Summary
The differentiation and activation of T cells are critically modulated by MAP kinases, which are in turn feed-back regulated by dual-specificity phosphatases (DUSPs) to determine the duration and magnitude of MAP kinase activation. DUSP4 (also known as MKP2) is a MAP kinase-induced DUSP member that is dynamically expressed during thymocyte differentiation. We generated DUSP4-deficient mice to study the function of DUSP4 in T-cell development and activation. Our results showed that thymocyte differentiation and activation-induced MAP kinase phosphorylation were comparable between DUSP4-deficient and wild type mice. Interestingly, activated DUSP4−/− CD4 T cells were hyperproliferative while DUSP4−/− CD8 T cells proliferated normally. Further mechanistic studies suggested that the hyperproliferation of DUSP4−/− CD4 T cells resulted from enhanced CD25 expression and IL-2 signaling through increased STAT5 phosphorylation. Immunization of the DUSP4−/− mice recapitulated the T-cell hyperproliferation phenotype in antigen recall responses, while the profile of Th1/Th2-polarized antibody production was not altered. Combined, these results suggest that other DUSPs may compensate for DUSP4 deficiency in T-cell development, MAP kinase regulation, and Th1/Th2-mediated antibody responses. More importantly, our data indicate that DUSP4 suppress CD4 T-cell proliferation through novel regulations in STAT5 phosphorylation and IL-2 signaling.
Keywords: DUSP4/MKP2, Knockout mice, Signal transduction, Cytokines, Immune regulation
Introduction
The activation of T lymphocyte requires signals transmitted from its surface antigen receptor, TCR. Upon TCR recognition of specific antigenic peptides, a collection of kinases, phosphatases, and adaptor molecules in various signaling pathways is mobilized to transmit and translate the variegated surface signals to unique nuclear genetic programs that govern further differentiation and expansion of naïve T cells into defined effector T cells [1]. Among the signaling pathways implicated in T-cell activation, NF-AT, NF-κB, and MAP kinase are shown to be essential for the activation of IL-2, the prototypic cytokine for T-cell proliferation and survival [2–3]. In addition, these pathways also regulate the differentiation of T cells in the thymus, which is marked by the progressive expression of co-receptor CD4/CD8 from CD4−CD8− (double-negative, DN), to CD4+CD8+ (double-positive, DP), and eventually to CD4+ (CD4 single-positive, CD4SP) or CD8+ (CD8 single-positive, CD8SP) thymocytes [4].
While valuable information is provided by the lack-of-function mutations of signaling molecules in the MAP kinase pathways [5], studying MAP kinase regulations is also important because these regulations may be tied to the delicate fine-tuning of immune cell polarization or immune response [6]. In this regard, the family of dual-specificity phosphatases (DUSPs), which is capable of dephosphorylating both the tyrosine and threonine residues of activated MAP kinases, has been demonstrated to be an important regulator of MAP kinases [7–8]. Indeed, DUSP1/MKP1-, DUSP2/PAC1-, and DUSP10/MKP5-deficient mice exhibit phenotypes in cytokine secretion, adaptive and innate immune regulation, susceptibility to sepsis, and resistance to autoimmune induction [9–11].
DUSPs can be categorized into typical DUSPs that contain an N-terminal kinase binding motif (KBM) and atypical ones that do not. Typical DUSPs can be further classfied based on their subcellular locations, in which the closely related DUSP1, DUSP2, DUSP4/MKP2, and DUSP5/hVH3 are all located predominantly in the nucleus [12]. While all these nuclear DUSPs are highly expressed in various immune cell types, DUSP4 is the major DUSP expressed in Th1, Th2, NK, and LPS-stimulated dendritic cells [12]. The activity of DUSP4 is thought to be regulated mainly at the transcription level, in which signals downstream of MAP kinases, as well as transcription factors such as p53 and HoxA10, are all involved [13–15]. In vitro results show that DUSP4 preferentially dephosphorylates ERK and JNK but not p38 MAP kinases [16], and is induced by growth factors or stress signals [13–14, 17]. Further studies suggest that DUSP4 is involved in fibroblast replication senescence [18] and macrophage apoptosis induction [19] through the regulation of ERK activity. Recently, a report using mice with target mutation of DUSP4 suggests that DUSP4 may regulate MAP kinase activity to alter inflammatory cytokine secretion in macrophage and induce resistance to sepsis [20], while another independent DUSP4 mutant mice showed altered macrophage JNK phosphorylation, Th1/Th2 responses, and susceptibility to L. mexicana [21]. However, the role of DUSP4 in T-cell development and activation has not been reported. Here we have generated DUSP4-deficient mice, and our results suggest that DUSP4 is dispensable for thymocyte differentiation as well as MAP kinase regulation in activated T cells. Nevertheless, DUSP4 appeared to play a non-redundant role in regulating IL-2 signaling and CD4 T-cell proliferation.
Results
Generation of DUSP4-deficient mice
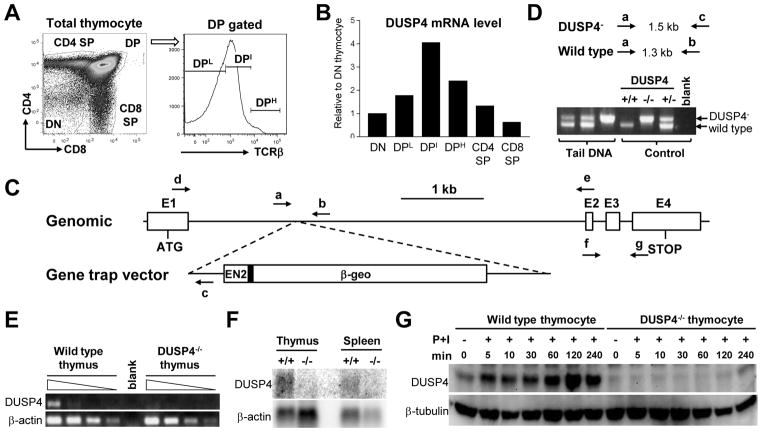
To study whether DUSP4 may be involved in thymocyte development, we sorted C57BL/6 thymocytes into DN, DP:TCRβLow (DPL), DP:TCRβIntermediate (DPI), DP:TCRβHigh(DPH), CD4SP, and CD8SP subsets to determine the relative levels of DUSP4 mRNA in these cells (Fig. 1A). The level of DUSP4 was lower in DN, CD4 SP, and CD8 SP cells, but was the highest in DPI thymocytes (Fig. 1B), in which thymic positive and negative selection occurred. To study the functions of DUSP4 during thymocyte development and T-cell activation, we generated DUSP4-deficient mice (hereafter referred to as DUSP4−/− mice) using gene trap mouse embryonic stem cells (Fig. 1C and 1D). Intercross of heterozygous DUSP4 mice revealed expected Mendelian ratio as well as normal growth curves (data not shown). Semi-quantitative RT-PCR and northern analysis suggested that the gene trap cassette efficiently blocked the formation of mature DUSP4 transcripts (Fig. 1E and 1F). Western analysis of PMA/ionomycin-stimulated thymocytes confirmed DUSP4 protein deficiency in DUSP4−/− cells (Fig. 1G). Interestingly, in WT thymocytes the level of DUSP4 protein was increased as quickly as 5 min following PMA/ionomycin stimulation and was further enhanced up to 2 h later (Fig. 1G), suggesting that, similar to other DUSPs [22–23], DUSP4 may be induced stepwise through both post-translational and transcriptional regulation in activated thymocytes.
