Introduction
RNA interference (RNAi) is an evolutionary conserved mechanism in which a double-stranded RNA (dsRNA) inhibits gene expression by degrading messenger RNA or by blocking the translation pathway of a specific gene.[1] During RNAi, exogenous (e.g., viral RNA) or endogenous (e.g., microRNA) dsRNA is processed and cut to 21- to 23-nucleotide RNA fragments (short interfering RNAs [siRNA]) by a ribonuclease called Dicer and assembled into a RNA-induced silencing complex (RISC). Components of the RISC destroy the passenger strand and recognize and cleave the target mRNA. Since RNAi was first discovered in Canorhabditis elegans a decade ago,[2] it has inspired researchers worldwide to explore more fundamental biological issues and possible biomedical applications The RNAi phenomenon was applied in mammalian cells by two research teams, demonstrating that siRNA can efficiently knock down genes in animal cells.[3, 4] Since then, researchers have been ardently pursuing approaches to manipulating this powerful tool in therapeutics.
Nanovehicle delivery of siRNA to mammalian systems
The potential advantage of RNAi in medical applications is that it may provide a cure for diseases that cannot be treated by conventional small molecular medicines. By introducing siRNA to the cell, specific genes can be silenced, resulting in either decreased translational product of the silenced gene, or increased protein levels of a gene that is downregulated by the silenced sequence. However, in vivo delivery of siRNA has been a challenge due to the instability of siRNA in blood (in the case of systemic delivery), its relatively large molecular size, and its highly negative charge
Depending on the type of vector used to present a potential gene silencer, siRNA can generally be delivered into cells or tissues by two vehicles: viral vectors and non-viral carriers. In the case of viral vectors, a DNA or RNA sequence containing one or multiple copies of an siRNA sequence is assembled into the genome of a virus, which infects the targeted cell and goes through the life cycle of a real virus, including transcription of the siRNA sequence to target the host cell mRNA. Despite the advantage of an enhanced sustained production of siRNA, this approach raises concerns about the potential dangers of infecting cells with a live virus, including acute toxicity such as competition of overexpressed shRNA for the endogenous miRNA, and unexpected long-term effects such as integration of the viral genome into the host cell DNA.[5, 6]
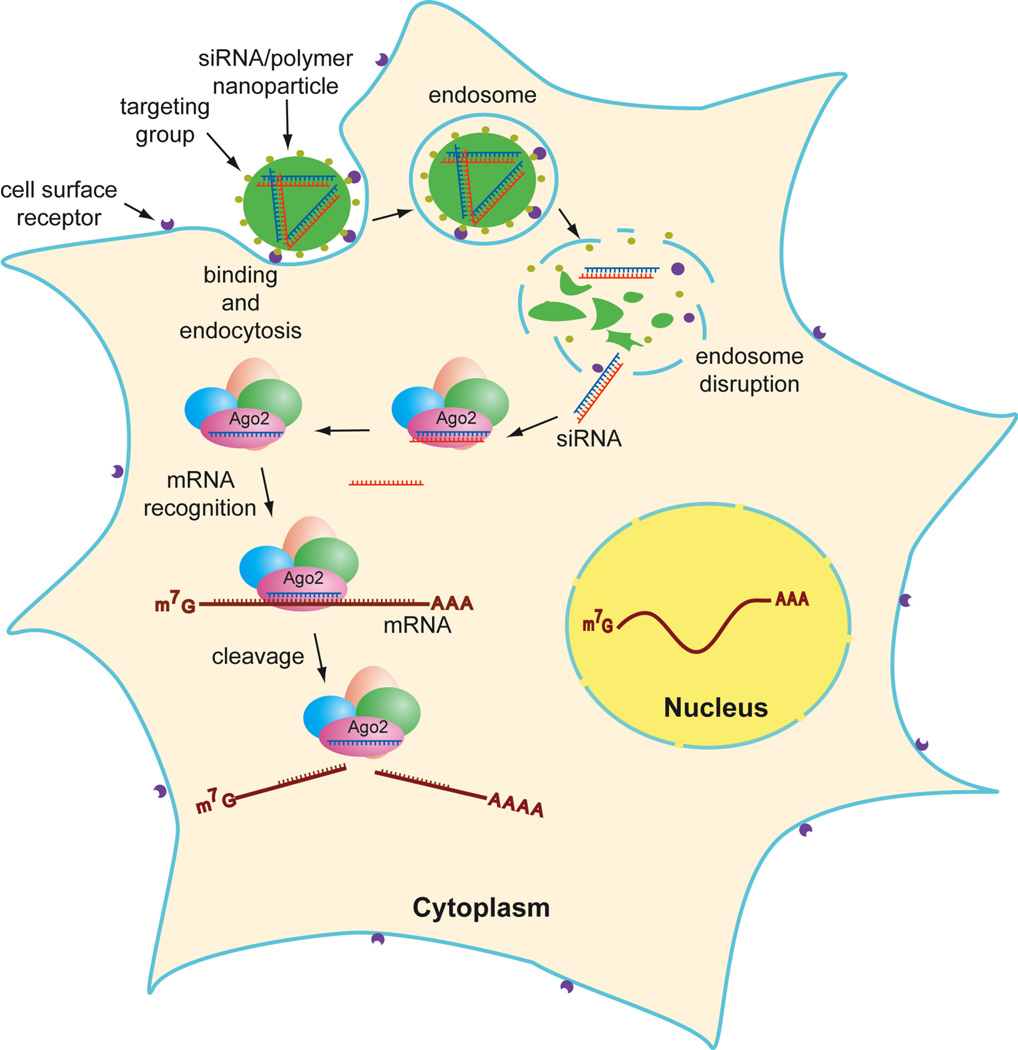
