Abstract
Since little is known regarding osteocytes, cells embedded within the mineralized bone matrix, a proteomics approach was used to discover proteins more highly expressed in osteocytes than in osteoblasts to determine osteocyte specific function. Two proteomic profiles obtained by two different proteomic approaches using total cell lysates from the osteocyte cell line MLO-Y4 and the osteoblast cell line MC3T3 revealed unique differences. Three protein clusters, one related to glycolysis, (Phosphoglycerate kinase 1, fructose-bisphosphate aldolase A, hypoxia up-regulated 1 [ORP150], triosephosphate isomerase), one to protein folding (Mitochondrial Stress-70 protein, ORP150, Endoplasmin), and one to actin cytoskeleton regulation (Macrophage-capping protein [CapG], destrin, forms of lamin A and vimentin) were identified. Higher protein expression of ORP-150, Cap G, and destrin in MLO-Y4 cells compared to MC3T3 cells was validated by gene expression, Western blotting, and in vivo expression. These proteins were shown to be selective in osteocytes in vivo using immuno-staining of mouse ulnae. Destrin was most highly expressed in embedding osteoid osteocytes, GapG in embedded osteocytes, and ORP150 in deeply embedded osteocytes. In summary, the proteomic approach has yielded important information regarding molecular mechanisms used by osteocytes for embedding in matrix, the formation of dendritic processes, and protection within a hypoxic environment.
Introduction
Several osteocyte selective/specific proteins have been recently identified, such as E11/gp38 (also known as podoplanin), Dentin Matrix Protein 1 (DMP1), Phosphate regulating neutral endopeptidase on chromosome X (PHEX), Matrix Extracellular Phosphoglycoprotein (MEPE), and Sclerostin [1–5]. The deletion of genes coding for these proteins has led to information regarding osteocyte function [6–10]. E11/gp38 was found to be a marker for early osteocytes, with expression occurring as osteoblasts differentiate into osteocytes, and shown to be regulated by mechanical loading in the form of fluid flow shear stress [6]. It appears that this molecule may play a role in dendrite formation of osteocytes in vitro [6]. The functions of DMP1, PHEX and MEPE are related to biomineralization and phosphate homeostasis [11–13]. Sclerostin is expressed in mature osteocytes and functions as an inhibitor of osteoblastic bone formation [14].
Transcriptional comparisons between osteocytes and osteoblasts have confirmed many of these previous identifications. Similar comparisons between osteocyte-like cells and osteoblast-like cells using genome wide transcription arrays showed major differences in actin cytoskeleton and cell communication systems [15]. By comparing immortalized human pre-osteocytic cells and pre-osteoblastic cells, transcription profiling showed increased levels of transcripts related to cytoskeleton, extracellular matrix, and cell adhesion during the differentiation of osteoblasts to osteocytes [16]. Another recent comparison between primary ostoecytes and osteoblasts collected from mouse bone showed differential transcription of genes associated with extracellular matrix proteins, plasma membrane proteins, transcription factors, and muscle function [17].
These differences during osteocyte development offer insight to new functions and requirements for osteocytes in bone. However, relative transcription levels rarely reflect protein expression levels accurately. Variations resulting from alternative splicing, translational regulation, post-translational modifications, proteolytic processing and protein stability ultimately control expression level. These processes also potentially change the function and activity of a protein. Therefore, proteomics provides directly relevant information regarding protein identification and potential structure and function. The premise for the present study was that additional osteocyte-specific or selective proteins remain to be discovered. The discovery of these proteins by proteomic profiling could improve our understanding of the function of osteocytes during bone mineralization and remodeling.
MC3T3-E1 and 2T3 cells are established models for osteoblasts [18, 19], and MLO-Y4 cells represent an osteocyte-like cell line [20]. These cells have been widely used in numerous mechanical loading and apoptosis studies [6, 21–23]. Both MC3T3 and 2T3 cells represent the early osteoblast precursor. In this study, we begin to define the osteocyte proteome compared to the osteoblast proteome, gaining insights into the biology of mature osteocytes in bone matrix.
Materials and Methods
Cell Culture
The MLO-Y4 cell line was used as a model of osteocytes [24]. This cell line was derived from a transgenic mouse in which the immortalizing T-antigen was expressed under control of the osteocalcin promoter. MLO-Y4 cells exhibit properties of osteocytes including high expression of osteocalcin, connexin 43, the antigen E11/gp38. In contrast, expression of the osteoblast marker, alkaline phosphatase, is low. Also, the dendritic morphology of MLO-Y4 cells is similar to that of primary osteocytes. MC3T3 and 2T3 cell lines were used as models of osteoblasts. There are many cell lines established as osteoblast cell models. Of them, MC3T3-E1 is a clonal non-transformed cell line established from newborn mouse calvaria [18]. The 2T3 osteoblast-like cell line is a clonal cell line derived from mouse calvaria immortalized by transgenic expression of the T-antigen driven by the BMP2 promoter [19]. Both of them are well-characterized murine osteoblast models which express osteoblast-specific proteins including alkaline phosphatase, collagen, osteocalcin, and both are capable of mineralization.
The MLO-Y4 cell line was cultured in alpha MEM containing ribonucleosides, deoxyribonucleosides and L-glutamine, (MediaTech, Manassas, VA, USA), supplemented with 2.5% fetal bovine serum and 2.5% calf serum, and the MC3T3 and 2T3 cell lines were cultured in alpha MEM with 10% fetal bovine serum. The fetal bovine serum and calf serum were from Hyclone, Logan, UT, USA. Different media are necessary to maintain biological and morphological features for each cell line. In the last passage before harvesting the cells, media for all the three cell lines were changed to the same alpha MEM with 7% fetal bovine serum to ensure identical culturing conditions before harvesting. All cells were harvested at 70–80% confluency, when cells start to contact each other but are not overlapping.
Protein Collection from Total Cell Lysates
To collect proteins for 2D gels, 1.4 ml lysis buffer (9 M urea, 4% NP40, 2% ampholytes pH 8–10.5, and 1% dithiothreitol [DTT]) was used to lyse the cells in 10 cm dishes. Ampholytes and DTT were added after measurement of protein concentration. After 30 min of 37 °C incubation with agitation or shaking, the lysates were collected and sonicated 3 seconds for 3 times with an interval of 15 minutes.
To collect proteins for Western Blotting, 1ml RIPA buffer (50 mM Tris-HCl, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, and 20 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1μg/ml pepstatin) was used to lyse the MLO-Y4, MC3T3 and 2T3 cells in each 10 cm dish. After 15 minutes of gentle shaking at 4 °C, the lysates were collected and sonicated. Homogenization was performed to increase solubility if necessary.
