Abstract
Enterococcus faecalis is a gram-positive bacterium that is part of the indigenous microbiotica of humans and animals as well as an opportunistic pathogen. In this study we have fractionated the membrane proteome of E. faecalis and identified many of its constituents by mass spectrometry. We present BN-/SDS-PAGE reference maps that contain 102 proteins. These proteins are important for cellular homeostasis, virulence, and antibiotic intervention. Intriguingly, many proteins with no known function were also identified, indicating that there are substantial gaps in knowledge of this organism’s biology. On a more limited scale we also provide insight into the composition of membrane protein complexes. This study is a first step toward elucidating the membrane proteome of E. faecaliswhich is critical for a better understanding of how this bacterium interacts with a host and with the extracellular milieu.
Keywords: Enterococcus faecalis, membrane proteome, protein complex, BN-PAGE, nano LC-ESI-MS/MS
Enterococci are gram-positive, facultatively anaerobic bacteria that inhabit the gastrointestinal tract of humans and animals [1, 2]. Although they are part of the indigenous microbiotica, enterococci are also opportunistic pathogens that can cause a variety of diseases (e.g. endocarditis, bacteremia, meningitis, wound infections, and urinary tract infections) and are one of the leading causes of nosocomial infections [1–3]. In the United States alone, an estimated 800,000 enterococcal infections occur each year [2]. These infections are predominantly caused by Enterococcus faecalis and Enterococcus faecium [1]. While relatively little is known about the pathogenic mechanisms of these organisms, it is known that many virulence proteins are localized in the cell envelope [4]. Some of these proteins can facilitate adherence to the host tissue, colonization, biofilm formation, evasion of the immune system, and antibiotic resistance [1, 2]. Some proteins embedded in the cell envelope are also essential for importing nutrients and maintaining cellular homeostasis [4–6] and, therefore, enable enterococci to colonize a wide range of niches. The importance of many cell envelope proteins for cellular homeostasis, pathogenesis and drug resistance makes them an obvious target for the development of novel therapeutics [7]. For these reasons, proteomics characterization of the E. faecalis cell envelope is of great interest. To this end, recent studies on secreted and surface proteins of E. faecalis have only identified 9 membrane embedded proteins (~1.5% of the predicted membrane embedded proteome) [8, 9].