In a non-viral delivery approach, a double-stranded siRNA, either “naked”, chemically modified,[7, 8] or covalently attached, is complexed with a carrier material that binds to siRNA through electrostatic interactions to form a particle. These particles usually range in size from nano- to micrometers, depending on the physical characteristics of the material and formulation conditions. The resulting particle protects the siRNA from degradation in the circulatory system, brings the siRNA into the target tissue, enters the cell through endocytosis, and releases the siRNA in the cytoplasm (Fig 1). The formulation of nanoparticles for in vitro and in vivo delivery of siRNA (systemic and/or local administration, respectively) has exploited various kinds of materials, including cationic lipids (liposomes), polymers, covalently conjugated polymers, peptides, and proteins (including antibodies), and inorganic nanoparticles such as quantum dots.
Figure1.
Functionalized nanoparticle enters cells by receptor-mediated endocytosis. Double-strand RNA is processed (by Dicer), assembled into RISC, which hydrolyzes mRNA.
siRNA loading to nanovehicles through electrostatic forces
Complexing siRNA to a delivery material is accomplished in most circumstances through electrostatic interactions between a positively charged material and the negatively charged siRNA. Positive charges come from amino groups on the backbone or side chains of a polymer, and the electrostatic strength and density of positive charges are among the factors that most influence the binding and releasing of the resulting nanoparticles. Primary, secondary, and tertiary amino groups are preferred over quaternary amines, due to their relative flexibility in binding and releasing in different pH environments. However, tertiary amines need to be acidified to be charged prior to being complexed with nucleic acids, limiting their ability to transfer their cargo under neutral physiological conditions.[9, 10]
The binding and releasing behaviors of siRNA/polymer complexes are often tested by simple agarose gel electrophoresis. The intensity of the free siRNA band usually decreases with increasing amounts of polymer to be complexed providing a quantitative method for RNA-polymer assembly. In the case of strong binding between siRNA and polymer, it is necessary to perform a replacement experiment, in which an anionic lipid (SDS) or polymer (heparin) is added to exclude siRNA from the complex. However, this method is limited to a certain pH range because electrophoresis is conducted in weak basic buffer (pH 8.0).
Polyethylenimine (PEI) polymers have been extensively investigated for their in vitro and in vivo siRNA transfection efficiency. PEIs bind strongly to siRNA through electrostatic forces due to the abundant amino groups present in their backbone (secondary amines of leaner or less branched PEIs) and on both the surface and inner layers of branched PEIs (primary and tertiary amines of branched PEIs). Although PEIs show remarkable ability to deliver genes (plasmids), their capacity for siRNA transfection has proved to be moderate, depending on more restricted conditions and formulations. Branched PEIs are favored over leaner ones, due to their abundant surface amino groups which afford strong binding as well as flexibility for various chemical modifications to reduce toxicity.