Protein concentration was measured using a BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL, USA).
2D Polyacrylamide Gel Electrophoresis (PAGE)
For quantitative comparison between large gels, 300 μg of protein from lysates of each cell line was mixed with rehydration buffer (8 M urea, 2% 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) (BioRad, Hercules, CA, USA), 15 mM DTT, 0.2% biolytes 40% pH3–10, 0.1% Orange O) to reach a final volume of 400 μl. Then, the mixture was loaded onto a 17 cm immobilized pH gradient (IPG) pH3–10 strip, and incubated at room temperature for 24 hours. After rehydration, the strip was focused on a Protean IEF Cell for 80 K volt hours (21–28 hours) to complete the first dimension. Before the second dimension, the strip was equilibrated with 12 ml of equilibration buffer (6 M urea, 37.5 mM Tris pH8.8, 2% SDS, 20% glycerol) with 240 mg DTT for 15 minutes, followed by 12 ml of equilibration buffer with 300 mg idoacetamine for another 15 minutes. SDS-PAGE was performed using 8–16% (18.3 × 19.3 cm) precast gels (BioRad, Hercules, CA, USA), electrophoresed for 2300 volt hours. Gels were stained with colloidal Coomassie blue dye (BioRad, Hercules, CA, USA), and photographed using a Kodak Image Station 2000R (Kodak, Rochester, NY, USA). The images of gels were pseudo-colored into red or green, and qualitatively overlaid using ImageJ software [25] to observe the similarities and differences between gels. The images were also quantitatively analyzed by PDQuest 7.2 software to calculate the ratio of intensity of spots between two gels [26]. Spots having at least a 2-fold difference in this analysis were considered differentially expressed protein candidates.
Protein Identification Using Mass Spectrometry
The comparison of the two bone cell lines and identification of proteins were conducted using two different approaches. Experiment 1 was conducted with manual gel spot excision and MALDI MS for peptide mass fingerprinting, whereas experiment 2 incorporated robotic spot picking and LC-tandem MS analysis. Trypsin digestions were conducted in the same manner for both experiments. Excised gel plugs were dehydrated with acetonitrile, swollen in 50 mM ammonium bicarbonate, dehydrated again with acetonitrile, and then rehydrated in 50 mM ammonium bicarbonate containing 20ug per uL porcine modified trypsin (Promega). Extracted peptides were dried down and analyzed as follows.
In experiment 1, a Voyager DE-STR MALDI TOF MS system (Applied Biosystems, Foster City, CA, USA) was operated to identify proteins by their tryptic peptide fingerprints. Threshold parameters used to obtain extract peaklists were 10000 minimum intensity, resolution of 8000, signal-to-noise of 100. Protein Prospector/MS-FIT (UCSF, San Francisco, CA, USA) was used to search the SwissProt database (version August 2004). The NCBI database was also used when SwissProt did not provide identification. Searching was performed with the Applied Biosystems Protein Solution 1 (PS1) system using internal mass calibration with trypsin autolysis masses. Some searches were performed manually using the on-line version of Protein Prospector. Mass error for all accepted peptide “hits” was less than 10 ppm. In addition, criteria for accepting a “hit” were: 1) pI and MW agree approximately with migratory position on the gel, 2) ≥ 10% protein coverage, and 3) at least 5 peptide masses were present.
In experiment 2, selected spots were picked from gels automatically using the Investigator Propic Robotic Workstation (Genomic Solutions, Ann Arbor, MI, USA). After digestion, extracted peptides were analyzed by capillary LC tandem MS using a 50uM inner diameter, 8cm long Phenomenex Jupiter C18 capillary column eluted with a linear gradient of acetonitrile in 50 mM acetic acid (2–70% acetonitrile in 45 min) at a flow rate of 250 nl/min. A ThermoFinnigan LTQ linear ion trap mass spectrometer (San Jose, CA) was operated in data-dependent mode in which one mass spectrum and eight collision induced dissociation (CID) MS/MS spectra acquired per cycle as described [27]. Centered data was exported for searching using Extract_MSN (lcq_dta.exe) with no scan grouping. Data were searched using Mascot 2.1 protein identification software (Matrix Science) allowing 1.8 Da precursor mass and 0.9 Da fragment mass tolerances, against the MSDB database version MSDB_20050227.fasta (Feb 27th, 2005), using average peptide mass and average fragment mass calculation, with searches restricted to Mammalian entries (280764 sequences considered), allowing up to two missed cleavages per peptide, fixed carbamidomethylation of cysteine, with variable Methionine oxidation. All identifications were based on at least two identified peptides receiving ions scores above threshold for 95% confidence [28], with manual inspection of representative spectra to ensure accurate interpretation of MS/MS data.
The two approaches in experiment 1 and 2 were conducted independently, including cell culture, total protein collection, 2D gel electrophoresis, comparison, spot picking and protein identification.
Western Blotting and IHC Validations
CapG, destrin and ORP150 were highly or only expressed in MLO-Y4 cells compared to the osteoblast cells. Therefore, they were chosen for further examination based on their potential biological importance. Proteomic experiments were validated by Western Blotting with protein extracts from the MLO-Y4 and MC3T3 cells, and protein expression in primary osteocytes was determined by immunohistochemical staining (IHC) on ulna sections from a 1-month-old mouse of the C57/BL background.
Western Blotting was conducted following a standard protocol as previously published [6]. For each cell line, 10 μg of total protein per lane was loaded on the 10% Tris-HCl gel, catalog number 161–1155 (BioRad Lab, Hercules, CA, USA), for Western Blotting. After electrophoresis, the samples were transferred to nitrocellulose membranes and hybridized to primary antibodies against the target proteins. Peroxidase conjugated secondary antibodies against the primary antibodies were used to react with ECL™ Chemiluminescent Western Blotting Detection Reagents (Amersham Biosciences, Pittsburgh, PA, USA). The membrane was finally exposed and quantified using Kodak Image Station 200R (Kodak, Rochester, NY, USA). The membrane was also stained with Ponceau S, catalog number P-7170 (Sigma-Aldrich, St. Louis, MO, USA) to confirm equal amounts of protein loaded. Proper dilutions of the primary antibodies were 1:200 for CapG antibody, 1:500 for destrin antibody, and 1:2000 for ORP150 antibody.
