectra were recorded in HEPES buffer (10 mM, with 2 mM DTT and 0.1% Prionex®, pH 7.33) at 30 °C. Each vial contained 0.5 μM probe (or probe cocktail) and 625 U/mL of corresponding caspase enzyme.
Cellular assays
Cells were plated in chambered coverglass and allowed to reach 60% confluency. The growth medium was removed and new growth medium containing 2.5 μM caspase sensor along with 82 nM Exgen 500 was added to the cells and incubated at 37 °C under 5% CO2 for 1 h. Cells were then incubated with 125 μg/mL of anisomycin at 37 °C under 5% CO2 for varied times. See Supporting Information for details of experiments and microscopic imaging.
Footnotes
This work was supported by the U.S. National Institutes of Health (GM067201). YNT acknowledges an A*STAR NSS scholarship.
Supporting information for this article is available on the WWW under http://www.angewandte.org
References
- 1.a) Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]; b) Lavis LD, Chao TY, Raines RT. ACS Chem Biol. 2006;1:252. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Johnsson N, Johnsson K. ACS Chem Biol. 2007;2:31. doi: 10.1021/cb6003977. [DOI] [PubMed] [Google Scholar]; d) Rao J, Dragulescu-Andrasi A, Yao H. Curr Opin Biotechnol. 2007;18:17. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]; e) Sadaghiani AM, Verhelst SH, Bogyo M. Curr Opin Chem Biol. 2007;11:20. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]; f) Diamond SL. Curr Opin Chem Biol. 2007;11:46. doi: 10.1016/j.cbpa.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 2.a) Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]; b) Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat Biotechnol. 2003;21:47. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]; c) Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Nat Biotechnol. 2003;21:41. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 3.a) Hardman R. Environ Health Perspect. 2006;114:165. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Gao J, Strassler C, Tahmassebi D, Kool ET. J Am Chem Soc. 2002;124:11590. doi: 10.1021/ja027197a. [DOI] [PubMed] [Google Scholar]; b) Cuppoletti A, Cho Y, Park JS, Strassler C, Kool ET. Bioconjug Chem. 2005;16:528. doi: 10.1021/bc0497766. [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Watanabe S, Kool ET. J Am Chem Soc. 2004;126:12748. doi: 10.1021/ja046910o. [DOI] [PubMed] [Google Scholar]
- 6.a) Wilson JN, Gao J, Kool ET. Tetrahedron. 2007;63:3427. doi: 10.1016/j.tet.2006.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Teo YN, Kool ET. Bioconjug Chem. 2009;20:2371. doi: 10.1021/bc9003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teo YN, Wilson JN, Kool ET. Chem Eur J. 2009;15:11551. doi: 10.1002/chem.200901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teo YN, Wilson JN, Kool ET. J Am Chem Soc. 2009;131:3923. doi: 10.1021/ja805502k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai N, Teo YN, Kool ET. Chem Commun. 2010;46:1221. doi: 10.1039/b926338a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Chowdhury I, Tharakan B, Bhat GK. Comp Biochem Physiol B Biochem Mol Biol. 2008;151:10. doi: 10.1016/j.cbpb.2008.05.010. [DOI] [PubMed] [Google Scholar]; b) Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Cell Death Differ. 2007;14:44. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- 11.O’Brien T, Linton SD. Design of Caspase Inhibitors as Potential Clinical Agents. CRC Press; Boca Raton, FL: 2008. [Google Scholar]
