Abstract
Aptamers are single-stranded nucleic acids that fold into stable three-dimensional structures with ligand binding sites that are complementary in shape and charge to a desired target. Aptamers are generated by an iterative process known as in vitro selection, which permits their isolation from pools of random sequences. While aptamers have been selected to bind a wide range of targets, it is generally thought that these molecules are incapable of discriminating strongly alkaline proteins due to the attractive forces that govern oppositely charged polymers (e.g., polyelectrolyte effect). Histones, eukaryotic proteins that make up the core structure of nucleosomes are interesting targets for exploring the binding properties of aptamers because these proteins have positively charged surfaces that bind DNA through non-covalent sequence-independent interactions. Previous selections by our lab and others have yielded DNA aptamers with high affinity but low specificity to individual histone proteins. Whether this is a general limitation of aptamers is an interesting question with important practical implications in the future development of protein affinity reagents. Here we report the in vitro selection of a DNA aptamer that binds to histone H4 with a Kd of 13 nM and distinguishes other core histone proteins by 100 to 480-fold, which corresponds to a ΔΔG of up to 3.4 kcal/mol. This result extends our fundamental understanding of aptamers to include the ability to fold into shapes that selectively bind alkaline proteins.
Keywords: Aptamer, DNA, molecular evolution, histone proteins
Introduction
Aptamers are short nucleic acid polymers (DNA or RNA) that fold into well-defined three-dimensional structures whose surfaces include binding sites that are complementary in shape and charge to a desired target. Aptamers were first discovered in 1990 when two labs independently reported the generation of RNA molecules with specific ligand binding properties from pools of random sequences.[1] In the original papers, Ellington and Szostak called these RNA molecules ‘aptamers’ from the Latin aptus, to fit, while Tuerk and Gold labeled this process ‘SELEX’, which stands for systematic evolution of ligands by exponential enrichment. SELEX is sometimes referred to as in vitro selection or test tube evolution since this laboratory procedure mimics the natural process of Darwinian evolution.[2] In these experiments researchers create a survival-of-the-fittest environment in which individual molecules compete against one another to overcome a selective pressure that is predefined, but often requires binding to a desired target. The small fraction of molecules that meet this requirement are collected and amplified to restore the population to its original size and create progeny molecules that can be further challenged in subsequent rounds of in vitro selection and amplification. Progeny molecules have the ability to inherit genetic mutations, either by intentional mutagenesis or through random mistakes made by a polymerase that can improve the fitness of the molecule for its intended function or lead to deleterious effects that cause the sequence to be removed from the pool.
The ability to harness the power of evolution at the molecular level has led to the development of straightforward procedures for creating tailor-made affinity reagents in the laboratory.[3] Since those initial experiments aptamers have been shown to display a wide range of structural plasticity, and it is now clear that aptamers can be selected to bind almost any kind of molecular target from small molecules to whole cells.[2a, 2e, 4] One major hallmark of aptamers is their ability to bind discrete targets with high specificity. An aptamer generated to bind theophylline, for example, recognizes its cognate ligand 10,000 times better than caffeine, which differs from theophylline by only one methyl group.[5] More recently, our lab developed an aptamer that recognizes an acetyl-lysine post-translational modification in a polypeptide sequence with 2,400-fold specificity.[6]
The strong recognition properties of aptamers combined with the ease by which they can be produced has fueled strong interest in the use of aptamers as affinity reagents in many areas of biotechnology and molecular medicine.[7] Aptamers function efficiently in standard protein-binding assays, including ELISA,[8] western blots analysis,[9] microarrays,[10] and affinity chromatography.[11] In one example, an L-selectin aptamer was used to purify the human L-selectin receptor from Chinese hamster ovary cells.[11] In this case, pure protein was obtained in a single step with 15,000-fold enrichment and 83% recovery. Aptamers have also been used as recognition elements in a variety of biosensors and analytical devices.[12] For example, an aptamer-based dipstick assay was made to detect cocaine,[13] and a colorimetric assay now exists to monitor the levels of lead in the environment.[14] Aptamers are also gaining attention as therapeutic agents.[15] The aptamer, Macugen®, is now approved by the FDA for the treatment of patients affected by neovascular age-related macular degeneration.[16] This VEGF aptamer functions as a drug by inhibiting the binding of VEGF-165 to its receptor. In clinical trials, 80% of the patients treated with this aptamer showed stable or improved vision three months after treatment.[16]
Despite the success that aptamers have achieved in recent years, many basic questions remain about how these molecules fold into shapes with discrete ligand-binding functions.[33-35] The ability for aptamers to target alkaline proteins constitutes an important aspect of this general problem as many proteins have highly basics surfaces. Clearly a greater understanding of the binding properties of aptamers is needed if these molecules are to be used as affinity reagents on a scale as large as the human proteome.[17] Conventional wisdom suggests that aptamers should be incapable of folding into structures that selectively recognize positively charged proteins due to the attractive forces that govern polymers of opposite charge. This problem, commonly referred to as the polyelectrolyte effect, occurs when negatively charged polymers like DNA interact with positively charged polymers like protein to create a ligand binding interaction that releases water molecules and counter ions that previously solvated overlapping regions of both polymers.[18] The magnitude of the polyelectrolyte effect is an important constraint on the ability of aptamers to target alkaline proteins, as aptamers would need to first overcome the barrier that defines the complementary attraction of oppositely charged polymers in order to bind a basic protein with high specificity. While the polyelectrolyte effect has been the subject of previous computational studies,[19] very little experimental consideration has been given to the thermodynamic properties of aptamers and their ability to bind alkaline proteins.
