Abstract
Amidation is a post-translational modification found at the C-terminus of ~50% of all neuropeptide hormones. Cleavage of the Cα-N bond of a C-terminal glycine yields the α-amidated peptide in a reaction catalyzed by peptidylglycine α-amidating monooxygenase (PAM). The mass of an α-amidated peptide decreases by 58 Da relative to its precursor. The amino acid sequences of an α-amidated peptide and its precursor differ only by the C-terminal glycine meaning that the peptides exhibit similar RP-HPLC properties and tandem mass spectral (MS/MS) fragmentation patterns. Growth of cultured cells in the presence of a PAM inhibitor ensured the coexistence of α-amidated peptides and their precursors. A strategy was developed for precursor and α-amidated peptide pairing (PAPP): LC-MS/MS data of peptide extracts were scanned for peptide pairs that differed by 58 Da in mass, but had similar RP-HPLC retention times. The resulting peptide pairs were validated by checking for similar fragmentation patterns in their MS/MS data prior to identification by database searching or manual interpretation. This approach significantly reduced the number of spectra requiring interpretation, decreasing the computing time required for database searching and enabling manual interpretation of unidentified spectra. Reported here are the α-amidated peptides identified from AtT-20 cells using the PAPP method.
Keywords: α-amidated peptide, spectral pairing, post-translational modification
1 Introduction
Amidation at the C-terminus is an important post-translational modification common to many neuropeptides (Figure 1A). About half of the peptides found in the nervous and endocrine systems are α-amidated [1, 2], such as corticotropin-releasing hormone, thyrotropin releasing hormone, neuropeptide Y, substance P, oxytocin, vasopressin, and α-melanotropin (α-MSH). For most of these peptides, the presence of the C-terminal amide structure is essential for their biological activity and stability [3]. Furthermore, most known α-amidated peptides are bioactive. In fact, it has been argued that peptide amidation is a “signature of bioactivity” [4].
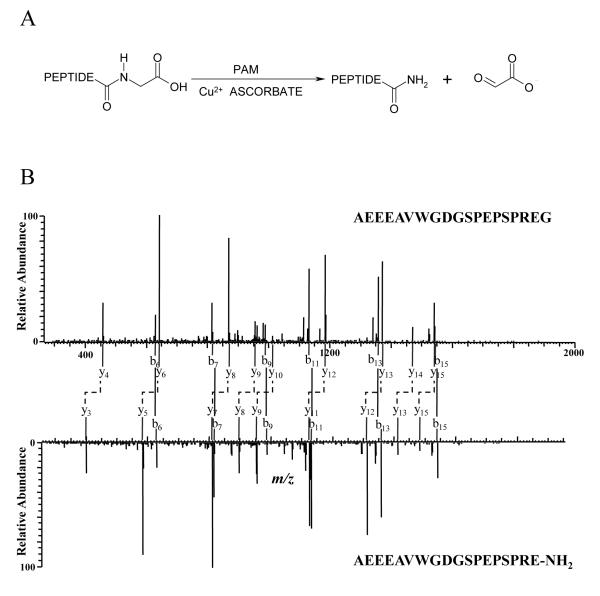
Figure 1. Detection of α-Amidated Peptides by Mass Spectrometry.
A, Peptide amidation reaction catalyzed by peptidylglycine α-amidating monooxygenase (PAM). In this reaction, the precursor peptide is cleaved, removing the C-terminal glycine and creating a new amidated C-terminus during PAM catalysis, resulting in a total difference of 58.0055 mass units (C2H2O2) between the glycine-extended and amidated forms. B, MS/MS spectra of mouse joining peptide (mJP) and its precursor. The two MS/MS spectra have similar fragment patterns. Corresponding b ions (connected with solid lines) have same mass values while y ions (connected with dashed elbow connectors) have a constant mass difference of 58 resulting from the cleavage of C2H2O2 group from the C-terminus by PAM.
Strategies for the discovery of novel peptide hormones dependent upon the presence of a C-terminal amide have been described. One chemical approach for identification of α-amidated peptides was to convert the amides to amines via the Hoffman rearrangement following acetylation of lysines and the N-terminus. Resultant amines were visualized with ninhydrin [5]. This assay has a limit of detection (LOD) in the mmol range and yields false positive results for peptides containing asparagine and glutamine residues. Tatemoto and Mutt [6] developed another chemical assay for the discovery of novel α-amidated peptides. The C-terminal amino acid amide was released enzymatically, converted to the dansyl derivative, and ultimately identified by two-dimensional thin layer chromatography (2D-TLC). A few α-amidated peptide hormones were originally identified using the Tatemoto and Mutt method, including galanin [7] and pancreastatin [8]. With improvements in separation and detection of the amino acid amide using HPLC [9-12] and capillary electrophoresis [13], the LOD of this method has been decreased into the picomole range.
