Summary
ER stress leads to upregulation of multiple folding and quality control components, known as the unfolded protein response (UPR). Glucose Regulated Protein 78 (GRP78) (also known as binding immunoglobulin protein, BiP, and HSPA5) and GRP94 are often upregulated coordinately as part of this homeostatic response. Given that endoplasmic reticulum (ER) chaperones have distinct sets of clients, we asked how cells respond to ablation of individual chaperones. The cellular responses to silencing BiP, GRP94, HSP47, PDIA6 and OS-9, were distinct. When BiP was silenced, a widespread UPR was observed, but when GRP94 was either inhibited or depleted by RNA interference (RNAi), the expression of only some genes was induced, notably those encoding BiP and protein disulfide isomerase A6 (PDIA6). Silencing of HSP47 or OS-9 did not lead to any compensatory induction of other genes. The selective response to GRP94 depletion was distinct from a typical ER stress response, both because other UPR target genes were not affected and because the canonical UPR signaling branches were not activated. The response to silencing of GRP94 did not preclude further UPR induction when chemical stress was imposed. Importantly, re-expression of wild-type GRP94 in the silenced cells prevented the upregulation of BiP and PDIA6, whereas re-expression of an ATPase-deficient GRP94 mutant did not, indicating that cells monitor the activity state of GRP94. These findings suggest that cells are able to distinguish among folding resources and generate distinct responses.
Introduction
Folding of secreted and membrane proteins, their post-translational modifications and their quality control are performed by endoplasmic reticulum (ER) resident chaperones, enzymes and co-factors. When these processes are compromised by accumulation of misfolded substrates, a signaling mechanism initiates the stress response known as the unfolded protein response (UPR), which aims to restore ER homeostasis (Ron and Walter, 2007; Walter and Ron, 2011). The UPR is initiated not only by pathological circumstances, but also in physiological situations like differentiation of secretory cells, in preparation for an increased demand on the ER folding capacity (van Anken et al., 2003).
In metazoa, the UPR comprises three signaling branches emanating from the transmembrane transducers inositol-requiring enzyme 1 (IRE1), activated transcription factor 6 (ATF6) and protein kinase RNA-activated ER kinase (PERK) (Ron and Walter, 2007). The mode of function of these pathways has been elucidated mostly by using chemically induced ER stress, such as with tunicamycin, thapsigargin or dithiothreitol (DTT) (Ron and Walter, 2007; Walter and Ron, 2011). Other mechanistic insights have come through the expression in the ER of misfolded proteins as models for various protein conformation diseases (Ron, 2002). These substrates are ‘proteotoxic’ because they are thought to occupy folding resources that in turn leads to the UPR (Balch et al., 2008). We sought to explore a complementary approach – limiting individual folding components of the ER by RNAi in order to assess the consequences to the cell.
In canonical UPR, hundreds of ER genes are co-induced, including many components of the protein folding machinery (Kamauchi et al., 2005; Murray et al., 2004; Travers et al., 2000). Nonetheless, because the ER fulfils multiple additional functions, such as calcium homeostasis and lipid synthesis, different physiological conditions may require distinct outcomes, characterized by the upregulation of selective subsets of ER genes. Indeed, recent work in yeast shows that UPR signaling can cause differential target gene expression depending on the nature of the stress (Thibault et al., 2011).
Two of the most inducible ER proteins are glucose-regulated protein 94, GRP94 (gp96 or HSP90B1) and BiP (immunoglobulin binding protein or GRP78), which are hallmarks of both pathological and physiological UPR (Chang et al., 1989; Shiu et al., 1977; Wiest et al., 1990). BiP functions as the ‘first encounter’ chaperone of the secretory pathway and interacts with many newly synthesized secretory proteins (Ma and Hendershot, 2004). BiP is also a negative regulator of the UPR, through its association with IRE1, ATF6 and PERK (Ron and Walter, 2007): its depletion induces ER stress signaling through all three UPR transducers (Paton et al., 2006). In contrast, less is known about the identities of GRP94's clients and interacting proteins, although for the few known clients GRP94 is essential (Yang et al., 2007). At least in some folding pathways, GRP94 acts later than BiP (Melnick et al., 1992; Melnick et al., 1994; Muresan and Arvan, 1997). Also in contrast to BiP, GRP94 has not been found to bind directly to the ER stress transducers.
Even though the two chaperones display no obvious genetic redundancy with each other, Link et al. described compensatory regulation in C. elegans: loss of either GRP94 or BiP upregulated expression of the other and activated IRE1 (Kapulkin et al., 2005). GRP94-deficient murine cells also expressed more BiP and other ER proteins, but unlike C. elegans, they were less responsive to ER stress because the level of spliced X-box binding protein 1 (XBP1) was substantially reduced (Mao et al., 2010). These reports suggest a compensatory network among at least some of the ER chaperones, but the underlying molecular mechanisms remain unclear.
The present study was designed to ask how the ER responds to limitation of individual folding resources such as BiP or GRP94. We identified a novel transcriptional network that appears distinct from the canonical UPR, by which the ER monitors the activity state of GRP94 and responds to the perturbation of this chaperone's function. This response does not preclude responses to other stress conditions, suggesting that the ER is able to distinguish among various metabolic stresses and respond to each adaptively.
Results
Silencing GRP94 produces a response distinct from the UPR
To determine the consequences of depleting individual ER chaperones we used RNAi knockdown (KD) approach and monitored the expression of individual proteins via western blot analysis. An example of the primary data is shown in Fig. 1A and the data are summarized in the subsequent panels. The effects of RNAi KDs were compared to the effects of tunicamycin (Tun) or thapsigargin (TG) treatments as reference points, since it is well known that when cells are treated with these agents they respond by upregulating a large battery of ER chaperones, enzymes and components of the quality control system. We measured the changes in protein levels of eight UPR target proteins, representing a few different classes of luminal ER stress response genes. In response to Tun, seven of the eight are upregulated (Fig. 1B) and in response to TG, six of eight were upregulated (supplementary material Fig. S1), as expected from coordinate UPR regulation.
Fig. 1.
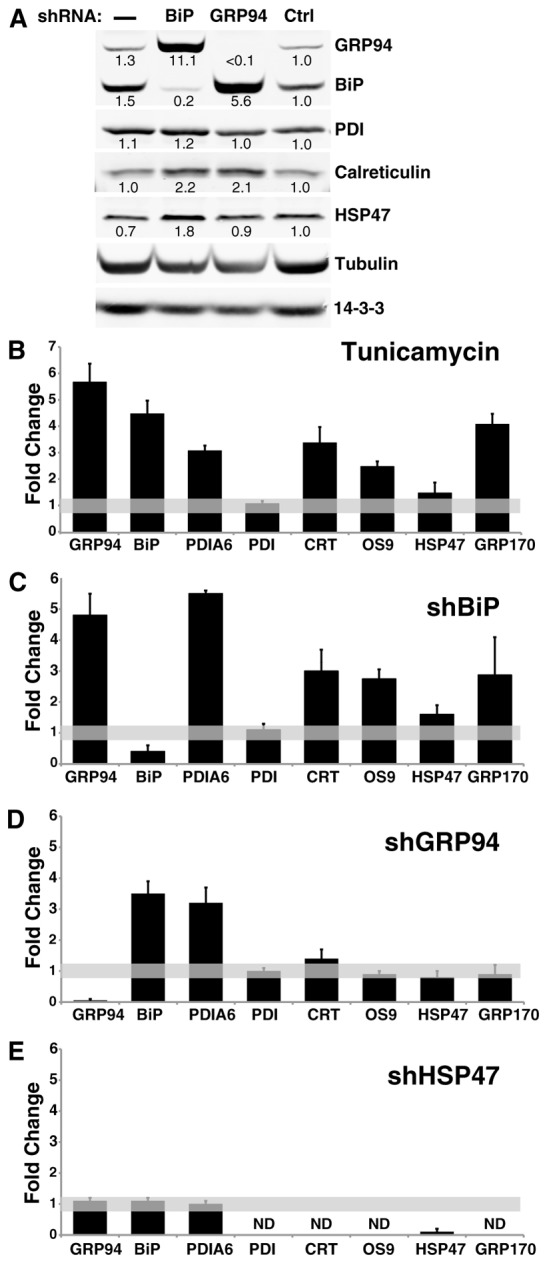
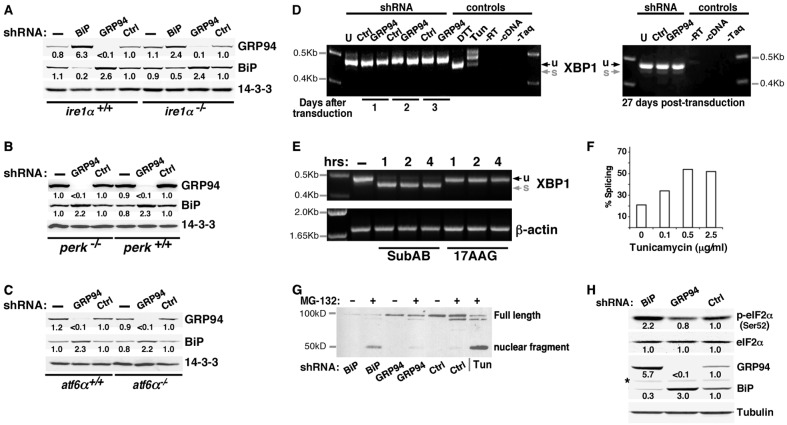
Silencing GRP94 produces a response distinct from a general UPR. (A) shRNA-mediated silencing of either BiP or GRP94 leads to upregulation of other ER components. In this representative experiment, murine 10T1/2 cells stably expressing shRNA against GRP94, BiP or an irrelevant shRNA (Ctrl) were lysed and analyzed by immunoblotting with antibodies to the proteins indicated on the right. Signals were quantified using a Li-Cor imager, because of its superior dynamic range. Levels of expression of the indicated proteins were determined by quantification of exposures at the linear range and normalized for protein load by comparison to the levels of tubulin and 14-3-3 in the same sample. The density of each band in the shCtrl lane is defined as 1.0. The relative fold-increase is indicated below each band. Similar results were obtained in various other cells, of either human of mouse origin, where the pattern was always consistent even if the amplitude of the effect differed. (B) Quantification of the response of the indicated eight ER proteins to tunicamycin treatment (2 µg/ml for 24 hours), as a typical chemical inducer of ER stress. Shown are means±s.d. of four independent experiments, performed and quantified as in A. (C) The protein levels of the same eight ER components as in B were determined by immunoblotting of lysates of cells stably expressing lentivirus shRNA against BiP. Shown are means±s.d. of four independent experiments. (D) The level of expression of the same protein set in cells silenced by shRNA against GRP94. Shown are means±s.d. of >4 independent experiments. (E) The level of expression of the same protein set in cells silenced by shRNA targeting HSP47. Shown are means±s.d. of three independent experiments. ND, not determined.