Figure 1.
Generation and validation of DUSP4−/− mice. (A) Schematic of thymocyte sorting for qPCR analysis. Total thymocytes were stained for CD4, CD8, and TCRβ followed by sorting for DN, CD4SP, and CD8SP cells. DP thymocytes were sorted separately based on their TCRβ expression levels. (B) cDNA was synthesized from sorted WT thymocytes in (A) for DUSP4 qPCR. The relative level of DUSP4 mRNA compensated by β-actin result was normalized to the level in DN thymocytes. (C) Schematic of the DUSP4 allele, gene trap vector, and locations of oligonucleotides. E1~E4, exon1~exon4. EN2, engrailed-2 intron. Filled rectangle, EN2 splice acceptor. β-geo, fusion gene of β-galactosidase and neomycin phosphotransferase. Arrows, oligonucleotides. ATG, start codon. STOP, stop codon. The schematic is drawn approximately to scale. (D) Expected PCR product sizes and representative PCR results of DUSP4 genotyping. The oligonucleotides used for PCR are indicated as in (C). PCR products corresponding to the DUSP4− or WT allele are also indicated. Blank, no DNA control. (E) Total thymocyte RNA was extracted from WT or DUSP4−/−thymocytes and used in cDNA synthesis and DUSP4 RT-PCR. Shown are RT-PCR results from 4-fold serial dilutions of cDNA. Control β-actin RT-PCR is also shown. Blank, no cDNA. (F) Total thymus or spleen RNA was extracted from WT (+/+) and DUSP4−/− (−/−) mice for northern analysis with a DUSP4-specific probe. The blot was then stripped and reprobed with a β-actin probe as RNA loading control. (G) Total thymocytes from WT or DUSP4−/− mice were activated with PMA and ionomycin (P+I) for the indicated time, followed by western analysis for DUSP4 protein. β-tubulin level as protein loading control is shown. Representative results from two independent experiments are shown (panel B, E, F and G).
DUSP4 is dispensable for thymocyte development and thymic positive selection
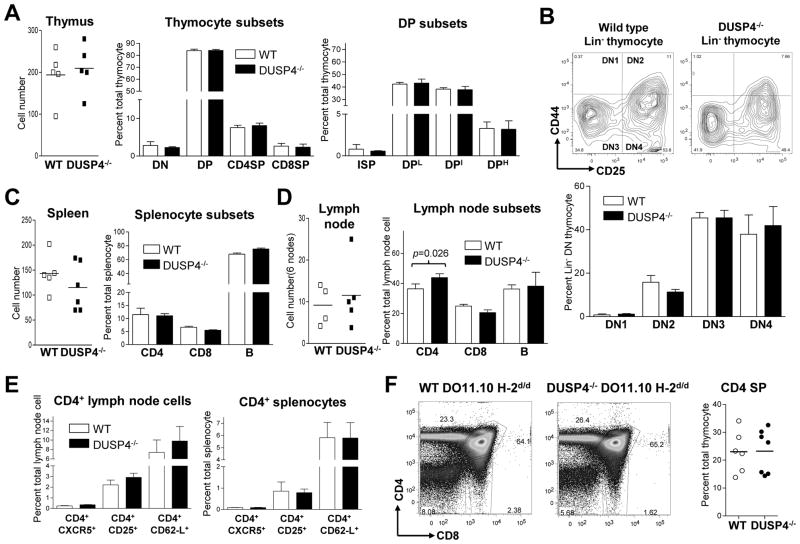
Littermates from DUSP4+/− heterozygous intercross were analyzed by flow cytometry to compare their T-cell development. The results showed that DUSP4−/− and WT littermates had similar numbers of DN, DP, CD4SP, and CD8SP thymocytes, as well as intermediate single-positive (ISP, CD4−CD8+TCRβIntermediate), DPL, DPI, and DPH cells (Fig. 2A). In addition, no significant difference was found when lineage-negative DN thymocytes were further divided based on their CD25 (IL-2Rα subunit) and CD44 expression (Fig. 2B top and bottom panel; p>0.2 for all four subset). In periphery lymphoid organs, splenocytes (Fig. 2C) and lymph node cells (Fig. 2D) from DUSP4−/− and WT littermates have similar number of CD4, CD8, and B cells, except for a marginal increase of CD4 T cells in DUSP4−/− lymph node (Fig. 2D, right panel, p= 0.026). Further analysis of CD4 T cells showed that in the lymph node this minor increase was equally distributed in various subsets including follicular T helper cells (CD4+CXCR5+), regulatory T cells (CD4+CD25+), and naïve T helper cells (CD4+CD62L+) (Fig. 2E, left panel) with no statistical significance. At the mean time no difference was found in the spleen (Fig. 2E, right panel). Finally, to assess thymocyte positive selection, thymocytes from WT or DUSP4−/− mice expressing MHC class II-restricted DO11.10 αβ TCR transgene on positive-selecting H-2d background were analyzed by flow cytometry (Fig. 2F, left panel). The results showed that both mice contained ~23% of CD4 SP thymocytes (Fig. 2F, right panel), suggesting that positive selection was not significantly altered by DUSP4 deficiency. From these results we conclude that, despite being dynamically regulated in the thymus, DUSP4 is dispensable for T-cell development and thymic positive selection.
Figure 2.
DUSP4−/− mice exhibit normal T-cell development and normal number of peripheral lymphocytes. (A), (C), and (D) Single cell suspension of thymus, spleen, and lymph node from 4 to 6 wk old WT and DUSP4−/− littermates (n=4~6) was stained and analyzed by flow cytometry. Total organ cell numbers are shown (left panel), as are the percentages of individual cell subsets (right panel). (B) Lineage-negative (Lin−, negative for CD3ε, CD4, CD8, B220, CD11b, CD11c, and Gr-1) cells from total thymocytes were gated for their relative expression of CD25 and CD44 (top panel). Percentages of individual DN subsets (DN1-DN4) in Lin− thymocytes are shown (bottom panel, n=6). See Supporting Information S1 for gating logics in (A) and (B). (E) CD4+ lymph node cells (left panel) or splenocytes (right panel) from (D) and (C) were further gated for their expression of CXCR5, CD25, and CD62-L. Percentages of the respective subsets in total lymph node cells or splenocytes are shown (n=5~6). (F) WT or DUSP4−/− thymocytes expressing the DO11.10 αβ TCR transgene on H-2d/d background were stained for CD4 and CD8. Shown are representive CD4-CD8 profiles (left panel) and percentages of CD4SP thymocytes (right panel; n=6~7). p > 0.05 for all panels unless specified.