IHC was performed on paraffin sections from the mid-shaft of mouse ulnae or dissected calvaria. Ulnae and calvaria were collected, fixed in commercial grade 10% formalin, and decalcified with 14% EDTA for 2 weeks, followed by paraffin embedding and sectioning. Primary antibodies against the target proteins and biotinylated secondary antibody were applied on the sections, using a Vectastain ABC kit (Vector Lab, Burlingame, CA, USA), catalog number PK6100, and PCNA DAB staining kit (Zymed Lab, San Francisco, CA, USA), catalog number 93–1143 according to the manufacture’s protocols. After counter-staining with methyl green (Vector Lab, Burlingame, CA, USA), catalog number H-3402, the slides were mounted and photographed with a Nikon E8000 microscope with a digital camera DXM1200 (Nikon, Melville, NY, USA). The imaging process was controlled by Analysis software version 3.1 (Soft Imaging System Corp, Lakewood, CO, USA). As a standard osteoblast marker, antibody against alkaline phosphatase (Santa Cruz Biotech, Santa Cruz, CA, USA), catalog number SC-23430, was used to distinguish osteoblasts from osteocytes. Non-specific IgGs from the same species as the primary antibodies were used as negative controls. Dilutions of the primary antibodies used were 1:300 for CapG antibody, 1:300 for destrin antibody, 1:100 for ORP150 antibody, and 1:200 for alkaline phosphatase antibody.
Statistical Analysis and Data Processing
Comparison between the proteomic results and mRNA array results was performed using Microsoft Access 2003 (Microsoft Corporation, WA, USA) by using Unigenes as the key attribute. Functional annotation was done with The Database for Annotation, Visualization and Integrated Discovery (DAVID) with EASE scores [29]. The cutoff for statistical significance was set to be p≤0.05.
Results
Proteomic Comparison between Osteoblastic and Osteocytic Cell Lines
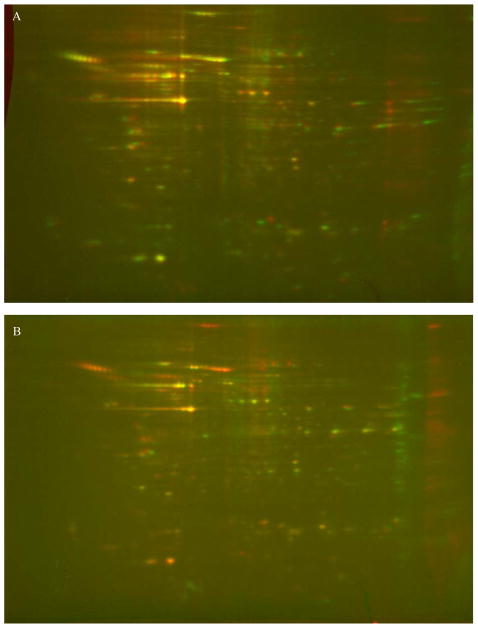
The proteomic profiles of the two osteoblast cell lines MC3T3 and 2T3 were nearly identical as measured by PDQuest, with 98–99% similarity (yellow spots) as shown in Fig. 1A. In contrast, comparison of MLO-Y4 cells with either of the osteoblast cell lines resulted in approximate 25% difference as reflected by more red and green spots in the overlaid images (Fig. 1B).
Fig. 1.
Overview of the proteomes of osteoblast and osteocyte cell lines. Two osteoblast cell lines showed a similar proteomic profile on qualitative 2D gels, which differs from the proteome of the osteocyte cell line. A. Overlay of 2D gels indicate that the profiles of the two osteoblast cell lines MC3T3 (green) and 2T3 (red) are nearly identical with 98–99% similarity as measured by PDQuest as shown by the prevailing yellow spots. B. In contrast, overlays of MLO-Y4 cells (green) with MC3T3 osteoblast cell line (red) gave a 25% difference as reflected by more red and green spots. The green and red are pseudo-colored Coomassie Blue staining.
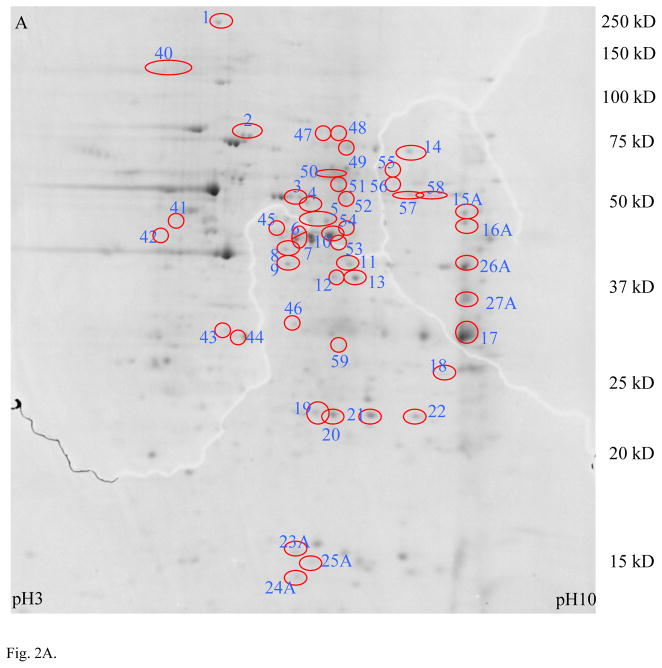
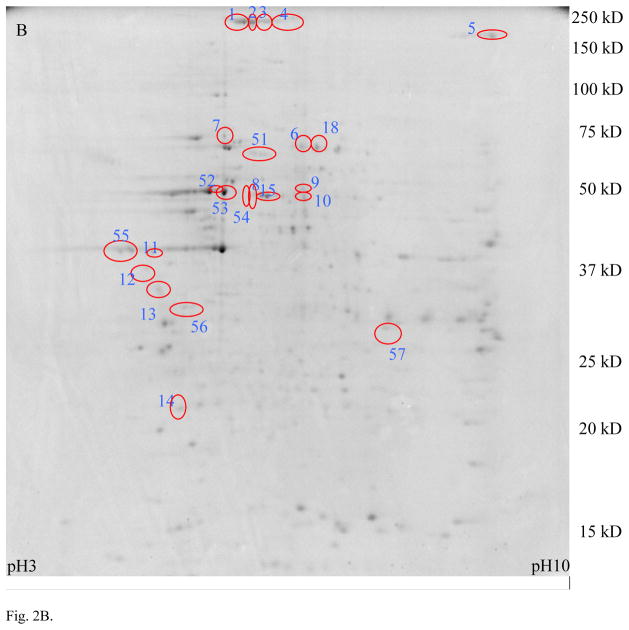
The results of the comparison between MLO-Y4 and MC3T3 cells in experiment 1 using MALDI MS are shown in Fig. 2 and the protein identifications are listed in Table S1 (Supporting Information). The spots with 2-fold or greater expression in MLO-Y4 compared to MC3T3 cells are shown in Fig. 2A, and those identified proteins preferentially expressed in MLO-Y4 cells are listed in Table S1A (Supporting Information). Spots exhibiting 2-fold or greater expression in MC3T3 compared to MLO-Y4 cells are shown in Fig. 2B, with protein identifications listed in Table S1B (Supporting Information).