BiP could generally be depleted only up to 60%, but even this partial depletion was sufficient to upregulate a similar pattern of target proteins as that caused by the chemical stress inducers (Fig. 1A–C). Thus, partial silencing of BiP is sufficient to cause UPR, as also noted by others (Ye et al., 2010). A more complete ablation of BiP is provided by using subtilase AB (subAB), a bacterial endopeptidase which cleaves BiP specifically at a di-leucine motif (L416–L417), rendering it nonfunctional (Paton et al., 2006). SubAB treatment depleted BiP acutely and completely, causing a wide spread ER stress response (Fig. 4E and Paton et al., 2006).
Fig. 4.
The response to GRP94 ablation is not mediated by UPR signaling. (A–C) The GRP94-specific response is observed in MEFs deficient for each of the three UPR transducers. (A) GRP94 and BiP were silenced individually, using lentiviral infections, in IRE1-deficient MEFs. Cell lysates were analyzed by western blotting. Relative band intensities, normalized to the loading control (14-3-3) are indicated. (B) Response of PERK-deficient MEFs to lentiviral-mediated silencing of GRP94. (C) Response of ATF6α-deficient MEFs to lentiviral-mediated silencing of GRP94. (D) XBP1 splicing is not triggered in GRP94-depleted cells. Total RNA was extracted from 293T cells that were either untransduced (U), transduced with a lentivirus with shRNA to GRP94 (GRP94) or with an irrelevant shRNA (Ctrl). Cells were harvested at the indicated times after viral transduction and XBP1 was amplified by RT-PCR. The unspliced (u, 473 bp) and spliced (s, 447 bp) forms of XBP1 differ by 26 nucleotides. Only the unspliced form is seen in all samples, whereas the positive controls for cells mounting active UPR show either efficient splicing (DTT treatment, 1 µM for 6 hours) or partial splicing (Tun, 10 µg/ml for 8 hours). (E) Inhibition of GRP94 does not trigger XBP1 splicing, whereas ablation of BiP does. XBP1 splicing in 10T1/2 cells that were treated with 100 ng/ml subAB, a toxin that cleaves BiP selectively and induces global UPR, or exposed to 10 µM 17AAG overnight. Cells were harvested at the indicated time points. RNA extracts were assayed as in panel D, with β-actin amplification serving as a loading control. (F) Sensitivity of detection of XBP1 splicing. 293T cells were subjected to the indicated doses of tunicamycin overnight and splicing was detected as shown in Fig. 4D,E. The percentage splicing is the ratio of the spliced band over the total amplicon [s/(u + s)] in each gel lane. Even though there is a higher level of basal activity in our 3T3 cells compared with that of the 293T cells or MEFs, splicing in response to as little as 0.1 µg/ml tunicamcyin was already detectable. Doses up to 2.0 µg/ml were used in the other figures. (G) ATF6 endo-proteolysis is induced in BiP-, but not in GRP94-depleted cells. Nuclear extracts of 293T cells stably co-expressing HA-tagged ATF6 with shRNA against BiP, GRP94 or Ctrl genes were analyzed by immunoblotting. The nuclear ATF6 fragment was detected with anti-HA antibody. Consistent with previous results (Haze et al., 1999), the nuclear ATF6 fragment was detected when cells were treated with tunicamycin (Tun; 2 µg/ml for 3 hours) to induce ER stress. The same fragment is induced in BiP-depleted cells, whereas the pattern of GRP94-depleted is similar to the control cells. Enhanced levels of the ATF6 fragment were detected when cells were treated with MG132 (10 µM for 3 hours) to prevent degradation. A longer exposure is presented in supplementary material Fig. S4C. (H) Depletion of BiP, but not of GRP94, triggers PERK activity. BiP, GRP94 or an irrelevant gene (Ctrl) were silenced with the appropriate shRNA-encoding lentivirus and then assayed by western blotting with the indicated antibodies. The anti-phospho-eIF2a antibody measures the phosphorylation site most indicative of PERK activation. The asterisk indicates a non-specific band. The data shown in all panels of this figure are representative of at least three independent experiments.
In contrast to the broad consequences of BiP silencing, when other chaperones are depleted by RNAi the cellular response is much more selective. For example, silencing GRP94 expression by lentiviral infection of short hairpin RNAs (shRNAs) triggered overexpression of BiP and of another KDEL-containing protein with an apparent molecular mass of 50 kDa (Fig. 1D), which we identified as PDIA6 (supplementary material Fig. S2). In contrast, calreticulin, HSP47, OS-9 and GRP170, other ER proteins that have been reported to be physically or functionally related to GRP94 (Christianson et al., 2008; Meunier et al., 2002), are marginally or not at all affected by silencing of GRP94 (Fig. 1D). These genes are not responsive to GRP94 depletion even though they are upregulated by chemical stimulation of ER stress (Fig. 1B) or in some cases by complete BiP ablation by subAB (data not shown). Particularly informative is the lack of response by GRP170 to GRP94 depletion, since GRP170 is highly induced by chemical ER stress (Fig. 1A). This underscores the selective nature of the response and argues against the possibility that all highly responsive genes are induced by GRP94 depletion. Therefore, the response to the silencing of GRP94 is different from the response to chemically induced ER stress. It is termed ‘the GRP94-specific response’ throughout this work.
The GRP94-specific response is unique also because it is not the common response to the depletion of any resident ER protein. Silencing of HSP47, a collagen chaperone in fibroblasts, or of OS-9, a component of ER-associated degradation, leads to little if any upregulation of the proteins in our test set (Fig. 1E and data not shown), and silencing of PDIA6 induces GRP94 and BiP only marginally (supplementary material Fig. S1B, Fig. S2D). Thus, the consequences of depleting individual ER chaperones are distinct rather than common, from a broad UPR through selective upregulation to very little response. On the basis of these data, we conclude that cells are able to distinguish between the ablation of particular chaperones in the ER, generating distinct outcomes.