DUSP4 deficiency does not impact MAP kinase and IKKβ phosphorylation in activated T cells
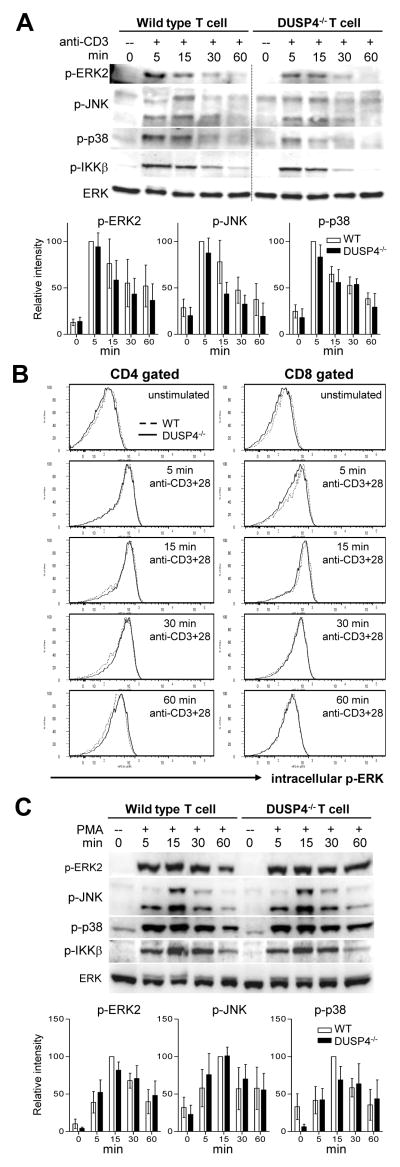
DUSP4 has been shown to dephosphorylate and inactivate ERK and JNK but not p38 in vitro [16]. To see if DUSP4 deficiency resulted in enhanced MAP kinase signaling in activated T cells, DUSP4−/− and WT total T cells were purified from pooled splenocytes and lymph node cells and stimulated with strepavidin-crosslinked anti-CD3. Western analysis results showed that the phosphorylation of ERK2, JNK, p38 and IKKβ was not increased in DUSP4−/− T cells (Fig. 3A, p>0.05 for all lanes). Purified total T cells were also stimulated with plate-bound anti-CD3 and anti-CD28, and the phosphorylation of ERK was examined by intracellular staining. The results showed no discernable difference between WT and DUSP4−/− CD4 (Fig. 3B, left column) or CD8 (Fig. 3B, right column) T cells. Lastly, purified T cells were also stimulated with PMA to bypass proximal TCR signaling, but again no enhanced ERK, JNK, p38, or IKKβ phosphorylation in DUSP4−/− T cells was found (Fig. 3C, p>0.05 for all lanes). Combined, these results suggest that DUSP4 deficiency does not significantly impact MAP kinase and NF-κB regulation in activated T cells.
Figure 3.
DUSP4 deficiency does not impact MAP kinase or IKKβ phosphorylation in activated T cells. (A) MACS-purified total T cells from pooled spleen and lymph node cells were stimulated with 2 μg/ml of anti-CD3/strepavidin mixture (anti-CD3) for the indicated time followed by western analyses. Top panel, representative western analyses results. One unrelated lane was cropped out as indicated by the dotted line without further modification of the image on either side. DUSP4 probing result is not shown because the level of DUSP4 protein in WT T cells was under the detection level in the specified condition. Bottom panel, densitometry quantification of p-ERK2, p-JNK, and p-p38 signals normalized to total ERK is shown as percent of WT signals at 5 min post stimulation (n=3~4). (B) Purified T cells as in (A) were stimulated with 5ug/ml plate-bound anti-CD3 and anti-CD28 for the indicated time, followed by CD4, CD8, and intracellular p-ERK staining. (C) Purified T cells were stimulated with 50 ng/ml PMA for the indicated time and analyzed by western. Top panel, representative western analyses results. Bottom panel, densitometry quantification as in (A) is shown as percent of WT signals at 15 min post stimulation (n=3~4). Representative results from three (panel B) or four (panel A and C) independent experiments are shown. p > 0.05 for all panels.
DUSP4−/− CD4 T cells are hyperproliferative
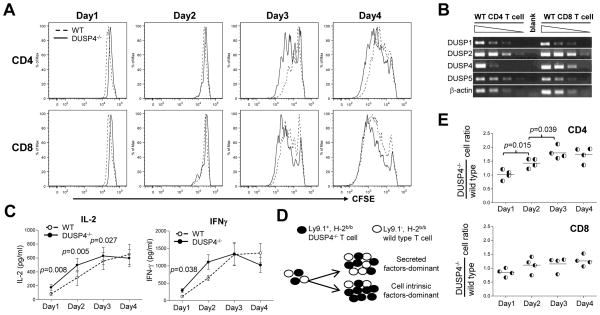
Regardless of the results for MAP kinase activation, purified total T cells from WT and DUSP4−/− mice were loaded with CFSE and stimulated by plate-bound anti-CD3 and anti-CD28 to test whether DUSP4 might regulate T-cell proliferation through MAP kinase-independent pathways. Daily analyses of the stimulated cells by 7AAD and AnnexinV revealed no alteration in cell death (see Supporting Information S2). However, CFSE dye dilution patterns reproducibly showed that DUSP4−/− CD4 T cells proliferated better than WT cells at day 2 and 3 following stimulation (Fig. 4A, top row), while DUSP4−/− CD8 T cells appeared to proliferate similarly as WT cells (Fig. 4A, bottom row). To further quantify cell proliferation, division indexes were calculated from the CFSE profile for each data point. The results showed that DUSP4−/− CD4 T cells proliferated nearly twice as well as WT cells on day 2 (p=0.0046), with the difference decreased to ~40% by day 3 (p=0.0003), while by day 4 both DUSP4−/− and WT CD4 cells were confluent (Table 1). The extent of this CD4 T-cell hyperproliferation varied between groups but the difference was reproducible, as indicated by the low p value. Similar quantification of CD8 T-cell proliferation revealed no significant difference between WT and DUSP4−/− CD8 T cells (Table 2, p>0.05 for all time points). These results thus indicate that DUSP4 deficiency induces hyperproliferation in CD4 but not CD8 T cells.
Figure 4.
DUSP4 deficiency induces CD4 T cells hyperproliferative by enhancing IL-2 signaling. (A) Purified total T cells as in Fig. 3A were loaded with CFSE and stimulated with 0.5, 1.6 or 5 μg/ml plate-bound anti-CD3 and anti-CD28. At each day cells were stained for CD4, CD8, CD25, 7AAD and AnnexinV, followed by analysis of CFSE histogram on gated live CD4+CD25+ (top row) or CD8+CD25+ (bottom row) T cells. See Supporting Information S3 for gating logics. (B) DUSP RT-PCRs were performed as in Fig. 1E with cDNA from MACS-sorted WT CD4 or CD8 T cells. Shown are RT-PCR results from 4-fold serial dilutions of cDNA. Control β-actin RT-PCR is also shown. Blank, no cDNA. (C) Culture supernatant from Fig. 4A was harvested daily and frozen at −80°C for simultaneous IL-2 (n=7, left panel) and IFN-γ (n=6, right panel) ELISA on all samples. (D) Schematic of the mixed T-cell proliferation assay. (E) Purified WT and DUSP4−/− T cells as in Fig. 3A were mixed at 1:1 ratio and stimulated in the same well with 0.5 or 1.6 μg/ml plate-bound anti-CD3 and anti-CD28, followed daily by staining with Ly9.1, CD4, and CD8. The ratios of Ly9.1+ DUSP4−/− T cells divided by Ly9.1− WT T cells are shown for CD4-gated (top) or CD8-gated (bottom) T cells (n=4). Representative results from two (panel B and E) or three (panel A and C) independent experiments are shown. p > 0.05 for all panels unless specified.
Table 1.
