Abstract
The current best serum marker for pancreatic cancer, CA 19-9, detects a carbohydrate antigen on multiple protein carriers. Better knowledge of the protein carriers of the CA 19-9 antigen in various disease states may lead to improved diagnostic tests. To identify proteins that carry the CA 19-9 antigen, we immunoprecipitated the CA 19-9 antigen from pooled sera and identified the associated proteins using mass spectrometry. Among the high-confidence identifications, we confirmed the presence of the CA 19-9 antigen on Apolipoprotein B-100 by antibody arrays and Western blot and on kininogen, ARVCF, and Apolipoprotein E by antibody arrays. We characterized the frequency and levels of the CA 19-9 antigen on the four proteins across various patient groups (pancreatic cancer, pancreatitis, and healthy controls) using antibody arrays. 10–25% of the subjects showed elevations of the antigen on each protein, but the elevations were not associated with disease state or total CA 19-9 levels. These results contribute to our knowledge of the carrier proteins of an important functional glycan and the rate at which the glycan is displayed. This work also demonstrates a strategy for using the complementary methods of mass spectrometry and antibody microarrays to identify protein carriers of glycans and assess the diagnostic value of measuring glycans on individual proteins.
Keywords: CA 19-9, pancreatic cancer, glycosylation, antibody arrays
Introduction
Improved diagnostic methods for pancreatic cancer are greatly needed. Pancreatic cancer often advances to an incurable stage prior to detection, leading to very short survival time for patients [1, 2]. Furthermore, difficulties in distinguishing benign from malignant disease [3] and predicting optimal treatment courses can lead to sub-optimal management of patients. Molecular diagnostics methods that can provide accurate detection of early-stage cancer or information about disease extent could lead to more effective treatment of patients and overall better outcomes [4, 5]. Thus far, serological tests that meet this need have been elusive.
The current best serological marker for pancreatic cancer is the CA 19-9 assay. The use of CA 19-9 for a wide variety of purposes has been extensively investigated, including early detection, diagnosis, prognostication, and monitoring of tumor responses and recurrence [6–9]. It is elevated in the blood of about 80% of pancreatic cancer patients [9], and its levels often follow the regression or progression of tumors in patients receiving therapy for pancreatic cancer. Accordingly, the marker is commonly used to follow patients receiving treatment for pancreatic cancer. Blood levels of CA 19-9 also can be elevated in conditions including liver damage, bile duct obstruction, and pancreatitis. Because of those additional causes of elevation, the CA 19-9 test is not specific enough to be used for pancreatic cancer detection or diagnosis. In addition, the sensitivity for cancer is not sufficiently high to rule out cancer upon a low reading. Improvements to the CA 19-9 assay are clearly needed for a better control of pancreatic cancer.
The CA 19-9 assay measures a carbohydrate antigen that is found on many different proteins. The repertoire of these “carrier” proteins is not well defined but is known to include mucins [10] and other adhesion molecules such as carcinoembryonic antigen [11]. It also is not known whether the composition of the CA 19-9 carrier proteins is different between disease states, for example whether the proteins bearing the CA 19-9 antigen in pancreatitis are different from those bearing the CA 19-9 antigen in pancreatic cancer. If the carrier proteins are different, improved discrimination between the disease states may be possible by measuring the CA 19-9 antigen on specific proteins, rather than on all proteins [12, 13]. The antibody-lectin sandwich array platform developed earlier is ideal for testing that concept [14], since the levels of a particular glycan epitope can be measured on many different proteins in parallel (Fig. 1a) and compared across many different samples [15]. In order to implement such experiments, one must first define the potential carrier proteins to target on the antibody array.
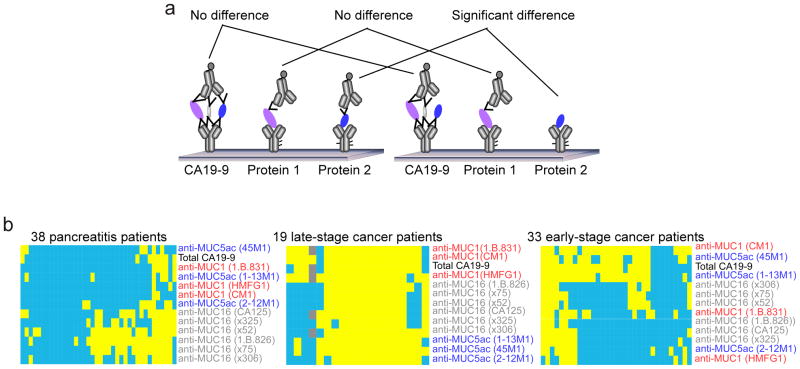
Figure 1. Detection of the CA19-9 antigen on individual protein carriers.
a) Schematic of antibody array measurements of total CA 19-9 and the CA 19-9 antigen on individual proteins, showing the theoretical basis for improvement using the latter. b) Antibody array measurements of the frequency of the CA 19-9 antigen on known protein carriers. Monoclonal antibodies targeting CA19-9 and individual proteins were immobilized on microarray slides and detected by the CA19-9 antibody. The capture antibodies are indicated by row labels (clone numbers in parentheses), and each column represents an individual sample from the indicated patient group. A yellow square indicates a high measurement, and blue indicates low. For the cancer groups, a measurement was defined as “high” if its CA19-9 level was above 90% of the benign samples (90% specificity). For the benign group, a measurement was defined as “low” if its CA19-9 level was below 90% of the cancer samples (90% sensitivity).