One example of successfully using PEI to deliver siRNA in vivo used a complex of low molecular weight PEI and siRNA targeting the human epidermal growth factor receptor (HER-2) in a mouse model.[11] The PEI/siRNA complex was administered intraperitoneally to mice bearing subcutaneous tumors; 12 d later HER-2 mRNA levels decreased ~50%, and tumor growth decreased significantly. In another application, an unmodified branched PEI was complexed with polyethylene glycol (PEG)-siRNA to give a nanoparticle with a diameter < 100nm, which effectively protected the siRNA from hydrolysis by serum nucleases, while preventing absorption by serum proteins. When these nanoparticles were injected into a mouse tumor model, the target gene, vascular endothelial growth factor (VEGF), was reduced by 86.4%, and microvessel formation was inhibited 77.5% compared to no-treatment control. Another application targeting the VEGF gene efficiently delivered siRNA to tumor tissue in nanoparticles functionalized by PEGylation of PEI with an integrin-binding Arg-Gly-Asp (RGD) peptide. Delivery of the siRNA to tumor tissue by intravenous administration resulted in its selective uptake by the integrin-expressing tumor and inhibition of VEGF expression.[12, 13]
Another ideal material for siRNA delivery is chitosan, a natural polysaccharide derived from the shells of shrimp and other sea crustaceans, because it is abundant, biodegradable, and has a unique structure. At neutral pH chitosan is strongly positively charged, which makes it readily complex with siRNA through electrostatic interactions upon simple mixing. When chitosan was complexed with siRNA targeting enhanced green fluorescent protein (EGFP), the resulting nanoparticle (N: P 57) knocked down EGFP expression in human lung carcinoma cells H1299 cells and mouse macrophages by 78% and 89%, respectively. The nanoparticle was also effective in vivo; its nasal administration to transgenic EGFP mice reduced the number of EGFP-expressing lung epithelial cells.[14]
siRNA loading to nanoparticles through covalent attachment
Complexes in which siRNA is covalently attached to a polymer have some advantages over complexes with traditional electrostatic binding between polymer and siRNA. The physical properties of covalently conjugated particles depend mostly on the characteristics of the polymer to be attached. One potential hurdle to this kind of coupling reaction is aggregation caused by electrostatic interactions, especially in the case of high molecular weight and highly positively charged polymers. A basic buffer (pH=9.0) may be feasible in this case, but the reaction time and other conditions such as temperature should be strictly controlled to minimize hydrolysis of siRNA under this alkaline condition.[15]
For covalent conjugation, the siRNA must have a reactive group, such as an amino or sulfhydryl group, attached at either the 3’ or 5’ end of the sense strand. For example, a thiolated apolipoprotein B (ApoB) siRNA was covalently coupled through a cleavable disulfide bond to an amine-containing amphipathic polyvinyl ether. After further modifications with PEG and N-acetylgalactosamine (NAG) groups, the conjugate formed 10nm particles which efficiently targeted asialoglycoprotein receptors (ASGPs) on hepatocytes and entered the cells via endocytosis. The acidic conditions of the endosomes led to hydrolysis of the PEG and NAG side chains from the polymer, enabling the polyconjugate to efficiently disrupt the endosomal membrane and enter the cytoplasm. The reducing environment of the cytoplasm cleaved the disulfide bond, releasing the siRNA and resulting in 84% knockdown of the target gene.[16] Making covalently siRNA-vehicle nanoparticle requires more tedious synthesis steps than for complexes joined by electrostatic forces, but better controls the particle size (usually less than 100nm). Smaller and more uniform articles result in better targeting and distribution inside the target tissue and reduced toxicity due to applying relatively lower doses of siRNA.