Division index of activated CD4+ cell from WT or DUSP4−/− mice
| Day 2 Div. indexa) | Day 3 Div. index | Day 4 Div. index | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| wt | DUSP4−/− | wt | DUSP4−/− | wt | DUSP4−/− | |
| Group 1 | 0.14 | 0.28 | 0.81 | 0.97 | 2.07 | 1.82 |
|
| ||||||
| Group 2 | 0.22 | 0.41 | 1.29 | 1.72 | 2.73 | 2.70 |
|
| ||||||
| Group 3 | 0.38 | 0.65 | 2.06 | 2.31 | 3.35 | 3.14 |
|
| ||||||
| Group 4 | 0.29 | 0.51 | 0.79 | 0.98 | 0.93 | 0.94 |
|
| ||||||
| Group 5 | 0.28 | 0.53 | 0.81 | 1.04 | 1.12 | 1.17 |
|
| ||||||
| Group 6 | 0.06 | 0.08 | 0.40 | 0.75 | 0.98 | 1.26 |
|
| ||||||
| Group 7 | 0.04 | 0.09 | 0.40 | 0.75 | 0.89 | 1.04 |
|
| ||||||
| Mean | 0.20 | 0.36 | 0.94 | 1.22 | 1.72 | 1.72 |
| SE | 0.05 | 0.08 | 0.22 | 0.22 | 0.38 | 0.33 |
|
| ||||||
| p valueb) | 0.0046 | 0.0003 | 1.0000 | |||
Div. index (division index) was calculated by FlowJo software based on CFSE dye-dilution pattern.
Obtained by paired two-tailed t-test.
Table 2.
Division index of activated CD8+ cell from WT or DUSP4−/− mice
| Day 2 Div. indexa) | Day 3 Div. index | Day 4 Div. index | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| wt | DUSP4−/− | wt | DUSP4−/− | wt | DUSP4−/− | |
| Group 1 | 0.28 | 0.28 | 1.23 | 1.12 | 2.77 | 2.14 |
|
| ||||||
| Group 2 | 0.46 | 0.47 | 1.76 | 1.94 | 3.59 | 3.06 |
|
| ||||||
| Group 3 | 0.87 | 0.99 | 3.01 | 3.16 | 4.33 | 4.12 |
|
| ||||||
| Group 4 | 0.63 | 0.51 | 1.30 | 1.24 | 1.27 | 1.60 |
|
| ||||||
| Group 5 | 0.61 | 0.57 | 1.04 | 1.23 | 1.86 | 1.39 |
|
| ||||||
| Group 6 | 0.24 | 0.30 | 0.74 | 1.05 | 1.04 | 1.38 |
|
| ||||||
| Group 7 | 0.11 | 0.21 | 0.74 | 0.99 | 0.98 | 1.15 |
|
| ||||||
| Mean | 0.46 | 0.48 | 1.40 | 1.53 | 2.26 | 2.12 |
| SE | 0.10 | 0.10 | 0.30 | 0.30 | 0.50 | 0.41 |
|
| ||||||
| p valueb) | 0.5770 | 0.0704 | 0.4018 | |||
Div. index (division index) was calculated by FlowJo software based on CFSE dye-dilution pattern.
Obtained by paired two-tailed t-test.
Hyperproliferation of DUSP4−/− CD4 T cells is correlated with enhanced IL-2/IL-2R expression
One possible explanation why enhanced proliferation is observed only in DUSP4−/− CD4 but not CD8 T cells is that DUSP4 may not be abundantly expressed in WT CD8 T cells, so that DUSP4 deficiency may have minimal impact on CD8 T-cell proliferation. However, semi-quantitative RT-PCR results showed that in WT CD8 T cells DUSP4 was expressed equally as, if not slightly higher than, that of WT CD4 T cells (Fig. 4B). In addition, DUSP1, DUSP2, and DUSP5 were also similarly expressed in WT CD4 and CD8 T cells (Fig. 4B). The mechanism responsible for the absence of hyperproliferation in DUSP4−/− CD8 T cells is currently unclear.
Since the early phase of T-cell proliferation is thought to be regulated by IL-2/IL-2R signaling from CD4 T cells [3, 24], we measured the amount of IL-2 in culture supernatant from stimulated T cells. As a control we also measured the concentration of IFN-γ, which was mostly produced by activated CD8 T cells as indicated by our intracellular stainings (data not shown). Day 1 ELISA results showed a ~115% increase of IL-2 from DUSP4−/− sample when compared with WT control, with the difference decreased to ~65% by day 2 and the amount of IL-2 gradually plateaued by day 3 and day 4 (Fig. 4C, left panel). In contrast the secretion of IFN-γ was only slightly increased in DUSP4−/− culture on day 1(Fig. 4C, right panel). Furthermore, the induction of CD25/IL-2Rα, the high-affinity subunit of IL-2 receptor, was also significantly enhanced in activated DUSP4−/− CD4 T cells on day 1 (p=0.0028, Table 3), but not in CD8 T cells (p>0.05, Table 4). It is worth noting that the kinetics of enhanced CD25 and IL-2 expression precedes the increased proliferation of DUSP4−/− CD4 T cells (Table 3, Fig. 4C, and Table 1). These results thus indicate that enhanced IL-2/IL-2R signaling may be responsible for the hyperproliferation in DUSP4−/− CD4 T cells.
Table 3.
Percentage of CD25+ cells in activated CD4+ cell from WT or DUSP4−/− mice
| Day 1 CD25+% | Day 2 CD25+% | Day 3 CD25+% | Day 4 CD25+% | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| wt | DUSP4−/− | wt | DUSP4−/− | wt | DUSP4−/− | wt | DUSP4−/− | |
| Group 1 | 40.80 | 58.60 | 70.60 | 80.10 | 94.70 | 93.20 | 99.20 | 98.20 |
|
| ||||||||
| Group 2 | 67.60 | 73.40 | 86.10 | 92.80 | 97.80 | 98.60 | 99.60 | 99.50 |
|
| ||||||||
| Group 3 | 14.40 | 18.40 | 30.70 | 21.50 | 70.40 | 77.80 | 81.50 | 85.40 |
|
| ||||||||
| Group 4 | 39.50 | 53.50 | 75.20 | 76.90 | 94.20 | 95.00 | 97.30 | 98.50 |
|
| ||||||||
| Group 5 | 15.80 | 29.10 | 42.70 | 57.80 | 57.50 | 87.40 | 87.90 | 94.00 |
|
| ||||||||
| Group 6 | 27.50 | 38.30 | 60.80 | 64.70 | 88.90 | 89.40 | 95.60 | 95.00 |
|
| ||||||||
| Group 7 | 29.00 | 37.00 | 63.80 | 67.30 | 83.70 | 89.70 | 93.90 | 94.30 |
|
| ||||||||
| Group 8 | 82.30 | 82.50 | 84.60 | 95.40 | 99.10 | 99.70 | 99.80 | 99.70 |
|
| ||||||||
| Mean | 39.61 | 48.85 | 64.31 | 69.56 | 85.79 | 91.35 | 94.35 | 95.58 |
| SE | 8.52 | 7.82 | 6.88 | 8.29 | 5.22 | 2.48 | 2.31 | 1.67 |
|
| ||||||||
| p valuea) | 0.0028 | 0.0818 | 0.1702 | 0.2067 | ||||
Obtained by paired two-tailed t-test.
Table 4.