The goals of this study were to identify potential blood-based carrier proteins of the CA 19-9 antigen and to investigate whether the prevalence of the CA 19-9 antigen on the protein carriers is different between pancreatic cancer and pancreatitis. We first examined whether the rate at which the CA 19-9 antigen is elevated on known mucin carriers is different between the two patient groups. This analysis gave insight into the concept that the protein carriers of the CA 19-9 antigen are distinct between disease states and provided the motivation for efforts to discover additional carriers of the CA 19-9 antigen. Next, to identify such carrier proteins, we immunoprecipitated the CA 19-9 antigen from the blood of patients from each of the disease groups and identified the captured proteins using mass spectrometry. Finally, we confirmed that selected proteins bear the CA 19-9 antigen and examined the prevalence of the antigen carried by these candidates in the patient groups. The value of using complementary MS-based and antibody array methods, as described earlier [14, 16], is highlighted in this work.
Materials and Methods
Serum and plasma samples
Serum samples from pancreatic cancer, pancreatitis and healthy subjects were collected at Evanston Northwestern Healthcare (ENH) and the University of Pittsburgh (UP) School of Medicine. All sample collection was performed under protocols approved by the local Institutional Review Boards, and written, informed consent was obtained from all participants. The healthy control samples from ENH were collected from high-risk individuals from pancreatic-cancer-prone families undergoing surveillance with Endoscopic Ultrasound (EUS) or Endoscopic Retrograde Cholangiopancreatography (ERCP). The control subjects had no pancreatic lesions. All samples were stored at −80°C and sent frozen on dry ice. Each aliquot had been thawed no more than three times before use.
Antibodies
The antibodies were obtained from various sources (Table s1). All antibodies were screened for reactivity and integrity, purified, and prepared at 0.25 mg/ml in pH 7.2 PBS. The steps of antibody purification included ultracentrifugation at 47,000g at 4 degree for 1 hour and dialysis (Slide-A-Lyzer Mini Dialysis Units, PIERCE) against pH 7.2 PBS at 4 degree for 2 hours.
Microarray fabrication
Approximate 175 pg (350 pl at 500 μg/ml or 700 pl at 250 μg/ml) of each antibody was spotted on the surfaces of ultra-thin nitrocellulose-coated microscope slides (PATH slides; GenTel Biosciences) by a piezoelectric non-contact printer (Biochip Arrayer; PerkinElmer Life Sciences) for the slides used in ENH sample set, and by a non-contact arrayer at GenTel Biosciences for the slides used in UP set. Forty-eight identical arrays containing triplicates of all selected antibodies were printed on each slide. The three spots in a triplicate were positioned adjacent to each other in the slides used in ENH and UM sets, and the position of each spot in triplicates was randomly assigned in the slides used in UP sample set. Hydrophobic borders were imprinted around each array using a stamping device (SlideImprinter, The Gel Company, San Francisco, CA).
Microarray assays
Microarray sandwich assays were performed to measure either the level of total CA19-9 or its level on the selected proteins captured by the immobilized antibodies (Fig. 1a). The sandwich assay consisted of four 1-hour-incubations in room temperature (RT) with the following reagents: 1) blocking buffer (PBS containing 0.5% Tween-20 (PBST0.5) and 1% BSA); 2) a serum sample, diluted two-fold in 1XTBS containing 0.08% Brij, 0.08 Tween-20, 50 μg/ml protease inhibitor cocktail (Complete Protease Inhibitor Tablet, Roche Applied Science), and a cocktail of IgG from mouse, goat, and sheep each at 100 μg/ml and rabbit IgG at 200 μg/ml (Jackson ImmunoResearch Laboratories, Inc.); 3) biotinylated detection antibody or lectin (2 μg/ml), diluted in PBST0.1 containing 0.1% BSA; 4) streptavidin-phycoerythrin (10 μg/ml, Roche Applied Science), diluted in PBST0.1 containing 0.1% BSA. After each step, the slides were rinsed in three baths of PBST0.1 and dried by centrifugation (Eppendorf 5810R, rotor A-4-62, 1500 × g). Fluorescence emission from the phycoerythrin was detected at 570 nm using a microarray scanner (LS Reloaded, Tecan). All arrays within one slide were scanned at a single laser power and detector gain setting. The images were quantified using the software program GenePix Pro 5.0 (Molecular Devices, Sunnyvale, CA). Spots were identified using automated spot-finding or manual adjustments for occasional irregularities. The local backgrounds were subtracted from the median intensity of each spot, and triplicate spots were averaged using the geometric mean. The coefficient of variation between replicate analyzed spots was 10–15%.
Immunoprecipitation
Serum samples pools from selected patients (Table s2) were incubated with biotinylated antibody against CA19-9 (1B.844, UsBiological) (1 μl serum with 2 μg antibody) and 50 μg/ml protease inhibitor cocktail (Complete Protease Inhibitor Tablet, Roche Applied Science) at 4° C overnight with gentle rotation. Superparamagnetic beads of 2.8 μm in diameter with a steptavidin monolayer covalently coupled to the surface was obtained commercially (Dynabeads M-280 Streptavidin, Invitrogen). Dynabeads (2 μl dynabeads to 1 μg antibody) were washed with 1X PBS containing 0.1% Tween-20 and 0.1mg/ml Dexton for three times before and after the 1 hour nutation at room temperature in the same buffer. The beads were next coupled with the mix of antibody and serum for 1.5 hour at room temperature under gentle rotation. After the completion of coupling, the beads were washed with 1XPBS with 0.1% Tween-20 for three times at room temperature. For western blotting, the antigens were eluted by being boiled with 1.5X SDS gel loading buffer (75 mM Tris, pH 6.8; 3% SDS; 15% Glycerol; 3.75% b-Mercaptoethanol; 0.03% Bromophenol blue) at 100° C for 5 min. For deglycosylation, the beads remained untouched after the final washed and processed as described in the deglycosylation section.