α-Amidated peptide identification methods based on mass spectrometry (MS) and tandem mass spectrometry (MS/MS) have also been reported. Mouls et al. [14] investigated the behavior of peptides with C-terminal amides and free carboxylic acids upon low energy collision-induced dissociation (CID) and found that α-amidated peptides produced an abundant fragment ion corresponding to the loss of ammonia from the protonated molecule. The side chain amides of Asn and Gln were more stable than the C-terminal amide under CID conditions. Therefore, C-terminal amidation can be identified by inspection of the peptide MS/MS spectra, even with the presence of asparagine or glutamine in the peptide chain. The other MS based method relied on chemical derivatization to convert free carboxyl groups (COOH) into methylamides (CONHCH3) [15]. Peptides with a free C-terminal carboxylate exhibit doublet peaks in MS spectra separated by 13 Da (the difference between COOH and CONHCH3) resulting from incomplete derivatization, while α-amidated peptides show singlet peaks as a consequence of the protected C-terminal carboxyl group. Immunological detection of the C-terminal amide was also developed utilizing antibodies specific to certain α-amino acid amides. The Grimmelikhuijzen group generated antisera against dipeptide amides Arg-X-NH2 (where X was Ala, Asn, Phe, Pro,Val, etc.) and have discovered a number of novel amidated peptides with these C-terminal sequences [16]. The methods summarized above for the identification of α-amidated peptides are most effective when the peptide is pure or is included in a relatively simple mixture. However, these methods are ineffective, inefficient, or laborious when applied to the discovery of low abundance or novel α-amidated peptides in a complex biological sample.
Liquid chromatography-tandem mass spectrometry peptide sequencing (LC-MS/MS) followed by database searching has emerged as a powerful tool for peptide identification due to its advantages of speed, sensitivity, and applicability to complex peptide mixtures. Many α-amidated peptides have recently been discovered using this method, such as the C-terminal fragments of chromogranin A, ER-20amide, and AR-28amide [17], as well as neuroendocrine regulatory peptides NERP-1 and NERP-2 [18]. However, peptide sequences cannot always be assigned for tandem mass spectra by database searching [19]. Peptide hormones, which are usually less concentrated and undergo post-translational modifications, are more likely to yield tandem mass spectra that require manual interpretation.
A few tools have been developed to detect general peptide/protein post-translational modifications (PTMs) based on spectral pair finding strategies, including ModifiComb [20], Mass Distance Fingerprint [21], and others [22, 23]. These methods were designed for the detection of general PTMs without prior assumption of their chemical composition and attachment sites, rendering these tools particularly useful for the detection and characterization of unanticipated PTMs. These methods can only detect relatively abundant PTMs because the spectra of modified and unmodified peptides must be repeatedly identified for the search to perform well. These methods also work under the assumption that both modified and unmodified peptides are present in the sample, which may not be true, especially for irreversible PTMs.
Reported herein is a novel mass spectrometry-based strategy for the discovery of α-amidated peptides that is dependent on their in vivo biosynthetic pathway. α-Amidated peptides are typically generated from larger, inactive precursors. Proteolytic cleavage of the precursor yields the final α-amidated peptide with a C-terminal glycine extension [24]. Generally, the glycine-extended precursor is inactive [3], one exception being glycine-extended gastrin [25]. The mature, bioactive α-amidated peptide is generated by the PAM-mediated oxidative cleavage of the glycyl Cα-N bond. The new C-terminus is amidated with a total difference of 58.0055 mass units (C2H2O2) between the precursor and the amidated product. PAM is the only known mammalian enzyme responsible for the conversion of the precursor glycine-extended peptide to the final α-amidated product. Since an α-amidated peptide and its precursor have identical amino acid sequences with the exception of the C-terminal glycine of the precursor, these peptide pairs should have similar hydrophobicities, reverse phase chromatographic retention times, and similar MS/MS fragmentation patterns. We demonstrate that α-amidated peptides can be detected in complex biological matrices by screening LC-MS/MS data using precursor and α-amidated peptide pairing (PAPP). This method is applied to datasets produced by LC-MALDI-MS/MS and LC-ESI-MS/MS; results from PAPP are compared to database searching strategies.
2 Material and Methods
2.1 Mammalian cell growth
Mouse pituitary AtT-20 cells were purchased from the ATCC and were cultured in the presence of 5% CO2 at 37°C using Hams F-12K culture medium supplemented with 15% (v/v) horse serum and 2.5% (v/v) fetal bovine serum. In order to accumulate precursor peptides, cells were collected and resuspended in fresh media with 5 μM disulfiram and incubated for 20 hrs prior to harvest. The same procedure was used for the growth of the untreated cells. Disulfiram is a known PAM inhibitor and has been used to inhibit PAM activity in cultured mammalian cells [26].
2.2 Sample preparation
Cell pellets were homogenized at 4° C in 8 M urea solution or an acid extraction solution containing 0.1 M HCl, 5% (v/v) formic acid, 1% (w/v) NaCl and 1% (v/v) trifluoroacetic acid (TFA). After centrifugation to remove cellular debris, the supernatant was passed through 10 kDa molecular weight ultrafilter to remove proteins. The extract was desalted and concentrated by solid phase extraction on a C18 cartridge (Sep-Pak Plus, Waters, Milford, MA, USA) and then further purified and concentrated using a Ziptip (Millipore, Billerica, MA, USA).