We chose to explore this problem by attempting to evolve DNA aptamers with high specificity to histone H4. Histones are eukaryotic proteins that package DNA into nucleosomes. The core proteins that make up the nucleosome are histones H2A, H2B, H3, and H4.[20] We hypothesized that histone H4 represented an ideal target for this investigation as a previous study by our lab produce a histone-binding aptamer with high affinity but low specificity.[21] Similar results were also achieved by Gonzalez and co-workers in their generation of DNA aptamers to Leishmania infantum histone proteins H2A and H3.[22] Collectively, these examples led us to wonder whether this was a general problem of aptamer binding or a specific problem related to the previous selection strategies. To explore this question in greater detail, we carried out an in vitro selection using counter selection steps to determine whether aptamers could be generated that distinguished histone H4 from the three other core histone proteins. The best aptamer identified in this selection binds to histone H4 with low nanomolar affinity and discriminates against histone proteins H2A, H2B, and H3 by ~100-500-fold. By comparison, all previous selections yielded aptamers with only 2-5-fold specificity. This result demonstrates that aptamers have the ability to fold into structures that distinguish highly basic proteins of similar structure and function.
Results and Discussion
Selection for single-stranded DNAs that Bind to Histone H4
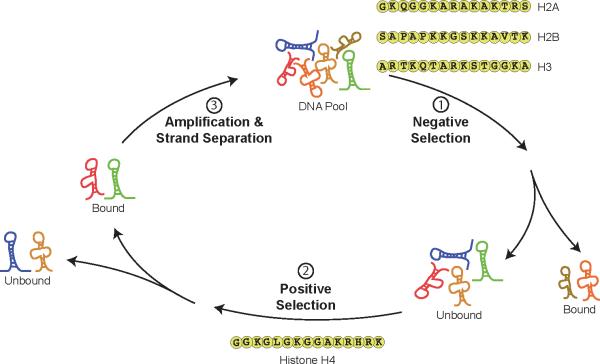
DNA sequences that bind to histone H4 were isolated by iterative rounds of in vitro selection and amplification. The initial pool contained 1012 unique single-stranded DNA molecules with a central random region of 50 unbiased nucleotide positions flanked on both sides with distinct primer-binding sites. To isolate molecules with affinity to the N-terminal region of histone H4, peptides reflecting the N-terminal tail of histone proteins H2A, H2B, H3, and H4 were used in place of the whole proteins. This substitution was feasible because this region of the protein remains natively unstructured when DNA threads itself around the histone octamer to form the nucleosome core.[20] The selection strategy (Figure 1) included a negative selection step to remove molecules that bound the off-target histone sequences of H2A, H2B, and H3, followed by a positive selection step to isolate molecules with affinity to the desired histone H4 target sequence. For each round of selection, the pool was incubated with the off-target peptides H2A, H2B, and H3, which were modified with a C-terminal biotin residue to enable their capture on a streptavidin-affinity matrix. Molecules that remained in the pool were incubated with the desired H4 peptide, and functional aptamers were separated from the unbound pool by injecting the mixture onto a neutral coated capillary. Five injections were made for each round of selection and 1011 molecules were sampled in the first round of the selection.
Figure 1.
In vitro selection strategy used to generate DNA aptamers with high affinity and specificity to histone H4 protein. For each round of selection and amplification, the pool of DNA was pre-cleared by removing DNA sequences with affinity to the N-terminal tails of histone proteins H2A, H2B and H3. Molecules collected in the flow-through were incubated with the N-terminal tail of histone H4 and the bound fraction was separated from the unbound pool by capillary electrophoresis. The set of bound molecules was recovered, amplified and used to generate a new pool of sequences for input into the next round of selection.
Capillary electrophoresis (CE) was chosen for the positive selection step because this technique leads to a higher partitioning efficiency than is commonly observed for traditional gravity filtration.[25] This in turn reduces the number of selection cycles required to generate high quality aptamers from ≥10 to just three or four rounds of selection and amplification. In the case of IgE, for example, an aptamer was generated after four rounds of CE-based selection that exhibited similar binding properties to an aptamer produced after 15 rounds of traditional selection.[26] A second major advantage of CE is that aptamer binding occurs free in solution, which obviates the need for complicated conjugation chemistry that can occlude surface binding sites or alter the native protein structure. Collectively, these advantages are making CE a popular separation technique for the in vitro selection of aptamers that bind peptides and proteins.[27] We have previously used this method to generate a DNA aptamer with >2000-fold specificity to an acetylated lysine residue in a short polypeptide sequence.[6]
To favor the selection of aptamers with high specificity to histone H4, the ratio of the DNA pool to the different histone tails was adjusted in the negative and positive selection steps to maintain high selective pressure on the pool of evolving molecules. In rounds one and two, the ratio of the DNA pool to the off-target histones was 100:1, which was stringent enough to remove DNA sequences that bound the off-target peptides, but permissive enough to allow desirable molecules to remain in the pool. The stringency was then increased in rounds three and four by reducing the ratio to 1:1, which favored the removal of molecules with weaker affinity to the H2A, H2B, and H3 peptide sequences. For each round of positive selection, the ratio was reversed such that the H4 peptide was present at limiting amounts relative to the DNA pool (1:1000). By limiting the target peptide, we aimed to increase competition between the pool and desired histone tail. After four rounds of selection, the DNA pool was cloned and sequenced. We obtained 23 clones and analyzed their sequences by calculating their predicted secondary structures using the computer program mFold (Figure S1). Five of the clones are predicted to fold into structures that are dominated by a simple stem-loop or internal bulge motif. The remaining clones adopt more complicated structures that contain tandem stem-loop motifs. The presence of many highly structured sequences suggests that sophisticated functions, such as the ability to discriminate subtle differences between peptides of similar sequence and composition, require molecules with significant structural complexity.
