Abstract
High-resolution top-down mass spectrometry was used to characterize eleven integral and five peripheral subunits of the 750 kDa Photosystem II (PSII) complex from the eukaryotic red alga, Galdieria sulphuraria. The primary separation used liquid chromatography mass spectrometry with concomitant fraction collection (LC-MS+) yielding around 40 intact mass tags (IMTs) at 100 ppm mass accuracy on a low-resolution electrospray-ionization mass spectrometer, whose retention and mass were used to guide subsequent high-resolution top-down nano-electrospray Fourier-transform ion-cyclotron resonance mass spectrometry experiments (FT-MS). Both collisionally activated and electron capture dissociation (CAD, ECD) were used to confirm the presence of eleven small subunits to mass accuracy within 5 ppm; PsbE, PsbF, PsbH, PsbI, PsbJ, PsbK, PsbL, PsbM, PsbT, PsbX and PsbZ. All subunits showed covalent modifications that fall into three classes including retention of initiating formyl-methionine, removal of methionine at the N-terminus with or without acetylation, and removal of a longer N-terminal peptide. Peripheral subunits identified by top-down analysis included oxygen evolving complex (OEC) subunits PsbO, PsbU, PsbV, as well as Psb28 (PsbW) and Psb27 (‘PsbZ-like’). Top-down high-resolution mass spectrometry provides the necessary precision, typically less than 5 ppm, for identification and characterization of polypeptide composition of these important membrane protein complexes.
Keywords: FT-MS, top-down proteomics, membrane proteins, Photosystem II, PTMs, Red algae
1 Introduction
Top-down proteomics is dependent upon high resolution and accurate mass measurements for analysis of intact proteins and their dissociation-derived product ions and can provide complete primary structure and post-translational modification assignment [1, 2]. In recent years, with the maturation of FT-MS coupled with electrospray ionization and fragmentation options including collisionally activated (CAD), electron capture (ECD), electron transfer (ETD) and infrared multi-photon (IRMPD) dissociation, top-down mass spectrometry has been developed into an extremely powerful method in protein science and proteomics and has gained high acceptance among the community, culminating with the award of the Biemann medal of the American Society for Mass Spectrometry to Neil Kelleher in 2009. For wider applicability, top-down mass spectrometry must address the entire proteome including the membrane proteins that constitute 20 to 30% of the open reading frames (ORF’s) in fully sequenced genomes [3], with their important functions ranging from photosynthesis, respiration and cellular communication to small molecule transport. Integral membrane proteins have transmembrane regions that pose major challenges to mass spectrometry as they are deficient in amino acids that carry charge and are insoluble in aqueous solvents, without detergents. Thus it can be argued that the most effective way to characterize these domains is to include them as part of the intact protein in a top-down experiment [4]. To date, studies have shown that integral membrane proteins can be analyzed by ESI-MS with mass accuracies comparable to those achievable for soluble proteins, and top-down high resolution FT-MS has been demonstrated for both polyhelix bundle and transmembrane β-barrel motifs [5–7].
Galdieria sulphuraria is a photosynthetic unicellular red alga that grows in acidic (pH 0.5 to 3.0), higher temperature (50 to 55 °C) environments [8]. Along with Cyanidium caldarium and Cyanidioschyzon merolae, these are the only photosynthetic eukaroyotes populating the Cyanidiales. Molecular phylogenetic studies have revealed that the Cyanidiales are one of the most ancient groups of algae, presumed to be at the base of eukaryotic lineages [9]. Galdieria is unique among other Cyanidiales as it propagates by endospores, has a cell wall and is highly tolerant of toxic metal ions (aluminium, cadmium and mercury) [10]. Galdieria can survive on more than fifty different carbon sources with the ability to grow photoautotrophically, heterotrophically and mixotrophically. The conditions under which Galdieria performs photosynthesis are at the extreme and it is of interest to study the proteins involved.
PSII is a large membrane protein complex that catalyzes the light driven electron transfer from water to plastoquinone, thereby oxidizing two water molecules to produce 4H+, 4e− and molecular oxygen. Because of its importance, several biochemical, biophysical, proteomic and structural studies of PSII from cyanobacteria, green algae and higher plants have been published. PSII is comprised of more than 20 subunits with most of these encoded in the chloroplast and the remainder in the nucleus [11]. Core integral subunits D1 (PsbA) and D2 (PsbD) bind most of the redox co-factors forming the electron transport chain assisted by antenna proteins CP43 (PsbC) and CP47 (PsbB) as well as the alpha and beta subunits of cytochrome b559 (PsbE and PsbF), and are present in PSII of all organisms. Along with the conserved core, there are several small integral membrane subunits (< 10 kDa) that are somewhat more diverse. Fully functional PSII requires assembly of the oxygen-evolving complex (OEC) which is stabilized by peripheral subunits. While the nature of the water-splitting reaction is conserved across organisms, the subunits that stabilize the OEC differ. In cyanobacteria, the OEC stabilizing proteins consist of PsbO, PsbU and PsbV subunits, while PsbO, PsbP and PsbQ are used in green algae and higher plants [12]. The eukaryotic Cyanidiales differs in their OEC composition: C. caldarium has a special subunit PsbQ’ along with cyanobacterial PsbO, PsbU and PsbV subunits, while C. merolae is reported to have PsbO, PsbU, PsbP and PsbQ [13, 14]. Considering Galdieria’s divergence from other Cyanidiales, it is important to characterize their peripheral subunits. Recently, others have shown that there are additional peripheral subunits associated with PSII (Psb27 and Psb28) though they are not thought to be associated with stabilization of the active OEC [15, 16].
Mass spectrometry has now made major contributions to the structural characterization of the PSII complexes from cyanobacteria, green algae and higher plants [5, 15, 17–20]. Recently, high-resolution FT-MS was applied to a number of different integral membrane proteins including the cyanobacterial cytochrome b6f complex [7]. The extension of this work to an ever-increasing array of membrane proteins is exemplified here where we have systematically applied FT-MS to study the composition of PSII from the eukaryotic red alga Galdieria. A total of sixteen peripheral and small integral subunits were analyzed by high resolution FT-MS using both CAD and ECD, allowing assignment of precursor and product ions to mass accuracies typically better than 5 ppm. The results show that while the Galdieria PSII integral core complex is very similar to that of other eukaryotes, the peripheral OEC composition is more similar to cyanobacteria. The detection of five peripheral subunits including Psb27 and Psb28 in the red algal preparation suggests they contain a functional and structurally specialized population of PSII complexes that may be important for survival in the harsh environments in which this algal species thrives.