siRNA stability in serum
When delivering siRNAs systemically, the most important factors to consider are serum influences and the circulation half-life of the siRNA/polymer complex. A positively charged siRNA/polymer in blood accumulates negatively charged serum proteins, which shield the surface, resulting in a larger particle, decreased zeta potential, and even aggregation, which could considerably decrease uptake by the target tissues. The circulation half-life of highly positively charged siRNA/polymers has proven to be short, less than 5 min in most cases. The reason for this quick blood clearance is not fully understood, but is considered as most likely due to degradation by serum enzymes, aggregation, and accumulation in capillary beds.[17, 18]
The half-life of unmodified siRNA in plasma is only a few minutes. Although siRNA in the circulatory system can be protected by cationic polymers or other carrier materials, chemical modifications have been shown to stabilize siRNA to resist hydrolysis and reduce off-target effects.[1, 7, 8] These chemical modifications can be made on the 2’ ribose, phosphodiester, 3’ or 5’ end of either the passenger or guide strand. For instance, phosphorothiolation on the backbone and/or 2’-O-halogenation of the passenger strand could protect siRNA from hydrolysis. 3’-end capping and 2’-fluoronation are tolerated in the guide strand, but the 5’-end of the guide strand should be unmodified.[19–24]
Another important polymer characteristic is hydrophobicity, especially in terms of solubility, serum compatibility, and particle size upon complexing with siRNA. A moderate hydrophobicity is necessary to maintain better binding, while keeping an appropriate particle size which may dramatically limit transfection efficiency. On the other hand, the molecular weight of a polymer, which may be anywhere between 1 to a few hundred kDa, is a key consideration to obtain suitable hydrophobicity. Polymers with higher molecular weight may give better binding to siRNAs, but their solubility may decrease while the viscosity increases, which may lead to poor handling properties for mixing and injecting in the case of systemic delivery.
These pitfalls can be avoided by incorporating PEG into the siRNA/polymer complex, which was found to effectively prevent siRNA interactions with serum proteins, protect siRNA degradation by serum nucleases, and prolong siRNA circulation time. PEG can be covalently coupled to siRNA, to polymer, or to a functional molecule that can be incorporated into the siRNA/polymer complex through non-covalent interactions. The amount of PEG chains in the complex should be controlled to adjust the hydrophobicity of the whole particle, with a cleavable linkage between the PEG and polymer or siRNA usually desirable. For example, when a siRNA-PEG-Cy5.5 /PEI complex was intravenously administered to tumor-bearing mice, their tissues, including kidney and blood, showed higher fluorescence intensity than tissues from mice injected with a siRNA/PEI complex, indicating that PEG can stabilize siRNA and prolong the circulation half-life. Meanwhile the siRNA complex with PEG knocked down the target gene, VEGF, by 86%, while a complex lacking PEG showed only reduced the target by 43%.[12]
Targeting organs and internalizing siRNA
siRNA can be delivered to a specific organ by functionalizing siRNA or polymers. For example, the liver can be targeted by nanoparticles generated from various kinds of lipidic or polymeric materials. By conjugating a small molecule to the 3’ or 5’ end of siRNA, its stability and tissue specificity can be increased. Indeed, when a single cholesterol molecule was conjugated to siRNA and systemically delivered to mice, liver ApoB mRNA was targeted and decreased by 55%.[23] Similarly, a-tocopherol-conjugated siRNA was efficiently delivered to mouse liver.[25] Another example is provided by siRNA/liposomes, which have been used to target hepatocytes. The liposome components include cationic lipid, neutral lipid, cholesterol and PEGylated lipid. When stable nucleic acid-lipid particles (SNALPs) containing ApoB siRNA were intravenously injected into cynomolgus monkeys, target ApoB mRNA was reduced (90%), resulting in significant decreased serum cholesterol and low-density lipoproteins.[21]
Another promising siRNA delivery technology is receptor-mediated delivery, especially for siRNA delivery to organs that reject siRNA by ordinary methods. In this approach, a targeting functional group is often assembled on the surface of the siRNA/polymer nanoparticle. The functional group could be a small molecule, such as folic acid (to target cancer cells), peptides, or monoclonal antibodies. The functional group could be conjugated to the polymer by covalent attachment or non-covalent interactions such as electrostatic forces. The functional group recognizes and binds to a specific cell surface receptor, after which the whole nanoparticle is internalized through endocytosis.