2.3 LC-MS/MS
Samples were analyzed by liquid chromatography-matrix assisted laser desorption ionization – tandem time of flight (MALDI-TOF/TOF) mass spectrometry. In these experiments, samples from disulfiram treated cells and the control untreated cells were analyzed separately. This was done to obtain peptide intensity information so that the accumulation filter can be applied when processing data to verify the identities of the precursor peptides. Each sample was fractionated and spotted into 192-well plates using a microfraction collector (LC Packings Probot, Dionex, Sunnyvale, CA, USA) interfaced with a capillary liquid chromatograph (Agilent 1200, Agilent, Santa Clara, CA, USA). The sample was loaded on a C18 column (100 mm × 150 μm ID, Vydac MS C18, Grace, Deerfield, IL, USA) and washed (at 1 μL/min) for 20 min. with 95% solvent A (2% (v/v) ACN and 0.1% (v/v) TFA) and 5% solvent B (80% (v/v) ACN and 0.1% (v/v) TFA). The 65 min. gradient was programmed as: 5% to 15% B in 10 min., 15% to 50% B in 40 min., 50% to 90% B in 5 min., 90% B for 5 min., and 90% to 5% B in 5 min. MALDI-TOF/TOF MS was performed on a 4700 Proteomics Analyzer (ABSciex, Foster City, CA, USA) using positive mode with reflected ion detection. The data was externally calibrated with mass standards provided by the manufacturer (4700 Calmix, ABSciex Foster City, CA, USA).
For LC-MS/MS using an electrospray hybrid linear ion trap-orbital ion trap, samples from the disulfram-treated and control sample were combined and analyzed together. A nanoflow liquid chromatograph (U3000, Dionex, Sunnyvale, CA, USA) was coupled to an LTQ-Orbitrap (Thermo, San Jose, CA, USA). The LC system was equipped with a trapping column (5 mm × 300 μm ID packed with C18 reversed-phase resin, 5 μm, 100Å) and an analytical column (C18, 150 mm × 75 μm ID, Pepmap 100, Dionex, Sunnyvale, CA, USA). The sample was first loaded onto the trapping-column and washed for 8 min. with an aqueous solution of 2% (v/v) ACN and 0.04% (v/v) TFA. Trapped peptides were then eluted onto the analytical column. The 120 min. gradient was programmed as follows: 95% A and 5% C for 8 min., 5-50% C in 90 min., 50-90% C in 7 min., 90% C for 5 min., 90-5% C in 1 min., and 95% A and 5% C for 10 min. to re-equilibrate the column, with solvent C being 90% (v/v) ACN and 0.1% (v/v) formic acid. The flow rate on analytical column was 300 nL/min. Survey scans were performed in Orbitrap to obtain accurate peptide mass measurement setting the resolving power to 60,000 at m/z 400. Each survey scan was followed by acquisition of five data dependent tandem mass spectra acquired in the linear ion trap with normalized CID energy of 30% and 60 sec. dynamic exclusion of previously sampled peaks. Mass measurement accuracy was monitored between runs using digests of bovine serum albumin.
2.4 Data processing
Mass lists from each MS1 spectrum (from both disulfiram treated and control groups) obtained by LC-MALDI-MS were exported and combined into one Microsoft Excel spreadsheet. The complete list was then imported into Microsoft Visual FoxPro 8.0. Peptides with mass difference smaller than m/z = 0.1 and retention time difference under 2 min. were considered to be the same. The peptide with the highest intensity was kept and all others were removed from the list. After removal of the redundant data, the list was scanned for peak pairs with 58 ± 0.1 m/z difference and a retention time difference less than 2 min. Based on the comparison of the control and disulfiram treated samples, the accumulation filter, defined as:
was used. Only peptides showing higher intensity in the disulfiram-treated group were kept. A signal-to-noise (S/N) filter was also used to eliminate peptides with S/N < 20. Peaks in peak-pairs suggestive of the presence of an α-amidated peptide were subjected to MS/MS analysis. The MS/MS spectra of each pair were inspected manually for similar fragment pattern and sequences were assigned to paired MS/MS spectra by database search and manual interpretation.
For LC-MS/MS, information from Scan Headers was extracted from the RAW data obtained from LTQ-Orbitrap using RawXtract1.93 (Finnigan Corp., San Jose, CA, USA). The list, which contains detailed information of each tandem mass spectrum such as scan number, charge state, monoisotopic m/z, and retention time, was imported to Microsoft Visual FoxPro 8.0. After removal of redundant data, the list was scanned for peak pairs with a mass difference of 58.006 ± 0.01 (for singly charged ions), 29.003 ± 0.01 (for doubly charged ions), and 19.335 ± 0.01 (for triply charged ions). The MS/MS spectra of each pair were inspected manually and paired spectra were sequenced by database search and/or manual interpretation.
2.5 Database search
MALDI MS and MS/MS data were extracted for database searches using the Launch Peaks to Mascot feature in the 4000 Series Explorer software (ABSciex, Foster City, CA). Briefly, 5 peaks (S/N > 3) per 200 Da were selected from the tandem mass spectra and searched against mammalian entries in the NCBInr database using Mascot (www.matrixscience.com). Peptide mass tolerance was set to 200 ppm and fragment ion mass tolerance was 0.8 Da. No enzyme was selected and oxidation of methionine and C-terminal amidation were selected as variable modifications.
LC-MS/MS data were submitted for Mascot database searches using the extractmsn.exe program (Thermo, San Jose, CA). Peptide mass tolerance was set to 1.2 Da and fragment ion mass tolerance was 0.8 Da. Enzyme specificity, modifications, and database searching were the same as described above for MALDI MS and MS/MS data. In addition, database searches were performed with Sequest (Thermo, San Jose, CA), extracting data for peptides between 600 and 4,500 Da. Mass tolerances and search settings were similar to those described for Mascot.
All sequence assignments for amidated peptides were manually verified by inspection of the tandem mass spectra.