The GRP94-specific response is also obtained by means other than RNAi
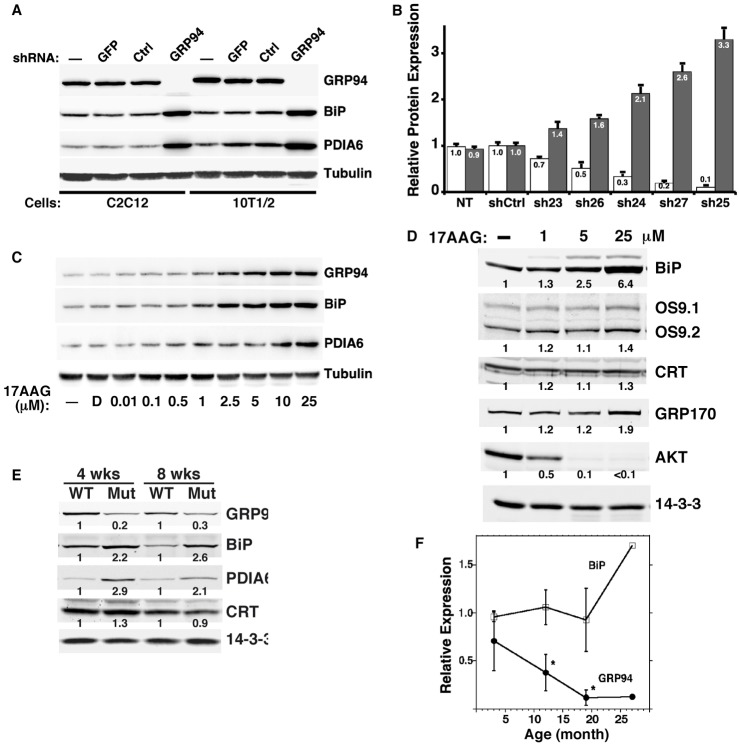
The upregulation of BiP and PDIA6 is not a peculiarity of the method of lentiviral delivery and is not due to the shRNA method of silencing, since (1) infection with a lentivirus expressing a non-targeting sequence (shRNA-Ctrl) or green fluorescence protein (GFP) does not induce the two genes (Fig. 2A); (2) BiP and PDIA6 are upregulated also when the effective shRNAs are introduced by transfection (data not shown); (3) the GRP94-specific response is observed in multiple cell lines from different species and tissue types (two shown in Fig. 2A); and (4) the extent of BiP induction is inversely proportional to the level of GRP94 expression [we used five shRNA sequences, with different knock-down efficiencies and observed that the lesser the expression of GRP94, the higher the induction of BiP (Fig. 2B)].
Fig. 2.
The GRP94-specific response occurs when GRP94 is ablated by several means in vitro and in vivo. (A) The GRP94-specific RNAi response is observed in several cell lines. Murine myoblasts (C2C12) and embryonic fibroblasts (10T1/2) were infected with a lentivirus encoding shRNA targeting GRP94 or with a control, non-targeting, shRNA (Ctrl) or a GFP-expressing lentivirus (GFP). Lysates were analyzed by immunoblotting with anti-KDEL antibody to detect GRP94, BiP and PDIA6 simultaneously; α-tubulin served as the loading control. (B) BiP induction is proportional to the degree of silencing of GRP94. GRP94 expression in C2C12 was knocked down to different extents with the indicated shRNAs. Levels of expression of GRP94 (white) and BiP (gray) were determined by densitometry of immunoblots. The means±s.d. of three independent experiments are shown. The numbers indicate the relative expression of BiP or GRP94 with each of the indicated shRNAs. (C) Pharmacological inhibition of GRP94 also induces BiP and PDIA6 expression. 10T1/2 cells were treated with 17AAG at the indicated concentrations for 24 hours [D, 0.1% (v/v) DMSO]. Lysates were analyzed by immunoblotting. Data are from a representative experiment out of three. (D) Pharmacological inhibition of GRP94 does not induce upregulation of calreticulin (CRT) or either OS-9 isoform, and only marginally induces GRP170 expression (in contrast to tunicamycin treatment, see Fig. 1B). 10T1/2 cells were treated with 17AAG at the indicated concentrations for 24 hours. Lysates were analyzed as in C. Note that unlike the shRNA treatment, 17AAG causes destabilization of AKT, indicating that the compound affects cytosolic HSP90 as well as GRP94. The numbers below the bands are the relative protein levels, determined by densitometry and normalized to the 14-3-3 loading control. (E) BiP and PDIA6, but not calreticulin (CRT) are induced in GRP94 knocked-out muscle. Gastrocnemius muscles were obtained from transgenic floxed-GRP94 mice expressing MCK-Cre recombinase (Mut) or from littermate controls (WT). 40 µg of muscle protein extracts were analyzed by immunoblotting as in A. 14-3-3 served as loading control. Note that the comparisons are normalized within each pair with the value of each protein in the WT mouse defined as 1.0. (F) Selective decline in GRP94 expression occurs during aging. WT (C57Bl/6) mice were killed and dissected at the indicated ages. Protein extracts from dissected tibialis anterior muscles were subjected to immunoblotting with anti-KDEL antibody to determine simultaneously the levels of GRP94 (black circles) and BiP (white squares). Means of relative expression levels (±s.d.) of 2–4 animals are shown, except the 25-month-timepoint, which is based on only one mouse. *timepoints when GRP94 expression values are significantly different from those of BiP in the same samples; P<0.002; two-tailed Student's t-test.
Silencing of GRP94 expression in fibroblastic cell lines is long-lived; it persists for months, and when it subsides, BiP expression returns to the basal level of expression (data not shown). This is in contrast to the tolerance of lymphoid or myogenic cells, where silencing of GRP94 expression is shorter-lived (Kropp et al., 2010; Ostrovsky et al., 2010)
To ask whether the induction is responsive to levels of GRP94 transcripts, protein or activity, we took advantage of 17-N-allylamino-17-demethoxygeldanamycin (17AAG), which inhibits the ATPase activity of GRP94 (and HSP90). The treatment with 17AAG induces a response comparable to the RNAi, in dose-dependent fashion: BiP and PDIA6 were induced but GRP170 expression increased only slightly and OS-9 expression did not change (Fig. 2C,D). Interestingly, the level of expression of GRP94 itself increases, suggesting that cells respond to limit of the activity, rather than of the amount of GRP94 mRNA or protein.
Although the GRP94-specific response was observed consistently in a variety of cell lines, we asked if it was also evident in vivo. To test this, we examined a conditional GRP94 knockout mouse, where GRP94 is deleted postnatally in skeletal muscle, due to Cre recombinase driven by the skeletal muscle-specific promoter of muscle creatine kinase (Barton et al., manuscript submitted). As shown in Fig. 2E, GRP94-deleted muscle displays similar induction of BiP and PDIA6 in murine skeletal muscle. In contrast, calreticulin expression remains steady. Importantly, distinct regulation of GRP94 and BiP is also observed in normal physiology: during aging, GRP94 expression, but not BiP expression in skeletal muscles declines (Fig. 2F). The selective reduction in GRP94 during aging is correlated with the reduced expression of insulin-like growth factors in aging muscle (Musarò et al., 2001), and are thought to be contributing factors to aging-related sarcopenia (Bartke, 2009). We showed that the activity of GRP94 is essential for production of IGF (Barton et al., 2012; Ostrovsky et al., 2010; Wanderling et al., 2007).
Another example of divergent chaperone regulation is provided by the androgen-responsive prostate carcinoma cell line LnCap (supplementary material Fig. S3). When treated with the androgen analog 5α-dehydroepiandrosterone (DHEA), there is a selective increase in GRP94 expression, without a concomitant increase in BiP in these cells. This increase appears to be cell-type specific, since PC-3, another prostate cancer cell line that is androgen-insensitive, as well as the embryonic kidney fibroblast 293T, do not respond in this fashion. These data indicate that GRP94 expression can be increased considerably even without a general UPR.