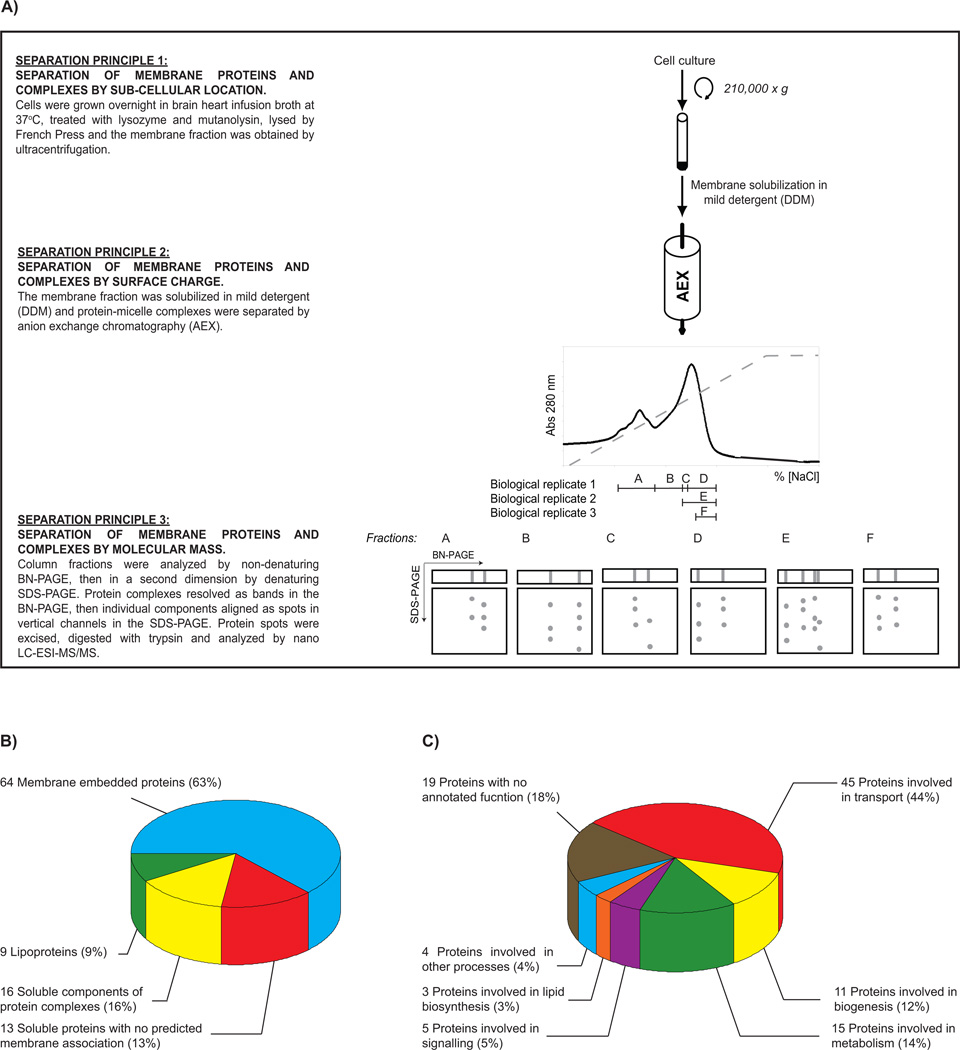
The aim of this study was to gain insight into the composition of the membrane proteome of E. faecalis. We used E. faecalis OG1X, a laboratory strain commonly used as a model system [10]. The study was carried out using a proteomics platform that separates membrane proteins by three orthogonal principles, enabling identification by mass spectrometry [11] (Figure 1A). From three biological replicate experiments we detected 294 protein spots (Supplementary Figure 1). Analysis of these spots by nano LC-ESI-MS/MS (Supplementary Table 1 and 2) indicated that they corresponded to 102 unique proteins (Table 1). Bioinformatics analysis of the amino acid sequences for these proteins was carried out with SignalP3.0 [12] (to identify cleavable signal peptides), SCAMPI [13] (to identify transmembrane proteins), and PRED-LIPO [14] (to identify lipoproteins). This analysis predicted that 64 membrane embedded proteins, 9 lipoproteins and 29 soluble proteins (Figure 1B). A large fraction of the soluble proteins (16/29) were known from the literature as components of membrane protein complexes. Indeed, 11 of these aligned in a vertical channel with a membrane embedded protein in our study, thus justifying their localization at the membrane. To our knowledge, all other proteomics studies on E. faecalis have focused on the soluble sub-proteomes (i.e. cytoplasmic and surface proteins) [8, 9, 15–18]. This study therefore represents a first-step towards a reference map of the membrane embedded proteome of E. faecalis and complements current knowledge on the cytoplasmic and surface proteomes of this important nosocomial bacterium.
Figure 1. A birds-eye view of the membrane proteome of E. faecalis OG1X.
(A) An overview of the methodology used in this study. A detailed description of all methodology is available in Supplementary Materials and Methods. Identified proteins were classified by (B) cellular location and (C) function.
Table 1.
Unique proteins found in this study. Salient features of mass spectrometry experiments are contained in Supplementary Table 1. Peptides matched in Mascot searches are listed in Supplementary Table 2. Fragmentation spectra for single-peptide protein indentifications are shown in Supplementary Figure 2.
