The initial stages of influenza A virus infection are mediated by the binding of the viral hemagglutinin (HA) to sialylated glycan receptors on host epithelial cells.[1] The specificity of the HA is believed a key determinant of viral host range.[2] While all 16 influenza HA subtypes are found in avian viruses, only three are found in viruses adapted to humans (H1, H2 and H3), each resulting in a major pandemic. HAs from avian and human viruses are characterized by their preference for α2–3 and α2–6 linked sialic acids, respectively. Studies now suggest that other elements of sialoglycan sequence are also important factors of HA specificity that contribute to the species barrier.[3] Recently, human and swine respiratory epithelial cells were shown to express sialylated N-glycans with extended poly-N-acetyllactosamine (poly-LacNAc) chains.[4] Poly-LacNAc chains are Galβ1–4GlcNAcβ1–3 tandem repeats that extend N- and O-linked glycans of glycoproteins and contribute to the biology mediated by glycan binding proteins.[5] Sasisekharan and coworkers have suggested that human HAs bind preferentially to extended α2–6 sialosides and may be critically important for viral adaptation to humans.[4a, 6]
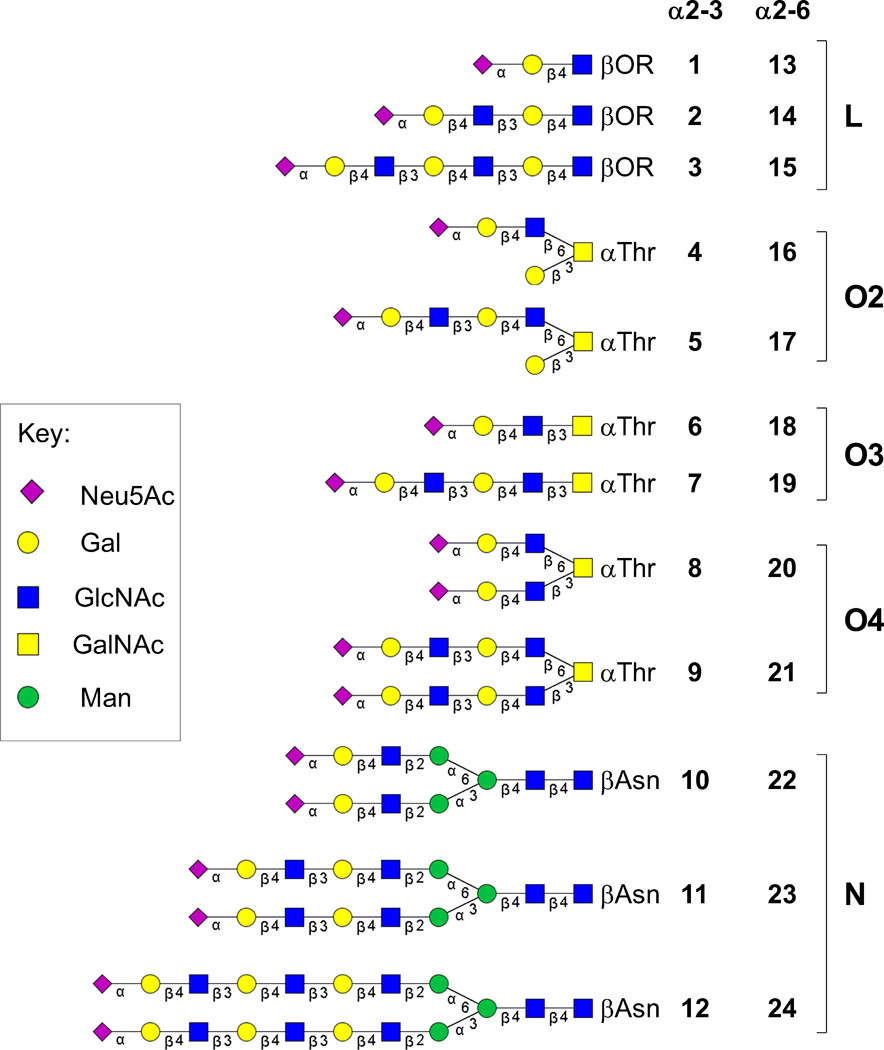
Studies on the preference of influenza HAs for extended glycans have employed synthetic sialosides that are linear terminal fragments of natural N- and O-linked glycans, which differ in their core structure and are often branched.[4a, 7] To more fully address the influence of poly-LacNAc chains on HA specificity in the context of natural glycans, we have synthesized a series of sialylated poly-LacNAc structures on intact O- (4–9, 16–21) and N-linked glycan (10–12, 22–24) cores (Figure 1). These sialosides were incorporated into a custom glycan microarray alongside the linear terminal fragments (1–3, 13–15) for analysis of specificities of human and avian influenza HAs.