3 Results
3.1 Proof of concept
The mouse pituitary corticotropic tumor cell line, AtT-20, is known to express high levels of PAM and two amidated peptides: mouse joining peptide (mJP) and α-MSH [27]. PAM activity in AtT-20 cells can be inhibited by disulfiram, resulting in the accumulation of α-amidated peptide precursors [26]. For proof of concept, we investigated the relative levels of these peptides in cells grown with and without a PAM inhibitor using LC-MALDI-TOF-MS. The relative intensity of the precursors (in comparison with intensity of the α-amidated forms) increased by 2.8-fold for mJP and 9.2-fold for α-MSH when the cells were incubated with the PAM inhibitor. Reverse phase retention times of these two α-amidated peptides were similar to their precursors. In this LC-MALDI experiment, the α-amidated peptides were detected in the same fraction or a neighboring fraction to the precursor peptide (i.e. elution within 20 sec.). In online LC-MS/MS analysis, retention time differences between α-amidated and precursor peptides were 52 sec. and 39 sec. for mJP and α-MSH, respectively. MS/MS spectra of mJP and its precursor are shown in Figure 1B. These two peptides exhibit very similar fragmentation patterns. They show b ions with the same mass value and y ions that show the mass difference of 58 Da associated with truncation of the glycine residue and amidation of the C-terminus. Furthermore, in the tandem mass spectra, the modification at the C-terminus of the peptide should enable detection of precursor and α-amidated peptides by the fact that they have identical b-ion series, but y ions that differ by 58 Da in mass.
3.2 LC-MALDI-MS/MS data set
Using the mass difference and retention time filters as described above, the PAPP method was compared to database searching for the identification of additional α-amidated peptides. Mascot database searching of LC-MALDI-MS/MS data reported one α-amidated peptide as shown in Table 1. This peptide is the C-terminus of chromogranin A (CgA) following a dibasic processing site.
Table 1.
Identification of α-Amidated Peptides by LC-MALDI and PAPP.
|
m/z of precursor |
m/z of amidated |
δm/z a | δRTb (s) |
Sequence | Mascot score |
|---|---|---|---|---|---|
| 3234.247 | 3176.261 | 57.986 | < 20 | (R)AEDQELESLSAIEAELEKVAHQLQALRr(G) | 67 |
| 1999.111 | 1941.091 | 58.020 | < 20 | (R)AEEEAVWGDGSPEPSPRe(G) | M.I.c |
| 1681.006 | 1622.971 | 58.035 | < 20 | (R)SYSMEHFRWGKPv(G) | M.I. |
δm/z is the difference in the m/z between the precursor peptide (with the C-terminal glycine) and the α-amidated peptide: (m/z for the precursor) - (m/z for the α-amidated peptide).
δRT is the difference in the RP-HPLC retention times between the precursor peptide (with the C-terminal glycine) and the α-amidated peptide: (RT for the precursor) - (RT for the α-amidated peptide).
M.I.: sequence assigned via manual interpretation.
Surprisingly, the two known α-amidated peptides (mJP and α-MSH) were not reported by database search. The peak list was then exported using Data Explorer (version 4.0, ABI, Foster City, CA, USA) and screened for peptide pairs as described previously. Considering external calibration was used in MALDI assay, the mass measurement accuracy obtained was relatively low. A wide window 58 ± 0.1 was applied to screen for peptide pairs, which resulted in a list of over three hundred peptide pairs. An accumulation filter was used to eliminate some of the false positive pairs. Pairs containing signals with low S/N values were eliminated since such pairs were unlikely to yield MS/MS spectra of sufficient quality for manual interpretation. After the use of these filters, the number of peptide pairs decreased to ~40. Manual inspection on MS/MS spectra of these peptide pairs found 4 pairs with similar fragmentation patterns. After database search and manual interpretation, 3 peptide pairs were identified, including the C-terminal fragment of CgA, mJP and α-MSH (Table 1).
3.3 High Resolution LC-MS/MS Data Set
Over 15,000 tandem mass spectra were obtained in each LC-MS/MS experiment. Nine α-amidated peptides were identified through Sequest and Mascot database searches (Table 2). Two sequences received low scores due to poor spectrum quality.
Table 2.
Identification of α-Amidated Peptides by LC-MS/MS and PAPP.