2 Materials and Methods
2.1 Cell culture and PSII isolation
Galdieria sulphuraria were grown at 42° C in 11 L capacity flasks containing a 10X medium at pH 2.0, with a constant supply of air, CO2 and light irradiances of 25 μmol photon m−2 s−1 (Li-cor, model LI-189). Cells were harvested by centrifugation (2000 x g; 2 minutes; 25° C). Cells were homogenized in 20 mM MES pH 6.0, 10 mM CaCl2, 10 mM MgCl2, 500 mM mannitol (MMCM buffer) with 1 mM PMSF added before cell breakage. Following breakage of the cells, PSII was isolated as described previously (Fig. 1) [21]. The original specimen, Galdieria sulphuraria strain 074, collected at Mount Lawu, Java, Indonesia was provided by Dr. Christine Oesterhelt, Max-Planck-Institut für Molekulare Pflanzenphysiologie, Am Mühlenberg 1, 14476 Golm, Germany. It was thought to be a single strain but growth capacity tests revealed that there were two strains in the sample [22]; under heterotrophic conditions, strain 074W (W-white) lost pigment whereas 074G (G- green) remained green on all substrates. Galdieria sulphuraria 074W was used to isolate PSII, which is the same strain that was used for the genome sequence information. The small differences in the sequences that were revealed by our study may be due to a diversification of the initial strain due to different growth conditions in the laboratories. The genome was sequenced from the laboratory strain that had been grown under photoheterotrophic conditions, while the Photosystem II was isolated from the strain that had been maintained under photoautrotrophic growth conditions since it was originally isolated.
Figure 1.
Flow chart summarizing the key steps in non-denaturing PSII membrane protein purification from Galdieria sulphurararia
2.2 Reversed phase chromatography with online electrospray-ionization mass spectrometry and fraction collection (LC-MS+)
Samples were precipitated using acetone. The suspension was split into two centrifuge tubes (125 μL each) and 1 mL 80% acetone in water (−20 °C stock) added to each tube, prior to vortex mixing (1 minute) and incubation at −20 °C for 1 hour. Precipitated protein was recovered by centrifugation (10,000 x g) and the supernatant removed. Pellets were dried briefly to allow evaporation of residual acetone (5 minutes, room temperature and pressure) and dissolved in 90% formic acid (total 100 μL) for immediate injection onto an HPLC system prepared for reversed phase chromatography [23, 24]. A column (5 μm, 300 Å PLRP/S, 2 × 150 mm; Varian) is previously equilibrated in 95% buffer A (0.1 % TFA in water), 5% B (0.05% TFA; 50% acetonitrile; 50% isopropanol) at 100 μL/minute at 40 °C for 30 minutes prior to sample injection. The column is eluted with a stepped linear gradient of increasing buffer B as previously described [23] and the eluent passed through a UV detector (280 nm) prior to a liquid flow splitter delivering 50 μL/minute to a low resolution mass spectrometer and 50 μL/minute to a fraction collector (1 minute/fraction). Fractions collected into microcentrifuge tubes were stored at −80 °C prior to off-line high-resolution nanospray analysis. Mass spectrometry was performed using a triple quadrupole instrument (AP III+, Applied Biosystems) tuned and calibrated using a PEG mixture as previously described [5]. Mass spectra were recorded by scanning from m/z 600 – 2300 with orifice voltage ramped with mass (60 – 120) using a 0.3 Da step size and a scan speed of 6 sec. Data were processed using BioMultiview 1.3.1 software (Applied Biosystems).
2.3 High resolution FT-MS mass spectrometry
Selected HPLC fractions collected during LC-MS+ were subjected to direct infusion nanospray analysis. Samples were individually loaded into 2 μm i.d. externally-coated nanospray emitters (Proxeon, Cambridge, MA) and desorbed using spray voltage of between 1.7 – 1.9 kV (versus the inlet of the mass spectrometer) using the nanospray source supplied by the manufacturer. These conditions produced a flow rate of 20 – 50 nL/min. All samples were analyzed using a hybrid linear ion-trap/FT-ICR mass spectrometer (7 T, LTQ FT Ultra, Thermo Scientific, Bremen, Germany) operated with standard (up to m/z 2000) or extended mass range (up to m/z 4000). Ion transmission into the linear trap and further to the FT-ICR cell was automatically optimized for maximum signal. The ion count targets for the full scan FT-ICR and MS2 FT-ICR experiments were 2 × 106. The m/z resolving power of the FT-ICR mass analyzer was set at 100,000 (defined by m/Δm50% at m/z 400) unless otherwise stated. Individual charge states of the multiply protonated protein molecular ions were selected for isolation and CAD in the linear trap followed by the detection of resulting fragments in the ICR cell. The precursor ions were activated using normalized collision energy settings in the range 10 – 15 at the default activation q-value of 0.25. For ECD studies precursor ions were transmitted to the FT-ICR cell and electrons introduced with normalized energy in the range 5 – 10, 10 ms duration and 50 ms delay. Spectra were derived from an average of between 50 – 200 transient signals. In addition, the analysis of Psb27 was performed on a hybrid linear ion-trap Fourier-transform orbitrap mass analyzer (LTQ XL Orbitrap ETD; Thermo Scientific, Bremen, Germany) similarly to the LTQ-FT experiments. High-energy collisional dissociation (HCD) was performed in the C-trap and ETD was performed using fluoranthene anions produced in a chemical ionization source according to the manufacturer’s instructions. Orbitrap product ion spectra were recalibrated using prominent y- and z- ions.
FT-ICR and FT-Orbitrap mass spectra were processed using ProSightPC software (ProSightPC 2.0, Thermo Scientific, Bremen, Germany) to produce monoisotopic mass lists (s/n = 1.1, fit 0%, remainder 0%, average table set to averagine) that were then assigned to protein sequences with various post-translational modifications. Protein identifications were achieved by generating sequence tags (sequence tag compiler and sequence tag searching tool) and matching these tags to a Galdieria database. Product ion assignments for known proteins were made using the software operated in single protein mode with a 10 ppm mass accuracy threshold and with the Deltamass feature deactivated. ProSightPC reported mass accuracy of assigned monoisotopic mass values of precursor and product ions and the probability that some other database entry might match the same dataset (P score).
All raw data files (.raw) and ProSightPC files (.puf) will be deposited at tranche (https://proteomecommons.org/) upon manuscript acceptance.
2.4 Annotation of PSII subunit genes from Galdieria sulphuraria
For the identification and annotation of chloroplast-endcoded PSII subunits, a collection of all open reading frames (ORFs) identified on the two supercontigs stig_35 and stig_158 from the Galdieria sulphuraria Genome Project [8, 10] (http://genomics.msu.edu/galdieria) was prepared after closure of all gaps (K. Krause, A. Weber et al., publication in preparation). This ORF collection was searched for sequences with homology to the amino acid sequences of PSII subunits from T. elongatus (Thermosynechococcus elongatus) and Arabidopsis thaliana and other higher plants. The nuclear-encoded PSII subunit genes were identified by searching the translated Galdieria nucleotide databases for sequences orthologous to the remaining subunits. The presence of predicted target peptides was analysed using the TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP) [25]. All nucleotide sequences were conceptually translated and nucleotide as well as protein sequences were prepared for annotation in Genbank by using Sequin Version 9.50 (provided by Genbank). The relevant accession numbers are: psbA - GU474525; psbB - GU474526; psbC -GU474527; psbD-GU474528; psbE - GU474529; psbF - GU474530; psbH - GU474531; psbI - GU474532; psbJ - GU474533; psbK - GU474534; psbL - GU474535; psbM -GU474520; psbN - GU474536; psbO - GU474521; psbP - GU474522; psbQ -GU971652; psbQ’ – GU721104; psbT - GU474537; psbU - GU474523; psbV -GU474538; psbW - GU474539; psbX - GU474540; psbY - GU474541; psbZ - GU474542; psb27 - GU474524. Nuclear encoded subunits include PsbM, PsbO, PsbP, PsbQ, PsbQ’, PsbU, Psb27; chloroplast encoded subunits include PsbA, PsbB, PsbC, PsbD, PsbE, PsbF, PsbH, PsbI, PsbJ, PsbK, PsbL, PsbN, PsbT, PsbV, Psb28 (PsbW), PsbX, PsbY and PsbZ.