Affinity and Specificity of the DNA Aptamers
Of the 23 sequences, eight representative clones with different secondary structures were chosen for further analysis. The eight sequences were constructed by solid-phase DNA synthesis, purified by gel electrophoresis, and assayed for affinity to histone H4 whole protein by dot blot analysis.[24] Close inspection of the dissociation constants (Kd's) reveal that all eight clones bind to histone H4 with Kd's of 1 to 10 nM, indicating these sequences are all capable of high affinity binding (Table 1). To examine the specificity of the selected aptamers, dissociation constants were measured for the four strongest binders to the off-target whole proteins H2A, H2B, and H3. In keeping with the literature.[28] this study defined specificity as the ratio of the off-target Kd to the on-target Kd, and aimed to produce aptamers with at least 100-fold specificity to each of the off-target proteins. Results from our initial specificity study demonstrate that the selected clones are relatively specific against histone proteins H2A and H2B (50-150-fold), but fail to discriminate histone H3 by more than 10-fold (Table 2). Creating aptamers that distinguish the N-terminal tail of histone H4 from the N-terminal tail of histone H3 is a challenging problem as previous selections performed in the absence of counter selection methods yielded aptamers with only 2-5-fold selectivity.[21-22]
Table 1.
Sequence and dissociation constants of round 4 aptamers to histone H4 protein.[a]
| Clone | Sequence | Kd (nM) |
|---|---|---|
| 4.33 | TGGTGGGGTTCCCGGGAGGGCGGCTACGGGTTCCGTAATCAGATTTGTGT | 1.3 ± 0.3 |
| 4.60 | TTGGCCCCGCGTGATCATTGAGGGGAGGAGCCGAGGCGGGTCCAAGATTG | 2.6 ± 0.4 |
| 4.32 | ATCACGATATGCCCGGCTCATCGGGGTTCAGTTGGGCGGTCACATGGAAA | 2.8 ± 0.6 |
| 4.36 | TTCAAGGCGGCGAGATTTAGTGGTTGGGAGGCTGTACGCCCTACGTGAAC | 2.9 ± 1.0 |
| 4.58 | TTTAACGTAACCTGCAAGGCGGGGAGGTGCGAGCCCCGTGTGTGGCTTGC | 3.8 ± 1.6 |
| 4.51 | GGTGCTCAGGGAACTGTCTGAGGGATCAGGCTTAAGCCTGTCGAGCAGTT | 5.6 ± 0.7 |
| 4.65 | TCGCATGGAGGGCAGAGCCGCCTGCCGGGATCCGGCCCTCTTGGGCGGGC | 7.4 ± 0.5 |
| 4.57 | TAGTCCCAAGGCACATAAGGGCCGGAGGTCTAGCGTCAGGGATACAGAGA | 8.8 ± 1.9 |
binding condition: 100 mM NaCl, 5 mM MgCl2, 10 mM HEPES (pH 7.5).
Table 2.
Affinity and specificity of representative aptamers.[a]
| H4 | H3 | H2A | H2B | |||||
|---|---|---|---|---|---|---|---|---|
| Clone | Kd (nM) | Sp. | Kd (nM) | Sp. | Kd (nM) | Sp. | Kd (nM) | Sp. |
| 4.33 | 1.3 ± 0.3 | -- | 14 ± 4 | 11 | 204 ± 22 | 157 | 84 ± 5 | 65 |
| 4.60 | 2.6 ± 0.4 | -- | 17 ± 5 | 7 | 332 ± 137 | 128 | 270 ± 117 | 104 |
| 4.32 | 2.8 ± 0.6 | -- | 18 ± 5 | 6 | 287 ± 44 | 103 | 153 ± 32 | 55 |
| 4.36 | 2.9 ± 1.0 | -- | 21 ± 3 | 7 | 249 ± 46 | 86 | 213 ± 70 | 73 |
Specificity (Sp.) is defined as the ratio of Kd (off-target) to Kd (on-target).
Salt Effects
Because electrostatic attraction between the negatively charged DNA backbone and positively charged histone protein might account for the low selectivity observed for the selected aptamers, we decided to examine the role of metal ions on ligand binding affinity. By increasing the concentrations of monovalent and divalent metal ions in the binding buffer, we aimed to stabilize the tertiary structure of the aptamer fold and simultaneously satisfy competing charges on the protein surface. To test this possibility, we chose clone 4.33 for further analysis as this sequence showed the highest degree of specificity to histone H3. Raising the salt concentration from 100 mM NaCl and 5 mM MgCl2 to 500 mM NaCl and 10 mM MgCl2 increased the binding specificity of clone 4.33 for histone H4 versus histone H3 from 10-fold to nearly 30-fold (Table 3). A similar increase in specificity was observed against histones H2A and H2B (up to 422- and 86-fold, respectively). We noticed that increasing the salt concentration beyond this level did not translate into further increases in specificity, indicating that all of the metal binding sites on the aptamer and protein were saturated under the higher salt conditions (data not shown). We speculate that the change in specificity is due to the formation of new intramolecular contacts within the aptamer structure. This hypothesis is consistent with the observation that clone 4.33 adopts a third stem-loop motif when its predicted secondary structure is calculated under conditions that simulate the higher salt concentration (Figure S2).
Table 3.