Figure 1.
Structures of sialylated poly-LacNAc linear (L) fragments (1–3, 13–15) and the same sequences elaborated on O-linked (O2, O3, O4) (4–9, 16–21) and N-linked (N) glycan cores (10–12, 22–24).
Several groups have reported chemical and chemo-enzymatic syntheses of poly-LacNAc structures.[8] For synthesis of extended natural N- and O-linked glycans, our strategy relied on enzymatic elaboration of advanced core structures. The α2–3 a n d α 2–6 sialoside targets comprised O-linked (Cores 2–4) and N-linked cores with up to two and three LacNAc repeats, respectively. Representative syntheses for N- (11 and 23) and Core-2 O-linked glycans (5 and 17) are described in Scheme 1. Key LacNAc extensions were attained by alternating reactions using recombinant Helicobacter pylori β1–3-N-acetylglucosaminyltransferase (β1–3GlcNAcT)[8b] and the bacterial β1–4-galactosyltransferase/UDP-4′-Gal-epimerase fusion protein (GalT-GalE).[9] Reaction of N-glycan 25 with UDP-GlcNAc (4 eq.) using β1–3GlcNAcT followed by treatment with UDP-Glc (4 eq.) and GalT-GalE allowed efficient construction of LacNAc on both antennae affording 27 (Scheme 1a). Divergent sialylation of 27 using rat α2–3-sialyltransferase (rST3Gal-III) or human α2–6-sialyltransferase (hST6Gal-I), with CMP-Neu5Ac gave the desired α2–3 11 and α2–6 23 products, respectively. The synthesis of O-linked cores 3–4 and the tri-LacNAc N-linked glycans were conducted following similar conditions (Schemes S1–S6 in the Supporting information).
Scheme 1.
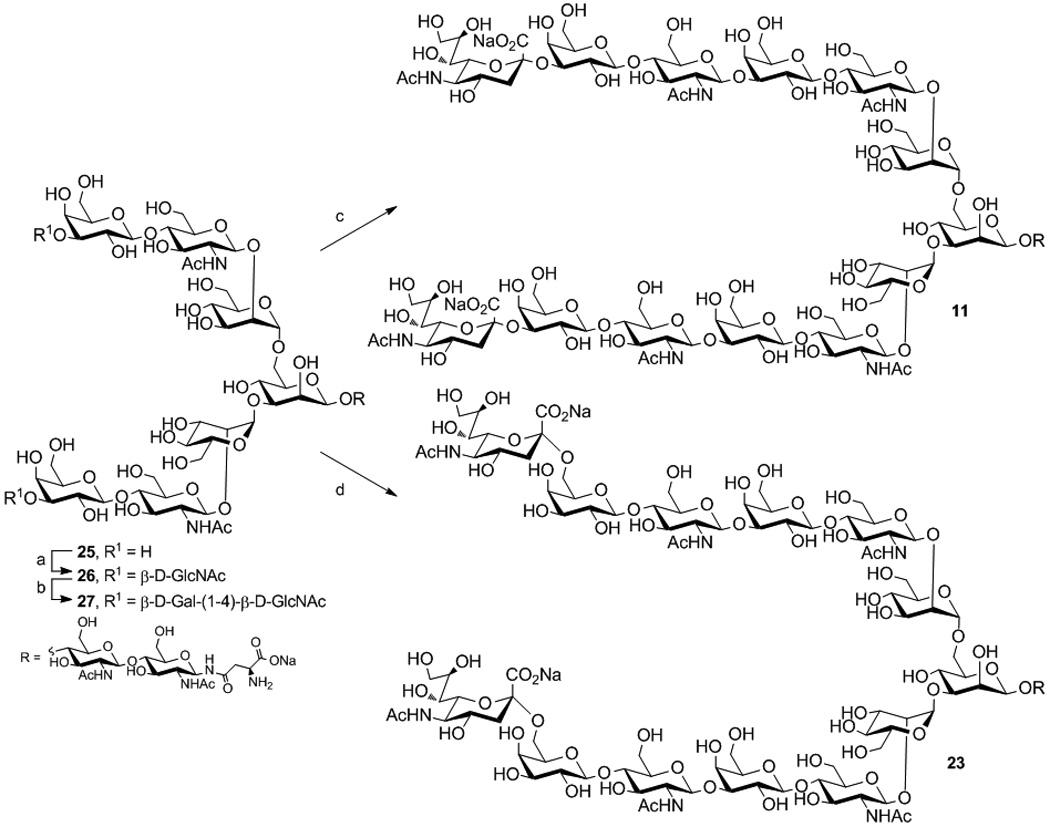
a. Enzymatic transformations of 25 to 11 and 23 a) β1–3GlcNAcT, UDP-GlcNAc; b) GalT-GalE, UDP-Glc; c) rST3Gal-III, CMP-Neu5Ac; d) hST6Gal-I, CMP-Neu5Ac.
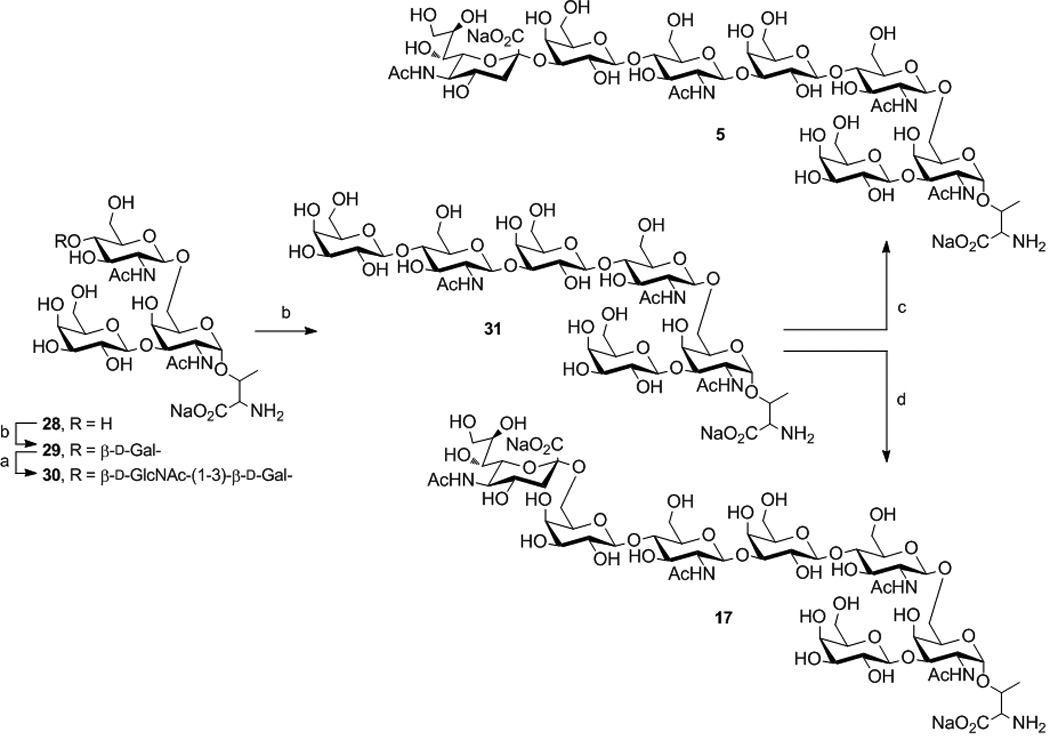
b. Enzymatic transformation of 28 to 5 and 17. See Scheme 1a for conditions.