| Protein name | Locus in Enterococcus faecalis OG1RF (a) |
Cellular location (b) |
TMs (c) | Type (d) |
|---|---|---|---|---|
| Proteins involved in transport | ||||
| Peptide ABC transporter | Contig 35 (24586–25509) | M | 6 | ABC1 |
| Peptide ABC transporter | Contig 35 (26499–27443) | S | 0 | ABC1 |
| Peptide ABC transporter | Contig 35 (27451–28450) | S | 0 | ABC1 |
| Cobalt import ATP-binding protein CbiO 1 | Contig 10 (31446–32285) | S | 0 | ABC2 |
| Cobalt import ATP-binding protein CbiO 2 | Contig 10 (30601–31476) | S | 0 | ABC2 |
| Cobalt transport family protein | Contig 10 (29807–30604) | M | 5 | ABC2 |
| Amino acid ABC transporter | Contig 5 (209776–210513) | S | 0 | ABC3 |
| Amino acid ABC transporter | Contig 5 (211684–212683) | M | 6 | ABC3 |
| ABC transporter, ATP-binding protein | Contig 10 (72643–73276) | S | 0 | ABC4 |
| ABC transporter, permease protein | Contig 10 (71091–72090) | M | 7 | ABC4 |
| ABC transporter | Contig 27 (10556–10969) | M | 5 | ABC5 |
| ABC transporter | Contig 27 (10974–11849) | M | 6 | ABC5 |
| ABC transporter | Contig 36 (11529–12271) | L | 0 | ABC6 |
| ABC transporter | Contig 36 (12552–13460) | M | 5 | ABC6 |
| ABC transporter | Contig 36 (13487–14425) | M | 4 | ABC6 |
| Amino acid ABC transporter | Contig 10 (23469–24468) | M | 3 | ABC7 |
| ABC transporter | Contig 5 (242535–243096) | M | 5 | ABC8 |
| ABC transporter | Contig 5 (243812–244811) | M | 5 | ABC8 |
| Peptide ABC transporter | Contig 4 (9909–10908) | L | 0 | ABC9 |
| Phosphate import ATP-binding protein pstB 1 | Contig 8 (185123–185881) | S | 0 | ABC10 |
| Phosphate import ATP-binding protein pstB 2 | Contig 8 (184303–185112) | S | 0 | ABC10 |
| Cell division ABC transporter | Contig 8 (180445–181329) | M | 4 | ABC11 |
| ABC transporter | Contig 27 (23767–24766) | M | 5 | ABC12 |
| ABC transporter | Contig 27 (25491–26490) | M | 6 | ABC12 |
| Glycine betaine/L-proline ABC transporter | Contig 32 (97401–99107) | M | 7 | ABC13 |
| PTS system, mannose-specific IIAB components | Contig 11 (5988–6947) | S | 0 | PTS1 |
| PTS system, mannose-specific IIC component | Contig 11 (7015–7818) | M | 7 | PTS1 |
| PTS system, mannose-specific IID component | Contig 11 (7841–8752) | M | 5 | PTS1 |
| PTS system, fructose-specific family, IIABC components | Contig 5 (165638–166637) | M | 8 | PTS2 |
| PTS family glucose porter, IICBA component | Contig 3 (52672–53671) | M | 9 | PTS3 |
| PTS system, IIC component | Contig 7 (40824–41823) | M | 8 | PTS4 |
| PTS system, IIC component | Contig 3 (73328–74327) | M | 10 | PTS5 |
| PTS system, IID component | Contig 8 (141368–142177) | M | 5 | PTS6 |
| C4-dicarboxylate transporter, putative | Contig 11 (97235–98159) | M | 8 | IT1 |
| Citrate transporter | Contig 23 (2321–3320) | M | 10 | IT2 |
| Major facilitator family transporter | Contig 11 (65570–66569) | M | 11 | MFS1 |
| Amino acid permease family protein | Contig 37 (11438–12437) | M | 12 | MFS2 |
| Amino acid permease family protein | Contig 5 (76782–77781) | M | 11 | |
| Formate/nitrite transporter family protein | Contig 41 (2254–3057) | M | 5 | |
| Mn2+/Fe2+ transporter, NRAMP family | Contig 13 (12302–13301) | M | 12 | |
| Na+/H+ antiporter | Contig 26 (63390–64389) | M | 11 | |
| Cation-transporting ATPase, E1–E2 family | Contig 8 (39552–40551) | M | 8 | |
| Large-conductance mechanosensitive channel | Contig 35 (74751–75200) | M | 2 | |