- 12.Boatright KM, Salvesen GS. Curr Opin Cell Biol. 2003;15:725. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 13.a) Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]; b) Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. Nature. 1992;356:768. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 14.a) Stennicke HR, Salvesen GS. Cell Death Differ. 1999;6:1054. doi: 10.1038/sj.cdd.4400599. [DOI] [PubMed] [Google Scholar]; b) Li J, Yuan J. Oncogene. 2008;27:6194. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 15.a) Marks N, Berg MJ, Guidotti A, Saito M. J Neurosci Res. 1998;52:334. doi: 10.1002/(SICI)1097-4547(19980501)52:3<334::AID-JNR9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]; b) Ekert PG, Silke J, Vaux DL. Cell Death Differ. 1999;6:1081. doi: 10.1038/sj.cdd.4400594. [DOI] [PubMed] [Google Scholar]; c) Hug H, Los M, Hirt W, Debatin KM. Biochemistry. 1999;38:13906. doi: 10.1021/bi9913395. [DOI] [PubMed] [Google Scholar]; d) Liu J, Bhalgat M, Zhang C, Diwu Z, Hoyland B, Klaubert DH. Bioorg Med Chem Lett. 1999;9:3231. doi: 10.1016/s0960-894x(99)00566-1. [DOI] [PubMed] [Google Scholar]
- 16.Bedner E, Smolewski P, Amstad P, Darzynkiewicz Z. Exp Cell Res. 2000;259:308. doi: 10.1006/excr.2000.4955. [DOI] [PubMed] [Google Scholar]
- 17.Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Nat Med. 2009;15:967. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Korngold EC, Jaffer FA, Weissleder R, Sosnovik DE. Heart Fail Rev. 2008;13:163. doi: 10.1007/s10741-007-9068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Leyva MJ, Degiacomo F, Kaltenbach LS, Holcomb J, Zhang N, Gafni J, Park H, Lo DC, Salvesen GS, Ellerby LM, Ellman JA. Chem Biol. 2010;17:1189. doi: 10.1016/j.chembiol.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Boeneman K, Mei BC, Dennis AM, Bao G, Deschamps JR, Mattoussi H, Medintz IL. J Am Chem Soc. 2009;131:3828. doi: 10.1021/ja809721j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Prasuhn DE, Feltz A, Blanco-Canosa JB, Susumu K, Stewart MH, Mei BC, Yakovlev AV, Loukov C, Mallet JM, Oheim M, Dawson PE, Medintz IL. ACS Nano. 2010;4:5487. doi: 10.1021/nn1016132. [DOI] [PubMed] [Google Scholar]
- 20.a) Kawai H, Suzuki T, Kobayashi T, Mizuguchi H, Hayakawa T, Kawanishi T. Biochim Biophys Acta. 2004;1693:101. doi: 10.1016/j.bbamcr.2004.05.009. [DOI] [PubMed] [Google Scholar]; b) Bardet PL, Kolahgar G, Mynett A, Miguel-Aliaga I, Briscoe J, Meier P, Vincent JP. Proc Natl Acad Sci USA. 2008;105:13901. doi: 10.1073/pnas.0806983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Chem Biol. 2008;15:1116. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. J Biol Chem. 1997;272:9677. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 23.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. J Biol Chem. 1997;272:17907. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 24.McStay GP, Salvesen GS, Green DR. Cell Death Differ. 2008;15:322. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 25.Stennicke HR, Renatus M, Meldal M, Salvesen GS. Biochem J. 2000;350:563. [PMC free article] [PubMed] [Google Scholar]
- 26.Curtin JF, Cotter TG. Br J Cancer. 2002;87:1188. doi: 10.1038/sj.bjc.6600612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mawji IA, Simpson CD, Gronda M, Williams MA, Hurren R, Henderson CJ, Datti A, Wrana JL, Schimmer AD. Cancer Res. 2007;67:8307. doi: 10.1158/0008-5472.CAN-07-1687. [DOI] [PubMed] [Google Scholar]
- 28.Pucci B, Indelicato M, Paradisi V, Reali V, Pellegrini L, Aventaggiato M, Karpinich NO, Fini M, Russo MA, Farber JL, Tafani M. J Cell Biochem. 2009;108:1166. doi: 10.1002/jcb.22345. [DOI] [PubMed] [Google Scholar]
- 29.Ren RX-F, Chaudhuri NC, Paris PL, SR, Kool ET. J Am Chem Soc. 1996;118:7671. doi: 10.1021/ja9612763. [DOI] [PMC free article] [PubMed] [Google Scholar]