Core-2 O-linked glycans are commonly extended with poly-LacNAc off the β1–6 branch. Initial galactosylation of 28 added Galβ1–4 to GlcNAc giving 29 (Scheme 1b). As both branches of 29 present terminal Gal, two sites were potentially reactive for GlcNAc addition. Regioselective reaction on the β1–6 branch was anticipated as β1–3GlcNAcT demonstrates higher selectivity for Galβ1–4GlcNAc substrates. Thus, under controlled conditions using UDP-GlcNAc (2 eq.), selective elongation of the β1–6 branch was achieved to afford 30.[10] NMR and MS analysis confirmed addition of a single GlcNAc unit. The asialo di-LacNAc structure 31 was prepared by reaction of 30 with UDP-Glc catalyzed by GalT-GalE. Finally, selective sialylation of 31 was performed with either rST3Gal-III or hST6Gal-I and CMP-Neu5Ac (2 eq). Both sialyltransferases show preference for Galβ1–4GlcNAc substrates and gave 5 and 17, respectively. The mono-sialylated products were confirmed by NMR and MS analysis.
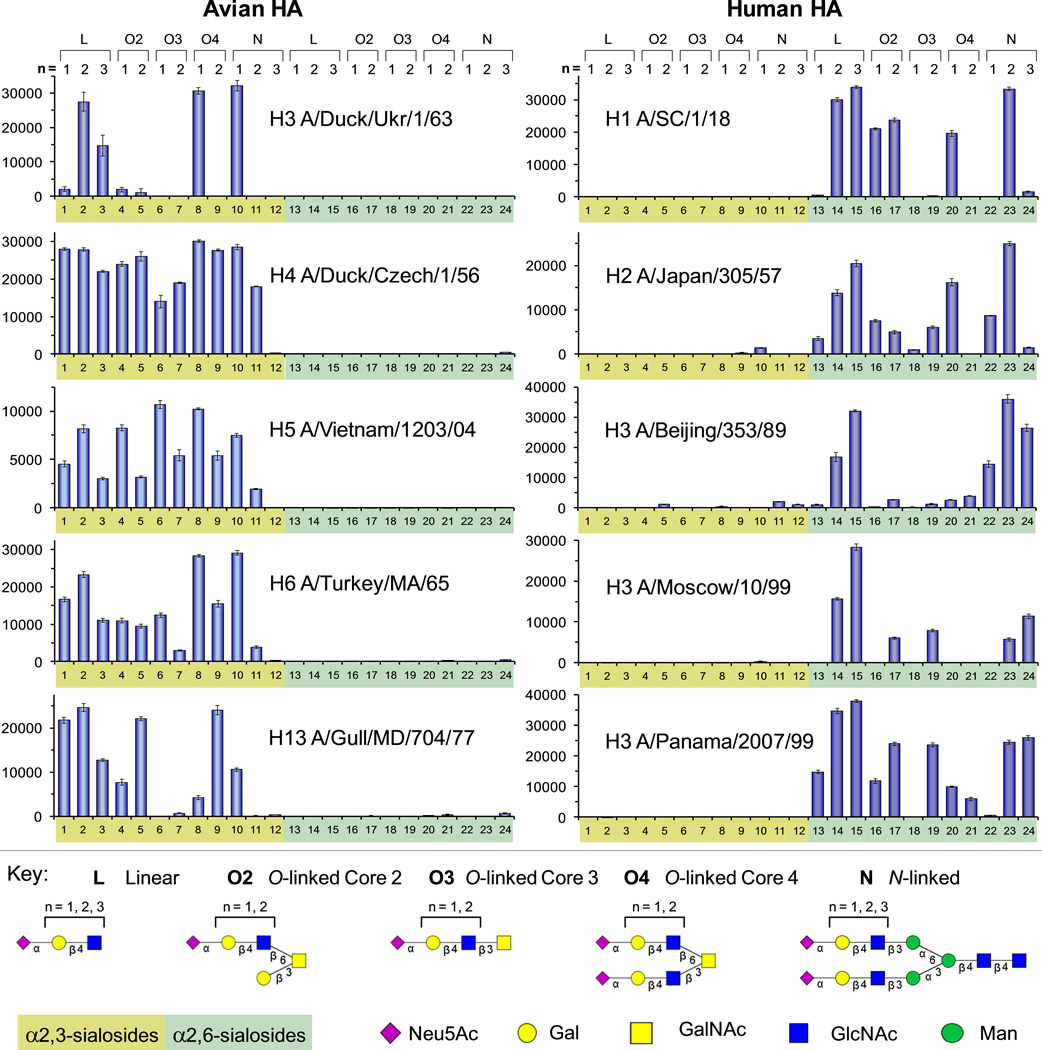
The 24 glycans in the sialoside library (Figure 1) contain either the terminal Neu5Acα2–3Gal (1–12) or Neu5Acα2–6Gal (13–24) sequence. A glycan microarray was constructed from this library to study the binding properties of influenza A virus HA.[7a, 11] The aglycone of each sialoside was equipped with a free amine for direct printing on N-hydroxy succinimde-activated slides (Figure S1 in the Supporting information). Recombinant HAs from selected avian and human influenza A viruses were then screened to assess the effects on HA binding of both length and presentation of sialylated poly-LacNAc.