Percentage of CD25+ cells in activated CD8+ cell from WT or DUSP4−/− mice
| Day 1 CD25+% | Day 2 CD25+% | Day 3 CD25+% | Day 4 CD25+% | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| wt | DUSP4−/− | wt | DUSP4−/− | wt | DUSP4−/− | wt | DUSP4−/− | |
| Group 1 | 38.30 | 51.80 | 51.60 | 56.60 | 92.40 | 88.40 | 99.20 | 98.40 |
|
| ||||||||
| Group 2 | 53.60 | 62.70 | 75.50 | 84.50 | 96.70 | 98.40 | 99.80 | 99.60 |
|
| ||||||||
| Group 3 | 76.90 | 79.40 | 91.40 | 95.40 | 99.50 | 99.80 | 99.90 | 99.80 |
|
| ||||||||
| Group 4 | 6.73 | 7.29 | 14.30 | 16.10 | 53.10 | 46.90 | 85.00 | 83.70 |
|
| ||||||||
| Group 5 | 3.48 | 5.00 | 4.38 | 5.94 | 13.00 | 26.70 | 60.30 | 67.90 |
|
| ||||||||
| Group 6 | 5.99 | 7.58 | 11.60 | 9.55 | 51.40 | 43.30 | 86.70 | 72.40 |
|
| ||||||||
| Group 7 | 5.86 | 8.14 | 15.50 | 15.90 | 51.90 | 59.20 | 90.00 | 88.90 |
|
| ||||||||
| Mean | 27.27 | 31.70 | 37.76 | 40.57 | 65.43 | 66.10 | 88.70 | 87.24 |
| SE | 11.10 | 12.04 | 13.22 | 14.28 | 12.10 | 11.09 | 5.31 | 4.99 |
|
| ||||||||
| p valuea) | 0.0539 | 0.0817 | 0.8264 | 0.5729 | ||||
Obtained by paired two-tailed t-test.
In addition to being induced in early T-cell proliferation, CD25 is also considered as a marker for regulatory T cells. To test whether the increase of CD25 expression in DUSP4−/− cells resulted from enhanced regulatory T-cell development, we performed similar T-cell proliferation assay using WT or DUSP4−/− T cells expressing the FOXP3-GFP reporter knock-in gene. The results showed that, in both WT and DUSP4−/−T samples, the majority (>90%) of CD25+ cells did not express FOXP3 (see Supporting Information S4), suggesting that CD25 served as a marker for activation but not regulatory T cell differentiation in these cells. Aslight increase in CD25+GFP+ cell percentage and IL-10 production hinted a role of DUSP4 in regulatory T-cell differentiation (see Supporting Information S4). However, under regulatory T-cell-polarizing condition, WT and DUSP4−/− T cell induced similar FOXP3-GFP expression (see Supporting Information S4). Suppression assays using purified wt or DUSP4−/− FOXP3-GFP+ regulatory T cells also did not reveal any functional difference (see Supporting Information S4). These results, together with a report showing that IL-10 may also be produced by activated Th1 and Th2 cells [25], thus suggest that the enhanced CD25 expression and IL-10 secretion in DUSP4−/− T cells likely reflect enhanced proliferation but not altered regulatory T-cell differentiation or function.
IL-2 and CD25 normally induce reciprocal transcriptional activation through positive feedback regulation, in which IL-2 induces CD25 expression and the formation of high affinity IL-2Rαβγ complex by CD25 helps to amplify IL-2 production [2]. To test whether CD25 or IL-2 served as the proximal target of DUSP4-mediated regulation in activated CD4 T cells, we performed mixed T-cell proliferation assay, in which WT and DUSP4−/− T cells that could be distinguished by the lineage marker Ly9.1 were mixed at 1:1 ratio and stimulated in the same well (Fig. 4D). The flow cytometry results showed that the ratio of DUSP4−/− versus WT CD8 cells did not change significantly (Fig. 4E, right panel), recapitulating the previous CFSE analyses. More importantly, the ratio of DUSP4−/− versus WT CD4 T cells increased gradually on day 2 and day3 (Fig. 4E, left panel, p<0.05), suggesting that cell-intrinsic mechanisms, such as the enhanced CD25 expression, may be the dominant factors responsible for the hyperproliferation of DUSP4−/− CD4 T cells.
DUSP4−/− CD4 T-cell hyperproliferation may be induced by enhanced CD25 expression through increased STAT5 phosphorylation
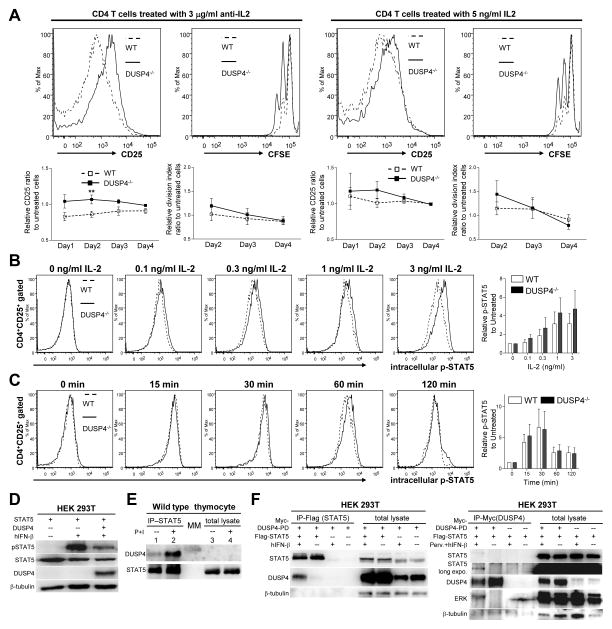
To further study how DUSP4 ablation altered CD25 induction and IL-2 signaling in activated T cells, WT and DUSP4−/− T cells were stimulated in the presence of neutralizing anti-IL-2 antibody or exogenous IL-2. The resulted CD25+ percentages and division indices were normalized to untreated samples to reveal the effects of the treatment. While anti-IL-2 antibody clearly decreased the percentage of CD25+ cells and blocked proliferation in the WT samples as indicated by the smaller-than-one ratio, DUSP4−/− CD4 T cells were more resistant to IL-2 neutralization (Fig. 5A, p<0.1 or 0.05 for all data points in the left panel; p>0.1 in the right panel). Meanwhile the addition of low-dose IL-2 also appeared to be slightly more effective in enhancing CD25 expression and proliferation in DUSP4−/− CD4 T cells than in WT cells, particularly in early time points (Fig. 5A, right panel, p>0.1). Combined with the mixed proliferation results, these data suggest that DUSP4 ablation decreases the threshold of IL-2 signaling, possibly through enhancing CD25 expression. Of the transcription factors reported to regulate CD25 induction, STAT5 is unique for the CD25 locus but is not involved in regulating IL-2 expression [2]. To test if STAT5 is regulated by DUSP4, WT or DUSP4−/− T cells were activated to induce CD25 expression, rested, and then treated with IL-2 to induce STAT5 phosphorylation. Intracellular staining results showed that DUSP4−/− CD4 T cells exhibited stronger STAT5 phosphorylation at various doses of IL-2 (Fig. 5B), but had relatively normal kinetics of p-STAT5 induction and recession (Fig. 5C). When overexpressed in HEK 293T cells, DUSP4 could reduce IFN-β-induced STAT5 phosphorylation (Fig. 5D). Furthermore, DUSP4 could be co-precipitated with endogenous STAT5 in WT thymocytes, with their interaction enhanced by PMA/ionomycin stimulation (Fig. 5E). Lastly, reciprocal co-precipitation of DUSP4 and STAT5 could also be observed in 293T cell over-expressing STAT5 and phosphatase-dead DUSP4 (DUSP4-PD), with IFN-β treatment increasing the efficiency of co-precipitation (Fig. 5F). As a control, ERK could also be co-precipitated with DUSP4 (Fig. 5F). Combined, these results suggest that STAT5 phosphorylation is modulated by DUSP4 and implicate this function to be responsible for DUSP4-mediated regulation of IL-2 signaling in activated T cells.