2.5 Experimental design
Separate Galdieria PSII preparations were analyzed by LC-MS+ on four different occasions over a twelve-month period (four biological replicates). Each top-down mass spectrometry experiment typically included CAD analyses of at least two different charge states and one ECD and/or ETD experiment. Results from the experiment that yielded the most product ions are presented.
3 Results
3.1 Chromatographic separation of PSII subunits
The primary reversed phase LC-MS+ separation yielded a retention map that was annotated with protein identities where they are known, and intact masses where they remain unknown (Fig. 2). Mass accuracy on the low-resolution quadrupole instrument was typically 100 ppm (1 Da at 10,000 Da) and the spectra were used to guide targeted top-down high-resolution experiments on selected fractions from the LC-MS+ experiment in order to fully define primary structure and post-translational modifications of PSII subunits. The physical basis of the separation relies upon hydrophobicity, and thus the first proteins to elute are the hydrophilic peripheral PSII subunits PsbO, PsbU, PsbV, Psb28 (PsbW) and Psb27 (‘PsbZ-like’). These were followed by a set of proteins in the 16 kD class, identified as phycobiliproteins (ApcA, ApcB, ApcD, ApcF, CpcA, CpcB, CpcG) and a pair of 55 kD proteins identified as AtpA and AtpB of ATP synthase (data not shown). Association of phycobiliproteins with PSII is predictable, while ATP synthase is probably a contaminant of the preparation. The remainder of the chromatogram was dominated by elution of the small and large integral subunits of PSII spanning a substantial range of hydrophobicity, with the PsbE and PsbF subunits that bind cytochrome b559 eluting around 70 minutes, and PsbZ not eluting until 133 minutes. Several of the small subunits (PsbF, PsbM, PsbT, PsbI, PsbH) had singly oxidized isoforms that eluted a little earlier than the unmodified population. The intensities of different peaks in the total ion chromatogram shown in Fig. 2 are dependent on abundance and ionization efficiency and thus stoichiometry should not be inferred. Table 1 lists intact mass tags (IMTs) that were assigned to PSII subunits based upon coincidence of measured average mass with calculated average mass based upon primary structure and limited post-translational modification. These assignments remain coincidental, with little statistical confidence until a high-resolution top-down analysis is completed (Table 2). Out of around 40 detected IMT’s, 20 PSII subunits were assigned with different PTMs. The 3409 Da IMT eluting at 78 minutes is potentially a PSII subunit but so far remains unidentified.
Figure 2.

Reversed phase total ion chromatogram elution profile of PSII from the red alga, Galdieria sulphuraria. PSII proteins were precipitated with acetone and dissolved in formic acid for loading onto a reversed phase column as described in the experimental procedures. The column was eluted with a compound gradient of increasing organic concentration with eluent directed to a flow splitter for online electrospray-ionization MS and fraction collection. Total ion current was summed across the m/z range 600 – 2300 every 6 s scan (arbitrary units, linear scale). Peak heights are related to both ionization and elution efficiency of individual proteins. PSII proteins identified at different elution times are labeled. Masses of other proteins including some phycobiliproteins and subunits of the ATP synthase (intact mass tags) are given in Dalton (Da).
Table 1.
Average masses of PSII subunits from low resolution LC-MS+
| PSII protein subunit | Genbank Accession # | # TMHa | Elution Time (min.) | Calculated average mass (Da) | Experimentally determined average mass (Da) | Proposed post-translational modification |
|---|---|---|---|---|---|---|
| Peripheral (extrinsic) subunits | ||||||
| PsbOb (33 KDa) | GU474521 | 0 | 39.4 | 28816.4 | 28821 | 1 – 21 removed, one disulfide bond |
| PsbUb | GU474523 | 0 | 36.1 | 10585.9 | 10587 | 1–81 removed |
| PsbV (Cyt c 550) | GU474538 | 0 | 38.4 | 15704.2 | 15704 | 1–30 removed 168–177 removed + heme |
| Psb27b (‘PsbZ-like’) | GU474539 | 0 | 38.4 | na | 12820 | na |
| Psb28 (PsbW) | GU474524 | 0 | 38.9 | 13246.0 | 13247 | -Met1 |
| Smaller integral membrane subunits | ||||||
| PsbE | GU474529 | 1 | 74.8 | 9392.5 | 9393 | -Met1 |
| PsbF | GU474530 | 1 | 68.8 | 4971.8 | 5015 | -Met1 + acetylation |
| PsbH | GU474531 | 1 | 111.1 | 7483.9 | 7485 | -Met1 |
| PsbI | GU474532 | 1 | 93.2 | 4476.3 | 4505 | Formyl-Met1 |
| PsbJ | GU474533 | 1 | 82.8 | 3917.6 | 3960 | -Met1+acetylation |
| PsbK | GU474534 | 1 | 95.1 | 4255.2 | 4256 | 1–8 removed |
| PsbL | GU474535 | 1 | 93.2 | 4247.9 | 4290 | -Met1 + acetylation |
| PsbMb | GU474520 | 1 | 80.2 | 4599.3 | 4600 | 1–38 removed |
| PsbT | GU474537 | 1 | 97.8 | 4217.4 | 4245 | Formyl-Met1 |
| PsbX | GU474540 | 1 | 102.7 | 4534.5 | 4563 | Formyl-Met1 |
| PsbZ | GU474542 | 2 | 132.9 | 6826.4 | 6855 | Formyl-Met1 |
| Larger integral membrane subunits | ||||||
| PsbA (D1) | GU474525 | 5 | 95.5 | 38160.7 | 38184 | -Met1+ acetylation, 345–360 removed |
| PsbB (CP 47) | GU474526 | 5 | 88.7 | 56558.8 | 56551 | -Met1+acetylation |
| PsbC (CP 43) | GU474527 | 6 | 97.8 | 50588.3 | 50927 | 1–14 removed |
| PsbD (D2) | GU474528 | 6 | 106.0 | 39344.1 | 39346 | -Met1+acetylation |
Transmembrane helices
nuclear encoded genes
not available
Table 2.