Affinity and specificity of aptamer 4.33 under low and high salt conditions.
| H4 (pI = 11)[a] | H3 (pI = 11)[a] | H2A (pI = 11)[a] | H2B (pI = 10)[a] | |||||
|---|---|---|---|---|---|---|---|---|
| Condition | Kd (nM) | Sp. | Kd (nM) | Sp. | Kd (nM) | Sp. | Kd (nM) | Sp. |
| Low sal[b] | 1.3 ± 0.3 | -- | 14 ± 4 | 11 | 204 ± 22 | 157 | 84 ± 5 | 65 |
| High salt[c] | 9.6 ± 6.2 | -- | 273 ± 78 | 28 | 4050 ± 70 | 422 | 825 ± 3 | 86 |
pI: isoelectric point.
low salt condition: 100 mM NaCl, 5 mM MgCl2, 10 mM HEPES (pH 7.5).
high salt condition: 500 mM NaCl, 10 mM MgCl2, 10 mM HEPES (pH 7.5).
Directed Evolution
In an effort to isolate aptamers with greater specificity for histone H4, we used directed evolution to optimize clone 4.33 for improved ligand binding affinity and specificity. We created a second-generation library based on the parent sequence of clone 4.33 in which each nucleotide position in the aptamer sequence was doped with a 15% mixture of the other three nucleotides. This level of mutagenesis was intended to optimize contacts within the aptamer structure and produce mutations that would lead to greater discrimination between histone H4 and the other three histone proteins. As a precaution new primer binding sites were added to the flanking regions to avoid the unwanted enrichment of aptamers from the original library. The doped library was subjected to three iterative rounds of directed evolution using two different selection strategies. The first strategy was performed in a manner identical to the original in vitro selection with a negative selection step performed on streptavidin-coated beads to remove molecules with affinity to the off-target sequence followed by a CE-based positive selection step to recover molecules that bound the N-terminal tail of histone H4. For each selection round, the ratio of the off-target to pool and on-target to pool was maintained at 1:1 and 1:1000, respectively. In the second selection strategy, both the negative and positive selection steps were performed using traditional affinity chromatography methods to separate the bound molecules from the unbound pool. After three rounds of directed evolution, both libraries were cloned and sequenced to examine the diversity of molecules that remained in each pool.
Eight clones from the CE-based selection and nine clones from bead-based selection were aligned with clone 4.33 (Table S2). The average number of mutations per sequence was 7.6, which closely approximates the number of mutations expected for a library of 50-nucleotides that was doped at a level of 15% per nucleotide position. Close inspection of the aligned sequences reveals several small patches of conserved nucleotides that are distributed among numerous single-point mutations. To examine the extent to which any of the selected sequences showed higher selectivity for histone H4, we randomly chose three sequences from the output of each selection and measured their affinity and specificity for histone H4. Each sequence was synthesized by solid-phase DNA synthesis, purified by gel electrophoresis, and assayed for affinity to histone proteins H2A, H2B, H3, and H4. Surprisingly, only clone 3.13 isolated from the CE-based selection showed high selectivity to histone H4 (Table 4). The remaining clones were unable to distinguish histone H4 from histone H3 by more than ~20-fold, which is less than the parent sequence (clone 4.33). Aptamer CE-3.13, however, binds histone H4 with a Kd of 13 nM and discriminates against histone proteins H2A, H2B, and H3 by 477-, 165-, and 100-fold, respectively (Figure 2). This result emphasizes the challenge of isolating aptamers with reasonable selectivity to highly basic proteins, but provides evidence that such sequences are not so rare that they cannot be discovered by in vitro selection.
Table 4.
Affinity and specificity of representative aptamers isolated by directed evolution.[a]
| H4 | H3 | H2A | H2B | |||||
|---|---|---|---|---|---|---|---|---|
| Clone | Kd (nM) | Sp. | Kd (nM) | Sp. | Kd (nM) | Sp. | Kd (nM) | Sp. |
| Parental sequence | ||||||||
| 4.33 | 9.6 ± 6.2 | -- | 273 ± 78 | 28 | 4050 ± 70 | 422 | 825 ± 3 | 86 |
| CE-based directed evolution | ||||||||
| CE-3.1 | 55 ± 5 | -- | 318 ± 16 | 6 | 5500 ± 800 | 100 | 5400 ± 2100 | 98 |
| CE-3.5 | 71 ± 8 | -- | 490 ± 95 | 7 | 10500 ± 700 | 148 | 5500 ± 800 | 77 |
| CE-3.13 | 13 ± 1 | -- | 1300 ± 50 | 100 | 6200 ± 100 | 477 | 2150 ± 70 | 165 |
| Bead-based directed evolution | ||||||||
| BB-3.12 | 22 ± 1 | -- | 415 ± 12 | 19 | 5950 ± 70 | 270 | 3900 ± 400 | 177 |
| BB-3.13 | 11 ± 2 | -- | 264 ± 7 | 23 | 4800 ± 300 | 425 | 3800 ± 500 | 332 |
| BB-3.16 | 20 ± 2 | -- | 121 ± 2 | 11 | 3750 ± 70 | 332 | 1800 ± 40 | 159 |
binding condition: 500 mM NaCl, 10 mM MgCl2, 10 mM HEPES (pH 7.5).
Figure 2.

Solution binding affinity and specificity of aptamer CE-3.13. Equilibrium binding affinity was measured by dot blot analysis for histone proteins H4 (circles), H3 (squares), H2B (diamonds) and H2A (triangles).