Western blot
The samples were fractionated on Bis-Tris 4–12% XT precast gels (Bio-Rad) and transferred to nitrocellulose (0.45μm, Bio-Rad). We used 5% milk in TBST0.05 for overnight blocking. Primary detection was completed by a 60 min incubation of 3 μg/ml mouse monoclonal antibodies, and secondary detection was accomplished by incubating HRP conjugated goat anti-mouse IgG (1:200,000) (ImmunoPure, Pierce Biotechnology) for another 60 min. The nitrocellulose membrane was developed using Supersignal West Femto substrate (Pierce Biotechnology) and visualized on a ChemiDoc (Bio-Rad).
Deglycosylation
Deglycosylation was performed using a commercial enzymatic kit (Enzyme CarboRelease Kit, QAbio). Washed beads from the immunoprecipitation were incubated with reaction buffer and denaturant from the kit and boiled for 5 min at 100° C. After chilled on ice, triton-X and enzymes were added to the beads (1 μl of each enzyme for 20 μl serum immunoprecipitated) followed by 4.5 hours incubation at 37° C. Next, 1.5X SDS gel loading buffer (75 mM Tris, pH 6.8; 3% SDS; 15% Glycerol; 3.75% b-Mercaptoethanol; 0.03% Bromophenol blue) (1 μl buffer for 2 μl initial beads slurry) was added, and samples were boiled at 100° C for 5 min to ensure full elution of the proteins from the beads.
Mass spectrometry analysis
Deglycosylated samples with SDS were initially separated by migrating into Bis-Tris 4–12% XT precast gel (Bio-Rad) for approximately 2 cm. The entire gel was fixed for 2 hours at room temperature in the solution containing 40% methanol, 10% acetic acid and 50% water. Mass spectrum identification was performed by the Michigan Proteome Consortium (Ann Arbor, MI). For each sample, two gel slices were obtained, one for proteins above 250kDa, the other for proteins between 75kDa to 250kDa. The samples were digested in-gel trypsin and were analyzed by capillary reversed-phase HPLC coupled offline to a MALDI TOFTOF tandem mass spectrometer (ABI model 4800). The proteins were identified by searching a genome database using Protein Pilot software.
Statistical analysis and data preparation
The area-under-the-curves (AUC) in receiver-operator characteristic (ROC) analysis were calculated using SAS, and the Mann-Whitney rank-sum tests and the box plots were performed using OriginPro 8. Clustering and visualization were performed using the programs Cluster and Treeview and MultiExperiment Viewer.
Results
Variation in the carriers of the CA 19-9 antigen in patient subpopulations
We began by examining known carriers of the CA 19-9 antigen, which included the mucin proteins MUC1, MUC5AC, and MUC16 [11], to investigate how often they displayed the CA 19-9 antigen in each group. In particular, we wanted to know whether certain proteins are more likely to be a carrier in one patient group relative to another. If that is the case, we may be able to distinguish the patient groups more accurately by measuring the CA 19-9 antigen on those particular proteins. We used antibody arrays to explore this question, by which the CA19-9 glycan could be measured for its total level as well as its level on individual proteins simultaneously (Fig. 1a). For the measurement of total CA19-9, a monoclonal antibody against CA19-9 was used as both the capture and detection antibody. To measure CA19-9 on specific proteins, antibodies targeting the potential carrier proteins were immobilized on the microscopic slide (see Table s1 for antibody information), and the CA 19-9 antibody was used for detection. In a standard antibody array platform, which can easily incorporate dozens of capture antibodies within one array [17], both the total amount of CA 19-9 and the specific levels of CA19-9 on particular proteins can be measured all in one assay. We took measurements on samples from three disease groups, including late-stage pancreatic cancer, early-stage pancreatic cancer, and pancreatitis patients (Fig. 1b–d).
To look at the prevalence of the CA 19-9 antigen on each of the mucins in each group, we set a threshold for each antibody to determine which of the individual measurements were elevated, relative to a control group (Fig. 1b–d). The specificity and reproducibility of these assays had been confirmed in previous work [11, 18]. In the pancreatitis group (Fig. 1b), about 25% of the patients showed elevations in CA 19-9 on all three mucins, and slightly less than half showed no or sporadic elevations. About a third of the patients showed elevations only on MUC16. In early-stage cancer, again about 25% of the patients have CA 19-9 elevations on all mucin proteins and another 25% have no or few elevations. In contrast to the pancreatitis group, a subset of about a third of the patients showed elevations only on two of the MUC5AC antibodies and one of the MUC1 antibodies, and another subset of about 15% (5 of 33) of the patients have elevations on those antibodies plus half of the MUC16 antibodies. Late-stage cancer showed yet another pattern. Over half (11 of 19) showed elevations on all mucins, and another ~25% (5 patients) showed variable elevations on each of the mucins. A distinct subset of three patients showed elevations only on MUC1. These results support the concept that the carrier proteins may shift between disease states, in this case from MUC16 being the most prevalent carrier in pancreatitis to MUC5AC and MUC1 being preferential carriers in cancer.