Monoisotopic masses of PSII subunits from high resolution Fourier-transform mass spectrometry (FT-MS)
| PSII protein subunit | Calculated Monoisotopic mass (Da) | Experimental Monoisotopic mass (Da) | Mass difference Delta (ppm) | P-score | Confirmed post-translational modification |
|---|---|---|---|---|---|
| Peripheral (extrinsic) subunits | |||||
| PsbO | 28798.6898 | 28796.7520 | 2.6387 | 1.41E-26 | 1 – 21 removed, one disulfide bond |
| PsbU | 10579.4347 | 10579.4648 | 2.8361 | 5.26E-75 | 1 – 81 removed |
| PsbV (Cyt. c550) | 15692.8033 | 15692.7162 | 5.1594 | 3.32E-45 | 1 – 30 removed, heme (+615.16947), Thr80 to Val (−1.972) |
| Psb27 (PsbZ-like) | 12811.4000 | 12811.3742 | 0.2939 | 8.93E-42 1.9E-100a |
168–177 removed unknown N-terminus 1375.65 Da on Ala19 |
| Psb28 (PsbW) | 13237.6972 | 13237.7755 | 4.6250 | 9.55E-28 1.0E-152a |
-Met1 |
| Smaller integral membrane subunits | |||||
| PsbE (α-cyt. b559) | 9386.7896 | 9386.8317 | 4.4238 | 2.81E-16 | -Met1 |
| PsbF (β-cyt. b559) | 5010.6243 | 5010.6386 | 3.8488 | 6.95E-88 | -Met1 + acetylation |
| PsbH | 7479.1312 | 7479.1417 | 1.2749 | 1.53E-49 | - Met1 |
| PsbI | 4501.4234 | 4501.4367 | 2.8954 | 6.85E-66 | formylMet1 |
| PsbJ | 3957.1113 | 3957.1322 | 5.2626 | 3.25E-61 | -Met1 + acetylation |
| PsbK | 4252.5198 | 4252.5336 | 3.2322 | 2.64E-59 | 1 – 8 removed |
| PsbL | 4287.2771 | 4287.2888 | 2.6928 | 2.15E-45 | -Met1 + acetylation |
| PsbM | 4596.4123 | 4596.4282 | 3.4407 | 2.21E-68 | 1–38 removed |
| PsbT | 4242.3140 | 4242.3284 | 3.2844 | 2.23E-44 | formylMet1 |
| PsbX | 4559.5632 | 4559.5807 | 3.7467 | 3.11E-45 | formylMet1 |
| PsbZ | 6849.9577 | 6849.9597 | 0.0925 | 7.55E-42 | formylMet1 |
From ECD/ETD experiments.
3.2 Larger integral membrane subunits
The four large subunits (PsbA, PsbB, PsbC and PsbD), which account for twenty-two of the total thirty four transmembrane alpha helices in PSII, eluted between 88 and 106 minutes along with other smaller subunits (Table 1). The large subunits PsbA, PsbB, PsbC and PsbD measured 38184, 56551, 50927 and 39350 Da respectively, largely consistent with their gene sequences and known N- and C- terminal post-translational modifications (Table 1). The experimentally determined average mass of 38184 Da for PsbA was consistent with the removal of the initiating methionine at the N-terminus with acetylation of Thr 2, and removal of 15 amino acids at the C-terminus (Table 1) in conservation with other species. The difference of 23 Da relative to the calculated average mass (38160.7 Da) could be due to minor DNA sequence differences between the strain (074W) used for PSII preparation and sequencing. In PsbD, the difference between the calculated (39344.1 Da) and experimental (39346 Da) average masses is within experimental measurement error (Table 1). For subunit PsbB, the experimentally derived mass (56551 Da) is in good agreement with the calculated mass (56558.8 Da) taking into account the loss of initial methionine and N-terminal acetylation (Table 1). In PsbC, if 14 amino acids are removed to form a processed PsbC with a free N-terminus in conservation with higher plants, the calculated mass (50588.3) is lower than measured (50888 Da) by 300 Da, indicating either divergent N-terminal processing or some other DNA sequence discrepancy. Despite minor inconsistencies, the intact mass tags of the larger integral subunits can be assigned with confidence. Since the main focus of this study was to identify and characterize the smaller integral (< 10 kDa) and the peripheral subunits constituting Galdieria PSII further experiments on the large subunits were not performed.
3.3 Peripheral subunits1
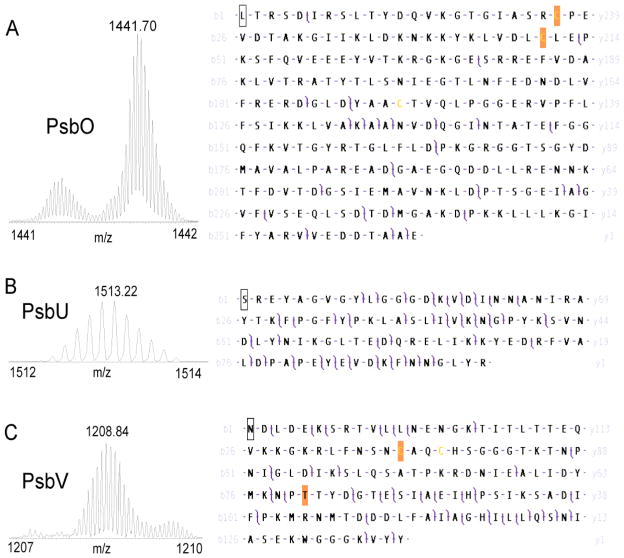
Of the three (PsbO, PsbU, PsbV) OEC stabilizing subunits identified, PsbO and PsbU are encoded by the nucleus and carry an N- terminal chloroplast target peptide. PsbO has a target peptide consisting of the first 21 amino acids so the mature form has 242 amino acids. The experimentally determined monoisotopic mass of 28796.7520 Da could be reconciled (Δ = 2.6387 ppm) with the calculated monoisotopic mass of 28796.67415 Da with inclusion of the formation of a single disulfide bond (−2.01565 Da) between the only two Cys residues, Cys23 and Cys47 (Figs. 2 and 3A). Assignment of product ions from a CAD experiment of the disulfide linked mature protein yielded 11 y-and 9 b- ion matches within a 10 ppm tolerance and a consequent P score of 1.41E-26 (Fig. 3A, Table 2). In the case of PsbU, the first 81 amino acids of the signal peptide are cleaved at the N terminus, forming a mature protein of 93 amino acids, with a calculated monoisotopic mass of 10579.4347 Da (Table 2). Top-down CAD analysis of the N-terminally truncated protein confirmed the primary structure of PsbU with a experimentally determined monoisotopic mass of 10579.4648 Da, within 5 ppm of the calculated monoisotopic mass (Δ = 2.8361 ppm), and a CAD experiment yielded 32 b- and 23 y- product ions for a P score of 5.26E-75 (Fig. 3B, Table 2). The third extrinsic subunit PsbV was assigned to an intact mass tag of 15704 Da with proposed modifications including removal of 30 amino acid residues from the N-terminus, removal of 10 residues from the C-terminus and attachment of a covalently bound heme group (615.16947 Da) (Fig. 2, Table 1). Analysis of the high-resolution CAD dataset for PsbV gave a mass difference of −2.0602 Da (−131.2860 ppm) with 11 b- and 15 y- ions matched. Further optimization of the match between measured and calculated mass was reached by changing Thr 80 to Val (mature protein numbering) significantly increasing coverage with 19 b- and 24 y- product ions matched and a P score of 3.32E-45 (Δ = 5.1595 ppm) (Fig. 3C, Table 2).