Studies of Site-directed Mutations
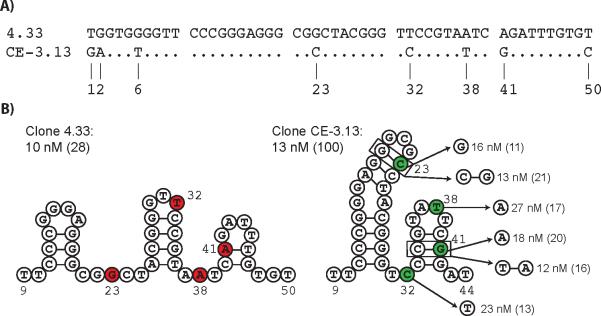
To examine the genetic changes that led to improved specificity, we compared the predicted secondary structure for the parent sequence to the evolutionary optimized variant. This analysis demonstrates that four of the eight single-point mutations occur in regions of the sequence that define the predicted secondary structure (Figure 3A). We therefore reverted each of the four point mutations individually back to their original nucleotide, and measured the solution binding affinity for histone proteins H3 and H4. While the four revertant clones recognized histone H4 with Kd's that are within 2-fold of aptamer CE-3.13, none of sequences were able to distinguish histone H3 by more than 20-fold (Figure 3B). This loss in selectivity suggests that each of the four point mutations play an important role in the folding and recognition properties of aptamer CE-3.13.
Figure 3.
Aptamer binding motif and analysis. (A) Sequence alignment of aptamers 4.33 and CE-3.13 showing the eight genetic mutations that gave rise to an evolutionary optimized variant with high specificity to histone H4. (B) The predicted secondary structures of aptamers 4.33 and CE-3.13 are shown with the mutated positions highlighted in red and green, respectively. Binding affinity and specificity (parentheses) of each reversion and compensatory mutation is shown next to the structure of aptamer CE-3.13.
To further explore the evolved mutations, we generated variants that contained compensatory mutations in stem-loop regions of the predicted secondary structure. Two clones were constructed that restore Watson-Crick base pairs to the C23G and G41A revertants by changing the G:G and C:A mismatches to C:G and T:A base pairs, respectively. These engineered clones bind histone H4 with Kd's equivalent to the evolutionary optimized aptamer, but again fail to restore selectivity to the aptamer sequence (Figure 3B). This result demonstrates that the selected mutations, G23C and A41G, which form G:C and C:G base pairs in aptamer CE-3.13, impart additional functionality beyond simply maintaining a contiguous helix in the stem-loop region of the secondary structure. One possibility is that these mutations form new contacts within the architecture of the aptamer that rigidify its structure and limit the amount of flexibility in and around the ligand-binding pocket.
To gain a better understanding of how the structure of aptamer CE-3.13 contributes to its recognition of the N-terminal tail of histone H4, we performed a hydroxyl radical footprinting analysis in the absence and presence of histone proteins H2A, H2B, H3, and H4. Consistent with the high specificity of aptamer CE-3.13 for histone protein H4, protection of the DNA backbone occurred to a greater extent for histone H4 than for any of the other three histone proteins (Figure S4). Careful analysis of the resulting gel indicates that histone proteins H2A, H2B, and H3 protect residues 34-38 of the aptamer. This relatively small region is likely due to weak electrostatic interactions between the DNA backbone and the three histone proteins. However, the case is quite different for histone H4, which protects a much larger region of the aptamer from hydroxyl cleavage. Here a clear footprint is observed for residues 31-45, which constitute a strong binding interface with histone H4. Combining this information with the predicted secondary structure suggests that the second stem-loop motif forms a binding pocket that is complementary in shape and charge to the N-terminal tail of histone H4. Indeed, three of the four genetic mutations observed in aptamer CE-3.13 occur in this region of the oligonucleotide, which implies that each of these mutations play an important role in the binding of histone H4. The fourth mutation, which occurs in the first stem-loop motif could help with aptamer stability by improving the packing interactions between the two stem-loop motifs. This result is consistent with the interpretation that the four selected mutations increase protein binding specificity by rigidifying the aptamer structure.
Discussion
We have applied the strategy of in vitro selection and directed evolution to isolate a single-stranded DNA molecule with high affinity and specificity to histone H4. When we began this study it was not obvious a priori that a random pool of DNA sequences would produce a nucleic acid molecule that folded itself into a shape that recognized an alkaline protein with high affinity and specificity. A previous study by our lab that aimed to produce a set of DNA aptamers to histone H4 yielded a number of high affinity sequences (Kd ~5-10 nM); however, the best sequence could only discriminate histone H3 by a factor of 5-fold.[21] Similar results were obtained by Ramos and co-workers on histone proteins H2A and H3, which produced aptamers with only 2-3-fold specificity.[22] Whether this was a general problem of aptamers (i.e., the potential inability of negatively charged polymers to fold into shapes that recognize positively charged polymers with high selectivity) or simply a limitation of the previous selection strategy was unclear. We therefore designed a new selection strategy that included the use of stringent counter selection steps between iterative rounds of selection and amplification, since this approach has been widely used to generate aptamers with specific ligand binding properties.[5-6, 29] The goal of this selection was to remove DNA molecules from the pool that exhibited high affinity to the off-target histone proteins H2A, H2B, and H3. In doing so, we aimed to address the broader question of whether aptamers could be used to bind alkaline proteins with high specificity.
Comparison of the binding properties of the aptamers isolated by directed evolution (Table 4) reveals a striking difference in the tolerance of each molecule for the off-target histone proteins. For example, aptamer CE-3.13, which was isolated using the capillary electrophoresis method, is significantly more fit in terms of its ability to bind histone H4 than all of the other DNA aptamers. This aptamer binds to histone H4 with a solution binding affinity of 13 nM and distinguishes the three remaining core histone proteins by a factor of 100-477-fold, which corresponds to a binding energy of up to 3.4 kcal/mol. In contrast, the less fit aptamers also bind to histone H4 with low nanomolar affinity, and are able to distinguish histone proteins H2A and H2B with high specificity (≥100-fold), but struggle with their ability to discriminate histone proteins H4 and H3. This problem was observed in our previous selection and likely stems from the fact that both proteins have similar sequence composition in their N-terminal tails, which was the protein region targeted in both selections. While it is exciting to wonder whether the isolation of aptamer CE-3.13 was due to the high partitioning efficiency of the capillary electrophoresis-based separation, many additional aptamers will need to be tested before this question can be answered.