Identification of protein carriers of the CA 19-9 antigen
Based on the above result, we sought to identify additional carriers of the CA 19-9 antigen in pancreatic cancer and pancreatitis patients. We used a glycoproteomics strategy based on immunoprecipitation and mass spectrometry (Fig. 2). For each of four groups, healthy controls, pancreatitis patients, early-stage pancreatic cancer patients, and late-stage pancreatic cancer patients, a sample pool was created from five to ten samples each containing a high level of CA19-9 (Table s3). Next, the CA 19-9 carrier proteins were isolated from each pool by immunoprecipitation with the CA 19-9 monoclonal antibody, followed by deglycosylation using a combination of glycosidases that cleaved both N- and O- linked glycans from the glycoproteins pulled down in the immuoprecipitation. Deglycosylation enabled us to obtain more protein hits identified in the mass spectrometry (data not shown). In the following steps, the pools of CA19-9 carrier proteins obtained from each sample group were further separated by one-dimensional sodium dodecyl sulfate gel electrophoresis (1D-SDS PAGE) for the identification of proteins by tandem mass spectrometry (MS), using matrix-assisted laser desorption/ionization with time-of-flight detection (MALDI/TOF/TOF). For selected identifications from the MS analysis, antibodies were obtained and used to measure the CA 19-9 levels on the target proteins in independent samples.
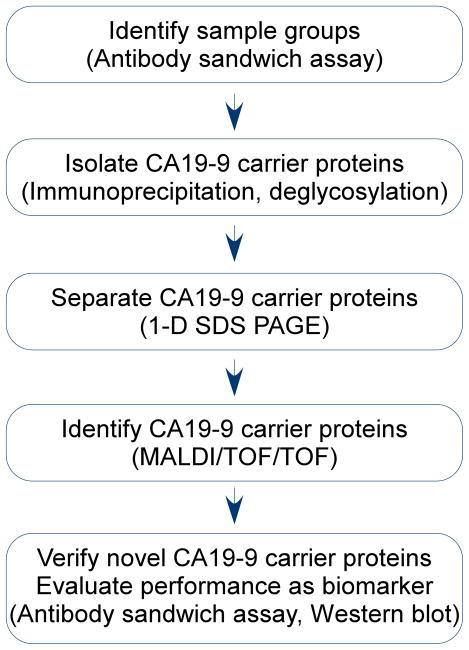
Figure 2. Experiment design.
The sequence of steps is shown with the methods used for each step in parentheses.
Three independent experiment sets were assembled. From 275 to 500 high-confidence protein identifications were achieved for the sample pools. Among these, 60 hits representing 21 distinct proteins (Table s3) were identified by more than one peptide and showed the greatest differences in the number of peptides between pancreatic cancer and non-cancer pools in at least one experiment. Four proteins targets were selected for further study (Table s4). Apolipoprotein B (ApoB) was one of the most abundant and consistent hits identified in all three experiments, and in two of the three experiments, ApoB was found with more hits identified in cancer than in benign or healthy (Table s5). Another protein from the apolipoprotein family, ApoE, was detected in the cancer pool in one experiment and not detected in any of the other pools. ApoE is closely associated with ApoB in function [19]. Peptides corresponding to ARVCF (armadillo repeat gene deleted in Velo-Cardio-Facial syndrome) were found in both the early-and late-stage cancer pools in one of the experiments but not detected in either the benign or healthy pools from all three experiments. Kininogen was found in just one experiment, although higher numbers of peptide hits were identified in cancer for both high- and low-molecular-weight isoforms, adding to the significance of the finding. Monoclonal antibodies targeting the above candidates were obtained and immobilized on microscopic slides for further validation using microarrays.
Confirmation and prevalence of the CA 19-9 antigen on newly-identified carrier proteins
We used antibody arrays (Fig. 1a) to verify that the CA 19-9 antigen is found on the selected proteins and to examine its frequency and levels across a population. Since the CA 19-9 antigen may be found on these proteins in highly varying levels between people, it was important to look at multiple, individual samples. Our ability to run antibody arrays in a high-throughput mode [15] facilitated this experiment. In examining the levels across 48 pancreatitis patients and 124 cancer patients, we observed significant diversity between the patients, with a subset showing elevations of the CA 19-9 antigen (Fig. 3a). ApoB showed strong signals in a subset of about 25% of the patients, while the other three proteins showed weaker elevations among about 10% of the patients (Fig. 3a). The non-random elevations in selected patients is consistent with these proteins carrying the CA 19-9 antigen in a subset of the population. These results also show the diversity that exists between people in glycosylation states and the importance of examining multiple samples.
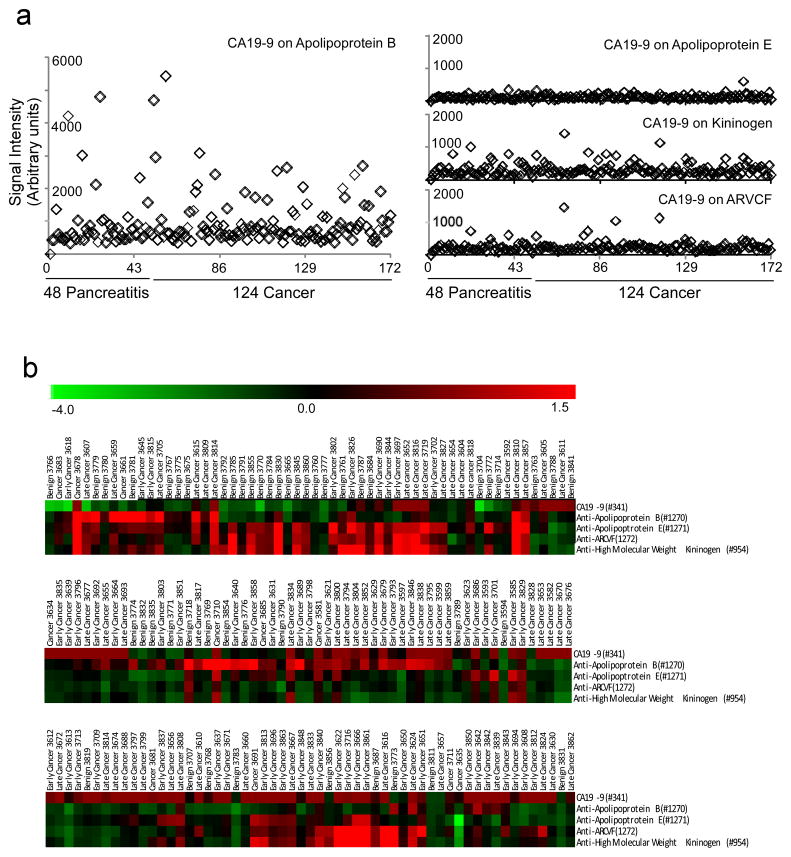
Figure 3. Detection of the CA19-9 antigen on individual protein carriers.