Figure 5.
DUSP4 ablation decreases IL-2 signaling threshold through sustained STAT5 phosphorylation. (A) WT or DUSP4−/− T cells were stimulated and analyzed as in Fig. 4A without additional treatment, or in the presence of neutralizing anti-IL-2 antibody or exogenous IL-2. Representative CD25 (day 1) or CFSE (day 2) flow cytometry results on gated CD4+ cells are shown (top row). Also shown are daily CD25+ percentage and the division index of viable CD4 T cells of treated samples normalized to untreated samples (bottom row, n=7). ** p<0.05. See Supporting Information S5 and S6 for raw data. (B-C) Purified T cells were stimulated as in Fig 4Afor 44 hrs, rested for 4 hrs, and treated with various doses of IL-2 for 15 min (B) or 3 ng/ml IL-2 for different time (C), followed by flow cytometry analyses of CD4, CD25, and p-STAT5 (Y694) (left panels). Mean fluorescence level of p-STAT5 in CD4+CD25+ cells was normalized to that of CD4 to compensate for staining variation, with the results shown as fold-induction to untreated samples (n=4) (right panels). (D) 293T HEK cells were transiently transfected with the indicated vectors. Twenty-four hr later, cells were treated with 10 ng/ml human IFN-β (hIFN-β) for 15 min or left untreated prior to western analyses for STAT5 Y694 phosphorylation (pSTAT5). STAT5, DUSP4, and β-tubulin loading control are also shown. (E) Total lysates from WT thymocytes treated or untreated with 50 ng/ml PMA and 500 ng/ml ionomycin (P+I) for two hours were immuoprecipitated with anti-STAT5 antibody. The precipitated proteins (IP-STAT5), together with aliquots of the original total lysate (total lysate) were analyzed by western analyses for DUSP4 and STAT5. Due to the amount of lysate loaded, DUSP4 was not detectable in lane 3 and 4. MM, molecular-weight marker. (F). 293T HEK cells were transfected and stimulated with 10 ng/ml human interferon-β (hIFN-β) for 1 hour as indicated. Cell lysates were analyzed by western blotting directly (total lysates) or following immunoprecipitation with anti-Flag (left panel) or anti-Myc (right panel) antibody. Perv., 25 μM pervanadate. Representative results from two (panel D and E) or three (panel A–C, and F) independent experiments are shown. p>0.05 unless specified.
Antigen-stimulated DUSP4−/− T cells are also hyperproliferative but induce normal Th1/Th2 hapten responses
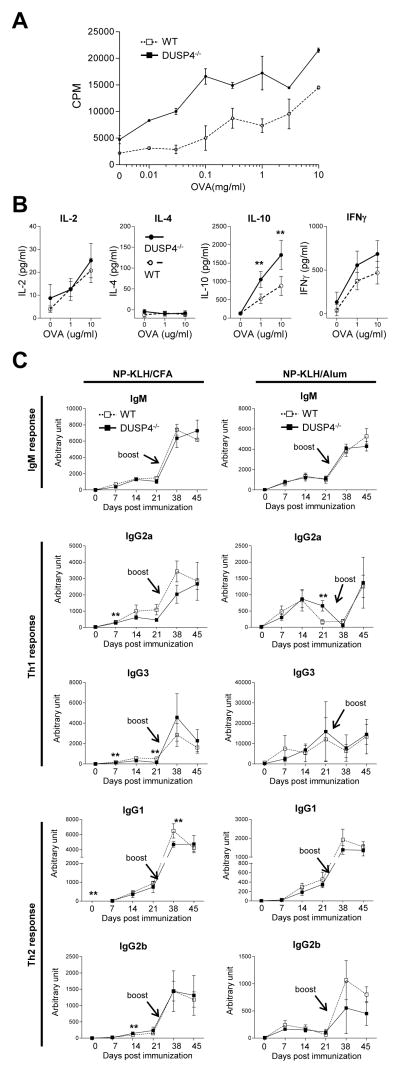
To test whether cognate antigen stimulation could also induce hyperproliferation of DUSP4−/− T cells, draining lymph node cells from OVA/CFA-immunized DUSP4−/− and WT mice were re-stimulated with titrating amount of OVA. Tritiated thymidine incorporation results showed a ~2–3 fold increase in the proliferation of DUSP4−/− T cells relative to WT cells at all OVA concentration (Fig. 6A, p<0.01), similar to the results from anti-CD3+anti-CD28- (Table 1) or ConA- (data not shown) stimulated T cells. OVA-activated DUSP4−/− T cells also showed enhanced IL-10 and, to a lesser extent, IL-2 or IFN-γ production than WT cells, while IL-4 production was undetectable (Fig. 6B). To see whether DUSP4 deficiency altered Th1 and Th2 T-cell functions in vivo, WT or DUSP4−/− mice were immunized and boosted with NP-KLH/CFA (Th1 adjuvant) or NP-KLH/alum (Th2 adjuvant) emulsion. ELISA measurements of serum NP-specific antibodies showed that, in WT and DUSP4−/− mice, similar primary and memory IgM, IgG2a/IgG3 (Th1 cytokine-biased) and IgG1/IgG2b (Th2 cytokine-biased) responses were induced by either immunization (Fig. 6C). The few data points reaching statistical significance are not substantiated by results from preceding or following time points, and likely represent experimental variation. These results are somewhat different from a previous report where decreased Th1 response was observed in DUSP4−/− mice following L mexicana immunization [21], although the difference in immunization protocol may contribute to this discrepancy. Regardless, our results suggested that, while DUPS4 is required for normal T-cell proliferation following antigen stimulation, it may not be essential for regulating hapten-specific antibody production.
Figure 6.
DUSP4−/− mice recapitulate T-cell hyperproliferation but have normal antibody production following immunization. (A) Mice were immunized with OVA/CFA, and the draining popliteal lymph node cells re-stimulated with titrating amount of OVA as described in the Method section for 3H thymidine incorporation (n=3 each). p < 0.01 for all data points. (B). Cells from (A) were stimulated with 0, 1, or 10μg/ml OVA. Supernatant was collected on day 3 for ELISA measurement of cytokine production (n=3 each). (C) Mice were immunized with NP-KLH/CFA or NP-KLH/alum emulsification i.p., followed by serum collection at 0, 7, 14, or 21 d later, antigen boost at day 31, and additional serum collection at day 38 and 45. NP-specific antibodies for the indicated immunoglobulin isotypes were measured by ELISA. Representative results from two (panel B and C) or three (panel A) independent experiments are shown. **, p<0.05.