Receptor-mediated siRNA delivery has been somewhat successful in the most difficult organ to target, the brain. Because of the blood-brain barrier, traditional lipid-based carriers have failed to deliver siRNAs to the brain and to other areas of the central nervous system.[26, 27]
One successful approach used a 29-amino-acid peptide, originally derived from the rabies virus glycoprotein (RVG), which binds specifically to acetylcholine receptors (AchR) on neuronal cells. When the RVG peptide was modified by conjugating 9 arginine residues (RVG-9R), it delivered a FITC-labeled siRNA into AchR-expressing mouse neuronal cells and efficiently reduced target GFP with limited toxicity. The RVG-9R/siRNA complex was also successfully delivered in vivo to the mouse brain, where it significantly reduced mRNA and protein levels of the targeted superoxide dismutase 1 (SOD1) gene. The RVG-9R/siRNA complex was also used to target viral FvEJ, thus protecting mice infected with Japanese encephalitis virus and significantly prolonging their survival time.[28]
Similarly, receptor-mediated siRNA delivery has been used in both brain tumor and leukocytes, another hard-to-transfect cell, by combining liposomal encapsulation and monoclonal antibody (mAb) technology. In addition, a mAb-protamine fusion protein, without any lipid components, efficiently delivered siRNA targeting the HIV-1 capsid gene gag into HIV-infected primary T cells.[29, 30]
siRNA release and cytotoxicity
After the siRNA/polymer nanoparticle has entered the cytoplasm, the siRNA needs to be completely separated from the complex. However, the mechanism of siRNA release from siRNA/polymer nanoparticles is not fully understood. The disassembly of nanoparticles and consequent release of siRNA may involve enzymatic hydrolysis. Some environmentally sensitive polymers have been designed to help siRNA escape from endosomes, which are small, acidic (<pH 5.5) membrane-enclosed compartments formed after the nanoparticle is taken up by cells through endocytosis. To release siRNA from endosomes, siRNA/polymer nanoparticles have incorporated acid-degradable or membrane-disruptive molecules. A ketalized PEI was shown to be hydrolyzed at pH 5.0 10 times faster than at pH 7.4, with a half-life of 2.6 h. The siRNA/ketalized PEI had lower toxicity in vitro and higher transfection efficiency than siRNA/unketalized PEI. Although this kind of polymer has shown promising activity in cell cultures, its serum stability and in vivo activity have not yet been reported.[31, 32]