The levels of the CA 19-9 antigen on the indicated proteins in serum samples from cancer patients and controls were measured using antibody microarrays. a) The signal is plotted for each patient sample for CA 19-9 on ApoB, ApoE, kiniogen, and ARVCF. b) A hierarchical cluster of the measurements of total CA 19-9 and the CA 19-9 antigen on each of the four indicated proteins. The columns indicate the samples, and the rows are the markers. The color bar indicates the scale, after log transformation of the raw fluorescence values and median-centering of each row. The cluster is split into three sections for clarity and shows the correlation between the measurements of CA 19-9 on ApoE, ARVCF, and kininogen, and a lack of correlation with total CA 19-9 and CA 19-9 on ApoB.
We examined the correlations between the individual measurements to gain insight into the coordinated regulation of the CA 19-9 antigen (Figure 3b). None of the measurements of CA 19-9 on individual proteins was significantly correlated with total CA 19-9 levels. CA 19-9 on ApoB was not correlated with any other measurement, but CA 19-9 on the other three proteins were highly correlated with each other. These relationships suggest that the regulation of CA 19-9 levels is divergent between certain proteins and similar among others. The lack of correlation with total CA 19-9 indicates that these proteins are not major contributors to the signal from the total CA 19-9 assay.
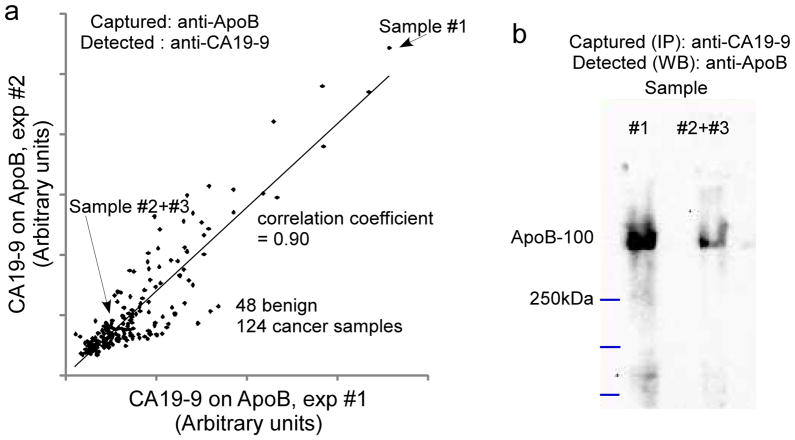
Additional analyses further confirmed ApoB as a carrier of the CA 19-9 antigen. Repeated measurements of CA 19-9 on ApoB (Figure 4a) correlated well (r = 0.9, Pearson’s r correlation coefficient), confirming that the signals were not due to random binding events. In addition, we immunoprecipitated CA 19-9-carrying proteins and probed for ApoB by Western blot (Figure 4b). This experiment is the reverse of the microarray assay, in which ApoB was captured first. One sample with high CA 19-9 levels on ApoB and two samples with low levels, as determined by antibody microarray, were selected. The Western blot results showed the same trend as the antibody microarray (Figure 4b). Western blot probes for the other candidate carriers were negative (data not shown), perhaps due to the much lower abundance of those proteins or their relative scarcity as CA 19-9 carriers.
Figure 4. Confirmation of Apo B as a CA19-9 carrier.
a) Correlation of measurements between duplicated antibody array assays. The level of CA 19-9 on ApoB was measured by antibody array (Fig 1a) in 172 serum samples in two independent experiments, and the measurements were plotted with respect to each other. b) Validation of the CA19-9 antigen on ApoB by immunoprecipitation using CA19-9 antibody and blotting with antibody targeting ApoB, using the three indicated samples identified from panel a.