Figure 3.
Top-down mass spectrometry of oxygen evolving complex subunits. Selected fractions were analyzed by static nanospray on a 7 Tesla LTQ-FT Ultra. Collisionally activated dissociation experiments on isolated precursor ions yielded complex tandem mass spectra that were matched to primary protein sequences using ProSightPC. The ion isolation used for each subunit is shown on the left while the b- and y- product ion assignments observed at 10 ppm tolerance are shown superimposed on the sequence to the right.
A. For subunit PsbO, an N-terminal signal sequence of 21 amino acids has been removed from the precursor sequence and highlighted Cys residues have been oxidized (−1.00782509 Da each) to account for disulfide bond formation. In the mature protein, 20% of product ions were matched to 19 b- and 10 y- ions giving a P score of 1.41E-26.
B. In PsbU, an 81 amino-acid signal peptide was removed from the precursor sequence. 25% of product ions were matched to 32 b- and 22 y- ions giving a P score of 5.26E-75.
C. In PsbV, 30 residues were removed from the N-terminus and 10 from the C-terminus of the precursor sequence to nominally match the measured intact mass tag. Change of Thr80 to Val optimized match of observed high-resolution precursor and product ion masses with 19 b- and 23 y- ions for a P score of 3.32E-45.
Besides the 3 OEC stabilizing subunits, 2 other peripheral subunits were identified, Psb27 and Psb28. In the primary LC-MS+ analysis, a hydrophilic protein eluted at 39 minutes with an intact mass tag of 13247 Da and was assigned as Psb28 with removal of Met1 (Fig. 2, Table 1). This protein is similar to the PsbW identified in Synechocystis and will be referred to as Psb28 hereafter (see discussion). Psb28 was assigned to a monoisotopic mass peak of 13237.7755 Da (Table 2), confirming the identity and processing with 4 b- and 17 y- product ions and the precursor matched below 5 ppm (Δ = 4.6250 ppm) for a P score of 9.55E-28 (Table 2). The identity of Psb28 was also confirmed by ECD (Fig. 4) with 57 c- and 39 z- product ions, and agreement between calculated and measured monoisotopic masses for the precursor ion within 5 ppm (Δ = 4.6250 ppm) for a P score of 1.02E-152 (Fig. 4A, Table 2). An IMT of 12820 Da was assigned to Psb27 (psbz-like) using top-down mass spectrometry after attempts to assign it with low-resolution data were unsuccessful. In Fig. 5 the assignments for the dissociation experiments performed on an LTQ-Orbitrap (HCD and ETD) are shown. Both experiments yield b- and y-, and c- and z- ions that support complete agreement with the C-terminal sequence over most of the polypeptide chain. However, the data strongly suggests that the sequence reported for Psb27 has some errors towards its N-terminus and we have therefore represented this region as an unknown modification on the Ala residue shown in Fig. 5 of mass 1375.65 Da to optimize b- and c- ion matches. Using this strategy we matched the experimental monoisotopic mass of 12811.3742 Da to the calculated value at <1 ppm, and matched 12 b- and 23 y- ions in the CAD (HCD) experiment and 36 c- and 44 z- ions in the ETD experiment. The measured P scores for each experiment (respectively) confirm the accuracy of the identification (Fig. 5). The nature of the N-terminus is under further study.
Figure 4.
Top-down mass spectrometry of subunit Psb28 (PsbW). A protein of average mass 13247 Da detected by LC-MS+, was analyzed by top-down FT-MS with ECD. The 14-charge m/z 947 precursor ion was isolated as shown inset left. The product spectrum is shown from m/z 650 – 1500, expanded between in the range 800 – 1200 and expanded again in the range m/z 960 – 990. As shown in the full range spectrum, the ion current is dominated by charge reduction to 13+ and to a lesser extent 12+ species. Careful inspection of lower abundance ions nevertheless reveals a very rich product ion spectrum across the m/z 800 – 1200 range, and at higher detail between m/z 960 and 990. After removal of the initiating Met residue, 61% of the product ions were matched to 57 c- and 39 z- ions for a final P score of 1.02E-152. Selected c- and z- ions are labeled. In a separate CAD experiment on a m/z 1020 precursor ion 23% of product ions were matched giving a P score of 9.55E-28 (not shown).
Figure 5.
Top-down mass spectrometry of subunit Psb27 (PsbZ-like).
A 12820 Da intact mass tag was confidently identified as Psb27 using top-down MS on an Orbitrap mass analyzer with HCD (CAD) and ETD (ECD).
A. A 15-charge precursor of m/z 855 (ion isolation inset top right) was isolated and fragmented using HCD generating the complex product ion mass spectrum shown (m/z 500 – 1400). The data was matched to the Psb27 sequence with the first Ala residue carrying a modification mass of 1375.65 Da to represent the as yet uncharacterized N-terminus of the protein. 26% of product ions were matched to 12 b- and 23 y-ions for a P score of 8.93E-42.
B. A 16-charge precursor of m/z 802 (ion isolation inset top right) was isolated and fragmented using ETD generating the complex product ion mass spectrum shown (m/z 300 – 1600). The data was matched to the Psb27 sequence with the first Ala residue carrying a modification mass of 1375.65 Da to represent the as yet uncharacterized N-terminus of the protein. 28% of product ions were matched to 36 c- and 44 z- ions for a P score of 1.9E-100.
Effort was expended finding the PsbQ, PsbQ’ and PsbP proteins, genes, which can be identified in the Galdieria genome. Attempts to match intact mass tags and top-down MS experiments in the 37 – 76 minute retention range to these sequences were unsuccessful so one minute fractions across this range were digested with trypsin and analyzed by nano-liquid chromatography with tandem mass spectrometry. While these experiments successfully identified many peptides from PsbO, PsbU, PsbV, Psb27, Psb28, ApcA, ApcB, ApcD, ApcF, CpcA, CpcB, AtpA, AtpB as well as some of the integral subunits there were no peptides detected for PsbQ, PsbQ’ or PsbP (data not shown).
3.4 Small integral membrane subunits
The analysis confirmed the presence of core subunits PsbE, PsbF and Psb I, and the remaining integral subunits PsbH, PsbJ, PsbK, PsbL, PsbM, PsbT, PsbX and PsbZ. The results of top-down CAD and, for PsbF and PsbL, ECD, are shown in Fig. 6, with coverage and P scores described in the legend and measured/calculated masses in Table 2. P scores ranged from 2.81E-16 for PsbE and 6.95E-88 for PsbF demonstrating the statistical confidence of the assignments achieved using high-resolution FT-MS with CAD. Even higher P scores were achieved for some of the ECD/ETD experiments. The small subunits could be grouped into three categories based upon post-translational processing. PsbI, PsbT, PsbX and PsbZ were unprocessed with initiating formyl-Met residue intact. PsbE, PsbF, PsbH, PsbJ and PsbL had Met1 removed though only PsbF, PsbJ and PsbL were subsequently acetylated. PsbK and PsbM had longer signal peptides removed from their N-termini and were not acetylated.