Fig. 2.
Proteomic comparison between MLO-Y4 and MC3T3 cells in experimental approach 1 (MALDI). Panel A shows a 2D gel of total protein lysate from MLO-Y4 cells, with spots exhibiting 2× or higher expression in MLO-Y4 compared to MC3T3 cells circled. Panel B shows a 2D gel of total protein lysate from MC3T3 cells, with spots showing 2× or higher expression in MC3T3 compared to MLO-Y4 cells circled. Identification of these spots by MALDI mass spectrometry are listed in Table S1 (Supporting Information).
Note the higher expression of CapG, destrin and ORP150 in MLO-Y4 cells compared to MC3T3 cells. These three proteins were chosen for further validation by Western Blotting and immunohistochemistry because CapG and destrin are actin regulators which may play a role in dendrite formation, and ORP150 may play a role in adaptation to hypoxia.
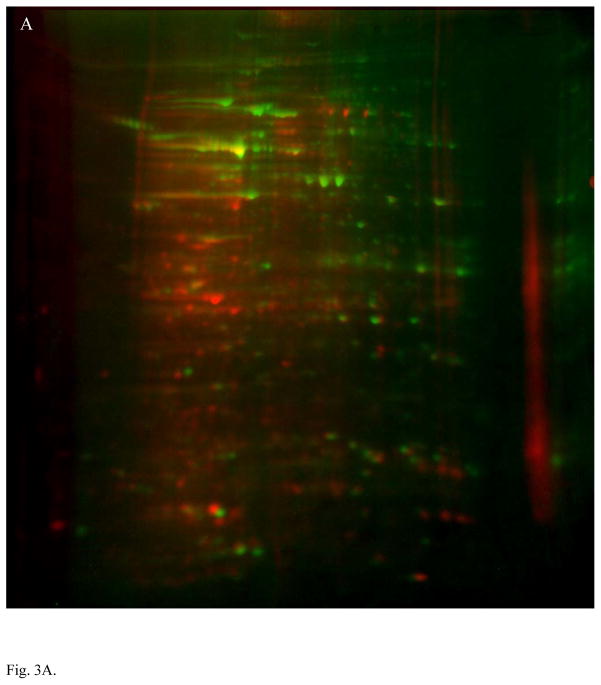
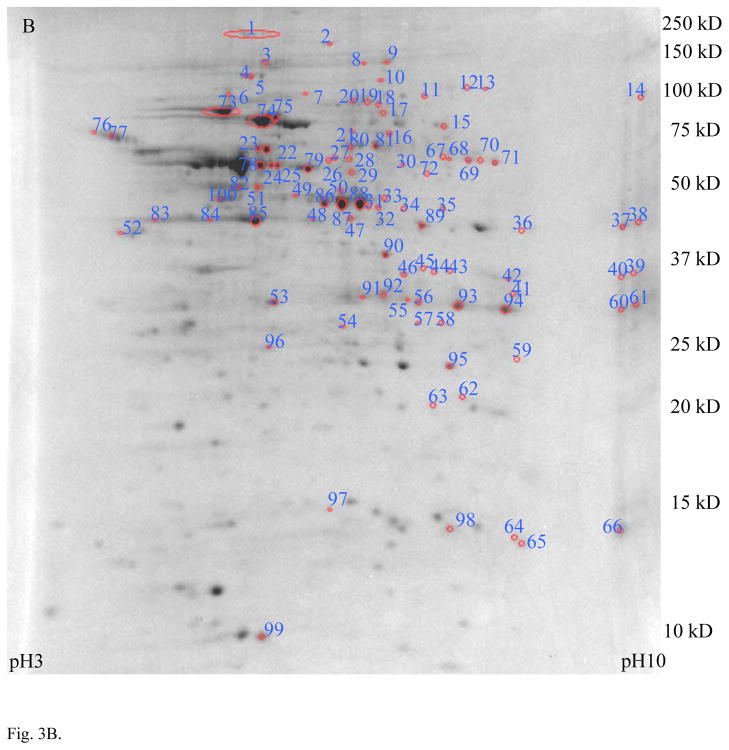
The comparison of MLO-Y4 and MC3T3 cells in experiment 2 is represented in the overlay of the 2D gel images, shown in Fig. 3A. Fig. 3B shows the protein spots that were found to be expressed at least two fold greater level in the osteocyte lineage MLO-Y4 cell. Identified proteins are listed in Table S2 (Supporting Information) with peptide identifications supporting these identifications summarized in Spreadsheet_1.xls (Supporting Information). Automation of the spot picking process allowed more spots to be picked, and the more sensitive LC-tandem MS instrumentation used in experiment 2 resulted in the confident identification of a much higher percentage of them.
Fig. 3.
Proteomic profile comparison between MLO-Y4 and MC3T3 cells in experiment approach 2 (LC-tandem MS). Panel A shows an overlay of MLO-Y4 cells (green) with MC3T3 osteoblast cell line (red) showing unique differences. The green and red are pseudo-colored Coomassie Blue staining. Panel B shows a 2D gel of total protein lysate from MLO-Y4 cells, with spots exhibiting 2× or higher expression in MLO-Y4 compared to MC3T3 circled. Identification of these spots by LC-tandem MS are listed in Table S2.
When results from experiment approaches 1 and 2 were compared, the two gels of MLO-Y4 showed a similarity of 91–98% as reported by PDQuest, indicating good reproducibility. In contrast, only 60–79% spots from MLO-Y4 and MC3T3 matched in experiment 2 using MS/MS, consistent with findings for experiment 1 using MALDI MS. Proteins that were consistently found to be preferentially expressed in MLO-Y4 cells (both experiments) are listed in Table 1. For each protein shown in Table 1, its corresponding ratio of mRNA level (MLO-Y4/MC3T3) as determined by gene arrays analysis [15] is also shown for comparison (if available). Of the 19 entries shown in Table 1, 10 were not previously identified as candidates in the transcription data, so these represent new osteocyte selection protein candidates. Of the remaining 9 entries, 7 were consistent with the transcription ratio determinations.
TABLE 1.