| V-type ATPase, subunit K | Contig 3 (30122–30595) | M | 3 | |
| V-type ATPase, subunit I | Contig 3 (28115–29114) | M | 8 | |
| Proteins involved in biogenesis | ||||
| Preprotein translocase, SecY subunit | Contig 10 (39044–40043) | M | 9 | |
| OG1RF_Possible O-antigen polymerase | Contig 30 (31004–32003) | M | 12 | |
| Septation ring formation regulator ezrA | Contig 26 (26980–27979) | M | 1 | |
| Cell division protein DivIVA | Contig 7 (57857–58558) | S | 0 | |
| Penicillin-binding protein 4 | Contig 32 (211716–212715) | S | 0 | |
| Penicillin-binding protein 2A | Contig 5 (126101–127100) | M | 1 | |
| Penicillin-binding protein 2B | Contig 18 (13934–14933) | M | 1 | |
| Preprotein translocase, YajC subunit | Contig 4 (20567–20944) | M | 1 | |
| Amidase, putative | Contig 5 (187646–188645) | S | 0 | |
| DltD protein | Contig 17 (5604–6603) | S | 0 | |
| Foldase protein prsA | Contig 5 (134096–134800) | L | 0 | |
| Proteins involved in metabolism | ||||
| ATP synthase subunit alpha | Contig 32 (122707–123706) | S | 0 | |
| ATP synthase subunit beta | Contig 32 (125269–126268) | S | 0 | |
| ATP synthase gamma chain | Contig 32 (124279–125187) | S | 0 | |
| ATP synthase epsilon chain | Contig 32 (126692–127111) | S | 0 | |
| ATP synthase subunit b | Contig 32 (121619–122149) | M | 1 | |
| Oxidoreductase, pyridine nucleotide-disulfide family | Contig 34 (7771–8770) | M | 5 | |
| Formate acetyltransferase | Contig 39 (81299–82298) | S | 0 | |
| Enolase | Contig 8 (18945–19944) | S | 0 | |
| Glucose-6-phosphate isomerase | Contig 3 (25333–26332) | S | 0 | |
| Ornithine carbamoyltransferase | Contig 11 (94338–95337) | S | 0 | |
| Decarboxylase | Ccontig 5 (74669–75668) | S | 0 | |
| 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | Contig 10 (58445–59131) | S | 0 | |
| Glyceraldehyde 3-phosphate dehydrogenases | Contig 8 (15688–16687) | S | 0 | |
| Thiamin biosynthesis lipoprotein ApbE | Contig 34 (5310–6309) | L | 0 | |
| Fumarate reductase flavoprotein subunit | Contig 32 (179927–180597) | L | 0 | |
| Proteins involved in signalling | ||||
| Probable protease eep | Contig 38 (3317–4016) | M | 4 | |
| Serine/threonine-protein kinase PrkC | Contig 35 (43308–44307) | M | 1 | |
| Sensor histidine kinase | Contig 7 (797–1796) | M | 2 | |
| Pheromone cAD1 lipoprotein | Contig 34 (6564–7272) | L | 0 | |
| Pheromone binding protein | Contig 28 (61901–62900) | L | 0 | |
| Proteins involved in lipid biosynthesis | ||||
| Glycerophosphoryl diester phosphodiesterase family protein | Contig 8 (75630–76629) | M | 9 | |
| (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase 1 | Contig 43 (1042–1476) | S | 0 | |
| (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase 2 | Contig 18 (35776–36201) | S | 0 | |
| Proteins involved in other processes | ||||
| Regulatory protein pfoR, putative | Contig 11 (86464–86967) | M | 8 | |
| Transcriptional regulator, PSR protein | Contig 39 (33224–33935) | M | 1 | |
| Transcriptional regulator lytR | Contig 1 (102661–103572) | S | 0 | |
| Bacterial sugar transferase | Contig 30 (42555–43341) | M | 5 | |
| Proteins with no annotated function | ||||
| hypothetical protein EF2929 | Contig 27 (32738–33728) | M | 11 | |
| Putative uncharacterized protein | Contig 39 (151365–151832) | M | 1 | |
| Membrane protein | Contig 11 (20435–21434) | M | 12 | |
| hypothetical protein OG1RF_0150 [Enterococcus faecalis OG1RF] | Contig 30 (18903–19902) | M | 10 | |