The GRP94-specific response is transcriptional
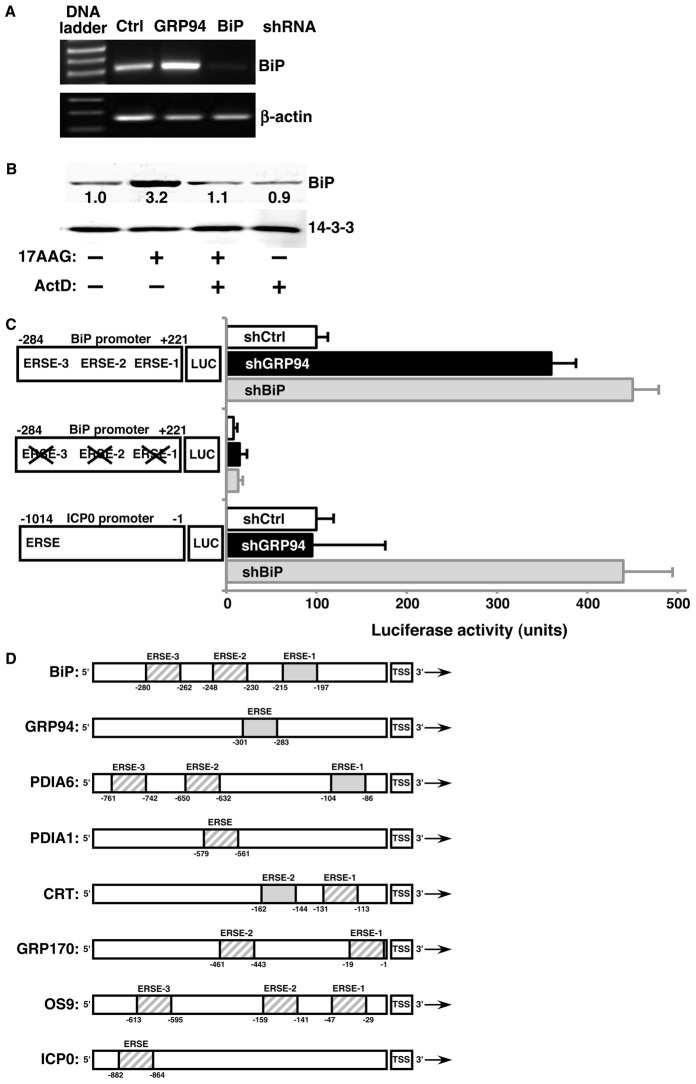
Besides an increase in steady state levels of BiP protein in GRP94-depleted cells, BiP mRNA levels are also elevated. (Fig. 3A). Moreover, the induction of BiP is abolished when cells are pre-treated with actinomycin D, to arrest transcription (Fig. 3B). In this experiment, we used inhibition of activity rather than RNAi, because under this condition the upregulation is fast and the time course of the experiment is compatible with cell viability. Yet a third line of evidence that the GRP94 specific response is transcriptional is provided by a luciferase reporter analysis. Using a construct where luciferase expression is driven by regulatory regions of the BiP gene, compared to controls, there is ∼3.5-fold induction of luciferase in 293T cells in which GRP94 was knocked down (Fig. 3C). The three ER stress elements (ERSEs) (Yoshida et al., 1998) of the BiP promoter are essential for the response to GRP94 silencing, since luciferase activity is abolished when all three ERSEs are mutated (Fig. 3C). Thus, ablation of GRP94 initiates a transcription response at the BiP promoter. While we did not check the transcription of PDIA6 explicitly, its promoter has the same organization of three ERSE elements as the BiP promoter and ERSE1 shows excellent correspondence to the canonical sequence (Fig. 3D; supplementary material Fig. S6). Not all ER stress-responsive promoters are induced by silencing of GRP94. One example is the promoter of PDIA1, which has only one ERSE and its sequence diverges from the canonical sequence (supplementary material Fig. S6). Furthermore, we tested another reporter, where luciferase is driven by the promoter of the Herpes Simplex Virus ICP0 gene (Burnett et al., 2012). This reporter is responsive to chemically induced ER stress (Burnett et al., 2012) and is also responsive to silencing of BiP (Fig. 3C). However, the ICP0-luciferase reporter is not induced by silencing of GRP94, supporting the conclusion that depletion of the two major ER chaperones is not equivalent and the GRP94-specific transcriptional response is mechanistically distinct from UPR.
Fig. 3.
The GRP94-specific response is transcriptional. (A) Total RNA derived from 293T cells stably expressing shRNAs against GRP94, BiP or an irrelevant shRNA (Ctrl) was subjected to RT-PCR. Amplicons of BiP and β-actin were then resolved on agarose gels. The gel shown is representative of two independent experiments. (B) 10T1/2 cells were treated with either 5 µM 17AAG, 0.5 µg/ml actinomycin D (ActD) or both. After 24 hours, cells were lysed and the indicated proteins resolved and quantified by immunoblotting. (C) 293T cells in which an irrelevant gene (white bars, shCtrl), GRP94 (black bars), or BiP (gray bars) were silenced, were transiently transfected with reporter plasmids carrying luciferase driven by: the minimal promoter of BiP; a mutant version of the BiP promoter where the ERSEs were disrupted; or the minimal promoter of HSV ICP0, a UPR-responsive viral gene. All cells were also co-transfected with a plasmid carrying Renilla luciferase. Luciferase activity was assayed 24 hours post-transfection and relative luciferase activity was calculated as a percentage after normalization against Renilla values. Results are mean±s.e.m.; n = 6 transfections for the wild-type and mutant BiP promoters; n = 2 for the ICP0 promoter. Left: schematic representation of intact or mutated promoter-luciferase constructs. Numbers indicate the nucleotide position from the transcription start site. (D) Schematic representation of the promoters of select murine UPR targets, as well as HSV ICP0 genes. The ERSEs in each promoter, are as defined previously (Yoshida et al., 1998), and are shown as by shaded boxes. The solid gray boxes are identical in sequence, as shown in supplementary material Fig. S6. The numbers below the ERSEs define the bp position relative to the transcription start site (TSS).
The response to GRP94 ablation is not mediated by UPR signaling
Transcriptional responses of ER quality control genes are usually initiated by the UPR signaling pathways, through activation of IRE1, PERK or ATF6. Therefore, we investigated whether depletion of GRP94 triggers these three transducers, using two approaches: (1) assaying the response to GRP94 deletion in cells where the signal pathways are genetically ablated (loss-of-function approach) and (2) measuring the activity of each transducer in GRP94-ablated cells.
The GRP94-specific response does not require IRE1, because it occurs in IRE1-deficient mouse embryonic fibroblasts (MEF) just as it does in wild-type MEFs (Fig. 4A). Similarly, the GRP94-specific response is normal in PERK-deficient and in ATF6α-deficient MEF (Fig. 4B,C). Therefore, none of the three transducers is needed individually for the response.
To test the possibility that in the absence of one transducer, signaling could occur via another UPR branch, we tested whether the main substrates mediating the signaling are activated. No splicing of XBP1 was observed at one, two or three days after infection with shRNA-GRP94-encoding virus, during the time frame needed to silence GRP94, and also not after 27 days, long after silencing was established (Fig. 4D). In contrast, tunicamycin and DTT treatments induce XBP1 splicing within a few hours, as expected, serving as positive controls for the assay. Since RNAi silencing requires days, we asked if there was an earlier XBP1 splicing event when GRP94 was inhibited by 17AAG, under conditions that induce BiP upregulation (Fig. 3). As shown in Fig. 4E, there was no detectable XBP1 splicing within 4 hours of 17AAG treatment. In contrast, efficient XBP1 splicing was observed already at the earliest time point of BiP ablation by subAB (Fig. 4E), which is known to induce UPR. The inability to detect XBP1 splicing was not because of low sensitivity of detection; as shown in Fig. 4F, splicing is easily detectable in our hands even at a dose of 100 ng/ml tunicamycin, 10-20×lower than the dose commonly used to induce UPR. Finally, the GRP94-specific response was intact in XBP1-deficient MEF (supplementary material Fig. S4A), showing that it is independent of either IRE1 or XBP1.
A second well-studied branch of the UPR machinery, ATF6, is activated by relocation to the Golgi complex, where proteolytic cleavage frees the N-terminal domain to become a transcription factor (Haze et al., 1999). We tested the activation of ATF6 by using 293T cells stably expressing an HA-tagged version of the protein (Wang et al., 2000). As shown in Fig. 4G, in GRP94-silenced cells, the basal level of the p50 active fragment of ATF6 is equal to that seen in control cells and there is no detectable accumulation of active ATF6. Note that the p50 fragment is unstable and is best detected when proteasome degradation is inhibited by MG-132 (Fig. 4G). In contrast, both tunicamycin treatment and BiP depletion are associated with accumulation of p50-ATF6.
The main substrate of the third UPR sensor, PERK, is the translation initiation factor 2 alpha (eIF2α) (Harding et al., 2000). Once phosphorylated, eIF2α activates the transcription factor ATF4 that in turn mediates the expression of ER proteins. Therefore, we assessed the phosphorylation of eIF2α in GRP94-silenced cells (Fig. 4H). Phospho-eIF2α levels do not appear significantly different in GRP94-depleted cells; in contrast, phosphorylation of eIF2α is induced by depletion of BiP (Fig. 4H and see also Wolfson et al., 2008). Further evidence against the involvement of PERK is that the GRP94-specific response occurs in ATF4-deficient MEF (supplementary material Fig. S4B).