Discussion
Our present data show that DUSP4 deficiency enhances IL-2 signaling and proliferation in activated CD4 T cells. While DUSP family proteins are thought to mediate negative-feedback regulation of MAP kinase pathways through transcriptional induction by MAP kinases followed by dephosphorylation and inactivation of MAP kinases [12], our STAT5 phosphorylation kinetics and STAT5/DUSP4 co-precipitation results suggested that DUSP4, and perhaps other DUSPs, may also be involved in setting the threshold for cytokine-activated genetic programs by constitutively dephosphorylating downstream transcription factors. This potential regulatory effect of DUSPs on cytokine signaling may also help to explain contradictory effects of DUSP4-deficiency on cytokine production [20, 21], which often cannot be rationalized by alterations in MAP kinase activation.
Several transcription factors are mapped to the CD25 promoter region to regulate its transcription, including NF-AT, NF-κB, AP-1, and CREB, which are shared between IL-2 and CD25 promoters, and others like GATA-1, Elf-1, SMAD3, and STAT5 that are unique to the CD25 locus [2]. Interestingly, the activity of GATA-1, Elf-1, SMAD3, and STAT5 are all regulated by serine/threonine or tyrosine phosphorylation [26–28], with SHP-1, SHP-2, PTP1B, and DUSP3/VHR all implicated in the regulation of STAT5 [29–32]. Furthermore, existing reports show that DUSP1 and DUSP22 dephosphorylate STAT1 and STAT3, respectively [33–34]. As a nuclear DUSP that is capable of dephosphorylating both serine/threonine and tyrosine residues, DUSP4 is certainly fit as a candidate regulator for GATA-1, Elf-1, and SMAD3. We are currently testing these possibilities, as well as using substrate-trap mutant of DUSP4 and proteomic analysis to systematically search for other transcription factors that may be regulated by DUSP4.
IL-2/IL-2R signaling promotes the proliferation of both CD4 and CD8 T cells. It is therefore intriguing that the enhanced proliferation is only observed in DUSP4−/− CD4 but not CD8 T cells. In this regard, CD8 T cells are reported to induce stronger S6K phosphorylation than CD4 T cells following IL-2 stimulation [35]. This implies that CD8 T cells may require a lower IL-2/IL-2R signaling threshold to induce proliferation, a possibility that is partly reflected on the faster proliferation kinetics of CD8 T cells as shown in our CFSE results. The lowered signaling threshold may thus nullify the effect of DUSP4 deficiency in increasing the effectiveness of IL-2/IL-2R signal in CD8 T cells. In parallel, The CD4-specific hyperproliferation may also result from the predominant production of IL-2 in activated CD4 T cells and the subsequent autocrine effect [3, 24]. The relative contribution of these two mechanisms remains to be determined.
We initially expected DUSP4−/− T cells to exhibit enhanced ERK or JNK phosphorylation because DUSP4 has been shown to dephosphorylate ERK and JNK in vitro [16], although two recent findings implicate contradictory results [19, 21]. However, our western and flow cytometry analyses of DUSP4−/− T cells showed no clear enhancement in MAP kinase phosphorylation. We believe that this may result from the redundancy of other DUSPs in regulating MAP kinase activities [11, 20]. The same redundancy may also operate to induce the grossly normal thymocyte development in DUSP4−/− mice, as DUSP5 and DUSP6 have both been demonstrated to regulate thymocyte differentiation [36, 37]. These possibilities may be further explored by breeding DUSP4−/− mice with other DUSP-deficient mice such as the DUSP1, -2, and -10 knockout mice [9–11].
Materials and methods
Mice
DUSP4 gene trap mouse embryonic stem cell clone XG164 was originally deposited by Baygenomics to Mutant Mouse Regional Resource Centers in the International Gene Trap Consortium database. Blastocyst injection and chimeric mouse generation were performed by the Transgenic Mouse Models Core of the National Research Program for Genome Medicine. Germline-transmitted DUSP4+/− mice on a mixed C57BL/6 : 129 background were intercrossed for experiments, as well as backcrossed to C57BL/6 for five more generations. DO11.10 αβ TCR transgenic and FOXP3-GFP mice has been described [38–39]. All experiments except for the analyses of DO11.10αβ TCR transgenic mice, which were analyzed on a mixed Balb/c : C57BL/6 : 129 background, were initially performed on F1 DUSP4−/− mice but repeated in F5 DUSP4−/− mice. Mice were housed under specific pathogen-free condition at the Laboratory Animal Center of the National Health Research Institutes (NHRI). The use of mice followed guidelines set forth by the NHRI’s Institutional Animal Care and Use Committee.
PCR, RT-PCR, northern analysis, and qPCR
Oligonucleotides for PCR, RT-PCR, qPCR, and northern probe generation are listed (see Supporting Information S7). All PCR conditions are available upon request. PCR and RT-PCR were performed with Phire Hot Start polymerase (Fynnzymes). RNA for northern, RT-PCR, and qPCR was extracted with TRIzol (Invitrogen). cDNA synthesis and northern analysis were performed as described previously [40]. Isotope signals were recorded on Storage Phosphor Screens (GE Healthcare) and read on Typhoon Trio with ImageQuant TL software (GE Healthcare). qPCR was performed with FastStart Universal Probe Master Rox (Roche Applied Science) on Realplex4 with Mastercycler ep realplex software (Eppendorf).
Statistical analyses
In all assays data were plotted as mean ± SEM or scattered with mean, with the p values calculated with unpaired or paired two-tailed Student’s t test. p values smaller than 0.05 were marked (**) or indicated.
Acknowledgments
This work was supported in whole or in part by NHRI Grant 99A1-IMPP01-014 (to T.-H. T.) and Grant 100A1-IMPP02-014 (to C. H.), by National Institutes of Health Grant 1R01-AI066895 (to T.-H. T.), and by National Science Council Grant 98-2320-B-400 -006 -MY3 (to C. H.). We thank Dr. Yi-Rong Chen and Dr. Yu-Wen Su for critically reviewing the manuscript, Dr. Toshio Kitamura for providing the STAT5 cDNA clone, Dr. Alexander Rudensky for providing the FOXP3-GFP mice, Dr. Mark Swofford for editing suggestions, and Transgenic Mouse Models Core for blastocyst injection.
Abbreviation
- CD4SP
CD4 single-positive
- CD8SP
CD8 single-positive
- DN
double-negative
- DP
double-positive
- DPH
DP:TCRβHigh
- DPI
DP:TCRβIntermediate
- DPL
DP:TCRβLow
- DUSP
dual-specificity phosphatase
- ISP
intermediate single positive
- KBM
kinase-binding motif
- KLH
keyhole limpet hemocyanin
- NHRI
National Health Research Institutes
- NP
nitrophenyl
- p-
phosphorylated-
- qPCR
quantitative PCR
Footnotes
Conflicts of interest
The authors declare no financial or commercial conflicts of interests.
Additional materials and methods descriptions can be found in Supporting Information S8.