| Membrane protein | Contig 30 (1279–1750) | M | 10 | |
| Putative uncharacterized protein | Contig 15 (106536–106931) | M | 3 | |
| YitT family protein | Contig 39 (20040–20923) | M | 6 | |
| Putative uncharacterized protein | Contig 27 (19139–19838) | M | 1 | |
| Putative uncharacterized protein | Contig 15 (54450–55181) | M | 3 | |
| Putative uncharacterized protein | Contig 18 (7102–7677) | M | 4 | |
| Putative uncharacterized protein | Contig 8 (194308–194714) | M | 1 | |
| conserved hypothetical protein [Enterococcus faecalis ATCC 29200] | Contig 32 (88786–89658) | M | 12 | |
| Phage infection protein | Contig 4 (65550–68213) | M | 6 | |
| PIN domain protein | Contig 11 (28478–29477) | M | 4 | |
| Cell wall surface anchor family protein | Contig 35 (105859–106858) | M | 1 | |
| Putative uncharacterized protein | Contig 30 (12932–13931) | S | 0 | |
| Putative uncharacterized protein | Contig 1 (61273–61944) | S | 0 | |
| Basic membrane protein family | Contig 10 (73934–74933) | L | 0 | |
| Basic membrane protein family | Contig 10 (75061–76060) | L | 0 | |
As the OG1X sequence is not publically available we verified the location of each protein in the closely related OG1RF strain (http://blast.hgsc.bcm.tmc.edu/blast.hgsc?organism=EfaecalisOG1).
Membrane (M), soluble (S), lipoprotein (L).
The number of predicted transmembrane helices for each protein was obtained by analyzing the amino acid sequence with SCAMPI (http://scampi.cbr.su.se/) and SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
Type of transporter: ABC, amino acid binding cassette; PTS, phosphoenolpyruvate:sugar phosphotransferase system; IT, ion transporter; MFS, major facilitator superfamily. Transporters were grouped according to the Transporter Classification Database (http://www.tcdb.org/superfamily.php) and numbered arbitrarily.
To approximate the depth-of-coverage we achieved, we downloaded the proteome of OG1RF [5] from the Pathosystems Resource Integration Centre (http://patricbrc.vbi.vt.edu/portal) and then generated a predicted membrane proteome using SCAMPI [13] (note that OG1RF contains essentially the same gene content as OG1X [10]). We then compared our list of experimentally identified membrane embedded proteins to the predicted membrane proteome. Using this measure, we estimate that we have experimentally identified ~10% of the membrane embedded proteome (~620 proteins). Retrospective analysis of replicate experiments (D, E and F in Figure 1A) indicated that the depth-of-coverage could be improved if narrower fractions from the anion exchange step were analyzed by BN-PAGE. This conclusion was based on the fact that most proteins were identified in the narrowest fraction and the fewest proteins from the largest fraction (61, 43, and 36 proteins in fractions F, D and E, respectively). The analysis also indicated that the experiment was reasonably reproducible, as fractions D and E gave 46% overlap (i.e. 25/54 proteins), even though they were not equal in size. Complete depth-of-coverage for bacterial membrane embedded proteomes is notoriously difficult to achieve [19–25]. Employing a single experimental approach usually identifies 10 – 20% of the predicted membrane embedded proteome (Supplementary Table 3). This can be improved using combinations of different approaches, however, no single study that we are aware of has exceeded 50% coverage of a membrane proteome.