On the basis of these combined data, we conclude that the GRP94-specific response does not cause activation of either IRE1, PERK or ATF6, while depletion of BiP by the same methods does, supporting that the GRP94-specific response is not a canonical UPR, but rather is mediated by a different mechanism.
The responsiveness to ER stress is not diminished by ablation of GRP94
If the GRP94-specific response involves upregulation of two proteins but not the activation of UPR sensors, is the UPR still functional under these conditions? To address this question we compared the responses to tunicamycin or thapsigargin of cells that are either GRP94-deficient or -sufficient.
In terms of viability, GRP94-deficient cells survived in increasing concentrations of Tun or TG less well than GRP94-sufficient cells, but the differences were not dramatic (Fig. 5A,B) when compared to the effect of BiP-deficient cells (see figure 5 in Morinaga et al., 2008) or to the GRP94-null ES cells (see figure 6 in Biswas et al., 2007). These observations indicate that despite the ubiquitous upregulation of GRP94 under ER stress conditions, this chaperone contributes to, but is not essential for coping with ER stress. A different conclusion was reached by Biswas and colleagues, and Ostrovsky and co-workers (Biswas et al., 2007; Ostrovsky et al., 2009), showing that GRP94 knockout (KO) ES cells are hypersensitive to stress, suggesting a pro-survival role for GRP94. The difference is likely due to the upregulation of BiP and PDIA6 seen in the KD cells but not in grp94−/− ES cells, arguing that the upregulation of the two compensates for some function of GRP94. Long-term loss of GRP94 function leads to adaptation at a cost of increased sensitivity to ER stress.
Fig. 5.
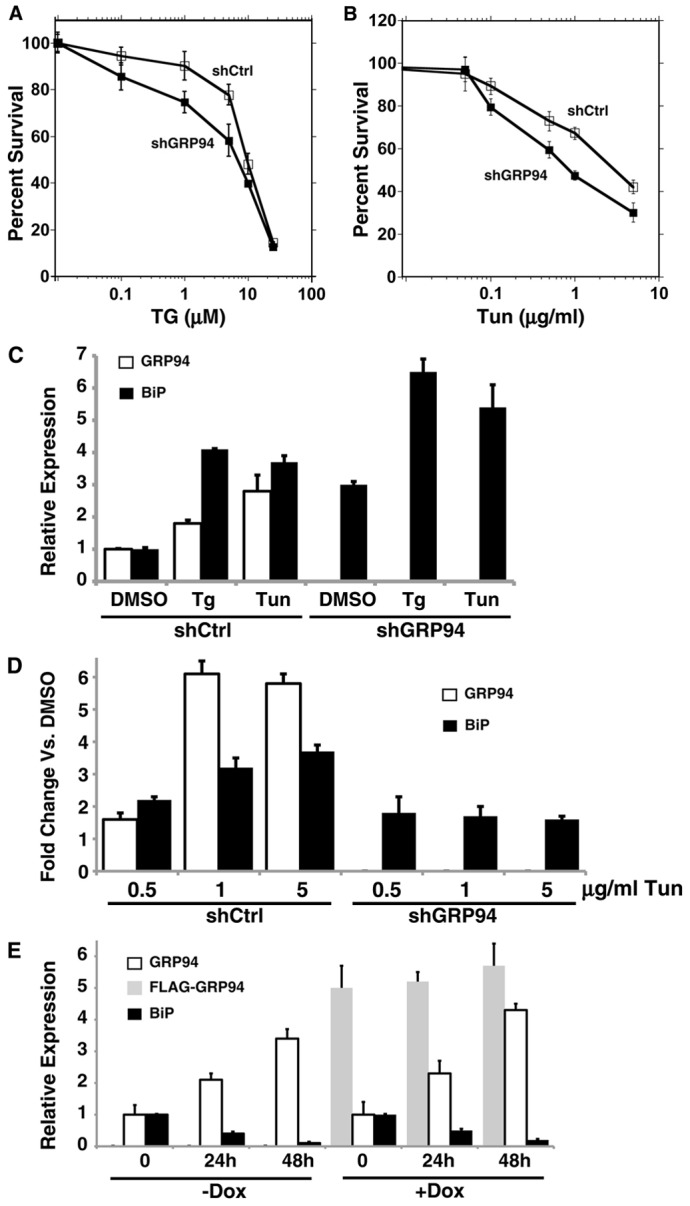
The responsiveness to ER stress is not diminished by ablation of GRP94. (A,B) HeLa cells stably expressing shRNA against GRP94 (shGRP94, black squares) or shRNA-Ctrl (white squares) were exposed to various doses of thapsigargin (TG) (A) or tunicamycin (Tun) (B). After 48 hours, cell viability was assayed by XTT and plotted as percentage relative to DMSO control. Results are the average ± s.d. of three (TG) and two (Tun) independent experiments performed in quadruplicate. (C) 293T cells stably expressing shRNA-CTRL or against GRP94 were treated with 10 µM TG, 10 µg/ml Tun or vehicle control (DMSO) for 18 hours. The protein levels were quantified by densitometry of immunoblots probed with anti-KDEL antibody to detect BiP and GRP94. The levels of expression of GRP94 (white) and BiP (black) were normalized against the DMSO levels and plotted in the bar graph. Values are means+s.d. of three experiments. (D) BiP response to titration of Tun doses. GRP94-silenced or control 293T cells were treated with Tun at the indicated concentrations for 18 hours. Cell lysates were analyzed as in C. The response values were renormalized to the response seen in each of cells without Tun, to separate the response to Tun from the inherent upregulation of BiP upon GRP94 KD. Note that BiP induction at the lower drug concentration is similar in the two cell lines, whereas they differ at higher concentrations of Tun. White bars, fold changes of GRP94 expression versus no Tun. Black bars, fold changes of BiP expression versus no Tun. Values are means+s.d. of three experiments. (E) CHO-tet cells were induced with 50 ng/ml doxycycline to express a Flag-tagged wild-type GRP94 cDNA. After 2 days, doxycycline or untreated cells were exposed to 10 ng/ml subAB to trigger ER stress. At the indicated time points after subAB addition, protein extracts were collected and analyzed as in A. White, level of GPR94; black, BiP; gray, FLAG–GRP94. Dox, doxycycline. The means±s.d. of four independent experiments are shown.
Fig. 6.
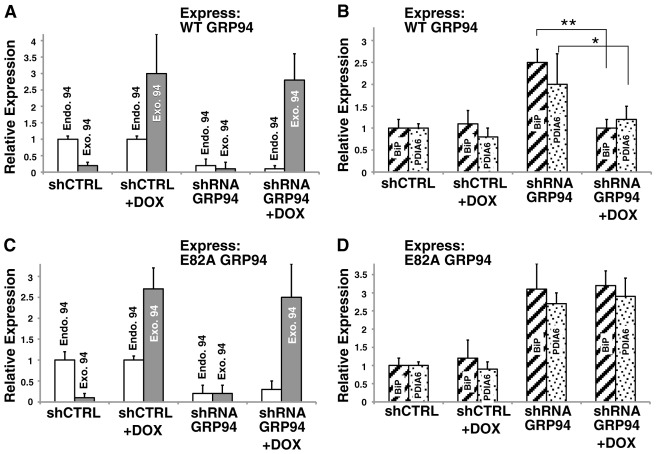
Cells monitor the level of active GRP94 in the ER. CHO-tet cells were induced with 50 ng/ml doxycycline to express Flag-tagged wild-type (WT) (A,B) or E82A (C,D) GRP94 cDNA, which is resistant to shRNA. Cells were then infected with shRNA against GRP94 or CTRL lentivirus and selected in presence of 2 mg/ml puromycin, to silence the endogenous GRP94. After 4 days, cells were harvested and subjected to immunoblotting to determine the level of expression of endogenous GRP94, Flag–GRP94, BiP and PDIA6. A and C show the level of expression of endogenous GRP94 (endo. 94, white bars) and of exogenous Flag–GRP94 (exo. 94, gray shaded bars) with and without doxycyclin (DOX). B and D show the level of BiP (striped bars) and of PDIA6 (spotted bars) before and after replacement of GRP94 expression. The means±s.d. of four independent experiments are shown. *P≤0.05; **P≤0.01.