TRANSCRIPTION AND GEL SPOT INTENSITY RATIOS OF OSTEOCYTE-SELECTIVE PROTEINS
| ID (2)a | Protein | Unigene | pIb | MWb (kDa) | Protein MLO-Y4 /MC3T3 (2)a | Protein MLO-Y4 /MC3T3 (1)a | mRNA MLO-Y4/2T3c |
|---|---|---|---|---|---|---|---|
| 1 | Hypoxia up-regulated 1 (Hyou1, ORP150) | Mm.116721 | 5.2 | 111 | 240 | 62 | n/a |
| 9 | Endoplasmin (Hsp90b1) | Mm.87773 | 4.74 | 92 | 19 | 17 | n/a |
| 16 | Pyruvate kinase isozymes M1/M2 (Pkm2) | Mm.326167 | 6.69 | 58 | 10 | 14 | n/a |
| 17 | Lamin-A/C (Lmna) | Mm.470666 | 6.54 | 74 | 44 | 79 | n/a |
| 24 | Vimentin (Vim) | Mm.268000 | 5.06 | 53 | 10 | 7 | 0.5 |
| 39 | Fructose-bisphosphate aldolase A (Aldoa) | Mm.275831 | 8.55 | 39 | 4 | 2.6 | 3.4 |
| 48 | Ornithine aminotransferase, mitochondrial (Oat) | Mm.13694 | 6.19 | 48 | 86 | 5.9 | n/a |
| 49 | Pyruvate kinase isozymes M1/M2 (Pkm2) | Mm.326167 | 7.42 | 58 | 13 | 14.8 | n/a |
| 50 | Aldehyde dehydrogenase, mitochondrial, (Aldh2) | Mm.284446 | 7.53 | 57 | 4.6 | 2.9 | n/a |
| 53 | Annexin A3 (Anxa3) | Mm.7214 | 5.33 | 36 | 24.8 | 13.6 | 1.7 |
| 56 | Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) | Mm.304088 | 8.44 | 36 | 8.4 | 2.6 | 3 |
| 67 | UDP-glucose 6-dehydrogenase (Ugdh) | Mm.344831 | 7.49 | 55 | 22 | 38 | 2 |
| 75 | Stress-70 protein, mitochondrial (Hspa9a) | Mm.209419 | 5.81 | 73 | 3.3 | 2.9 | n/a |
| 78 | Vimentin (Vim) | Mm.268000 | 5.06 | 53 | 7.8 | 7.2 | 0.5 |
| 79 | Protein disulfide-isomerase A3 (Pdia3) | Mm.263177 | 5.78 | 57 | 2.5 | 2.7 | n/a |
| 89 | Phosphoglycerate kinase 1 (Pgk1) | Mm.336204 | 8.02 | 44 | 2.8 | 3.3 | n/a |
| 90 | Macrophage-capping protein (CapG) | Mm.18626 | 6.47 | 39 | 3.3 | 4.1 | 2.5 |
| 95 | Triosephosphate isomerase (Tpi1) | Mm.4222 | 7.08 | 26 | 3.5 | 3.7 | 1.8 |
| 97 | Destrin (Dstn) | Mm.28919 | 8.2 | 18 | 3.6 | 5.2 | 5 |
(1) or (2) means data from experiment 1 or 2 of the proteomic comparisons.
pI and MW were estimated from the full length GenBank entries for the amino acid sequence of the identified protein. Actual pI and MW on gels may differ resulting from post-translational modifications (e.g. phosphorylation, glycosylation and truncation).
mRNA microarray result was from Yang [15]. “n/a” means the gene was not included in the microarray.
Gene ontology (GO) term enrichment analysis showed higher abundance of proteins involved in biological processes of glycolysis, protein folding, and actin polymerization depolymerization with statistical significance (Table 2). The repetitively identified ostoecyte-selective proteins were selected for input into DAVID (http://david.abcc.ncifcrf.gov) with the total mouse genome as background.
TABLE 2.
GO TERM ENRICHMENT OF OSTEOCYTE-SELECTIVE PROTEINSa
| Enriched GO Term | Proteins | EASE Scoreb |
|---|---|---|
| Glycolysis | Phosphoglycerate kinase 1 (Pgk1) | 0.00001 |
| Fructose-bisphosphate aldolase A (Aldoa) | ||
| Hypoxia up-regulated 1 (Hyou1, ORP150) | ||
| Triosephosphate isomerase (Tpi1) | ||
|
| ||
| Protein folding | Stress-70 protein, mitochondrial (Hspa9a) | 0.01 |
| Hypoxia up-regulated 1 (Hyou1, ORP150) | ||
| Endoplasmin (Hsp90b1) | ||
|
| ||
| Actin polymerization depolymerization | Macrophage-capping protein (CapG) | 0.03 |
| Destrin (Dstn) | ||
Unigenes of ostoecyte selective proteins listed in Table 3 were used as a input into DAVID. And, the total mouse genome was used as background. The option of GO term was set as GOTERM_BP_ALL (including all 5 levels of biological processes) following the DAVID instruction manual.
An EASE score (modified Fisher Exact P-Values) of less than 0.05 indicates siginificant enrichment of proteins of the corresponding GO term.
Validation of Osteocyte-Selective Proteins
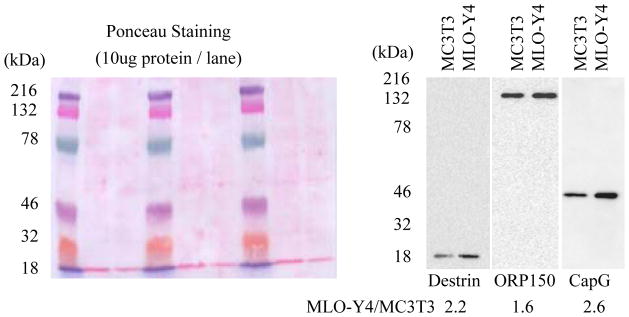
CapG, destrin and ORP150 were consistently present at higher levels in the MLO-Y4 cells in the proteomic comparisons (transcription data was not available for ORP150). They were chosen for further validation by Western Blotting because of 1) the availability of antibodies, 2) the potential role of CapG and destrin in dendrite formation, and 3) the potential role of ORP150 in adaptation to hypoxia. The results showed consistently higher abundance of the three proteins in MLO-Y4 cells than in MC3T3 cells (Fig. 4).
Fig. 4.
Western Blotting confirmed higher expression of osteocyte-selective proteins in MLO-Y4 cells. Antibodies against CapG, destrin and ORP150 were used to do Western Blotting on total proteins from MC3T3 osteoblasts and MLO-Y4 osteocytes. Equivalent loading of total proteins was shown by uniform Ponceau staining of the membrane on the left. On the right, these three proteins showed higher expression in MLO-Y4 than in MC3T3 cells. Ratios between the two cell lines from quantitative densitometry are labeled at the bottom.