Unlike long dsRNA, siRNA does not elicit RNA-activated protein kinase (PKR), which destroys RNA and inhibits protein synthesis. However, injecting siRNA/polymer nanoparticles may activate a non-specific immune response, leading to significant toxicity and apoptosis. Non-specific immune responses are indicated by the expression levels of interferon (IFN)-inducible genes (IFIT-1, STAT 1) and plasma IFN-a levels, respectively. The toxicity may come from the polymer or chemically modified siRNA.[33–35]
Delivery of siRNA via dendrimers
Dendrimers, or repeatedly branched polymers, bear many advantages over conventional leaner polymers in delivering nucleic acids. With their well-defined molecular structure, monodispersity, and densely located surface functionalities, dendrimers provide more surface volume for interactions and more condensed particles upon complexing with nucleic acids. For example, a generation-four polyamidoamine (PAMAM) dendrimer was complexed with a Cy3-labeled siRNA and used to deliver siRNA to HeLa cells. Although the target gene CDK9 was efficiently knocked down, in subsequent transfections of the CDK9 siRNA/PAMAM complex to HeLa cells, the range of working concentrations of the dendrimer proved to be relatively narrow.[36] In another study, the complex of a luciferase siRNA/PAMAM gave 60–110nm particles, which were highly efficient in silencing the target GL3 luciferase gene at higher dendrimer generations (G7).[37]
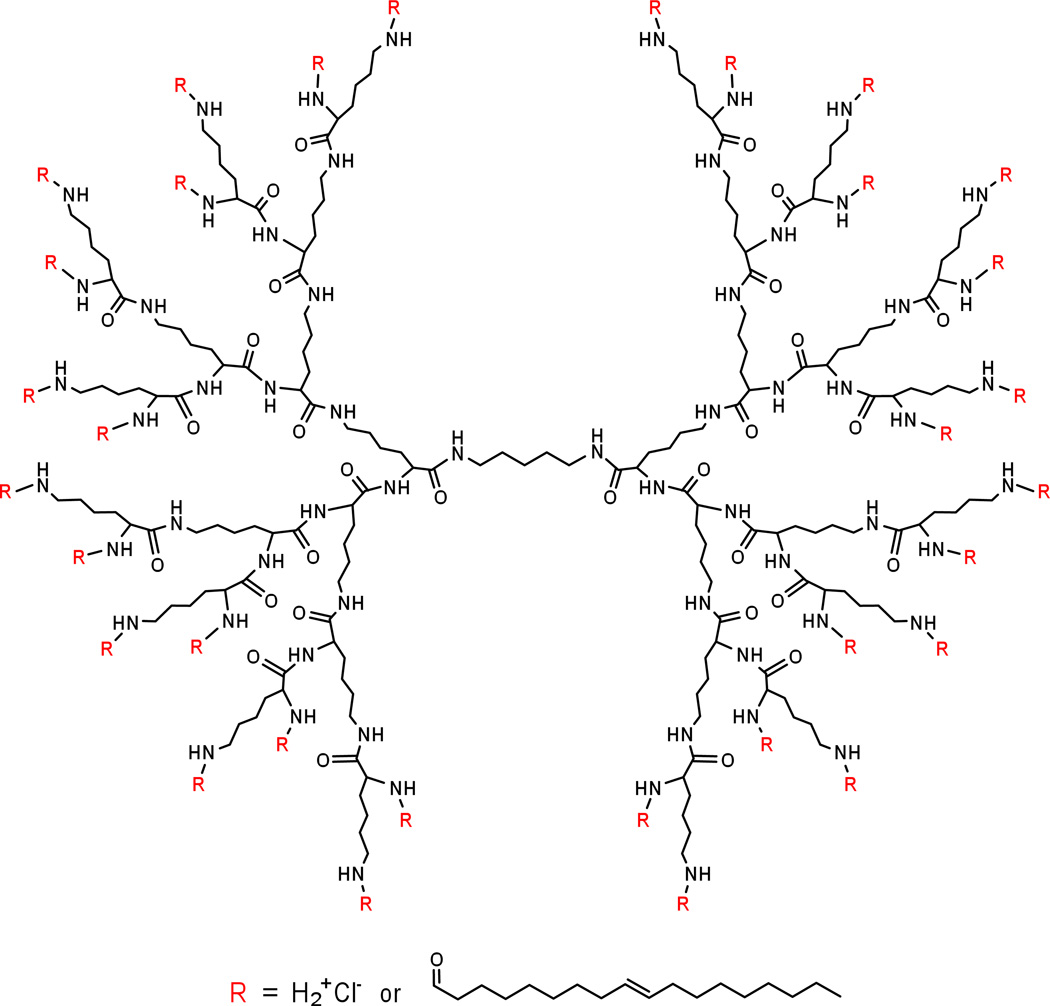
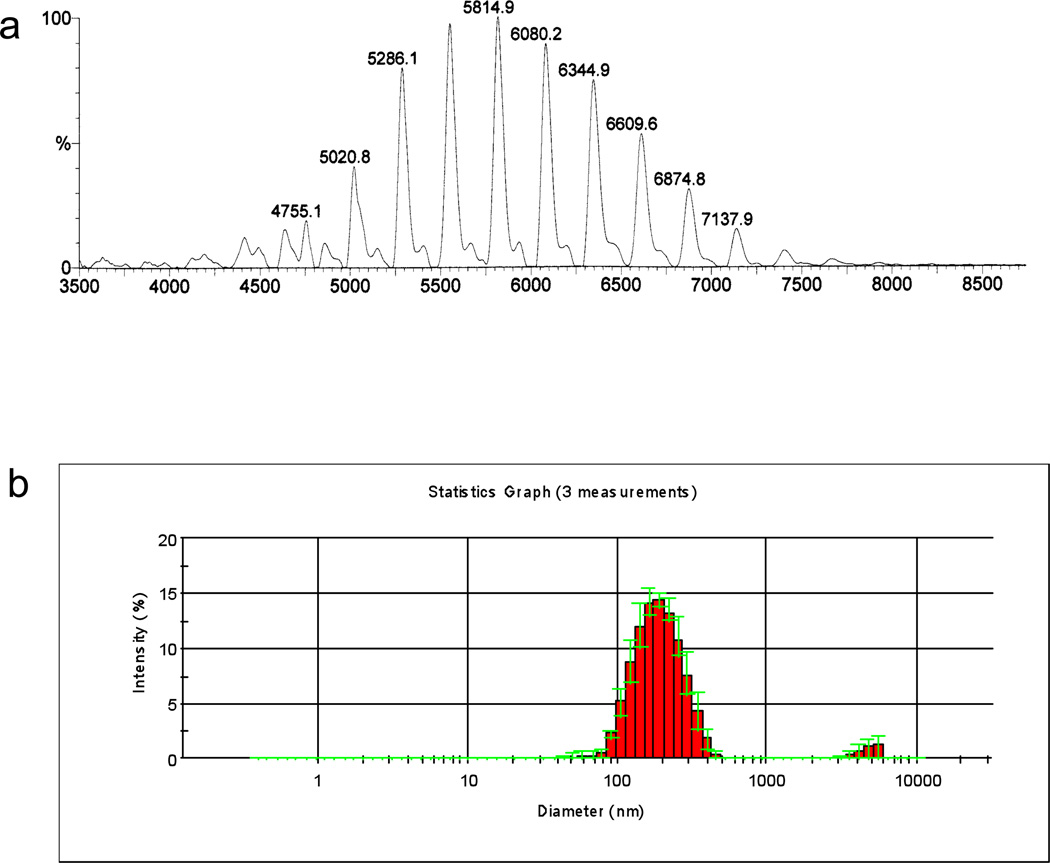
Dendrimers composed of naturally occurring small molecules have the advantage of biocompatibility and biodegradability. When a natural amino acid, lysine, was built up in branched way, it provided maximum surface charges, which could be utilized for binding to siRNA and conveniently conjugated with a variety of functional groups. Moreover, unlike linear polymers, dendrimers that are systemically administered resist immediate hydrolysis by serum proteases. A third-generation lysine dendrimer was conjugated in our lab to oleic acid.[38] By carefully controlling the number of lipids on each dendrimer molecule, we obtained a lipid-functionalized dendrimer, HB-OLD7, with 25 surface amino groups and 7 oleoyl moieties (Fig 2). This 14-nm particle readily complexes with siRNA upon mixing in HEPES buffer at pH7.4 to provide 180-nm nanoparticles (Fig 3).
Figure2.
Chemical structure of lipid-functionalized polylysine dendrimer HB-OLD7.
Figure 3.
MALDI-TOF MS spectrum of HB-OLD7 (a) and particle size distribution of siRNA/HB-OLD7 nanoparticle by dynamic light scattering (b).