|
m/z of precursor |
m/z of amidated |
CS a |
δm/zb | δRTc (min) |
Sequence | Mascot score |
|---|---|---|---|---|---|---|
| 999.9359 | 970.9304 | 2 | 58.0110 | -0.01 | (R)AEEEAVWGDGSPEPSPRe(G) | 94.6 |
| 964.4114 | 935.4109 | 2 | 58.0010 | 0.08 | (A)EEEAVWGDGSPEPSPRe(G) | 86.4 |
| 899.8951 | 871.8935 | 2 | 58.0032 | 0.72 | (E)EEAVWGDGSPEPSPRe(G) | 64.1 |
| 835.3716 | 806.3690 | 2 | 58.0052 | 1.22 | (E)EAVWGDGSPEPSPRe(G) | 59.7 |
| 770.8514 | 741.8502 | 2 | 58.0024 | 1.22 | (E)AVWGDGSPEPSPRe(G) | 32.4 |
| 735.3329 | 706.3295 | 2 | 58.0068 | 1.67 | (A)VWGDGSPEPSPRe(G) | 61.8 |
| 685.8043 | 656.8019 | 2 | 58.0048 | 1.10 | (V) WGDGSPEPSPRe(G) | 59.0 |
| 592.7625 | 563.7588 | 2 | 58.0074 | 1.22 | (W)GDGSPEPSPRe(G) | M.I.d |
| 564.2518 | 535.2484 | 2 | 58.0068 | 1.57 | (G)DGSPEPSPRe(G) | 31.2 |
| 506.7379 | 477.7352 | 2 | 58.0054 | 1.09 | (D)GSPEPSPRe(G) | M.I. |
| 840.8983 | 811.8932 | 2 | 58.0102 | 1.12 | (R)SYSMEHFRWGKPv(G) | 62.8 |
| 560.9345 | 541.5995 | 3 | 58.0050 | 0.50 | (R)SYSMEHFRWGKPv(G) | M.I. |
| 500.2572 | 471.2552 | 2 | 58.0040 | 1.42 | (R)ELEGERPl(G) | M.I. |
CS = charge state
δm/z is the difference in the m/z between the precursor peptide (with the C-terminal glycine) and the α-amidated peptide: (m/z for the precursor) - (m/z for the α-amidated peptide).
δRT is the difference in the RP-HPLC retention times between the precursor peptide (with the C-terminal glycine) and the α-amidated peptide: (RT for the precursor) - (RT for the α-amidated peptide).
M.I.: sequence assigned via manual interpretation.
The peptide list was then screened for peptide pairs with 58.006 ± 0.01 mass difference and similar retention time (within 2 min.) resulting in a list of 33 peptide pairs. After manual inspection of the tandem mass spectra for these peptides, we found 13 pairs of peptides (Table 2) which displayed the similar fragmentation pattern expected for an α-amidated peptide and its C-terminal glycine-extended precursor, including 9 α-amidated peptides reported by database searching. In addition, the PAPP method detected peaks corresponding to triply charged α-MSH and 3 more pairs, which were not identified by database searching.
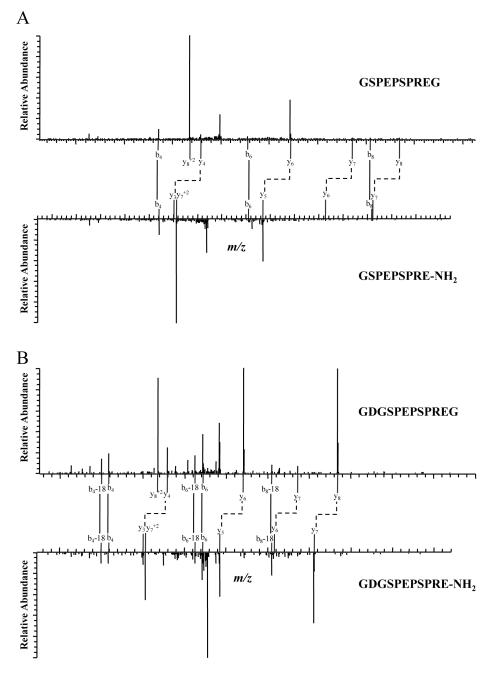
Shown in Figure 2A are MS/MS spectra of one interesting peptide pair. By looking for peaks with mass difference of 58 between the two spectra, we identified 4 pairs of y ions with the following m/z values: 458.44 and 400.30, 642.41 and 584.40, 771.46 and 713.23, and 868.66 and 810.51. These y ions are identical to the y4, y6, y7, and y8 ions in the mJP precursor and α-amidated peptide pair (Figure 1B), suggesting that all of these peptide pairs are mJP-related peptides. Based on the m/z values of the parent ions, the sequences are, most likely, GSPEPSPREG and GSPEPSPRE-NH2. Although the complete sequences could not be deduced from the fragments, accurate mass measurements of the intact peptides compared to the theoretical mass (with mass differences in the range of −0.0013 (2.56 ppm) to −0.0009 (1.88 ppm) respectively), as well as assigned b ions indicate that they were the correct sequences. Similarly, another pair of peptides was identified as GDGSPEPSPREG and GDGSPEPSPRE-NH2 (Figure 2B).
Figure 2. MS/MS Spectra of Two mJP Related Peptide Pairs.
These spectra have same the y ions as mJP and its precursor.
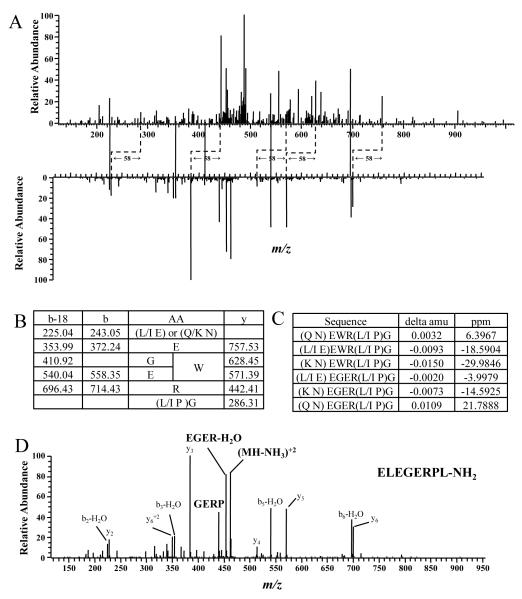
De novo sequencing was required to identify the sequence of the other peptide pair, tandem mass spectra are shown in Figure 3A. Through these peaks, we were able to get partial sequence of this peptide: EWR or EGER (Figure 2B). The fragment ions 243.05 and 286.31 were used to deduce the remaining compositions at N-terminus and C-terminus, which turned out to be (L/I, E) or (Q/K, N) for N-terminus and (L/I, P)G for C-terminus. Shown in Figure 3C are the error values in parts per million (ppm) for each of the possible sequences. Considering the mass measurement accuracy of mass analyzer (~5 ppm), (Q, N) EWR(L/I, P)G and (L/I, E) EGER(L/I, P)G are more likely to match the actual sequence of this peptide. A BLAST search using all possible combinations against the NCBI non-redundant protein sequences (NR) database revealed one match for sequence ELEGERPLG. This sequence was confirmed by MS/MS of the synthetic peptide, ELEGERPL-NH2, (Figure 3D) obtained under the same conditions.