As a second readout for UPR responsiveness we measured the extent of BiP induction by Tun or TG stress. As shown in Fig. 5C, GRP94-silenced cells can overexpress BiP when challenged with Tun or TG beyond the level of the GRP94-specific response, demonstrating that the stable induction of BiP when GRP94 is silenced is not maximal, and further increase of BiP is possible. GRP94-silenced cells responded by increasing the level of BiP in a dose-dependent fashion, similarly to the control cells, but BiP induction appeared saturated at lower concentration of Tun in comparison to the control cells (Fig. 5D). Interestingly, GRP94-silenced cells responded with similar amplitude to the control cells at the lowest concentration of Tun, indicating that the trigger is not compromised by the lack of GRP94. A third readout for UPR responsiveness was the XBP1-Venus reporter, where YFP fluorescence is induced by splicing of the upstream XBP1 sequence (Iwawaki and Akai, 2006). While GRP94-silenced cells displayed only background reporter levels, when treated with Tun, the reporter in these cells was activated (Eletto and Argon, data not shown). Together, these readouts show that the GRP94-specific response does not obviate the ability of cells to mount the UPR transcriptional response.
If depletion of GRP94 does not impair the ER stress response and since GRP94 is normally induced under such stress conditions, we also tested the effect of overexpressing GRP94 on the ER stress response. Taking advantage of doxycycline-inducible CHO cells, we expressed a controlled amount of Flag-tagged GRP94 in cells that were simultaneously treated with subAB (the time course of BiP ablation by this method is compatible with the time course of doxycycline induction). Depleting cells of BiP resulted in 3–4-fold induction of the endogenous GRP94, even in cells that already overexpressed Flag-tagged GRP94 (Fig. 5E). This indicates that the ER has a selective mechanism of sensing levels of BiP that fails to monitor the abundance of GRP94. Surprisingly, the overexpression of GRP94 is not counter-balanced by a reduction in the level of BiP, as mirrored by the loss-of-function experiment. Altogether, these results suggest that: (a) the relative abundance of GRP94 is monitored only below a minimal threshold; (b) cells employ a distinct mechanism to sense the relative abundance of GRP94; (c) although GPR94 is one of the major UPR targets, it is not required for mounting the response to chemical stresses.
Cells monitor the activity, not the protein level of GRP94 in the ER
Since RNAi was not the only treatment that induced BiP and PDIA6 and chemical inhibition led to the same response, we asked if cells sense the expression of GRP94 or its activity. To this end, we determined whether WT GRP94 or an ATPase-deficient (E82A) mutant can substitute for the endogenous protein when the latter is silenced in doxycycline-inducible CHO cells. The exogenous GRP94 genes were constructed to be shRNA-resistant (supplementary material Fig. S5). When an active version of GRP94 (Flag-tagged WT GRP94), replaced the silenced endogenous GRP94, the induction of BiP and PDIA6 was abolished. On the other hand, expression of E82A GRP94 failed to prevent the induction of the two genes (Fig. 6). Together with the effect of geldanamycin, these results strongly suggest that the mechanism that monitors the amount of GRP94 in the ER is dependent on the activity of the chaperone. Since GRP94 activity is much more client-selective than BiP activity, loss of this chaperone may necessitate a more restricted and mechanistically distinct response than UPR.
Discussion
The present work shows that limitation of folding resources in the ER has distinct outcomes, depending on which chaperone or enzyme activity is limited. When GRP94 is either inhibited or depleted by RNAi, a selective transcriptional response and de novo synthesis are initiated, resulting in upregulation of BiP and PDIA6. This response is different from the UPR that is induced by loss of BiP and is characterized by several functional and mechanistic features. Silencing of PDIA6, HSP74 or OS-9 does not cause compensatory protein upregulation, while depleting BiP causes a more global UPR (Wolfson et al., 2008; Ye et al., 2010 and this work). Such distinct ER stress responses illustrate the adaptability of the ER to various types of stress.
The response to GRP94 ablation is much more selective than standard UPR and does not include many UPR target genes, even those that are known to interact with GRP94 (such as OS-9, Christianson et al., 2008). Among all the genes we tested, only BiP and PDIA6 are very responsive to ablation of GRP94. This contrasts with the typical ER stress response, which is characterized by transcriptional activation of hundreds of ER proteins (Kamauchi et al., 2005; Murray et al., 2004). Consistent with this selectivity, not every UPR target promoter is activated by silencing of GRP94, even if it contains ER stress responsive elements. The second distinguishing feature of the transcriptional response to silencing of GRP94 is that it does not rely on either the IRE1, PERK or ATF6 pathways of ER stress signaling. At no time after inhibition or depletion of GRP94 was there activation of the proximal mediators of each of the UPR pathways – spliced XBP-1, phospho-eIF2α or cleaved ATF6. Furthermore, the transcriptional response to loss of GRP94 still occurs in MEFs deficient for IRE1, XBP-1, PERK, ATF4 or ATF6α.
The use of two luciferase reporter constructs show that the GRP94-specific transcriptional response is mediated by some promoter elements, and not only by the ERSE cis elements that are normally activated by ER stress signaling. We notice that there are genes that are strong responders to canonical ER stress (e.g. GRP170), but are not responsive to GRP94 depletion in spite of the presence of one or more ERSEs. Furthermore, the number of predicted ERSE does not correlate with the amplitude of either the GRP94-specific response or the general UPR.
We speculate that there is a different transcription factor, perhaps analogous to OASIS (Kondo et al., 2005), which binds to the promoters and mediates the activation of BiP and PDIA6. While the functional ERSEs in BiP and GRP94 promoters have been extensively studied (Yoshida et al., 1998), little is known about the regulation of PDIA6. It is regulated during ER stress by XBP1 (Lee et al., 2003; and our data) and we have found that the PDIA6 promoter has a perfectly homologous ERSE. The same motif is either poorly conserved or not present at all in other PDIAs, explaining why they are not induced and PDIA6 is highly responsive to ER stress.
Despite GRP94 being one of the main hallmarks of the ER stress response, its loss or inhibition do not impair the cells' ability to mount a stress response and cope with ER stress. GRP94-depleted cells, which have increased levels of BiP, can still further upregulate BiP when ‘insulted’ with chemical stress inducers. In contrast, a GRP94-null clone that does not show BiP induction, is dramatically more susceptible to ER stress (Biswas et al., 2007). We propose that ER stress responsiveness is not impaired without GRP94 as long as there is sufficient activity of BiP and PDIA6. This explanation fits with the cytoprotective effect of BiP overexpression (Dorner et al., 1992). Also consistent with this explanation is the inability of cells to survive simultaneous ablation of GRP94 and BiP (D.E., unpublished data). Furthermore, KD of GRP94 in myeloma lines, which are specialized secretory cells, is barely achievable and only lasts for few days (Kropp et al., 2010). In contrast, GRP94 gene remained silenced for indefinite period of time in non-professionally secretory cells, as reported here. Thus, GRP94 may have a central role in general UPR only when there is high demand on folding resources, when even mild accumulation of folding intermediates may be harmful.
It is unclear why BiP and PDIA6 in particular are overexpressed when GRP94 activity is lost. There is no obvious functional reason why upregulation of either one would compensate for the loss of another component. BiP and GRP94 do work in concert on the folding of some client proteins, such as immunoglobulins or thyroglobulin, but they are not redundant and work sequentially, apparently recognizing distinct folding intermediates (Melnick et al., 1994). We hypothesize that when GRP94 is ablated, BiP, which recognizes the less advanced folding intermediates, must be induced to deal with accumulation of GRP94 clients. PDIA6 is a BiP partner that binds to BiP clients (Jessop et al., 2009), and its level is co-regulated with BiP under chemically induced ER stress and in response to the accumulation of misfolded proteins (this work and Hartley et al., 2010). GRP94 and BiP may also be affected by each other due to other shared common functions. Both are calcium-binding proteins (Biswas et al., 2007; Lièvremont et al., 1997) and both are involved in the degradation of soluble substrates (Christianson et al., 2008; Kabani et al., 2002). In conclusion, the expression network formed by BiP, GRP94 and PDIA6 can be rationalized by several known functional parameters that can be tested in the future.
Cells must have developed mechanisms to monitor the abundance of chaperones in order to keep their levels within a certain range that is compatible with their cooperative mode of actions. For example, when cytosolic HSP90 is inhibited, HSP70 is upregulated, due to the cooperation of these two chaperones in the folding pathways of many kinases and transcription factors (Bagatell et al., 2000; Zou et al., 1998). Such compensatory mechanisms are likely based on sensing the loss of chaperone activity. We provide evidence that the presence of GRP94 is also detected based on its activity and not simply the abundance of the protein. First, the transcriptional response of BiP and PDIA6 is activated not only when GRP94 is depleted, but also when it is inhibited pharmacologically. Second, the upregulation of BiP and PDIA6 occurs even if an inactive GRP94 mutant is overexpressed, but is prevented when active GRP94 is supplied. Interestingly, the detection mechanism works only when GRP94 activity is reduced; overexpression of GRP94 does not perturb the normal functioning of the ER stress response machinery.