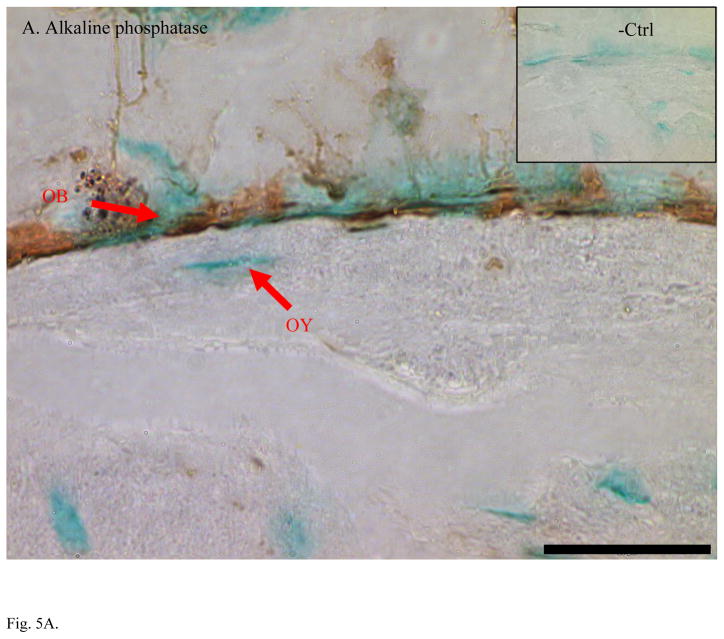
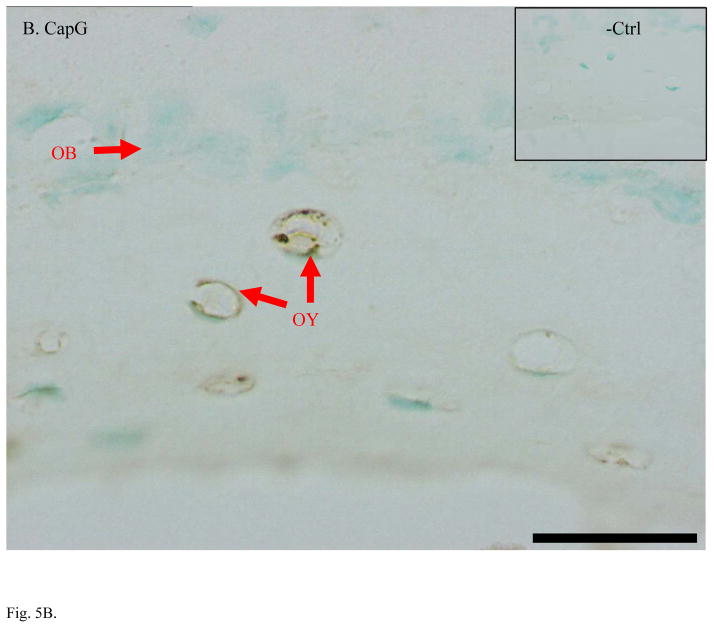
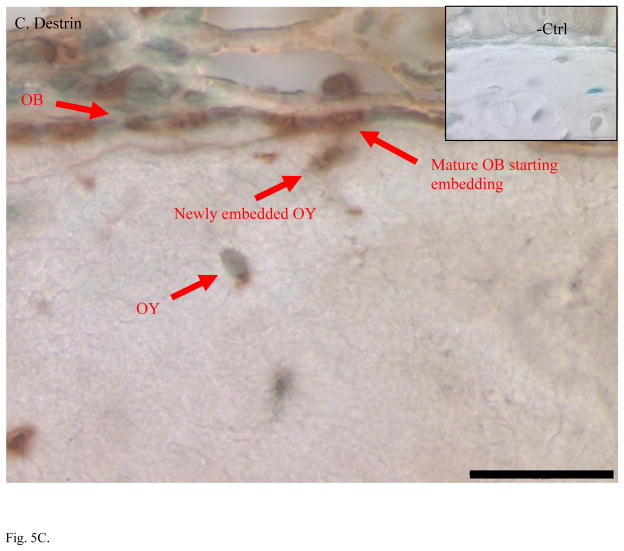
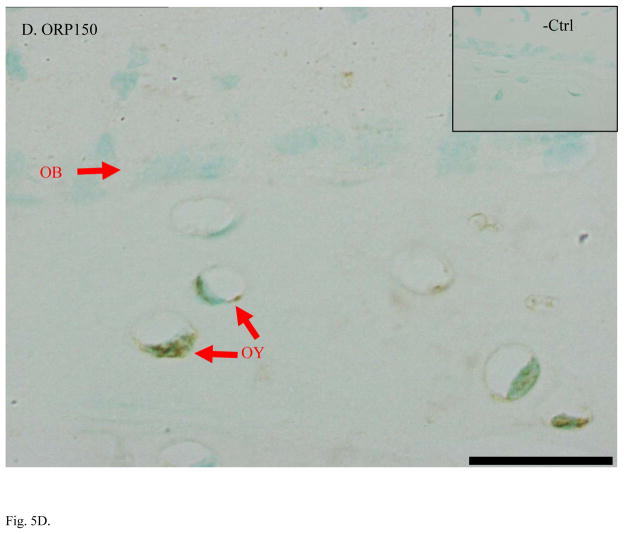
Higher expression of CapG, destrin, and ORP150 found in MLO-Y4 osteocytes was validated in bone tissue using immunohistochemical staining (IHC) of ulna sections (Figure 5). IHC on mouse ulna demonstrated stronger staining in osteocytes than in osteoblasts with antibodies against CapG and ORP150. Antibody against destrin stained both osteoblasts and osteocytes while the strongest signal of destrin staining was in the embedding osteocytes. As expected for an osteoblast specific marker, alkaline phosphatase antibody clearly stained osteoblasts but not osteocytes. All negative controls showed little background staining of non-specific IgG. Methyl green was used as counter-stain to label nuclei.
Fig. 5.
Immunohistochemical staining confirmed higher expression of osteocyte-selective proteins in mouse ulnae. A. Alkaline phosphatase is a specific marker of osteoblasts (OB) but not osteocytes (OY). B. CapG was only detected OY but not in OB. C. Destrin showed its polarized peak expression during the differentiation of OB to OY and embedding osteocytes. D. ORP150 showed higher expression in OY than in OB. The signal of immunohistochemical staining is in dark brown. Normal IgG from the same species of the primary antibody was used as the negative control. Counter-stains of Methyl green was used to indicate nuclei. Length of the scale bar is 20 μm. The mid-shaft ulnae cross-sections were from 1-month-old wild type mice.