One interesting observation to come from the aggregate set of binding data is that aptamers with high affinity are not automatically more specific for their target ligands. Although it has long been assumed that the easiest way to improve aptamer specificity is to increase its shape and charge complementary for a given target,[30] a recent study by Szostak and co-workers suggests that specificity is a physical property that emerges when biopolymers adopt folded structures that are reinforced with additional intramolecular contacts.[31] This revised aptamer binding theory takes into account the free energy term provided by intramolecular contacts that contribute to the overall stability of the tertiary structure. According to this model, it is expected that as an aptamer evolves from an initial simple motif to a more complex tertiary structure it will acquire addition structural elements that allow it to form a more rigid ligand-binding pocket that is less willing to accept target analogues. This hypothesis is consistent with the binding properties of our aptamers and suggests that aptamer CE-3.13 adopts a folded structure, either as a free molecule or in the bound state that is more rigid than the other aptamers that we tested.
Mutagenesis data supports the prediction that aptamer CE-3.13 represents a complex solution to the chemical problem of how a DNA molecule would fold itself into a tertiary structure with a ligand binding site that is capable of selectively recognizing the alkaline protein histone H4. Single-nucleotide revertants constructed for each of the four genetic mutations that occur in the region of the sequence that defines the predicted secondary structure maintain high affinity binding but abate specificity. Furthermore, specificity is not restored when the C23G and G41A revertants are modified with compensatory mutations that change the G19:G23 and C34:A41 mismatches to C19:G23 and T34:A41 base pairs, respectively. Since positions 23 and 41 occur in adjoining helices of the predicted secondary structure, successful resuscitation of specificity would have meant that these mutations were selected to maintain two contiguous helices in the aptamer structure. However, since neither compensatory mutation allowed the aptamer to recover specificity, it is reasonable to assume that both mutations play a greater role in aptamer folding. This prediction is supported by our footprinting analysis (Figure S4).
A second interesting observation to emerge from our results was that a limited sampling of aptamers (in this case six aptamers were examined after directed evolution) yielded a DNA molecule that was capable of achieving high specificity. One interpretation that is consistent with our results is that the counter selection method used to isolate these aptamers provided access to complex structures that are capable of folding into rigid shapes with well-defined ligand binding sites, but that these structures are still somewhat rare when compared to simpler structures that continue to dominate the pool. This scenario agrees with the long held belief that in vitro selection tends to produce the simplest solutions to a given biochemical problem. This hypothesis is evident from our previous selection for histone-binding aptamers, which selected for protein-binding affinity only and produced molecules with high affinity but low specificity. In contrast, the current selection strategy, which included a direct selection for specificity allowed us to favor the enrichment of aptamers with specific ligand binding properties by removing many of the simpler solutions from the pool. We speculate that our previous selection contained aptamers that were capable of high specificity but these molecules were so rare that random sampling of the selection output could not identify them.
Conclusion
In summary, we provide evidence that nucleic acid aptamers can be evolved by in vitro selection to fold into shapes that recognize alkaline proteins with high specificity. Because these aptamers are rare relative to simpler solutions that bind with high affinity but low specificity, their isolation requires strong counter selection measures that deplete the pool of low specificity binders. In the broader context of aptamer binding, these results suggest that aptamers could be used as affinity reagents to target a wide range of human proteins, including structures whose surfaces are dominated by an abundance of positive charge.
Experimental Section
General
DNA oligonucleotides were purchased from Integrated DNA Technologies and purified by denaturing polyacrylamide gel electrophoresis. Histone peptides (H4, GGKGLGKGGAKRHRK; H3, ARTKQTARKSTGGKA; H2A, GKQGGKARAKAKTRS; H2B, SAPAPKKGSKKAVTK) were purchased from Sigma-Aldrich in >95% purity. Histone peptides with a C-terminal biotin residue (H4, GGKGLGKGGAKRHRK-Biotin; H3, ARTKQTARK-STGGKAGKBiotin; H2A, GKQGGKARAKAKTR-SGK-Biotin; H2B, SAPAPKKGSKKAVTK-Biotin) were purchased from New England Peptide in >95% purity. Histone proteins H2A, H2B, H3, and H4 were purchased from New England BioLabs. The 100-mer DNA library containing a random region of 50 nucleotides flanked on both sides with constant PCR primer binding sites, and a second generation DNA library based on clone 4.33 were purchased from the Keck Facility at Yale University.
In vitro Selection
For each round of selection, the DNA library was amplified by PCR using a 6-carboxyfluorescein (6-FAM) labeled forward primer (5’-FAM-GAG CTA CGT ACG AGG ATC CGG TGA G-3’) and a biotin labeled reverse primer (5’-Biotin-GGA CCT GGG GCC GAA GCT TAG CAG T-3’). The pool was made single-stranded by immobilizing the dsDNA onto streptavidin-coated agarose beads and eluting the top strand with 0.15 M NaOH. The single-stranded library was neutralized, ethanol precipitated, and folded by heating to 95 °C for 5 minutes and cooling on ice for 10 minutes in selection buffer (100 mM NaCl, 5 mM MgCl2, 10 mM HEPES, pH 7.5). The DNA library was incubated for 1 hour at 24 °C with histone peptides H2A, H2B and H3 derivatized with a C-terminal biotin residue. After 1 hour, the solution was passed through a column of streptavidin-coated agarose beads. The unbound fraction was collected, concentrated by ethanol precipitation, and refolded. The DNA pool was incubated with the histone H4 peptide for 1 hour at 24 °C, and histone H4 aptamers were isolated by separating the bound molecules from the unbound library by capillary electrophoresis. After four rounds of in vitro selection and amplification, the library was cloned and sequenced to examine the diversity of molecules that remained in the pool.