Figure 6.
Top-down mass spectrometry of small integral membrane subunits A. PsbE, B. PsbF, C. PsbH, D. PsbI, E. PsbJ, F. PsbK G. PsbL H. PsbM I. PsbT J. PsbX and K. PsbZ. The ion isolation used for CAD/ECD for each subunit is shown on the left, while the product ion assignments (b- and y- ions for CAD; c- and z- ions for ECD) observed at 10 ppm tolerance are shown on the sequence to the right. A shaded box around a Met residue in position 1 indicates retention of the formyl group as translated. A shaded box around a different residue implies removal of one or more amino-acid residues from the N-terminus and N-acetylation. An open box around the first amino acid implies removal of one or more amino-acids with no further modification. Percentage of matched product ions for small subunits: PsbE – 6%; PsbF – 36/51%; PsbH – 59%; PsbI – 29%; PsbJ – 23%; PsbK – 29%; PsbL – 38%; PsbM – 40%; PsbT – 33%; PsbX – 35% and PsbZ – 44%. P scores are given in Table 2.
4 Discussion
4.1 Number of subunits/transmembrane helices
Top-down mass spectrometry has been used to characterize integral [23] and peripheral [26] thylakoid membrane proteins for several years now. Top-down high-resolution FT-MS was first applied to an integral thylakoid protein demonstrating the potential benefits of using ECD [6]. Recently, all 8 subunits of the cytochrome b6f complex from Nostoc were analyzed in this way for full characterization of post-translational modifications [7]. Here we have used top-down FT-MS and orbitrap FT-MS to characterize the smaller integral and the peripheral subunits of PSII from Galdieria. A significant challenge was the lack of a highly annotated Galdieria proteome database and considerable effort was devoted to the search for PSII homologues. We were able to define the identity of all the subunits with high confidence. In total Galdieria PSII appears to have 4 large subunits, 11 smaller integral subunits and 3 peripheral OEC subunits for a total of 18 subunits. This total gives 2 fewer subunits and 2 fewer transmembrane helices than the cyanobacterial complex in agreement with the 34 transmembrane α-helices identified in spinach [27] and in contrast to the 36 transmembrane helices identified in the PSII X-ray structure of T. elongatus [28], perhaps a little surprising given the similarity between the Galdieria OEC and that of the cyanobacteria. It remains possible that the small 3409 Da intact mass tag observed in the LC-MS+ experiment is a PSII subunit, but we were unable to match top-down FT-MS data to any sequence in our Galdieria database. PsbN and ycf12 of the cyanobacterial complex were found in the Galdieria genome by homology but were not found in the protein analysis despite their inclusion in the database used for interpretation of the data. Final resolution of the issue will require a crystal structure of sufficient quality to identify the number and subunit assignment of the transmembrane helices in Galdieria PSII.
4.2 Post-translational Modifications
Top-down mass spectrometry allowed characterization of PTMs with molecular detail. Modification of small subunits shows a distinct pattern based on the exposure of their N terminus to the stroma or lumen. Subunits PsbI, PsbK, PsbM, PsbT, PsbX and PsbZ have their N-termini exposed to the lumen with PsbI, PsbT, PsbX and PsbZ retaining their initiating formyl-Met residue unmodified. PsbK and PsbM had transit peptides removed (Table 2). The other subunits PsbE, PsbF, PsbH, PsbJ and PsbL have their N-termini exposed to the stroma and in all cases have their initiating Met residue removed. PsbF, PsbJ and PsbL have their N-terminii acetylated. The observed differences in modification for different subunits may be due to the availability of different processing enzymes in stroma and lumen [28] and accessibility of the N-termini to these enzymes.
4.3 Peripheral subunits
Inspection of the Galdieria genome allowed identification of genes for the OEC stabilizing subunits, which are important for the assembly of the lumenal side of PSII that catalyzes the water-splitting reaction (PsbO, PsbU, PsbV, PsbP, PsbQ, PsbQ’) from both prokaryotic and eukaryotic origins. PsbO is the only extrinsic protein conserved from cyanobacteria to higher plants. As in green algae and higher plants, PsbO is nuclear encoded in Galdieria with a transit peptide sequence of the first 21 residues, which is shorter than the 34 residue predicted by TargetP [25]. In the cyanobacterium T. elongatus 26 residues are removed [28] while transit peptide sequences of 52, 76 or 84 residues are predicted for Chlamydomonas reinhardtii (green alga), Cyanidium caldarium (red alga) and Spinacia oleracea (higher plant) respectively [29]. PsbO from Arabidopsis was shown by mass spectrometry to have 85 N-terminal residues removed during processing [26, 30]. Processing of Galdieria PsbO leaves the mature N-terminus conserved compared with other organisms. Finally, the two cysteine residues in Galdieria are conserved across all organisms, so it is likely that the disulfide bond is present in all organisms. PsbU was processed by cleavage of the first 81 residues of the transit peptide at the consensus sequence A-X-A (79 – 81), a recognition site for the thylakoid processing peptidase [29]. The Galdieria transit sequence is longer compared to that of T. elongatus (30 residues) and C. caldarium (61 residues). In Galdieria, the mature PsbV N-terminus is obtained after removal of the transit peptide (1 – 30 residues) in agreement with the other red algae, Cyanidium caldarium and Cyanidioschyzon merolae and the cyanobacterium, T. elongatus [13, 28]. Alignment of the Galdieria PsbV sequence with PsbV from Cyanidium caldarium (CCPsbV - Q9TLW2-1) and Cyanidioschyzon merolae (CMPsbV - Q85FS3) showed Galdieria to be extended by 10 amino acids at the C-terminus, and their removal allowed matches of more product ions. A change of Thr80 to Val further optimized measured and calculated masses to within 10 ppm and maximized the number of product ions matched. Such discrepancies might be explained by small differences between the laboratory strains that were used for genome sequencing and protein isolation (see methods).
Two other peripheral proteins were identified in the preparation, Psb27 and Psb28. Both are in the 12 – 13 kDa range and hydrophilic, being the first to elute in reversed phase chromatography. The gene for Psb28 was included in the Galdieria database based upon homology to cyanobacterial PsbW, also a hydrophilic 13 kDa protein. It is noted that this is different to the PsbW of higher plants, which is a hydrophobic 6.1 kDa protein. We prefer the Psb28 designation to avoid confusion with PsbW [11]. Top-down MS demonstrated removal of the initiating Met residue and no other PTMs. Psb27 was included in the Galdieria database based upon homology to cyanobacterial ‘psbZ-like’ protein. Similarly to Psb28, we note that Psb27 is not like PsbZ, which is a smaller hydrophobic integral subunit. Top-down MS using both CAD and ECD gave numerous y-and z- product ions matched to better than 5 ppm, confirming the accuracy of the C-terminal sequence. However, there appears to be disagreement toward the N-terminus that could be due to alternative splicing of the primary transcript, sequencing errors or differences between lines of the 074W strain that was used for PSII preparation versus sequencing. Only a sequence analysis of the mature psb27 transcript will be able to resolve the genetic basis of this discrepancy. The problem appears to be toward the N-terminus from Ala19 of the database sequence, and we therefore placed a modification of mass 1375.65 Da on that residue to account for the unmatched stretch of polypeptide. In this way many b- and c- ions could be matched confirming with confidence the sequence beyond Ala19. Psb27 is lipidated at the N-terminus in cyanobacteria [15] and our ongoing work will reveal the situation in Galdieria.