Discussion
In our study, we showed that the cytoskeletal network of an osteocyte differs from that of an osteoblast by differential expression of components including actin binding proteins such as CapG and destrin, as well as lamin A, and certain forms of vimentin. The discovery of abundant actin binding proteins in MLO-Y4 cells and primary osteocytes in bone suggests a relatively more active cytoskeleton in the osteocyte compared to the osteoblast cells. CapG and destrin indicate active cytoskeletal rearrangement in osteocytes especially in embedding cells. This may be essential for the embedding and dendrite elongation processes.
CapG is a member of the gelsolin family responsible for regulation of actin filament length by capping the barbed positive ends [30]. Expression of CapG has been localized to multiple types of tissues and cells. Its function has been linked to the motility and ruffling of macrophages, neutrophils, fibroblasts and endothelial cells [30–32]. Recently, it was also reported to be a potential transcriptional repressor in neurons [33]. In bone, CapG mRNA has been reported to be more highly expressed in immortalized human pre-osteocytic cells than pre-osteoblastic cells [16]. CapG has only 3 gelsolin-like domains, although other gelsolin members have all the 6 domains [34], and lacks the severing activity of the other gelsolin members [30]. CapG is the only gelsolin member showing a nuclear localization [35], consistent with its possible role as transcription regulator. Its nuclear localization may be essential in the embedding process of osteocytes into bone matrix because only nuclear import of over-expressed CapG induced invasion of epithelial cells into collagen matrix [36]. Surprisingly, CapG knockout mice showed no apparent developmental phenotype other than motility defects in macrophages [37]. This may be due to the compensatory effects of other gelsolin members, or lack of examination of other tissues such as bone. In our study, CapG protein was observed to be more highly expressed in osteocytes than in osteoblasts. We also observed CapG antibody staining of both the nuclei and cytoplasm of osteocytes. Cell morphology dramatically changes from polygonal to dendritic when osteoblasts differentiate into osteocytes as they embed into bone matrix. In osteocytes, binding of CapG may protect actin filaments from being severed by other gelsolins, and therefore elongate the cell protrusion counteracting the surface tension. Furthermore, CapG could be involved in the response to shear stress because it has been reported to be increased in endothelial cells after exposure to flow [38], probably by coupling with calcium channels since its activity depends on Ca2+ [39].
Destrin is a member of the actin depolymerizing factor/cofilin family which regulates assembly and disassembly of actin filaments at the pointed minus ends. It is essential in regulating actin filament turnover by severing and enhancing depolymerization of actin filaments [40]. It has been associated with cell motility and formation of cell processes in a variety of cells including fibroblasts, neurons, astrocytes, epithelial cells, and spermatogenic cells [40–44]. Although there is no explicit report of a skeletal defect in destrin knockout mice, deletion of its regulator, LIM kinase 1, caused a spine abnormality [45]. Over-expression of destrin enhances the invasiveness of glioblastoma tumor cells by regulating their motility [46]. Destrin displays a polarized expression in early osteocytes towards the matrix. In a similar way, destrin selectively localizes to lamellipodium, and therefore may direct the migration of fibroblasts [41]. Destrin increases the rate of filament turnover by disassembly of actin filaments from the minus ends, thereby providing sufficient actins for barbed-end growth by CapG. The overall treadmilling process could facilitate the embedding of osteocytes into bone matrix.
A traditional dogma is that house-keeping enzymes in the glycolysis pathway, such as GAPDH, should be constantly expressed in all cells. However, MLO-Y4 cells surprisingly showed a higher abundance of six out of the ten enzymes in glycolysis pathway. The six glycolysis enzymes identified are fructose-bisphosphate aldolase A, GAPDH, triosephosphate isomerase, enolase, pyruvate kinase, and phosphoglycerate kinase. Messenger RNA of the first four of these six proteins were consistently more highly transcribed in MLO-Y4 than in MC3T3 cells, consistent with microarray results. Greater glycolytic flux may provide needed energy for osteocyte activity under anaerobic conditions, especially for a dynamic cytoskeleton system.
Deep embedding of osteocytes within bone implies reduced access to oxygen and nutrition. However, instead of a short life span, osteocytes are known to be able to live for decades in the bone matrix. Together with elevated glycolysis, the detection of chaperone proteins such as ORP150, endoplasmin precursor, and heat shock protein 9 suggest a possible explanation for the osteocyte’s capacity to adapt to its environment. Most reports of ORP150 are in the neural system showing an anti-apoptotic function against hypoxia in cooperation with other stress proteins [47–50]. A similar protective function was also observed in renal tubular epithelium [51] and cardiomyocytes [52]. Human prostate cancer, bladder cancer and HeLa cells also synthesize ORP150 for self-protection against hypoxia, and VEGF has been reported to be correlated to ORP150 in prostate cancer and bladder cancer cells [53–55]. ORP150 also accelerated wound healing through increased VEGF transport [56]. ORP150 has also been detected in osteonecrotic bones [57]. An in vitro study showed hypoxia accelerated the differentiation of osteoblasts to osteocytes coupled with the expression of ORP150 and other osteocyte markers [58]. Similarly, annexin A3 was highly expressed in MLO-Y4 cells along with ORP150. Annexin A3 is believed to be involved in the secretion of VEGF through activation of the hypoxia-induced factor-1 (HIF-1) pathway [59]. It is reasonable to hypothesize that ORP150, annexin A3 and the heat shock proteins could support osteocyte survival within an adverse environment.
Together with the chaperone proteins involved in protein folding, many enzymes catalyzing post-translational modifications were detected with greater abundance in MLO-Y4 than in MC3T3 cells. UDP-glucose 6-dehydrogenase is the enzyme which converts UDP-glucose to UDP-glucuronate, a critical componentof the glycosaminoglycans, hyaluronan, chondroitin sulfate, andheparan sulfate, etc. Protein level and mRNA level were consistently higher in MLO-Y4 cells. To control the proper folding and disulfide connection of peptides, protein disulfide-isomerase A3 was also more abundantly expressed in MLO-Y4 cells. Many osteocyte-specific/selective membrane proteins, such as MEPE, E11 and DMP1, are heavily post-translationally modified. These enzymes involved in post-translational modifications in MLO-Y4 cells may provide necessary machinery for extensive post-translation modifications.