Capillary Electrophoresis
Capillary electrophoresis was performed on Beckman ProteomeLab PA 800 Protein Characterization System. Prior to use, the glass capillary (0.1 mm inner diameter, total length = 60 cm) was rinsed with water and equilibrated with selection buffer. A small portion (70 nl) of the library/peptide mixture was injected onto the capillary using pressure injection (0.5 psi for 5 seconds) and electrophoresis was performed under a constant voltage of 15 kV at 20 °C for 35 minutes. Laser-induced fluorescence (LIF) was used to monitor the separation of 6-FAM labeled DNA (excitation = 488 nm; emission = 520 nm). Five injections were performed for each round of in vitro selection.
Directed Evolution
Directed evolution was performed to optimize clone 4.33 (5’-CAC GAC TCT CAC CTC ATA GC tgg tgg ggt tcc cgg gag ggc ggc tac ggg ttc cgt aat cag att tgt gt CTG GTT CTG TAG ACG GCT TG-3’). A degenerate DNA library was constructed by solid-phase DNA synthesis using mixtures of phosphoramidite monomers that allowed for 15% mutagenesis to occur at each nucleotide position in the aptamer sequence. Lower case bases in clone 4.33 denote a region of the sequence that contains 85% of the wild-type nucleotide and 5% of each of the other three bases. New PCR primers were used to avoid possible contamination with the first-generation library. The DNA library was amplified by PCR and made single-stranded by denaturing on streptavidin-coated agarose beads. The pool of single-stranded DNA was split into two parts, and two separate selections were carried out in parallel. The first selection was performed as described above with the exception that only histone H3 peptide was used in the negative selection step. All other steps were the same, including the solution-phase separation of the bound aptamers by capillary electrophoresis. The second selection was perform in a similar manner with the exception that the positive selection step was performed by capturing the portion of DNA that remained bound to histone H4 peptide on a streptavidin-coated magnetic beads, washing to remove the unbound molecules, and amplifying the bound material by PCR. After three rounds of in vitro selection and amplification, both libraries were cloned and sequenced to examine the diversity of molecules that remained in the pool.
DNA sequencing and analysis
DNA sequences present in the output of each selection were amplified by PCR and cloned into a pJET DNA cloning vector (Fermentas). The vectors were transformed into E. coli TOP10 competent cells and grown on ampicillin agar plates at 37 °C with an overnight incubation. Individual colonies were randomly picked and checked by colony PCR to ensure that the vector contained the insert. Positive clones were grown in liquid media, mini-preped, and sequenced at the ASU Sequencing Facility. The predicted secondary structures were determined using the computer program mFold.[23]
Dot blot binding assay
DNA aptamers (150 pmole) were labeled with 32P by incubating with [] ATP and T4 polynucleotide kinase for 1 hour at 37 °C. The [32P]-labeled aptamers were desalted on a sephadex G-25 column, diluted with selection buffer and folded by heating at 95 °C for 5 minutes and cooling on ice for 10 minutes. The purified aptamers were then divided into 12 tubes and incubated for 1 h at 24 °C with the histone protein poised at concentrations that span the expected Kd (typically 0.1-100 nM). After 1 hour, the solutions were placed into a vacuum manifold dot blot apparatus and the bound aptamers were partitioned away from the free DNA by passing the solution through a layer of nitrocellulose and nylon membranes.[24] To reduce any nonspecific binding and retention of the free DNA, the nitrocellulose membrane was presoaked for 10 minutes in 0.4 M KOH and rinsed with water until the pH returned to neutral. Prior to analysis, both membranes were equilibrated in selection buffer for 30 minutes at 4 °C followed by passing selection buffer through the wells with vacuum. Samples were then loaded into the dot blot manifold and vacuum was applied to separate the bound aptamer from the unbound DNA. Aptamers that are bound to histone become captured on the surface of the nitrocellulose membrane (top layer), while unbound DNA passes through the nitrocellulose layer and becomes captured on the nylon membrane (bottom layer). The wells were then washed with selection buffer, dried, and the amount of radioactivity present on both membranes was determined by phosphorimaging. The protein-bound aptamer fraction and protein concentration were used to determine the Kd using the following equation: Ib/(Ib + Iu) = c1 +c2([protein]/([protein] + Kd)), where Ib and Iu are the intensity of protein-bound aptamer and free aptamer, respectively, c1 and c2 are constants. Dissociation constants were calculated using a nonlinear least-squares regression analysis performed with DeltaGraph program.
Structural probing by hydroxyl radical footprinting
Hydroxyl radical footprinting reactions were performed similar to previously described.[32] Briefly, [32P]-5’-end labeled aptamer was incubated in high salt conditions in the presence or absence of histone proteins and equilibrated for 1 hour at room temperature in a total volume of 10 μl. The hydroxyl radical cleavage reaction was prepared by carefully spotting 1 μl of a fresh Fe(II)-EDTA solution (3 mM Fe (II)/6 mM EDTA), 1 μl sodium ascorbate solution (30 mM), and 1 μl hydrogen peroxide solution (1.8% freshly diluted from a 30% stock) as three separate drops on the wall of the tube. The reaction was initiated by simultaneously mixing the three individual reagent drops together and immediately adding to the aptamer solution. The reaction was quenched by addition of 7 μl of stop solution (100 mM thiourea) after 1 min of digestion.