Figure 3. De novo Identification of an α-Amidated Peptide.
A, Spectral pair found by PAPP. Two spectra have peaks with a 58 mass difference (tentative y ions, marked with dashed elbow connectors) and peaks with the same m/z value (tentative b ions, marked with solid lines). B, Fragment ions found in the spectrum of the larger peptide and the corresponding possible amino acid residues. C, Mass errors of all possible sequences. D, MS/MS spectrum of the synthetic peptide ELEGERPL-NH2.
4 Discussion
Liquid chromatography (LC) coupled with tandem mass spectrometry (MS/MS) is one method useful in the screening of biological extracts for endogenous α-amidated peptides. Standard MS/MS data processing procedures use MASCOT, SEQUEST, or another database search engine to compare theoretical spectra generated from sequences within the database to those obtained experimentally. Although this approach has been successful, this strategy does not work well in identifying novel bioactive peptides. Cleavage of a glycyl residue from the C-terminus of a peptide and C-terminal amidation are difficult to account for when using database search engines as PAM is not usually included as a processing enzyme. The lack of a predictable N-terminus further complicates database searches for the presence of novel α-amidated peptides. The net effect of these two complicating factors means that “no enzyme” specificity must be employed to generate theoretical spectra from database sequences, meaning that all possible sequences from each peptide must be evaluated. For a peptide of N amino acid residues, [N(N+1)]/2 theoretical spectra must be considered. This translates into the generation and evaluation of 120 theoretical spectra for a peptide comprised of 15 amino acids. The number of theoretical spectra to consider increases as the length of the α-amidated peptide increases and long α-amidated peptides are known. Examples include a few α-amidated peptides containing 40-50 amino acids [3], a 72-amino acid α-amidated peptide from crustaceans [28], a 78-amino acid α-amidated peptide identified in the venom of the South American armed spider Phoneutria nigriventer [29], and porcine sorbin is 153-amino acids long with a C-terminal alaninamide [30]. Given that modern LC-MS/MS instruments generate tens of thousands of MS/MS spectra per sample, database searching for endogenous peptides is time-consuming and requires considerable computing power. The database SwePep, which contains known endogenous peptides from different species, was constructed to alleviate this problem [31, 32]. The masses of experimental peptides are compared against the masses of database peptides using SwePep. Sequence identifications are then verified by comparing experimental MS/MS data with spectra stored in the database. Searches for endogenous peptides require significantly less time using SwePep relative to conventional databases, but SwePep is best for the identification of known peptides and is of limited use for the discovery of novel peptides.
Peptide identification using database searching methods is further hampered by the relatively low success rate in successfully interpreting MS/MS spectra within the database [33]. Reasons for the low identification rate include unknown modifications [19,20], poor spectral quality [34], and spectra that have fragments from multiple peptides [35]. For a small dataset, manual interpretation of unassigned spectra may identify more peptides of interest, as has been shown for both LC-MALDI-MS/MS and LC-MS/MS. However, manual interpretation of spectra is not feasible for the high throughput analysis of a large dataset containing tens of thousands of spectra. The PPAP method presented here enables the screening of a large data set for the LC-MS/MS spectra of interest by significantly reducing the number of spectra requiring interpretation. Our PAPP method filtered out 99.8% of the spectra from the 15,580 MS/MS spectra obtained in the LTQ-Orbitrap experiment yielding 33 paired tandem mass spectra for further analysis. Such a small sample enabled manual interpretation of the data for the presence of α-amidated peptides and significantly reduced the time required for the database search. Three additional α-amidated peptides were identified through manual interpretation.
Our method is designed to screen for peptides with a known modification based upon a well understood biosynthetic pathway. We inhibited PAM resulting in the accumulation of the glycine-extended precursor peptides. The coexistence of α-amidated peptides and their C-terminal glycine-extended precursors was assured by combining the peptides from the PAM inhibited sample and from the control sample without PAM inhibition. Because we specified the expected mass difference between an α-amidated peptide and its precursor (58.0 ± 0.1 Da), our method is sufficiently robust to identify a peptide pair represented by a single tandem mass spectrum. We identified the α-amidated peptide ELEGERPL-NH2 in this way.
The method detailed herein can be easily adapted to screen for peptides with other modifications as long as modified and unmodified peptides coexist in the sample. This condition is usually fulfilled for reversible modification due to their dynamic nature. Proper treatment of the sample can increase the concentration of the less abundant peptide; thus, increasing the likelihood of its detection. The detection of peptide S-glutathionylation/de-glutathionylation is one potential application of our technique. If the levels of the modified peptide(s) level are low, cells can be exposed to oxidative or nitrosative stress to induce S-glutathionylation [36]. If the levels of the unmodified peptide(s) level are low, treatment of the cells with glutaredoxin will increase peptide deglutathionylation [37].