Small integral subunits
In Galdieria, the larger subunit of cytochrome b559 PsbE has the intiating Met residue removed, while the smaller subunit, PsbF, loses Met1 and becomes acetylated as also seen in cyanobacteria (T. elongatus and Synechocystis sp. PCC 6803) and higher plants (Hordeum vulgare and Nicotiana tabacum) [20, 28, 31]. The PsbH subunit showed removal of Met1 with a free N-terminus as was reported for other organisms [20, 28, 31, 32]. Though light-sensitive phosphorylation at one or two sites close to the N-terminus has been identified in higher plants [33] a sequence comparison of PsbH between higher plant (Spinacia oleracea) and Galdieria revealed lack of conservation of these phosphorylation sites, as also seen in T. elongatus. No evidence of phosphorylation of PsbH (or any other subunit of Galdieria) was detected. PsbI retained the initating formyl-Met residue as also observed for cyanobacteria and higher plants [20, 28, 31]. Galdieria PsbJ contains 38 amino-acid residues and is located in a gene cluster containing PsbE, PsbF and PsbL. PsbJ has Met1 removed and the N-terminus acetylated consistent with cyanobacteria [28]. While PsbJ was confirmed as a part of PSII in Synechocystis sp. PCC 6803 and T. elongatus, it was not detected in PSII of T. vulcanus by N-terminal sequencing [28, 32], probably because of blockage of Edman degradation by N-acetylation. In Galdieria, the first 8 residues are removed from the N-terminus of PsbK while in T. elongatus 9 residues are removed and in N. tabacum the first 24 [28, 31]. A free amino terminus was retained in all cases. In Galdieria, PsbL is processed by removal of the initiating Met and N-terminal acetylation. In T. elongatus and H. vulgare, PsbL is unmodified while in N. tabacum, it has lost its initiating Met [20, 28, 31]. In cyanobacteria and higher plants, the PsbM subunit retains the initiating formyl-Met residue [20, 28, 31, 33] and in Chlamydomonas, Edman sequencing showed a blocked N-terminus [34]. While higher plant and green algal PsbM are chloroplast encoded, the Galdieria PsbM is nuclear encoded and has a 38 amino-acid transit peptide removed leaving a free amino-terminus. Galdieria PsbT retains the initiating formyl-Met residue in agreement with PsbTc in P. sativum and N. tabacum, and PsbT in T. elongatus [28, 31, 33]. In H. vulgare, N-terminal acetylation is reported however [20]. In recent structural models of dimeric PSII complexes, PsbT along with PsbL and PsbM are located at the PSII monomer interface [28]. It will be of high interest to know the oligomeric state of PSII in Galdieria as this organism performs photosynthesis under extreme conditions. Galdieria PsbX retains the initiating formyl-Met residue and is otherwise unmodified. Mass spectrometric analysis of a protein assigned as PsbX in T. elongatus, was concluded to show cleavage of the first 10 amino acids due to the presence of a lumen targeting pre-sequence [28, 35]. The current version of T. elongatus PsbX (Uniprot Q9F1R6) does not have this extension, requiring removal of just Met1 to achieve the match. PsbZ is conserved across photosynthetic organisms and our analysis confirmed retention of the initiating formyl-Met residue in Galdieria PsbZ in good agreement with T. elongatus, P. sativum and N. tabacum [20, 28, 31].
Conclusions
Top-down high-resolution MS is clearly the technology of choice for absolute characterization of protein identity and modification. The reversed phase LC-MS+ platform presents an attractive primary separation that avoids problems associated with gels. Low resolution MS data from the primary LC-MS+ experiment is used to drive user directed data-dependent top-down MS experiments. Interpretation of top-down data relies upon accurate annotation of high-quality genomic data though it is now very easy to see where actual primary structure diverges from that predicted by translation of DNA sequence. The small integral and the peripheral subunits of Galdieria PSII have been characterized with unprecedented detail, emphasizing the amenability of membrane proteins to analysis by top-down MS.
Acknowledgments
The authors congratulate Neil Kelleher and Fred McLafferty for the development of top-down mass spectrometry and NK for his award of the 2009 Biemann medal from the American Society of Mass Spectrometry. NIH is thanked for funds for instrument purchase (S10 RR023045). This work was supported by funds from the National Science Foundation NSF award 0417142 to PF. APMW acknowledges support from DFG SFB TR1. We thank Dr. C. Oesterhelt for providing us with the photoautotrophically grown strains 074W and 074G.
Abbreviations
- CAD
collision activated dissociation
- ECD
electron capture dissociation
- IMT
intact mass tags
- OEC
oxygen evolving complex
- PSII
photosystem II
Footnotes
In this article, peripheral subunits refers to all of the subunits PsbO, PsbP, PsbQ, PsbQ’, PsbU, PsbV, Psb27 and Psb28, whereas OEC stabilizing subunits refers specifically to all these subunits, excluding Psb27 and Psb28.