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Cancer Institute [CA125510]. We thank members of the Chaput lab for helpful comments and suggestions.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.a Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; b Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.a Wilson DS, Szostak JW. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]; b Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]; c Bittker JA, Phillips KJ, Liu DR. Curr. Opin. Chem. Biol. 2002;6:367–374. doi: 10.1016/s1367-5931(02)00321-6. [DOI] [PubMed] [Google Scholar]; d Famulok M, Mayer G, Blind M. Acc. Chem. Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]; e Mayer G. Angew. Chem. Int. Ed. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 3.Bunka DHJ, Stockley PG. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 4.Jayasena SD. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 5.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 6.Williams BAR, Lin L, Lindsay SM, Chaput JC. J. Am. Chem. Soc. 2009;131:6330–6331. doi: 10.1021/ja900916p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Mukhopadhyay R. Anal. Chem. 2005;77:115–118. doi: 10.1021/ac053500p. [DOI] [PubMed] [Google Scholar]; b Famulok M, Hartig JS, Mayer G. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 8.Drolet DW, Moon-McDermott L, Romig TS. Nat. Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 9.Murphy MB, Fuller ST, Richardson PM, Doyle SA. Nuc. Acids Res. 2003;31:e110. doi: 10.1093/nar/gng110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock C, Coleman M, Collins B, Davis J, Foulds G, Gold L, Greef C, Heil J, Heilig JS, Hicke B, Hurst MN, Husar GM, Miller D, Ostroff R, Petach H, Schneider D, Vant-Hull B, Waugh S, Weiss A, Wilcox SK, Zichi D. Proteomics. 2004;4:609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 11.Romig TS, Bell C, Drolet DW. J. Chromatrgr. B. 1999;731:275–284. [PubMed] [Google Scholar]
- 12.Tombelli S, Minunni M, Mascini M. Biosens. Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Mazumdar D, Lu Y. Angew. Chem. Int. Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Lee JH, Lu Y. Adv. Mat. 2008;20:3263–3267. [Google Scholar]
- 15.White RR, Sullenger BA, Rusconi CP. J. Clin. Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui MA, Keating GM. Drugs. 2005;65:1571–1577. doi: 10.2165/00003495-200565110-00010. [DOI] [PubMed] [Google Scholar]
- 17.Taussig MJ, Stoevesandt O, Borrebaeck CAK, Bradbury AR, Cahill D, Cambillau C, de Daruvar A, Dübel S, Eichler J, Frank R, Gibson TJ, Gloriam D, Gold L, Herberg FW, Hermjakob H, Hoheisel JD, Joos TO, Kallioniemi O, Koegl M, Konthur Z, Korn B, Kremmer E, Krobitsch S, Landegren U, van der Maarel S, McCafferty J, Muyldermans S, Nygren P, Palcy S, Plückthun A, Polic B, Przybylski M, Saviranta P, Sawyer A, Sherman DJ, Skerra A, Templin M, Ueffing M, Uhlén M. Nat. Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- 18.Lohman TM. CRC Crit. Rev. Biochem. 1986;19:191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- 19.Jayaram B, DiCapua FM, Beveridge DL. J. Am. Chem. Soc. 1991;113:5211–5215. [Google Scholar]
- 20.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 21.Lin L, Hom D, Lindsay SM, Chaput JC. J. Am. Chem. Soc. 2007;129:14568–14569. doi: 10.1021/ja076488m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a Ramos E, Piñeiro D, Soto M, Abanades DR, Martín ME, Salinas M, González VM. Laboratory Investigation. 2007;87:409–416. doi: 10.1038/labinvest.3700535. [DOI] [PubMed] [Google Scholar]; b Ramos E, Moreno M, Martin ME, Soto M, Gonzalez VM. Oligonucleotides. 2010;20:207–213. doi: 10.1089/oli.2010.0240. [DOI] [PubMed] [Google Scholar]
- 23.Zuker M. Nuc. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong I, Lohman TM. Proc. Natl. Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berezovski MV, Musheev MU, Drabovich AP, Jitkova JV, Krylov SN. Nat. Protoc. 2006;1:1359–1369. doi: 10.1038/nprot.2006.200. [DOI] [PubMed] [Google Scholar]
- 26.Mendonsa SD, Bowser MT. J. Am. Chem. Soc. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 27.a Mendonsa SD, Bowser MT. J. Am. Chem. Soc. 2005;127:9382–9383. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]; b Mosing RK, Mendonsa SD, Bowser MT. Anal. Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]; c Berezovski M, Musheev M, Drabovich A, Krylov SN. J. Am. Chem. Soc. 2006;128:1410–1411. doi: 10.1021/ja056943j. [DOI] [PubMed] [Google Scholar]; d Tang J, Xie J, Shao N, Yan Y. Electrophoresis. 2006;27:1303–1311. doi: 10.1002/elps.200500489. [DOI] [PubMed] [Google Scholar]
- 28.Carothers JM, Oestreich SC, Davis JH, Szostak JW. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sazani PL, Larralde R, Szostak JW. J. Am. Chem. Soc. 2004;126:8370–8371. doi: 10.1021/ja049171k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaton BE, Gold L, Zichi DA. Chem. Biol. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 31.Carothers JM, Oestreich SC, Szostak JW. J. Am. Chem. Soc. 2006;128:7929–7937. doi: 10.1021/ja060952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain SS, Tullius TD. Nat. Protoc. 2008;3:1092–1100. doi: 10.1038/nprot.2008.72. [DOI] [PubMed] [Google Scholar]
- 33.Patel DJ, Suri AK. Rev. Mol. Biotechnol. 2000;74:39–60. doi: 10.1016/s1389-0352(99)00003-3. [DOI] [PubMed] [Google Scholar]
- 34.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Proc. Natl. Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long SB, Long MB, White RR, Sullenger BA. RNA. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.