References
- 1.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–366. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Burchill MA, Yang J, Vang KB, Farrar MA. Interleukin-2 receptor signaling in regulatory T cell development and homeostasis. Immunol Lett. 2007;114:1–8. doi: 10.1016/j.imlet.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberola-Ila J, Hernandez-Hoyos G. The Ras/MAPK cascade and the control of positive selection. Immunol Rev. 2003;191:79–96. doi: 10.1034/j.1600-065x.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 5.Kuida K, Boucher DM. Functions of MAP kinases: insights from gene-targeting studies. J Biochem (Tokyo) 2004;135:653–656. doi: 10.1093/jb/mvh078. [DOI] [PubMed] [Google Scholar]
- 6.Germain RN, Stefanova I. The dynamics of T cell receptor signaling: complex orchestration and the key roles of tempo and cooperation. Annu Rev Immunol. 1999;17:467–522. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 7.Kondoh K, Nishida E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim Biophys Acta. 2007;1773:1227–1237. doi: 10.1016/j.bbamcr.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Camps M, Nichols A, Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- 9.Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, Flavell RA. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc Natl Acad Sci U S A. 2006;103:2274–2279. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffrey KL, Brummer T, Rolph MS, Liu SM, Callejas NA, Grumont RJ, Gillieron C, et al. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat Immunol. 2006;7:274–283. doi: 10.1038/ni1310. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Blattman JN, Kennedy NJ, Duong J, Nguyen T, Wang Y, Davis RJ, et al. Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature. 2004;430:793–797. doi: 10.1038/nature02764. [DOI] [PubMed] [Google Scholar]
- 12.Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- 13.Brondello JM, Brunet A, Pouyssegur J, McKenzie FR. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J Biol Chem. 1997;272:1368–1376. doi: 10.1074/jbc.272.2.1368. [DOI] [PubMed] [Google Scholar]
- 14.Shen WH, Wang J, Wu J, Zhurkin VB, Yin Y. Mitogen-activated protein kinase phosphatase 2: a novel transcription target of p53 in apoptosis. Cancer Res. 2006;66:6033–6039. doi: 10.1158/0008-5472.CAN-05-3878. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Lu Y, Huang W, Papoutsakis ET, Fuhrken P, Eklund EA. HoxA10 activates transcription of the gene encoding mitogen-activated protein kinase phosphatase 2 (Mkp2) in myeloid cells. J Biol Chem. 2007;282:16164–16176. doi: 10.1074/jbc.M610556200. [DOI] [PubMed] [Google Scholar]
- 16.Chu Y, Solski PA, Khosravi-Far R, Der CJ, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271:6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 17.Teng CH, Huang WN, Meng TC. Several dual specificity phosphatases coordinate to control the magnitude and duration of JNK activation in signaling response to oxidative stress. J Biol Chem. 2007;282:28395–28407. doi: 10.1074/jbc.M705142200. [DOI] [PubMed] [Google Scholar]
- 18.Torres C, Francis MK, Lorenzini A, Tresini M, Cristofalo VJ. Metabolic stabilization of MAP kinase phosphatase-2 in senescence of human fibroblasts. Exp Cell Res. 2003;290:195–206. doi: 10.1016/s0014-4827(03)00309-4. [DOI] [PubMed] [Google Scholar]
- 19.Ramesh S, Qi XJ, Wildey GM, Robinson J, Molkentin J, Letterio J, Howe PH. TGF beta-mediated BIM expression and apoptosis are regulated through SMAD3-dependent expression of the MAPK phosphatase MKP2. EMBO Rep. 2008;9:990–997. doi: 10.1038/embor.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornell TT, Rodenhouse P, Cai Q, Sun L, Shanley TP. Mitogen-activated protein kinase phosphatase 2 regulates the inflammatory response in sepsis. Infect Immun. 2010;78:2868–2876. doi: 10.1128/IAI.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Mutairi MS, Cadalbert LC, McGachy HA, Shweash M, Schroeder J, Kurnik M, Sloss CM, et al. MAP Kinase Phosphatase-2 Plays a Critical Role in Response to Infection by Leishmania mexicana. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanzola MB, Kersh GJ. The dual specificity phosphatase transcriptome of the murine thymus. Mol Immunol. 2006;43:754–762. doi: 10.1016/j.molimm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Katagiri C, Masuda K, Urano T, Yamashita K, Araki Y, Kikuchi K, Shima H. Phosphorylation of Ser-446 determines stability of MKP-7. J Biol Chem. 2005;280:14716–14722. doi: 10.1074/jbc.M500200200. [DOI] [PubMed] [Google Scholar]
- 24.Long M, Adler AJ. Cutting edge: Paracrine, but not autocrine, IL-2 signaling is sustained during early antiviral CD4 T cell response. J Immunol. 2006;177:4257–4261. doi: 10.4049/jimmunol.177.7.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 26.Yu YL, Chiang YJ, Chen YC, Papetti M, Juo CG, Skoultchi AI, Yen JJ. MAPK-mediated phosphorylation of GATA-1 promotes Bcl-XL expression and cell survival. J Biol Chem. 2005;280:29533–29542. doi: 10.1074/jbc.M506514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juang YT, Solomou EE, Rellahan B, Tsokos GC. Phosphorylation and O-linked glycosylation of Elf-1 leads to its translocation to the nucleus and binding to the promoter of the TCR zeta-chain. J Immunol. 2002;168:2865–2871. doi: 10.4049/jimmunol.168.6.2865. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci U S A. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao W, Hong H, Kawakami Y, Kato Y, Wu D, Yasudo H, Kimura A, et al. Tumor suppression by phospholipase C-beta3 via SHP-1-mediated dephosphorylation of Stat5. Cancer Cell. 2009;16:161–171. doi: 10.1016/j.ccr.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyt R, Zhu W, Cerignoli F, Alonso A, Mustelin T, David M. Cutting edge: selective tyrosine dephosphorylation of interferon-activated nuclear STAT5 by the VHR phosphatase. J Immunol. 2007;179:3402–3406. doi: 10.4049/jimmunol.179.6.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki N, Matsuda T. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000;275:39718–39726. doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- 32.Yu CL, Jin YJ, Burakoff SJ. Cytosolic tyrosine dephosphorylation of STAT5. Potential role of SHP-2 in STAT5 regulation. J Biol Chem. 2000;275:599–604. doi: 10.1074/jbc.275.1.599. [DOI] [PubMed] [Google Scholar]
- 33.Venema RC, Venema VJ, Eaton DC, Marrero MB. Angiotensin II-induced tyrosine phosphorylation of signal transducers and activators of transcription 1 is regulated by Janus-activated kinase 2 and Fyn kinases and mitogen-activated protein kinase phosphatase 1. J Biol Chem. 1998;273:30795–30800. doi: 10.1074/jbc.273.46.30795. [DOI] [PubMed] [Google Scholar]
- 34.Sekine Y, Tsuji S, Ikeda O, Sato N, Aoki N, Aoyama K, Sugiyama K, et al. Regulation of STAT3-mediated signaling by LMW-DSP2. Oncogene. 2006;25 :5801–5806. doi: 10.1038/sj.onc.1209578. [DOI] [PubMed] [Google Scholar]
- 35.Yu A, Zhu L, Altman NH, Malek TR. A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells. Immunity. 2009;30:204–217. doi: 10.1016/j.immuni.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bettini ML, Kersh GJ. MAP kinase phosphatase activity sets the threshold for thymocyte positive selection. Proc Natl Acad Sci U S A. 2007;104:16257–16262. doi: 10.1073/pnas.0705321104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovanen PE, Bernard J, Al-Shami A, Liu C, Bollenbacher-Reilley J, Young L, Pise-Masison C, et al. T-cell development and function are modulated by dual specificity phosphatase DUSP5. J Biol Chem. 2008;283:17362–17369. doi: 10.1074/jbc.M709887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 39.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol. 2005;6:331–337. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 40.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]