Disease associations of the CA 19-9 antigen on ApoB
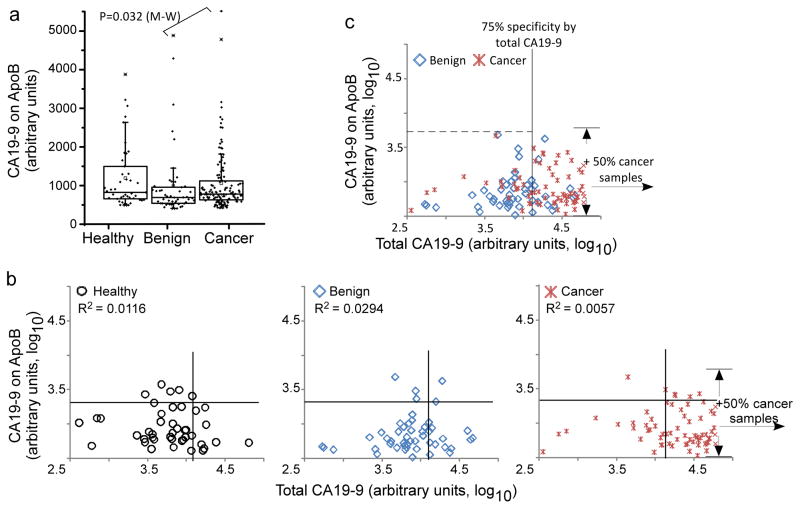
We examined more closely the disease-associations of the CA 19-9 antigen on ApoB and whether those measurements have implications for the detection of pancreatic cancer. The level of CA 19-9 on ApoB was not significantly different between the patient groups (including healthy subjects), with only a moderate difference between the cancer and pancreatitis groups (p=0.044, Mann-Whitney test, Fig. 5a). This result indicates that ApoB is not a major cancer-associated carrier of the CA 19-9 antigen. Because measurements of total CA 19-9 are composed of many individual proteins, we wanted to know whether signals from ApoB contributed to total CA 19-9 levels in any of the patient groups. If so, ApoB may be a confounding factor contributing to the non-specificity of total CA 19-9, since ApoB itself is not a disease-associated carrier. Comparisons of the total levels and the levels on ApoB in each of the patient groups revealed a general lack of correlation (Fig. 5b). The patients that were elevated in CA 19-9 were not more or less likely to be elevated in CA 19-9 on ApoB, indicating that other proteins besides ApoB were responsible for the total CA 19-9 elevations. Selected patients with moderate CA 19-9 levels were elevated in CA 19-9 on ApoB, suggesting that in certain cases, ApoB can be the dominant carrier in the blood.
Figure 5. Apo B as a CA19-9 carrier in healthy, pancreatitis and pancreatic cancer groups.
A) Box-plot of CA19-9 level on ApoB from 43 healthy, 48 pancreatitis (benign) and 124 cancer individuals. Each point represents an individual sample. The boxes indicate the quartiles, with the median indicated by the solid horizontal lines and the ranges marked by the vertical lines. B) Scatter plots comparing CA19-9 on ApoB (y-axis) to total CA19-9 (x-axis). Each point represents a sample from 124 pancreatic cancer or 48 benign patients. The cancer patients with total CA19-9 signal in the higher half were omitted in the figure for concision purpose, and the range of CA19-9 signal on ApoB from the 50% patients were marked by the arrows. R2, Pearson correlation coefficient.
We also asked whether the CA19-9 level on ApoB provides complementary information to total CA19-9, so that the two measurements could be combined in a biomarker panel. A complementary relationship between markers would be characterized by an increased ability to selectively detect certain patients with one marker and other patients with the other marker. As noted above, selected patients that were not highly elevated in total CA 19-9 showed relatively high levels of the CA 19-9 antigen on ApoB. However, the frequency of that occurrence was similar between the patient groups, so that no additional, complementary information that enhances the discrimination of cancer from pancreatitis is provided. This result further confirms that the level or rate of the CA 19-9 antigen on ApoB is not influenced be the presence of pancreatic cancer or pancreatitis in the patient, in contrast to the CA 19-9 antigen on MUC1, MUC5AC, and MUC16 (Figure 1b).
Discussion
Improved serological tests for pancreatic cancer could be useful for detecting early-stage disease, for the differential diagnosis of pancreatic cancer from pancreatitis, and for monitoring the response to treatment in drug trials. This work demonstrated an approach for developing new markers based on both protein and glycan measurements. We previously showed that the measurement of glycans on specific proteins can yield improved biomarker performance relative to measuring just protein levels, as in standard immunoassays [11]. That work took the approach of enhancing the detection of known cancer-associated proteins by identifying and measuring cancer-associated glycans on those proteins. This work takes a complementary approach by starting with a known cancer-associated glycan, the CA 19-9 antigen, and identifying the proteins that might carry the antigen more often in cancer than in other states. We used immunoprecipitation followed by tandem MS to identify such carrier proteins, followed by antibody arrays with glycan detection to examine the rate at which the proteins carried the glycan. We found several candidate carrier proteins and characterized the prevalence of CA 19-9 on four of the newly-found carriers.
We found that the frequency with which these proteins carried the CA 19-9 antigen was not associated with pancreatic diseases. Although these findings did not uncover a new biomarker, other valuable results were achieved. More information about which proteins in the blood carry biologically-important glycans such as the Lewis blood group antigens is valuable both for the biological implications and for the use of those molecules as biomarkers, as discussed below. In addition, we demonstrate here a valuable strategy using complementary MS-based and antibody array methods [16] (Figure 2). The MS protocol enables the identification of carriers of a specific glycan, and the profiling of many samples using antibody arrays allows exploration of the diversity between people and associations with disease.
This study adds to the knowledge base of proteins that are carriers of the CA 19-9 or related glycan structures in the blood. Kininogen was not previously known to be a carrier of the CA 19-9 antigen, although it was identified as a carrier of the Lewis X carbohydrate epitope [20]. Lewis X is closely related to the CA 19-9 antigen, which is the sialyl Lewis A blood group antigen, so it is reasonable that any particular protein could carry both epitopes. ApoB previously had not been directly demonstrated as a CA 19-9 antigen carrier, although glycolipids in general were thought to be carriers of Lewis antigens. ApoE would belong to the same category of Lewis antigen carrier, although at a lower abundance. ARVCF and other proteins identified here had not been previously found to be CA 19-9 antigen carriers. Additional experimentation will be required to confirm that other identified proteins (Table s3) are carriers of the CA 19-9 antigen.