In conclusion, a new strategy for screening of C-terminal α-amidated peptides is described. This approach is based on the biosynthetic pathway for peptide amidation at the C-terminus and the high mass measurement accuracy of LC-MS/MS. The coexistence of α-amidated peptides and their precursors was guaranteed by the use of a PAM inhibitor. Peptides were scanned for pairs with a mass difference of 58 and a retention time difference of less than 2 minutes. Selected pairs were further validated by comparing their fragmentation patterns in MS/MS. This method significantly reduces the workload of spectral interpretation and increases the identification rate for α-amidated peptides. Thirteen α-amidated peptides were identified from cultured AtT-20 cells using this method; four of these α-amidated peptides could only be assigned by PAPP with manual interpretation of the MS/MS data.
Acknowledgments
This study was supported by NIH grant R44-DK063812. All mass spectrometry experiments were done in Moffitt Proteomics which is supported by the US Army Medical Research and Materiel Command under Award No. DAMD17-02-2-0051, continuing as W81XWH-08-2-0101, for a National Functional Genomics Center, the National Cancer Institute under Award No. P30-CA076292 as a Cancer Center Support Grant, and the Moffitt Foundation.
Abbreviations
- CgA
chromogranin A
- CS
charge state
- disulfiram
1,1′,1″,1‴-[disulfanediylbis(carbonothioylnitrilo)]tetraethane
- ESI
electrospray ionization
- LOD
limit of detection
- MALDI
matrix-assisted laser desorption/ionization
- mJP
mouse joining peptide
- α-MSH
α-melanotropin
- PAM
peptidylglycine α-amidating monooxygenase
- PAPP
precursor and α-amidated peptide pairing
- RP-HPLC
reverse-phase high pressure liquid chromatography
- TOF
time of flight
Footnotes
Conflicts of Interest The authors have no financial or commercial interests resulting from this research.
5 References
- [1].Eipper BA, Mains RE. Peptide α-amidation. Annu. Rev. Physiol. 1988;50:333–344. doi: 10.1146/annurev.ph.50.030188.002001. [DOI] [PubMed] [Google Scholar]
- [2].Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide α-amidation. Annu. Rev. Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- [3].Merkler DJ. C-Terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extened peptides and the importance of the amide to bioactivity. Enzyme Microb. Technol. 1994;16:450–456. doi: 10.1016/0141-0229(94)90014-0. [DOI] [PubMed] [Google Scholar]
- [4].Cuttitta F. Peptide amidation: signature of bioactivity. Anat. Rec. 1993;236:87–95. doi: 10.1002/ar.1092360112. [DOI] [PubMed] [Google Scholar]
- [5].Hill JC, Flannery GM, Fraser BA. Identification of α-carboxamidated and carboxy-terminal glycine forms of peptides in bovine hypothalamus, bovine pituitary and porcine heart extracts. Neuropeptides. 1993;25:255–264. doi: 10.1016/0143-4179(93)90110-v. [DOI] [PubMed] [Google Scholar]
- [6].Tatemoto K, Mutt V. Chemical determination of polypeptide hormones. Nature. 1978;75:4115–4119. doi: 10.1073/pnas.75.9.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tatemoto K, Rökaeus Å , Jörnvall H, McDonald TJ, Mutt V. Galanin - a novel biologically-active peptide from porcine intestine. FEBS Lett. 1983;164:124–128. doi: 10.1016/0014-5793(83)80033-7. [DOI] [PubMed] [Google Scholar]
- [8].Tatemoto K, Efendić S, Mutt V, Makk G, et al. Pancreastatin, a novel pancreatic peptide that inhibits insulin secretion. Nature. 1986;324:476–478. doi: 10.1038/324476a0. [DOI] [PubMed] [Google Scholar]
- [9].Simmons WH, Meisenberg G. Separation of DNS-amino acid amides by high-performance liquid chromatography. J. Chromatogr. A. 1983;266:483–489. [Google Scholar]
- [10].Schmidt WE, Conlon JM, Mutt V, Carlquist M, et al. Identification of the C-terminally α-amidated amino acid in peptides by high-performance liquid chromatography. Eur. J. Biochem. 1987;162:467–472. doi: 10.1111/j.1432-1033.1987.tb10663.x. [DOI] [PubMed] [Google Scholar]
- [11].Bennett HPJ, Solomon S. Use of Pico-Tag methodology in the chemical analysis of peptides with carboxyl-terminal amides. J. Chromatogr. A. 1986;359:221–230. doi: 10.1016/0021-9673(86)80076-0. [DOI] [PubMed] [Google Scholar]
- [12].Treston AM, Vicchio D, Mulshine JL, Yergey AL. High-performance liquid chromatography with thermospray mass spectrometric detection of α-carboxyamido amino acids. J. Chromatogr. A. 1989;474:187–195. [Google Scholar]
- [13].Feng L, Mitchell ME. Selective fluorescence derivatization and capillary electrophoretic separation of amidated amino acids. J. Chromatogr. A. 1999;832:211–224. [Google Scholar]
- [14].Mouls L, Subra G, Aubagnac J-L, Martinez J, Enjalbal C. Tandem mass spectrometry of amidated peptides. J. Mass Spectrom. 2006;41:1470–1483. doi: 10.1002/jms.1118. [DOI] [PubMed] [Google Scholar]