To investigate protein functions in the membrane proteome, we examined the functional annotations of the identified proteins (taken from the homologous protein in the V583 strain [4] or the OG1RF strain [5]) (Supplementary Table 1). The majority of the identified proteins are predicted to be involved in transport processes (Figure 1C, Table 1) and include 13 ABC-type transporters, 6 PTS-type transporters, 2 IT-type transporters, and 2 MFS-type transporters (data not shown). The genome sequence of E. faecalis V583 contains a large number of transport systems compared to other bacteria [4], and it was therefore not surprising that we detected so many. Intriguingly, a large fraction of the proteins that we identified had no known function (Figure 1C, Table 1). The presence of these proteins indicates that there is biology in the membrane of E. faecalis that we do not yet understand. We also identified 17 proteins that are potentially of biomedical interest, as they have been implicated by other studies to play roles in biofilm formation (Supplementary Table 4) and virulence (Supplementary Table 5). Together with approaches that address soluble and surface proteomes, our platform will be useful for understanding proteomes in pathogenic and antibiotic resistant strains of E. faecalis.
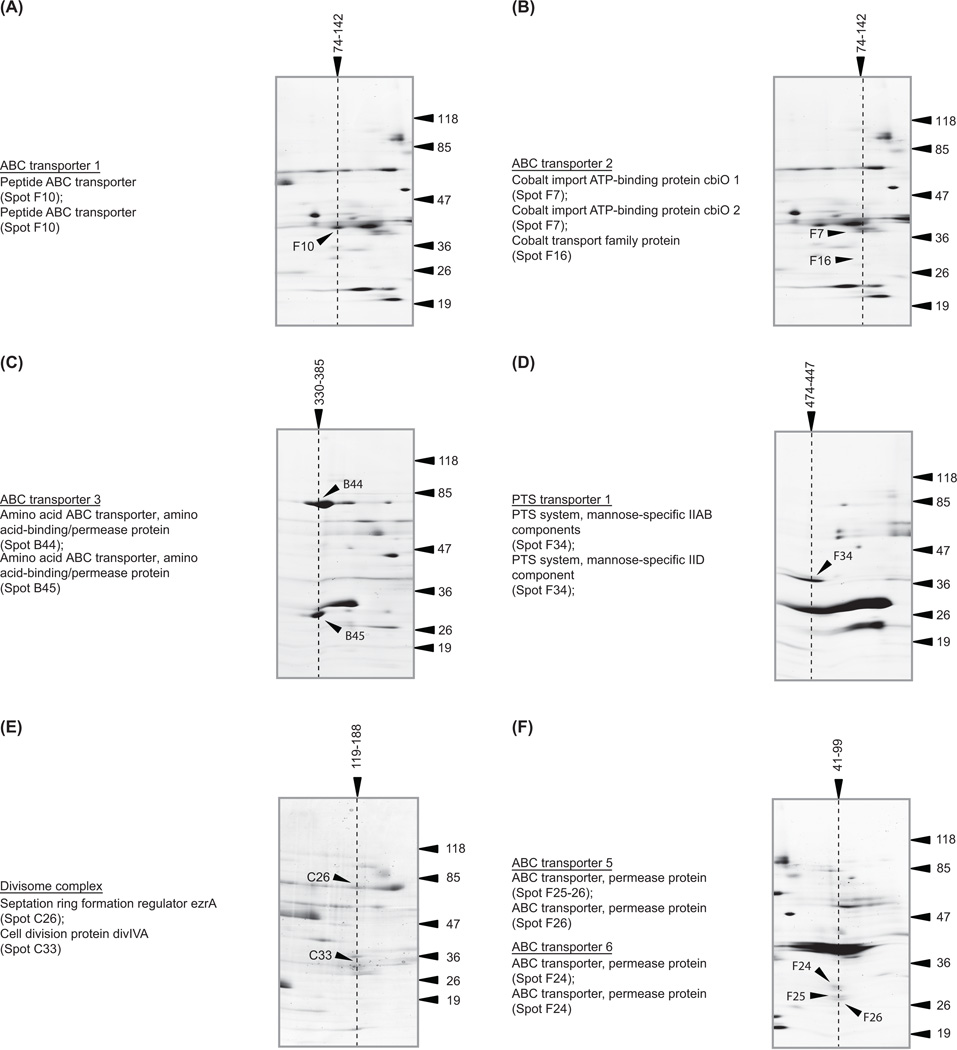
In addition to identification of single membrane constituents we were able to resolve membrane protein complexes. This was possible because we had utilized a mild detergent to extract the proteins from the membrane and native conditions to separate them during anion exchange chromatography and BN-PAGE. Membrane complexes resolved as a band in the non-denaturing BN-PAGE, and constituent proteins resolved as spots in a vertical channel in the denaturing BN-/SDS-PAGE (Figure 1A). For example, we could clearly recognize three ABC transporter complexes (Figure 2A – C), a mannose PTS transporter (Figure 2D), and the DivIVA-EzrA cell divison complex (Figure 2E). Unfortunately the composition of many complexes was difficult to assign from the gels without prior biological knowledge, as some vertical channels were clearly over-lapping (Figure 2F). The molecular mass of these complexes in the BN-PAGE indicated that the two complexes had the same electrophoretic mobility rather than being super-complexes. However, the data show that most proteins resolved at a higher molecular mass in the BN-PAGE than in the denaturing SDS-PAGE, indicating that they exist in a high molecular mass complex (Supplementary Figure 1). If no prior biological knowledge is available, then the vertical