HB-OLD7 at low concentration (1µM) efficiently delivered apoB siRNA to cultured mouse hepatocytes (FL83B cells) and knocked down 95% of the corresponding gene without significant toxicity. When HB-OLD7 was complexed with apoB siRNA at 1:10 w/w ratio and injected into mice, the apoB gene was knocked down by 50% after a single injection of a clinically feasible dose (1.25mg/kg). Analysis of apoB protein levels in mouse plasma by Western blot showed that both apoB100 and apoB48, two major products of the apoB gene, were significantly reduced (>70%). Moreover, 24 h after injection, the total plasma cholesterol level of mice decreased by approximately 35%.
Discussion
One of the advantages of non-viral siRNA delivery by nanovehicles is that the formulation needs minimal procedures and the dose of RNAi can be controlled. In most cases, creation of a nanovehicle is accomplished by a simple step of mixing siRNA with delivery agent followed by a short incubation before in vivo administration. However, a successful in vivo siRNA delivery needs many technical issues to be considered during this simplified scenario. For example, the size of the nanovehicle that should ideally be between 50 nm to 200 nm, could easily be affected by the ingredients in the formulation and the mixing methods. A quick mixing of siRNA and delivery agent in a buffered solution usually gives a larger particle (less clear suspension) than in an isotonic glucose solution does, which could result in a less knockdown of a target gene in the target tissue.
While smaller particles are advantageous over larger ones, challenges still exist for the nanovehicles to evenly distribute in the target tissue. It is not well-understood how nanovehicles, especially the ones formed from polymeric materials which most probably have a cell entering mechanism different from those of liposomes, to ‘travel’ further and ‘spread’ deeper into the tissue against the resisting networks of various tissue structures. Histological slides or live animal imaging could assess the distribution profiles of the nanovehicles.
A higher ratio between siRNA and delivery agent usually gives a better knockdown. However, balance between the knockdown and the toxicity should be carefully evaluated because an increased amount of delivery agent, especially positively charged polymers, usually causes toxicity. Although the strong interactions of amine groups with cell membrane and other cell components is believed to be the reason of cytotoxicity,[39–41] other possibility such as induction of immune responses still remains to be elucidated. Designing non-toxic nanovehicle is another challenge to in vivo RNAi.