References
- 1.Kelleher NL, LH, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J Am Chem Soc. 1999;121:806–807. [Google Scholar]
- 2.McLafferty FW, Breuker K, Jin M, Han XM, et al. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. Febs Journal. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 3.Nam HJ, Jeon J, Kim S. Bioinformatic approaches for the structure and function of membrane proteins. Bmb Reports. 2009;42:697–704. doi: 10.5483/bmbrep.2009.42.11.697. [DOI] [PubMed] [Google Scholar]
- 4.Whitelegge J, Halgand F, Souda P, Zabrouskov V. Top-down mass spectrometry of integral membrane proteins. Expert Rev Proteomics. 2006;3:585–596. doi: 10.1586/14789450.3.6.585. [DOI] [PubMed] [Google Scholar]
- 5.Whitelegge JP, Gundersen CB, Faull KF. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998;7:1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zabrouskov V, Whitelegge JP. Increased coverage in the transmembrane domain with activated-ion electron capture dissociation for top-down Fourier-transform mass spectrometry of integral membrane proteins. J Proteome Res. 2007;6:2205–2210. doi: 10.1021/pr0607031. [DOI] [PubMed] [Google Scholar]
- 7.Whitelegge J. Confident Assignment of Post-Translational Modifications Using Top-Down Mass Spectrometry. Molecular & Cellular Proteomics. 2009:S15–S15. [Google Scholar]
- 8.Weber APM, Oesterhelt C, Gross W, Brautigam A, et al. EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Molecular Biology. 2004;55:17–32. doi: 10.1007/s11103-004-0376-y. [DOI] [PubMed] [Google Scholar]
- 9.Ciniglia C, Yoon HS, Pollio A, Pinto G, Bhattacharya D. Hidden biodiversity of the extremophilic Cyanidiales red algae. Molecular Ecology. 2004;13:1827–1838. doi: 10.1111/j.1365-294X.2004.02180.x. [DOI] [PubMed] [Google Scholar]
- 10.Barbier G, Oesterhelt C, Larson MD, Halgren RG, et al. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiology. 2005;137:460–474. doi: 10.1104/pp.104.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi LX, Schroder WP. The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochimica Et Biophysica Acta-Bioenergetics. 2004;1608:75–96. doi: 10.1016/j.bbabio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Roose JL, Wegener KM, Pakrasi HB. The extrinsic proteins of photosystem II. Photosynthesis Research. 2007;92:369–387. doi: 10.1007/s11120-006-9117-1. [DOI] [PubMed] [Google Scholar]
- 13.Enami I, Iwai M, Akiyama A, Suzuki T, et al. Comparison of binding and functional properties of two extrinsic components, Cyt c550 and a 12 kDa protein, in cyanobacterial PSII with those in red algal PSII. Plant and Cell Physiology. 2003;44:820–827. doi: 10.1093/pcp/pcg106. [DOI] [PubMed] [Google Scholar]
- 14.Thornton LE, Ohkawa H, Roose JL, Kashino Y, et al. Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechopystis 6803. Plant Cell. 2004;16:2164–2175. doi: 10.1105/tpc.104.023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowaczyk MM, Hebeler R, Schlodder E, Meyer HE, et al. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell. 2006;18:3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobakova M, Sobotka R, Tichy M, Komenda J. Psb28 Protein Is Involved in the Biogenesis of the Photosystem II Inner Antenna CP47 (PsbB) in the Cyanobacterium Synechocystis sp PCC 6803. Plant Physiology. 2009;149:1076–1086. doi: 10.1104/pp.108.130039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma J, Panico M, Barber J, Morris HR. Purification and determination of intact molecular mass by electrospray ionization mass spectrometry of the photosystem II reaction center subunits. J Biol Chem. 1997;272:33153–33157. doi: 10.1074/jbc.272.52.33153. [DOI] [PubMed] [Google Scholar]
- 18.Premstaller A, Oberacher H, Walcher W, Timperio AM, et al. High-performance liquid chromatography-electrospray ionization mass spectrometry using monolithic capillary columns for proteomic studies. Anal Chem. 2001;73:2390–2396. doi: 10.1021/ac010046q. [DOI] [PubMed] [Google Scholar]
- 19.Zolla L, Timperio AM, Walcher W, Huber CG. Proteomics of light-harvesting proteins in different plant species. Analysis and comparison by liquid chromatography-electrospray ionization mass spectrometry. Photosystem II. Plant Physiol. 2003;131:198–214. doi: 10.1104/pp.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ploscher M, Granvogl B, Zoryan M, Reisinger V, Eichacker LA. Mass spectrometric characterization of membrane integral low molecular weight proteins from photosystem II in barley etioplasts. Proteomics. 2009;9:625–635. doi: 10.1002/pmic.200800337. [DOI] [PubMed] [Google Scholar]
- 21.Kern J, Loll B, Luneberg C, DiFiore D, et al. Purification, characterisation and crystallisation of photosystem II from Thermosynechococcus elongatus cultivated in a new type of photobioreactor. Biochimica Et Biophysica Acta-Bioenergetics. 2005;1706:147–157. doi: 10.1016/j.bbabio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Gross W, Schnarrenberger C. Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiology. 1995;36:633–638. [Google Scholar]
- 23.Whitelegge JP, Zhang HM, Aguilera R, Taylor RM, Cramer WA. Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein - Cytochrome b6f complex from spinach and the cyanobacterium Mastigocladus laminosus. Molecular & Cellular Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Whitelegge J. In: HPLC of Peptides and Proteins:Methods and Protocols. Aguilar M-I, editor. Humana Press; New Jersey: 2004. pp. 323–340. [Google Scholar]
- 25.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 26.Zabrouskov V, Giacomelli L, van Wijk KJ, McLafferty FW. A new approach for plant proteomics: characterization of chloroplast proteins of Arabidopsis thaliana by top-down mass spectrometry. Mol Cell Proteomics. 2003;2:1253–1260. doi: 10.1074/mcp.M300069-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Yakushevska AE, Jensen PE, Keegstra W, van Roon H, et al. Supermolecular organization of photosystem II and its associated light-harvesting antenna in Arabidopsis thaliana. Eur J Biochem. 2001;268:6020–6028. doi: 10.1046/j.0014-2956.2001.02505.x. [DOI] [PubMed] [Google Scholar]
- 28.Guskov A, Kern J, Gabdulkhakov A, Broser M, et al. Cyanobacterial photosystem II at 2. 9-angstrom resolution and the role of quinones, lipids, channels and chloride. Nature Structural & Molecular Biology. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 29.Tohri A, Suzuki T, Okuyama S, Kamino K, et al. Comparison of the structure of the extrinsic 33 kDa protein from different organisms. Plant and Cell Physiology. 2002;43:429–439. doi: 10.1093/pcp/pcf053. [DOI] [PubMed] [Google Scholar]
- 30.Gómez SM, Bil KY, Aguilera R, Nishio JN, et al. Transit peptide cleavage sites of integral thylakoid membrane proteins. Mol Cell Proteomics. 2003;2:1068–1085. doi: 10.1074/mcp.M300062-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Granvogl B, Zoryan M, Plocher M, Eichacker LA. Localization of 13 one-helix integral membrane proteins in photosystem II subcomplexes. Analytical Biochemistry. 2008;383:279–288. doi: 10.1016/j.ab.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 32.Kashino Y, Koike H, Yoshio M, Egashira H, et al. Low-molecular-mass polypeptide components of a photosystem II preparation from the thermophilic cyanobacterium Thermosynechococcus vulcanus. Plant and Cell Physiology. 2002;43:1366–1373. doi: 10.1093/pcp/pcf168. [DOI] [PubMed] [Google Scholar]
- 33.Gómez SM, Nishio JN, Faull KF, Whitelegge JP. The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Molecular & Cellular Proteomics. 2002;1:46–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Devitry C, Diner BA, Popot JL. Photosystem-II particles from Chlamydomonas-reinhardtii - purification, molecular-weight, small subunit composition, and protein-phosphorylation. Journal of Biological Chemistry. 1991;266:16614–16621. [PubMed] [Google Scholar]
- 35.Katoh H, Ikeuchi M. Targeted disruption of psbX and biochemical characterization of photosystem II complex in the thermophilic cyanobacterium Synechococcus elongatus. Plant and Cell Physiology. 2001;42:179–188. doi: 10.1093/pcp/pce024. [DOI] [PubMed] [Google Scholar]