channels can simply be used to suggest a list of ‘putative’ interacting partners. As these data were generated from natively expressed proteins that are not tagged or over-expressed, it is complementary to other protein-protein interaction methodologies. We therefore believe that it will be useful to the scientific community for the prediction and/or confirmation of novel protein interactions at the membrane.
Figure 2. Examples of membrane protein complexes that were identified.
Cropped sections from the BN-/SDS-PAGE showing ‘vertical channels’ that contain proteins in known complexes. Only spots in a precise vertical channel can be part of the same complex. For the sake of clarity we have omitted annotations for proteins that are in close (but unrelated) channels. These spots were identified and are shown in Supplementary Figure 1. Soluble and membrane proteins were used to calibrate the protein complexes in the BN-PAGE (Supplementary Materials and Methods). As these two independent sets of proteins give significantly different calibration curves, we have calculated the molecular mass for each protein complex using both curves and reported a molecular mass range. All molecular mass markers are in kDa.
In conclusion, we have presented a first reference map of the membrane proteome of E. faecalis OG1X. This map contains proteins important for cell homeostasis, virulence, and antibiotic intervention. It also contains a large number of proteins with no known function, indicating that much is left to learn about the biology of this important sub-proteome. On a more limited scale, we were also able to provide insight into membrane protein complexes. Taken together, the information we have presented is a first step towards understanding how E. faecalis uses its membrane proteome to interact with the host and with the extracellular milieu.
Supplementary Material
Acknowledgements
We thank Scott Hultgren (Washington University in St. Louis) and Birgitta Henriques-Normark (Karolinska Institute) for technical assistance and support. This work was supported by grants from the Swedish Research Council and the NIH (R01 GM081827-01) to DOD.
Non-standard Abbreviations
- ABC
ATP binding cassette
- AEX
anion exchange
- BN-PAGE
blue native-polyacrylamide gel electrophoresis
- DDM
n-dodecyl-β-D-maltoside
- IT
ion transport
- MFS
major facilitator superfamily
- PTS
phosphoenolpyruvate-dependent transferase system
Footnotes
The mass spectrometry data set has been deposited into the PRIDE database (accession no. 17167, username: review18709, password: Kn2smN#B).
The authors declare no conflict of interests.