Conclusion
Targeted delivery is one of the main goals for the design of nanoparticles in future. In most cases, siRNA is to be delivered to the pathological sites rather than the normal tissues. To do this, the detailed analysis of microenvironment of the tissue would be necessary. By conjugating a peptide or antibody which specifically binds to cell surface receptor of a specific tissue, siRNA can be delivered to the pathologic sites. To eliminate non-specific absorption, nanoparticles are often modified with PEG, and provide a neutral or even negative surface to prolong the circulation time and improve the binding to the targeting cells.
Most of the reports on RNAi therapeutics focus on the direct knockdown of an overexperssed gene, or disease-causing gene. What can siRNA do when a disease happens due to an insufficient expression of a gene? One way is to employ gene (plasmid) delivery and express the desired gene. One could also imagine that knocking down a gene that down-regulates the disease-causing gene by RNAi would be an interesting alternative approach.
References
- 1.Rana TM. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Proc Natl Acad Sci U S A. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 5.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 6.Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y, McCaffrey AP. Mol Ther. 2009;17:538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu YL, Rana TM. Mol Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 8.Chiu YL, Rana TM. Rna. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Bioconjug Chem. 2008;19:1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Lim YB, Choi JS, Lee Y, Kim TI, Kim HJ, Yoon JK, Kim K, Park JS. Bioconjug Chem. 2003;14:1214–1221. doi: 10.1021/bc034095g. [DOI] [PubMed] [Google Scholar]
- 11.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. J Control Release. 2008;129:107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MO, Hovgaard MB, Schmitz A, Nyengaard JR, Besenbacher F, Kjems J. Mol Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Wakefield DH, Klein JJ, Wolff JA, Rozema DB. Bioconjug Chem. 2005;16:1204–1208. doi: 10.1021/bc050067h. [DOI] [PubMed] [Google Scholar]
- 16.Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hagstrom JE, Wolff JA. Proc Natl Acad Sci U S A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dash PR, Read ML, Barrett LB, Wolfert MA, Seymour LW. Gene Ther. 1999;6:643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- 18.Trubetskoy VS, Wong SC, Subbotin V, Budker VG, Loomis A, Hagstrom JE, Wolff JA. Gene Ther. 2003;10:261–271. doi: 10.1038/sj.gt.3301888. [DOI] [PubMed] [Google Scholar]
- 19.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choung S, Kim YJ, Kim S, Park HO, Choi YC. Biochem Biophys Res Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 22.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 23.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 24.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishina K, Unno T, Uno Y, Kubodera T, Kanouchi T, Mizusawa H, Yokota T. Mol Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 26.Xia CF, Zhang Y, Boado RJ, Pardridge WM. Pharm Res. 2007;24:2309–2316. doi: 10.1007/s11095-007-9460-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Zhang YF, Bryant J, Charles A, Boado RJ, Pardridge WM. Clin Cancer Res. 2004;10:3667–3677. doi: 10.1158/1078-0432.CCR-03-0740. [DOI] [PubMed] [Google Scholar]
- 28.Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 29.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 30.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paramonov SE, Bachelder EM, Beaudette TT, Standley SM, Lee CC, Dashe J, Frechet JM. Bioconjug Chem. 2008;19:911–919. doi: 10.1021/bc7004472. [DOI] [PubMed] [Google Scholar]
- 32.Shim MS, Kwon YJ. Biomacromolecules. 2008;9:444–455. doi: 10.1021/bm7007313. [DOI] [PubMed] [Google Scholar]
- 33.Williams BR. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 34.Marques JT, Williams BR. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 35.Robbins MA, Rossi JJ. Nat Med. 2005;11:250–251. doi: 10.1038/nm0305-250. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Wu J, Hafdi N, Behr J-P, Erbacher P, Peng L. Chem. Commun. 2006:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 37.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Chem Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Baigude H, McCarroll J, Yang CS, Swain PM, Rana TM. ACS Chem Biol. 2007;2:237–241. doi: 10.1021/cb7000582. [DOI] [PubMed] [Google Scholar]
- 39.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 40.Fogera F, Noonpakdee W, Loretz B, Joojuntr S, Salvenmoser W, Thaler M, Bernkop-Schnurch A. Int. J. Pharm. 2006;319:139–146. doi: 10.1016/j.ijpharm.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 41.Morgan DML, Larvin VL, Pearson JD. J. Cell Sci. 1989;94:553–559. doi: 10.1242/jcs.94.3.553. [DOI] [PubMed] [Google Scholar]