- [15].Kuyama H, Nakajima C, Nakazawa T, Nishimura O, Tsunasawa S. A new approach for detecting C-terminal amidation of proteins and peptides by mass spectrometry in conjunction with chemical derivatization. Proteomics. 2009;9:4063–4070. doi: 10.1002/pmic.200900267. [DOI] [PubMed] [Google Scholar]
- [16].Grimmelikhuijzen CJP, Williamson M, Hansen GN. Neuropeptides in cnidarians. Can. J. Zool. 2002;80:1690–1702. [Google Scholar]
- [17].Taylor SW, Andon NL, Bilakovics JM, Lowe C, et al. Efficient high-throughput discovery of large peptidic hormones and biomarkers. J. Proteome Res. 2006;5:1776–1884. doi: 10.1021/pr0600982. [DOI] [PubMed] [Google Scholar]
- [18].Yamaguchi H, Sasaki K, Satomi Y, Shimbara T, et al. Peptidomic identification and biological validation of neuroendocrine regulatory peptide-1 and -2. J. Biol. Chem. 2007;282:26354–26360. doi: 10.1074/jbc.M701665200. [DOI] [PubMed] [Google Scholar]
- [19].Ye D, Fu Y, Sun R-X, Wang H-P, et al. Open MS/MS spectral library search to identify unanticipated post-translational modifications and increase spectral rate. Bioinformatics. 2010;26:i399–i406. doi: 10.1093/bioinformatics/btq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Savitski MM, Nielsen ML, Zubarev RA. ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- [21].Potthast F, Gerrits B, Häkkinen J, Rutishauser D, et al. The Mass Distance Fingerprint: a statistical framework for de novo detection of predominant modifications using high-accuracy mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;854:173–182. doi: 10.1016/j.jchromb.2007.04.020. [DOI] [PubMed] [Google Scholar]
- [22].Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. Proc. Natl. Acad. Sci. USA. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fu Y, Jia W, Lu Z, Wang HP, et al. Efficient discovery of abundant post-translational modifications and spectral pairs using peptide mass and retention time differences. BMC Bioinformatics. 2009;10(Suppl. 1):S50. doi: 10.1186/1471-2105-10-S1-S50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kovac S, Shulkes A, Baldwin GS. Peptide processing and biology in human disease. Curr. Opin. Endocrinol. Diabetes Obes. 2009;16:79–85. doi: 10.1097/MED.0b013e3283202555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Seva C, Dickinson CJ, Yamada T. Growth-promoting effects of glycine-extended gastrin. Science. 1994;265:410–412. doi: 10.1126/science.8023165. [DOI] [PubMed] [Google Scholar]
- [26].Mains RE, Park LP, Eipper BA. Inhibition of peptide amidation by disulfiram and diethyldithiocarbamate. J. Biol. Chem. 1986;261:11938–11941. [PubMed] [Google Scholar]
- [27].Eipper BA, Park L, Keutmann HT, Mains RE. Amidation of joining peptide, a major pro-ACTH/endorphin-derived product peptide. J. Biol. Chem. 1986;261:8686–8694. [PubMed] [Google Scholar]
- [28].Montagné N, Desdevises Y, Soyez D, Toullec J-Y. Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans. BMC Evol. Biol. 2010;10:62. doi: 10.1186/1471-2148-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Martin-Moutot N, Mansuelle P, Alcaraz G, Gouvêa R, et al. Phoneutria nigriventer toxin 1: a novel, state-dependent inhibitor of neuronal sodium channels that interacts with μ conotoxin binding sites. Mol. Pharmacol. 2006;69:1931–1937. doi: 10.1124/mol.105.021147. [DOI] [PubMed] [Google Scholar]
- [30].Vagne-Descroix M, Pansu D, Jörnvall H, Carlquist M, et al. Isolation and characterization of porcine sorbin. Eur. J. Biochem. 1991;201:53–59. doi: 10.1111/j.1432-1033.1991.tb16254.x. [DOI] [PubMed] [Google Scholar]
- [31].Falth M, Skold K, Norrman M, Svensson M, et al. SwePep, a database designed for endogenous peptides and mass spectrometry. Molecular & Cellular Proteomics. 2006;5:998–1005. doi: 10.1074/mcp.M500401-MCP200. [DOI] [PubMed] [Google Scholar]
- [32].Falth M, Svensson M, Nilsson A, Skold K, et al. Validation of endogenous peptide identifications using a database of tandem mass spectra. Journal of Proteome Research. 2008;7:3049–3053. doi: 10.1021/pr800036d. [DOI] [PubMed] [Google Scholar]
- [33].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [34].Mujezinovic N, Schneider G, Wildpaner M, Mechtler K, Eisenhaber F. Reducing the haystack to find the needle: improved protein identification after fast elimination of non-interpretable peptide MS/MS spectra and noise reduction. BMC Genomics. 2010;11(Suppl. 11):S13. doi: 10.1186/1471-2164-11-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen X, Drogaris P, Bern M. Identification of tandem mass spectra of mixtures of isomeric peptides. J. Proteome Res. 2010;9:3270–3279. doi: 10.1021/pr100205k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- [37].Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]