References
- 1.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 2.Tendolkar PM, Baghdayan AS, Shankar N. Pathogenic enterococci: new developments in the 21st century. Cell Mol Life Sci. 2003;60:2622–2636. doi: 10.1007/s00018-003-3138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hidron AI, Edwards JR, Patel J, Horan TC, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention. 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 4.Paulsen IT, Banerjei L, Myers GS, Nelson KE, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 5.Bourgogne A, Garsin DA, Qin X, Singh KV, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott JR, Barnett TC. Surface proteins of gram-positive bacteria and how they get there. Annu Rev Microbiol. 2006;60:397–423. doi: 10.1146/annurev.micro.60.080805.142256. [DOI] [PubMed] [Google Scholar]
- 7.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benachour A, Morin T, Hebert L, Budin-Verneuil A, et al. Identification of secreted and surface proteins from Enterococcus faecalis. Can J Microbiol. 2009;55:967–974. doi: 10.1139/w09-052. [DOI] [PubMed] [Google Scholar]
- 9.Bohle LA, Riaz T, Egge-Jacobsen W, Skaugen M, et al. Identification of surface proteins in Enterococcus faecalis V583. BMC Genomics. 2011;12:135. doi: 10.1186/1471-2164-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ike Y, Craig RA, White BA, Yagi Y, Clewell DB. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci U S A. 1983;80:5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddalo G, Stenberg Bruzell F, Gotzke H, Toddo S, et al. Systematic analysis of native membrane protein complexes in Escherichia coli. J Proteome Res. 2011;10(4):1848–1859. doi: 10.1021/pr101105c. [DOI] [PubMed] [Google Scholar]
- 12.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Bernsel A, Viklund H, Falk J, Lindahl E, et al. Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci U S A. 2008;105:7177–7181. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. Prediction of lipoprotein signal peptides in gram-positive bacteria with a Hidden Markov Model. J Proteome Res. 2008;7:5082–5093. doi: 10.1021/pr800162c. [DOI] [PubMed] [Google Scholar]
- 15.Bohle LA, Faergestad EM, Veiseth-Kent E, Steinmoen H, et al. Identification of proteins related to the stress response in Enterococcus faecalis V583 caused by bovine bile. Proteome Sci. 2010;8:37. doi: 10.1186/1477-5956-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giard JC, Laplace JM, Rince A, Pichereau V, et al. The stress proteome of Enterococcus faecalis. Electrophoresis. 2001;22:2947–2954. doi: 10.1002/1522-2683(200108)22:14<2947::AID-ELPS2947>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 17.Heim S, Lleo MM, Bonato B, Guzman CA, Canepari P. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis as determined by proteome analysis. J Bacteriol. 2002;184:6739–6745. doi: 10.1128/JB.184.23.6739-6745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, He X, Jiang Z, Wang J, et al. Proteomic analysis of the Enterococcus faecalis V583 strain and clinical isolate V309 under vancomycin treatment. J Proteome Res. 2010;9:1772–1785. doi: 10.1021/pr901216e. [DOI] [PubMed] [Google Scholar]
- 19.Bernsel A, Daley DO. Exploring the inner membrane proteome of Escherichia coli: which proteins are eluding detection and why? Trends Microbiol. 2009;17:444–449. doi: 10.1016/j.tim.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Eichacker LA, Granvogl B, Mirus O, Muller BC, et al. Hiding behind hydrophobicity. Transmembrane segments in mass spectrometry. J Biol Chem. 2004;279:50915–50922. doi: 10.1074/jbc.M405875200. [DOI] [PubMed] [Google Scholar]
- 21.Fischer F, Poetsch A. Protein cleavage strategies for an improved analysis of the membrane proteome. Proteome Sci. 2006;4:2. doi: 10.1186/1477-5956-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Stenberg F, Daley DO. Exploring membrane proteomes. In: Hagen v., editor. Proteomics Sample Preparation. Wiley-VCH; 2008. pp. 303–312. [Google Scholar]
- 24.Weiner JH, Li L. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim Biophys Acta. 2008;1778:1698–1713. doi: 10.1016/j.bbamem.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Griffin NM, Schnitzer JE. Overcoming key technological challenges in using mass spectrometry for mapping cell surfaces in tissues. Mol Cell Proteomics. 2011;10:R110 000935. doi: 10.1074/mcp.R110.000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.