The tissue of origin may affect both the normal and disease-associated glycosylation. ApoB is produced mainly in the liver, whereas the other proteins studied here are produced in the liver and in a variety of other tissues. An interesting question is which types of tissue produce the version of the protein with the CA 19-9 antigen attached. A study to address that question could consist of analyzing the glycosylation on each of the proteins acquired separately from different tissue types. For the proteins produced in the liver, it is possible that the glycosylation changes when cancer or some other major stress is present elsewhere in the body, as indicated by a study of liver protein glycosylation in a mouse model of cancer [21]. An example of that occurrence in humans is the altered fucosylation of the major liver protein haptoglobin in many different inflammatory and malignant conditions [22–24]. Although the CA 19-9 antigen did not change on the proteins studied here, other features such as fucosylation may show disease-associated effects on these proteins. Regarding expression in the tumor itself, the resource Protein Atlas (www.proteinatlas.org) provides some information. According to Protein Atlas, ApoB, ApoE, and KNG are formed in moderate amounts in up to half of pancreatic tumors, and ARVCF is produced mainly in the stroma of over half the cases. It is possible that the glycosylation produced in the tumor environment was different elsewhere, but that the cancer-associated signals were not detected because of masking from normal variants. In addition, other glycans besides the CA 19-9 antigen may reveal the disease-associated changes. The mucin proteins MUC1, MUC5AC, and MUC16, which do show disease-associated alterations in glycosylation, are expressed in the great majority of pancreatic tumors. Since the normal levels in the blood of these proteins are low, the main source of elevation in disease is likely leakage from the pancreas, which would increase the ability to detect tumor-associated glycosylation over the background of normal variants. Future studies could examine the glycosylation of these proteins acquired specifically from tumor tissue, which may reveal whether the CA 19-9 antigen or other glycans are produced on these proteins in that environment.
We demonstrated the importance of not only identifying the protein carriers of the glycan but also characterizing how often each protein displays the glycan. For certain proteins, such as ARVCF, the prevalence is quite low at just about 10%. Low prevalence could make the identification of certain carrier proteins difficult using pooled sera. The ability to profile the carriers over populations also gives insights into the co-regulation of the synthesis of the glycan on each of the proteins and the association with disease state. The presence of CA 19-9 on three of the proteins, ApoE, ARVCF, and kininogen, was highly correlated, indicating that the modification of these proteins may be controlled by similar factors. The CA 19-9 antigen on ApoB was found on a completely uncorrelated set of patients, indicating a different regulatory system controlling the post-translational modification of that protein. Since most pancreatitis and pancreatic cancer patients have active and systemic inflammatory signals present, the levels are likely not affected by inflammation.
The presence of the CA 19-9 antigen on ApoB has physiological implications. ApoB has been intensively studied since late-1980s [25] for its central roles in lipoprotein metabolism and arteriosclerosis. The two isoforms of ApoB, ApoB-48 and ApoB-100, have distinct functions [26]. ApoB-48 is responsible for intestinal absorption of dietary fibers, while ApoB-100 is virtually the only protein component of low-density-lipoproteins (LDL). We found the CA 19-9 antigen only on ApoB-100 (Table s5 and Figure 4b), although that finding may be due to the much higher level in the blood (about 100-fold) of ApoB-100 than ApoB-48. The presence of the sialyl Lewis A glycan on ApoB-100 may affect inflammation and the development of atherosclerosis. P-selectin, an adhesion molecule expressed on platelets and endothelium, is expressed at sites of inflammation to promote leukocyte attachment and rolling. P-selectin interacts with its ligands through its C-type lectin domain which binds sialylated lewis structures. By tagging a subset of Apo-B-100’s with the sialyl Lewis A antigen, these molecules may be able to cluster at sites of inflammation to serve as signaling molecules for recruitment and maturation of additional immune cells [27]. Additional evidence supporting a role for sialyl Lewis A in inflammation is stimulation of FUT3 expression by the inflammatory modulators IL-6 and IL-8 [28], as FUT3 is the sole fucosyltransferase known to create the Lewis A structure.
Although mucins are highly-prevalent carriers of the CA 19-9 antigen in pancreatic cancer and pancreatitis, our MS analysis did not uncover mucins. This negative result likely stems from the difficulty in analyzing the large glycoproteins by MS and the fact that some mucins do not contain the trypsin cleavage site in their main extracellular domains. Studies targeted at very high molecular weight proteins may be fruitful in finding additional CA 19-9 carriers secreted from the tumor environment. Such approaches may be necessary since it appears from our study that a variety of liver-produced produced proteins can carry the CA 19-9 antigen in a subset of people, without regard to disease status. Future studies should focus on identifying the CA 19-9 carrier proteins specifically in the pancreatic cancer patients that are hard to classify using the standard CA 19-9 assay—the patients with CA 19-9 levels similar to those in the pancreatitis patients.
This study contributes to our knowledge of the proteins that carry an important functional glycan in the blood. In addition, we demonstrate a useful approach, using complementary MS-based and antibody microarray methods, for both the identification of protein carriers of particular glycans as well as the characterization of the prevalence of the display of the glycan on specific proteins. We found that the CA 19-9 antigen in found on ApoB in about 25% of people, independent of the presence of pancreatic disease, and it is found on other serum proteins in both healthy and diseased individuals at a lower prevalence. Further experimentation will be required to uncover the protein carriers of the glycan that are predominant in disease. The methods presented here should be useful in such studies and for the identification and study of other functional glycans.
Supplementary Material
Acknowledgments
We gratefully acknowledge support of this work from the National Cancer Institute (R33 CA122890, to B.B.H.) and the Van Andel Research Institute. We thank Vivien Wang and Mary Ann Regner (Evanston Northwestern Healthcare) for assistance with the sample collection and handling.