As expected, the avian HAs preferentially recognized α2–3-linked sialosides (Figure 2 and Figure S2 in the Supporting information). However, while H4 (A/duck/Czech/1/56) bound strongly to nearly all α2–3 structures, other avian HAs showed more selective binding patterns. For instance, H3 (A/duck/Ukr/1/63), a progenitor of the 1968 Hong Kong pandemic,[12] only bound the linear glycans (2, 3), and the O- (8) and N-linked (10) glycans. Remarkably, all avian HAs, including H5 (A/Vietnam/1203/04), a highly pathogenic human isolate of the bird flu,[13] showed strong preference for short N-linked structures, binding strongly to 10, and reduced or no binding to the longer glycans (11, 12).
Figure 2.
Glycan microarray binding analyses as measured by fluorescence intensity for avian and human influenza A recombinant hemagglutinins. All HAs were evaluated at 15 µg/ml except for A/SC and A/Beijing, which were evaluated at 150 µg/ml. See additional details in Supporting information.
Although human HAs demonstrated classic preference for α2–6-sialosides, they exhibited varied fine specificity for the extended N- and O-linked glycans (Figure 2 and Figure S2 in the Supporting Information). As previously reported, the human HAs bound best to the linear sialosides with di- and tri-LacNAc extensions (13–15).[4] Significantly, however, the same sequences were not uniformly recognized when presented on N- and O-linked glycan cores. For instance, while the H1 (A/SC/1/18) and the H2 (A/Japan/305/57) HAs bound strongly to the linear sialoside with the di-LacNAc extension (14), they bound poorly to the same sequence presented on Core 3 (19) and Core 4 (21) glycans. Surprisingly, these same two HAs exhibited strong binding to N-glycans with the di-LacNAc sequence (23), but dramatically reduced binding to the same sequence with the tri-LacNAc repeat (24).
In summary, we have synthesized a panel of novel glycans containing sialylated poly-LacNAc on intact N- and O-linked glycan cores as candidates of the natural glycan receptors of influenza viruses. While all avian and human virus HAs retained their basic α2–3 and α2–6 linkage specificity, respectively, the N- and O-linked glycan cores differentially impacted the ability of individual HAs to recognize the sialic acid as a receptor. The lack of a consistent recognition pattern for human HAs suggests that the fine specificity of the virus for receptor(s) may drift under antigenic selective pressure, while retaining the ability to bind to a subset of α2–6-sialosides sufficient to mediate infection and transmission. It should also be noted that the branched N-linked and Core 4 O-linked glycans produced with our synthetic strategy are symmetric di-sialylated glycans. However, glycans extended on a single branch also occur in nature.[4a, 4b] Thus, it will also be of interest to investigate the role of asymmetric glycans on influenza receptor biology.