These results suggest the following model: when GRP94 activity is insufficient, either because of a high demand of obligate clients that accumulate in the ER or because of another need, a dedicated sensor is activated, which reports on the limiting GRP94 through a signaling pathway, culminating in activation of transcription of BiP, PDIA6, and perhaps other genes.
Our results show that ER proteins are not necessarily upregulated as a cohort. Rather, multiple signal transduction pathways may be evocated in response to distinct stimuli or stresses. The plasticity of ER dynamics may be due to its ability to initiate such distinct stress responses, leading to distinct outcomes and different kinetics.
Materials and Methods
Ethics statement
All experiments were approved by the CHOP and the University of Pennsylvania animal care committees.
Chemicals and plasmids
Actinomycin D, doxycycline, tunicamycin and thapsigargin were purchased from Sigma Chemicals (St Louis, MO). MG-132 was from Calbiochem (San Diego, CA). Lipofectamine 2000 transfection reagent was from Invitrogen (Carlsbad, CA). 17-Allylamino-17-demethoxygeldanamycin (17AAG), puromycin and G418 were from InvivoGen (San Diego, CA); the XTT cell proliferation kit from Biotium, Inc. (Hayward, CA). DMEM was from Mediatech, Inc. (Manassas, VA), fetal bovine serum was from Gemini (West Sacramento, CA). Glutamine, penicillin/streptomycin supplement was from Gibco-Invitrogen (Grand Island, NY). Subtilase AB toxin was a kind gift of J. C. Paton (Univ. of Adelaide, Australia). A Flag-tagged GRP94 expressed into the pTRE Vector (Clontech, Mountain View, CA) was subjected to site-directed mutagenesis using the QuickChange Kit (Agilent, Santa Clara, CA) to generate the E82A GRP94 ATPase deficient mutant.
Cell culture and GRP94 conditional KO mice
C2C12, 10T1/2, NIH-3T3, 293T, and HeLa cells were from the ATCC. CHO-K1 Tet-On were from Clontech. 293T cells stably expressing a HA-tagged ATF6 were a kind gift of Drs H. Steiner and Haass (Ludwig Maximilians University, Germany). These cell lines were grown in DMEM in the presence of 10% FBS and Gln/Pen/Strept, and, when needed, the proper eukaryotic selection agent (puromycin or G418).
IRE1 and XBP1 KO, PERK and ATF4 KO MEFs were generous gifts from Dr D Ron (Univ. of Cambridge, UK). ATF6alpha KO MEFs from Dr K. Mori (Univ. of Kyoto, Japan), Mice containing a floxed allele of GRP94 (grp94flox) (Yang et al., 2007) were crossed with muscle creatine kinase (MCK)-Cre transgenic mice (on a C57Bl/6 background; Jackson Labs, stock 006475). Double heterozygous progeny were then bred to grp94flox/flox mice, in order to deplete GRP94 within skeletal and cardiac muscle. Tissue samples were collected and processed for immunoblotting as reported in Barton et al. (Barton et al., 2010).
Immunoblotting
Cells were lysed with a 0.5% NP-40/Igepal detergent solution as described in Ostrovsky et al. (Ostrovsky et al., 2009). Images were recorded using an Alpha Innotech (Santa Clara, CA) or Odyssey (Li-Cor, Lincoln, NE) imagers. Band intensities were normalized for total protein loads using house-keeping proteins (α–tubulin or 14-3-3). Fold changes were calculated relative to internal references (wt or control samples), as indicated in the figure legends.
Antibodies: rabbit anti-14-3-3 (C16) and mAb anti-myogenin (F5D) were purchased from Santa Cruz, Biotechnology, Santa Cruz, CA. mAb anti-desmin(D33) was from Imgenex (San Diego CA); mAb anti-KDEL was from StressGen (Vancouver, BC); anti-HSP90 was from BD Transduction Laboratories (San Jose, CA) and anti-caspase 3 and anti-cleaved caspase 3 (Asp175) were from Cell Signaling; the anti-GRP94 monoclonal antibody 9G10 (SPA-850) from Stressgen Biotechnologies (Victoria, BC). The monoclonal anti-HA antibody HA.11 (clone 16B12) was obtained from Covance (Princeton, NJ). Secondary antibodies conjugated to HRP were from Jackson ImmunoResearch Laboratories (West Grove, PA), secondary antibodies conjugated to near-infrared fluorophores were from Li-Cor.
RNAi silencing
GRP94, BiP, PDIA6, HSP47 or OS-9 were knocked-down, using the following shRNAs from Sigma Life Science (S Louis, MO): SHCLNG-NM_011631 (a set of five vectors referred to in this paper as shRNA23 to 27; Fig. 2B), TRCN0000008455, SHCLNG-NM_027959 (a set of five vectors, supplementary material Fig. S2D), TRCN0000008534, TRCN0000175937, respectively. Briefly, cells were transduced with lentiviral particles encoding the shRNA sequences, as in Ostrovsky et al. (Ostrovsky et al., 2010). The efficiency of knockdown was consistently >90% for GRP94, PDIA6, HSP47 and OS-9. BiP expression typically could only be reduced to 40% of control level.
Analysis of XBP1 mRNA splicing and ATF6 endoproteolysis
XBP1 and β-actin were PCR amplified from total RNA as in Calfon et al. (Calfon et al., 2002). 293T cells stably expressing HA-tagged ATF6 were analyzed by immunoblotting with anti HA-antibody HA.11 after the cells were stressed with Tun and treated with MG132, to block degradation of the ATF6 fragment. The levels of the ATF6 fragment nuclear fractions were enriched as described in Li et al. (Li et al., 2000).
Luciferase reporter assay
Constructs composed of nucleotides −284 to +221 of the human GRP78 promoter driving luciferase were described in Doroudgar et al. (Doroudgar et al., 2009). The construct composed of 1024 nucleotides upstream of the coding sequence of the herpes simplex virus ICP0 gene fused to luciferase was from Dr Liu (Burnett et al., 2012). 293T stably expressing shRNA targeting either GRP94, BiP or a non-relevant sequence were transfected and analyzed for luciferase activity as in (Doroudgar et al., 2009).
Statistical analysis
Statistical analysis was performed using a one-way analysis of variance followed by Student's Newman-Keul's post hoc analysis of variance (*, P<0.05, unless otherwise stated in the figure legends).
Supplementary Material
Acknowledgments
We are grateful to K. Mori (University of Kyoto, Japan), D. Ron (Cambridge University, UK), J. C. Paton (University of Adelaide), H. Steiner and C. Haass (Ludwig-Maximilians-University, Germany), R. Prywes (Columbia University) and R. Lu (University of Guelph) for generous gifts of cell lines, plasmids and other reagents. We also thank Yina Dong and Erikka Carr for technical support and T. Gidalevitz, M. Marzec, M. Chou and A. Gentilella for their comments and suggestions.