References
- 1.Maitra A, Hruban RH. Pancreatic cancer. Annual review of pathology. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hruban RH, Adsay NV. Molecular classification of neoplasms of the pancreas. Human pathology. 2009;40:612–623. doi: 10.1016/j.humpath.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Kloppel G, Adsay NV. Chronic pancreatitis and the differential diagnosis versus pancreatic cancer. Arch Pathol Lab Med. 2009;133:382–387. doi: 10.5858/133.3.382. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Etzioni R, Urban N, Ramsey S, McIntosh M, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 6.Pleskow DK, Berger HJ, Gyves J, Allen E, et al. Evaluation of a serologic marker, CA19–9, in the diagnosis of pancreatic cancer. Ann Intern Med. 1989;110:704–709. doi: 10.7326/0003-4819-110-9-704. [DOI] [PubMed] [Google Scholar]
- 7.Ko AH, Hwang J, Venook AP, Abbruzzese JL, et al. Serum CA19-9 response as a surrogate for clinical outcome in patients receiving fixed-dose rate gemcitabine for advanced pancreatic cancer. Br J Cancer. 2005;93:195–199. doi: 10.1038/sj.bjc.6602687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritts RE, Jr, Nagorney DM, Jacobsen DJ, Talbot RW, Zurawski VR., Jr Comparison of preoperative serum CA19-9 levels with results of diagnostic imaging modalities in patients undergoing laparotomy for suspected pancreatic or gallbladder disease. Pancreas. 1994;9:707–716. doi: 10.1097/00006676-199411000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266–270. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Akagi J, Takai E, Tamori Y, Nakagawa K, Ogawa M. CA19-9 epitope a possible marker for MUC-1/Y protein. Int J Oncol. 2001;18:1085–1091. doi: 10.3892/ijo.18.5.1085. [DOI] [PubMed] [Google Scholar]
- 11.Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, et al. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697–1707. doi: 10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, LaRoche T, Hamelinck D, Bergsma D, et al. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nature methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 14.Haab BB. Antibody-lectin sandwich arrays for biomarker and glycobiology studies. Expert review of proteomics. 2010;7:9–11. doi: 10.1586/epr.09.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester S, Kuick R, Hung KE, Kucherlapati R, Haab BB. Low-volume, high-throughput sandwich immunoassays for profiling plasma proteins in mice: identification of early-stage systemic inflammation in a mouse model of intestinal cancer. Molecular Oncology. 2007;1:216–225. doi: 10.1016/j.molonc.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Z, Hincapie M, Haab BB, Hanash S, et al. The development of an integrated platform to identify breast cancer glycoproteome changes in human serum. Journal of chromatography. 2009 doi: 10.1016/j.chroma.2009.09.029. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue T, Haab BB. Microarrays in glycoproteomics research. Clin Lab Med. 2009;29:15–29. doi: 10.1016/j.cll.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YM, Nowack DD, Omenn GS, Haab BB. Mucin glycosylation is altered by pro-inflammatory signaling in pancreatic-cancer cells. Journal of proteome research. 2009;8:1876–1886. doi: 10.1021/pr8008379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahley RW, Innerarity TL, Rall SC, Jr, Weisgraber KH. Plasma lipoproteins: apolipoprotein structure function. J Lipid Res. 1984;25:1277–1294. [PubMed] [Google Scholar]
- 20.Cho W, Jung K, Regnier F. Use of Glycan Targeting Antibodies To Identify Cancer-Associated Glycoproteins in Plasma of Breast Cancer Patients. Analytical chemistry. 2008;80:5286–5292. doi: 10.1021/ac8008675. [DOI] [PubMed] [Google Scholar]
- 21.Lee A, Chick JM, Kolarich D, Haynes PA, et al. Liver membrane proteome glycosylation changes in mice bearing an extra-hepatic tumour. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M900538-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson S, Kelly CA, Griffiths ID, Turner GA. Abnormally-fucosylated serum haptoglobins in patients with inflammatory joint disease. Clinica chimica acta; international journal of clinical chemistry. 1989;184:251–258. doi: 10.1016/0009-8981(89)90058-2. [DOI] [PubMed] [Google Scholar]
- 23.Thompson S, Dargan E, Turner GA. Increased fucosylation and other carbohydrate changes in haptoglobin in ovarian cancer. Cancer letters. 1992;66:43–48. doi: 10.1016/0304-3835(92)90278-4. [DOI] [PubMed] [Google Scholar]
- 24.Okuyama N, Ide Y, Nakano M, Nakagawa T, et al. Fucosylated haptoglobin is a novel marker for pancreatic cancer: A detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. International journal of cancer. 2006;118:2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 25.Young SG. Recent progress in understanding apolipoprotein B. Circulation. 1990;82:1574–1594. doi: 10.1161/01.cir.82.5.1574. [DOI] [PubMed] [Google Scholar]
- 26.Kane JP, Hardman DA, Paulus HE. Heterogeneity of apolipoprotein B: isolation of a new species from human chylomicrons. Proc Natl Acad Sci U S A. 1980;77:2465–2469. doi: 10.1073/pnas.77.5.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li AC, Glass CK. The macrophage foam cell as a target for therapeutic intervention. Nature medicine. 2002;8:1235–1242. doi: 10.1038/nm1102-1235. [DOI] [PubMed] [Google Scholar]
- 28.Groux-Degroote S, Krzewinski-Recchi MA, Cazet A, Vincent A, et al. IL-6 and IL-8 increase the expression of glycosyltransferases and sulfotransferases involved in the biosynthesis of sialylated and/or sulfated Lewisx epitopes in the human bronchial mucosa. The Biochemical journal. 2008;410:213–223. doi: 10.1042/BJ20070958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.