Footnotes
We thank the Otsuka Chemical Co., Ltd for the generous gift of N-linked glycans (10 and 22), and the Consortium for Functional Glycomics (GM62116) for providing the linear sialosides used in these studies. This work was supported in part by NIH grants AI058113 (to JCP and IAW), GM62116 (to JCP), a predoctoral fellowship from the Achievement Rewards for College Scientists Foundation (to DCE); grant GM080209 from the NIH Molecular Evolution Training Program (to DCE), and the Skaggs Institute (IAW).
Contributor Information
Corwin M. Nycholat, Department of Chemical Physiology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA)
Ryan McBride, Department of Chemical Physiology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Damian C. Ekiert, Department of Molecular Biology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA)
Rui Xu, Department of Molecular Biology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Janani Rangarajan, Department of Chemical Physiology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Wenjie Peng, Department of Chemical Physiology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Nahid Razi, Department of Chemical Physiology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Michel Gilbert, Institute for Biological Sciences, National Research Council Canada, Ottawa, ON K1A 0R6 (Canada).
Warren Wakarchuk, Institute for Biological Sciences, National Research Council Canada, Ottawa, ON K1A 0R6 (Canada).
Ian A. Wilson, Department of Molecular Biology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA)
James C. Paulson, Department of Chemical Physiology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
References
- 1.a) Neumann G, Kawaoka Y. Emerg Infect Dis. 2006;12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Salomon R, Webster RG. Cell. 2009;136:402–410. doi: 10.1016/j.cell.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 3.a) Matrosovich MN, Gambaryan AS, Klenk HD. In: Avian Influenza. Klenk HD, Matrosovich MN, Stech J, editors. Vol. 27. Basel: Basel; 2008. pp. 134–155. [Google Scholar]; b) Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Trends Microbiol. 2008;16:149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 4.a) Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]; b) Bateman AC, Karamanska R, Busch MG, Dell A, Olsen CW, Haslam SM. J Biol Chem. 2010;285:34016–34026. doi: 10.1074/jbc.M110.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Cho MJ, Cummings RD. Trends Glycosci Glyc. 1997;9:47–56. [Google Scholar]; b) Renkonen R, Niemela R, Natunen J, Majuri ML, Maaheimo H, Helin J, Lowe JB, Renkonen O. Journal of Biological Chemistry. 1998;273:4021–4026. doi: 10.1074/jbc.273.7.4021. [DOI] [PubMed] [Google Scholar]; c) Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. Proc Natl Acad Sci U S A. 1997;94:14294–14299. doi: 10.1073/pnas.94.26.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viswanathan K, Chandrasekaran A, Srinivasan A, Raman R, Sasisekharan V, Sasisekharan R. Glycoconjugate J. 2010;27:561–570. doi: 10.1007/s10719-010-9303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Stevens J, Blixt O, Paulson JC, Wilson IA. Nat Rev Microbiol. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Childs RA, Palma AS, Wharton S, Matrosovich T, Liu Y, Chai W, Campanero-Rhodes MA, Zhang Y, Eickmann M, Kiso M, Hay A, Matrosovich M, Feizi T. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Blixt O, Razi N. Methods Enzymol. 2006;415:137–153. doi: 10.1016/S0076-6879(06)15009-0. [DOI] [PubMed] [Google Scholar]; b) Sauerzapfe B, Krenek K, Schmiedel J, Wakarchuk WW, Pelantova H, Kren V, Elling L. Glycoconj J. 2009;26:141–159. doi: 10.1007/s10719-008-9172-2. [DOI] [PubMed] [Google Scholar]; c) Mong TK, Huang CY, Wong CH. J Org Chem. 2003;68:2135–2142. doi: 10.1021/jo0206420. [DOI] [PubMed] [Google Scholar]
- 9.Blixt O, Brown J, Schur MJ, Wakarchuk W, Paulson JC. J Org Chem. 2001;66:2442–2448. doi: 10.1021/jo0057809. [DOI] [PubMed] [Google Scholar]
- 10.Peng W, Pranskevich J, Nycholat CM, Gilbert M, Wakarchuk W, Paulson JC, Razi N. 2012. doi: 10.1093/glycob/cws101. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. Virology. 2003;309:209–218. doi: 10.1016/s0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 13.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]