Footnotes
Funding
This work was funded by the National Institutes of Health [grant numbers GM077480, AG18001 to Y.A., HL085577, HL75573, HL104535, EB011698 to C.G.G.]. D.D. was supported by a National Institutes of Health training grant [grant number GM008275]. S.D. was supported by the Rees-Stealy Research Foundation, the San Diego Chapter of the Achievement Rewards for College Scientists (ARCS) Foundation, an American Heart Association Predoctoral Fellowship and an Inamori Foundation Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.108928/-/DC1
References
- Bagatell R., Paine–Murrieta G. D., Taylor C. W., Pulcini E. J., Akinaga S., Benjamin I. J., Whitesell L. (2000). Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents. Clin. Cancer Res. 6, 3312–3318 [PubMed] [Google Scholar]
- Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008). Adapting proteostasis for disease intervention. Science 319, 916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Bartke A. (2009). The somatotropic axis and aging: mechanisms and persistent questions about practical implications. Exp. Gerontol. 44, 372–374 10.1016/j.exger.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E. R., DeMeo J., Lei H. (2010). The insulin-like growth factor (IGF)-I E-peptides are required for isoform-specific gene expression and muscle hypertrophy after local IGF-I production. J. Appl. Physiol. 108, 1069–1076 10.1152/japplphysiol.01308.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E R., Park S., James J. K., Makarewich C. A., Philippou A., Eletto D., Lei H., Brisson B., Ostrovsky O., Li Z., Argon Y. (2012). Deletion of muscle GRP94 impairs both muscle and body growth by inhibiting local IGF production. FASEB J. 26, 3691–3702 10.1096/fj.11-203026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas C., Ostrovsky O., Makarewich C. A., Wanderling S., Gidalevitz T., Argon Y. (2007). The peptide-binding activity of GRP94 is regulated by calcium. Biochem. J. 405, 233–241 10.1042/BJ20061867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett H. F., Audas T. E., Liang G., Lu R. R. (2012). Herpes simplex virus-1 disarms the unfolded protein response in the early stages of infection. Cell Stress Chaperones 17, 473–483 10.1007/s12192-012-0324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Chang S. C., Erwin A. E., Lee A. S. (1989). Glucose-regulated protein (GRP94 and GRP78) genes share common regulatory domains and are coordinately regulated by common trans-acting factors. Mol. Cell. Biol. 9, 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008). OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 10.1038/ncb1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner A. J., Wasley L. C., Kaufman R. J. (1992). Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 11, 1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroudgar S., Thuerauf D. J., Marcinko M. C., Belmont P. J., Glembotski C. C. (2009). Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response. J. Biol. Chem. 284, 29735–29745 10.1074/jbc.M109.018036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Hartley T., Siva M., Lai E., Teodoro T., Zhang L., Volchuk A. (2010). Endoplasmic reticulum stress response in an INS-1 pancreatic beta-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol. 11, 59 10.1186/1471-2121-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano T., Kikuchi M. (1995). Cloning and sequencing of the cDNA encoding human P5. Gene. 164, 377–378 [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Akai R. (2006). Analysis of the XBP1 splicing mechanism using endoplasmic reticulum stress-indicators. Biochem. Biophys. Res. Commun. 350, 709–715 10.1016/j.bbrc.2006.09.100 [DOI] [PubMed] [Google Scholar]
- Jessop C. E., Watkins R. H., Simmons J. J., Tasab M., Bulleid N. J. (2009). Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J. Cell Sci. 122, 4287–4295 10.1242/jcs.059154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Beckerich J. M., Brodsky J. L. (2002). Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22, 4677–4689 10.1128/MCB.22.13.4677-4689.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi S., Nakatani H., Nakano C., Urade R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272, 3461–3476 10.1111/j.1742-4658.2005.04770.x [DOI] [PubMed] [Google Scholar]
- Kapulkin W. J., Hiester B. G., Link C. D.2005). Compensatory regulation among ER chaperones in C. elegans. FEBS Lett. 5793063–3068 10.1016/j.febslet.2005.04.062 [DOI] [PubMed] [Google Scholar]
- Kondo S., Murakami T., Tatsumi K., Ogata M., Kanemoto S., Otori K., Iseki K., Wanaka A., Imaizumi K. (2005). OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7, 186–194 10.1038/ncb1213 [DOI] [PubMed] [Google Scholar]
- Kropp L. E., Garg M., Binder R. J. (2010). Ovalbumin-derived precursor peptides are transferred sequentially from gp96 and calreticulin to MHC class I in the endoplasmic reticulum. J. Immunol. 184, 5619–5627 10.4049/jimmunol.0902368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Baumeister P., Roy B., Phan T., Foti D., Luo S., Lee A. S. (2000). ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20, 5096–5106 10.1128/MCB.20.14.5096-5106.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvremont J. P., Rizzuto R., Hendershot L., Meldolesi J. (1997). BiP, a major chaperone protein of the endoplasmic reticulum lumen, plays a direct and important role in the storage of the rapidly exchanging pool of Ca2+. J. Biol. Chem. 272, 30873–30879 10.1074/jbc.272.49.30873 [DOI] [PubMed] [Google Scholar]
- Ma Y., Hendershot L. M. (2004). ER chaperone functions during normal and stress conditions. J. Chem. Neuroanat. 28, 51–65 10.1016/j.jchemneu.2003.08.007 [DOI] [PubMed] [Google Scholar]
- Mao C., Wang M., Luo B., Wey S., Dong D., Wesselschmidt R., Rawlings S., Lee A. S. (2010). Targeted mutation of the mouse Grp94 gene disrupts development and perturbs endoplasmic reticulum stress signaling. PLoS ONE 5, e10852 10.1371/journal.pone.0010852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J., Aviel S., Argon Y. (1992). The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J. Biol. Chem. 267, 21303–21306 [PubMed] [Google Scholar]
- Melnick J., Dul J. L., Argon Y. (1994). Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature 370, 373–375 10.1038/370373a0 [DOI] [PubMed] [Google Scholar]
- Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456–4469 10.1091/mbc.E02-05-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga N., Yahiro K., Matsuura G., Moss J., Noda M. (2008). Subtilase cytotoxin, produced by Shiga-toxigenic Escherichia coli, transiently inhibits protein synthesis of Vero cells via degradation of BiP and induces cell cycle arrest at G1 by downregulation of cyclin D1. Cell. Microbiol. 10, 921–929 10.1111/j.1462-5822.2007.01094.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan Z., Arvan P. (1997). Thyroglobulin transport along the secretory pathway. Investigation of the role of molecular chaperone, GRP94, in protein export from the endoplasmic reticulum. (published erratum appears in J. Biol. Chem. 272, 30590. J. Biol. Chem. 272, 26095–26102 10.1074/jbc.272.42.26095 [DOI] [PubMed] [Google Scholar]
- Murray J. I., Whitfield M. L., Trinklein N. D., Myers R. M., Brown P. O., Botstein D. (2004). Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15, 2361–2374 10.1091/mbc.E03-11-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò A., McCullagh K., Paul A., Houghton L., Dobrowolny G., Molinaro M., Barton E. R., Sweeney H. L., Rosenthal N. (2001). Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 27, 195–200 10.1038/84839 [DOI] [PubMed] [Google Scholar]
- Ostrovsky O., Ahmed N. T., Argon Y. (2009). The chaperone activity of GRP94 toward insulin-like growth factor II is necessary for the stress response to serum deprivation. Mol. Biol. Cell 20, 1855–1864 10.1091/mbc.E08-04-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky O., Eletto D., Makarewich C., Barton E. R., Argon Y. (2010). Glucose regulated protein 94 is required for muscle differentiation through its control of the autocrine production of insulin-like growth factors. Biochim. Biophys. Acta 1803, 333–341 10.1016/j.bbamcr.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A. W., Beddoe T., Thorpe C. M., Whisstock J. C., Wilce M. C., Rossjohn J., Talbot U. M., Paton J. C. (2006). AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 443, 548–552 10.1038/nature05124 [DOI] [PubMed] [Google Scholar]
- Ron D. (2002). Proteotoxicity in the endoplasmic reticulum: lessons from the Akita diabetic mouse. J. Clin. Invest. 109, 443–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Shiu R. P., Pouyssegur J., Pastan I. (1977). Glucose depletion accounts for the induction of two transformation-sensitive membrane proteinsin Rous sarcoma virus-transformed chick embryo fibroblasts. Proc. Natl. Acad. Sci. USA 74, 3840–3844 10.1073/pnas.74.9.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault G., Ismail N., Ng D. T. (2011). The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc. Natl. Acad. Sci. USA 108, 20597–20602 10.1073/pnas.1117184109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- van Anken E., Romijn E. P., Maggioni C., Mezghrani A., Sitia R., Braakman I., Heck A. J. (2003). Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity 18, 243–253 10.1016/S1074-7613(03)00024-4 [DOI] [PubMed] [Google Scholar]
- Walter P., Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wanderling S., Simen B. B., Ostrovsky O., Ahmed N. T., Vogen S., Gidalevitz T., Argon Y. (2007). GRP94 is essential for mesoderm induction and muscle development because it regulates insulin-like growth factor secretion. Mol. Biol. Cell 18, 3764–3775 10.1091/mbc.E07-03-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R. J., Prywes R. (2000). Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275, 27013–27020 [DOI] [PubMed] [Google Scholar]
- Wiest D. L., Burkhardt J. K., Hester S., Hortsch M., Meyer D. I., Argon Y. (1990). Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J. Cell Biol. 110, 1501–1511 10.1083/jcb.110.5.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J. J., May K. L., Thorpe C. M., Jandhyala D. M., Paton J. C., Paton A. W. (2008). Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell. Microbiol. 10, 1775–1786 10.1111/j.1462-5822.2008.01164.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liu B., Dai J., Srivastava P. K., Zammit D. J., Lefrançois L., Li Z. (2007). Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity 26, 215–226 10.1016/j.immuni.2006.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Jung D. Y., Jun J. Y., Li J., Luo S., Ko H. J., Kim J. K., Lee A. S. (2010). Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes 59, 6–16 10.2337/db09-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Haze K., Yanagi H., Yura T., Mori K. (1998). Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273, 33741–33749 10.1074/jbc.273.50.33741 [DOI] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R. (1998). Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.