Some of our discoveries were consistent with a recent study of MLO-Y4 proteome mapping. Protein disulfide isomerase A3, several vimentins, pyruvate kinase, lamin A, CapG, annexins, triosephosphate isomerase, and several chaperone proteins were identified as abundantly expressed proteins in MLO-Y4 cells [60]. In summary, our analysis of the osteocyte proteome suggests a dynamic cytoskeleton, enhanced glycolytic flux, enriched protective chaperone proteins, and abundant post-translationally modified proteins in osteocytes. It supports the emerging view in the field that osteocytes are actively embedding cells with capacity to extend dendrites, and that they are terminally differentiated long-lived adaptive cells.
Supplementary Material
Acknowledgments
Rabbit anti-CapG IgG was kindly provided by Dr. Helen Yin at the University of Texas Southwestern Medical Center, USA. Rabbit anti-destrin IgG was kindly provided by Dr. Jim Bamburg at the Colorado State University, USA. Rabbit anti-ORP150 IgG was kindly provided by Dr. Satoshi Ogawa at the Kanazawa University Medical School, Japan. We thank the technical support from Jie Zhao and Leonardo Barragan at the University of Missouri at Kansas City. This study was supported by NIH NIAMS PO1 AR046798.
Footnotes
Conflict of interest statement
The authors hereby declare no financial or commercial interests related to the content of this manuscript or the publication of this manuscript.
References
- 1.Wetterwald A, Hoffstetter W, Cecchini MG, Lanske B, Wagner C, Fleisch H, et al. Characterization and cloning of the E11 antigen, a marker expressed by rat osteoblasts and osteocytes. Bone. 1996;18:125–32. doi: 10.1016/8756-3282(95)00457-2. [DOI] [PubMed] [Google Scholar]
- 2.Feng JQ, Huang H, Lu Y, Ye L, Xie Y, Tsutsui TW, et al. The Dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. Journal of dental research. 2003;82:776–80. doi: 10.1177/154405910308201003. [DOI] [PubMed] [Google Scholar]
- 3.Westbroek I, De Rooij KE, Nijweide PJ. Osteocyte-specific monoclonal antibody MAb OB7.3 is directed against Phex protein. J Bone Miner Res. 2002;17:845–53. doi: 10.1359/jbmr.2002.17.5.845. [DOI] [PubMed] [Google Scholar]
- 4.Gluhak-Heinrich J, Pavlin D, Yang W, Macdougall M, Harris SE. MEPE expression in osteocytes during orthodontic tooth movement. Arch Oral Biol. 2007;52:684–90. doi: 10.1016/j.archoralbio.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. The EMBO journal. 2003;22:6267–76. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, et al. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Mol Cell Biol. 2006;26:4539–52. doi: 10.1128/MCB.02120-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling Y, Rios HF, Myers ER, Lu Y, Feng JQ, Boskey AL. DMP1 depletion decreases bone mineralization in vivo: an FTIR imaging analysis. J Bone Miner Res. 2005;20:2169–77. doi: 10.1359/JBMR.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strom TM, Francis F, Lorenz B, Boddrich A, Econs MJ, Lehrach H, et al. Pex gene deletions in Gy and Hyp mice provide mouse models for X-linked hypophosphatemia. Human molecular genetics. 1997;6:165–71. doi: 10.1093/hmg/6.2.165. [DOI] [PubMed] [Google Scholar]
- 9.Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, et al. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. The Journal of biological chemistry. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- 10.Balemans W, Cleiren E, Siebers U, Horst J, Van Hul W. A generalized skeletal hyperostosis in two siblings caused by a novel mutation in the SOST gene. Bone. 2005;36:943–7. doi: 10.1016/j.bone.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38:1310–5. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowe PS, de Zoysa PA, Dong R, Wang HR, White KE, Econs MJ, et al. MEPE, a new gene expressed in bone marrow and tumors causing osteomalacia. Genomics. 2000;67:54–68. doi: 10.1006/geno.2000.6235. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Zhou J, Tang W, Jiang X, Rowe DW, Quarles LD. Pathogenic role of Fgf23 in Hyp mice. Am J Physiol Endocrinol Metab. 2006;291:E38–49. doi: 10.1152/ajpendo.00008.2006. [DOI] [PubMed] [Google Scholar]
- 14.Poole KE, van Bezooijen RL, Loveridge N, Hamersma H, Papapoulos SE, Lowik CW, et al. Sclerostin is a delayed secreted product of osteocytes that inhibits bone formation. Faseb J. 2005;19:1842–4. doi: 10.1096/fj.05-4221fje. [DOI] [PubMed] [Google Scholar]
- 15.Yang W, Kalajzic I, Lu Y, Guo D, Harris MA, Gluhak-Heinrich J, et al. In vitro and in vivo study on osteocyte-specific mechanical signaling pathways. Journal of musculoskeletal & neuronal interactions. 2004;4:386–7. [PubMed] [Google Scholar]
- 16.Billiard J, Moran RA, Whitley MZ, Chatterjee-Kishore M, Gillis K, Brown EL, et al. Transcriptional profiling of human osteoblast differentiation. Journal of cellular biochemistry. 2003;89:389–400. doi: 10.1002/jcb.10514. [DOI] [PubMed] [Google Scholar]
- 17.Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, et al. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009 doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudo H, Kodama HA, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. The Journal of cell biology. 1983;96:191–8. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh-Choudhury N, Windle JJ, Koop BA, Harris MA, Guerrero DL, Wozney JM, et al. Immortalized murine osteoblasts derived from BMP 2-T-antigen expressing transgenic mice. Endocrinology. 1996;137:331–9. doi: 10.1210/endo.137.1.8536632. [DOI] [PubMed] [Google Scholar]
- 20.Bonewald LF. Establishment and characterization of an osteocyte-like cell line, MLO-Y4. Journal of bone and mineral metabolism. 1999;17:61–5. doi: 10.1007/s007740050066. [DOI] [PubMed] [Google Scholar]
- 21.Cheng B, Zhao S, Luo J, Sprague E, Bonewald LF, Jiang JX. Expression of functional gap junctions and regulation by fluid flow in osteocyte-like MLO-Y4 cells. J Bone Miner Res. 2001;16:249–59. doi: 10.1359/jbmr.2001.16.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Tan SD, Kuijpers-Jagtman AM, Semeins CM, Bronckers AL, Maltha JC, Von den Hoff JW, et al. Fluid shear stress inhibits TNFalpha-induced osteocyte apoptosis. Journal of dental research. 2006;85:905–9. doi: 10.1177/154405910608501006. [DOI] [PubMed] [Google Scholar]
- 23.Ponik SM, Triplett JW, Pavalko FM. Osteoblasts and osteocytes respond differently to oscillatory and unidirectional fluid flow profiles. Journal of cellular biochemistry. 2007;100:794–807. doi: 10.1002/jcb.21089. [DOI] [PubMed] [Google Scholar]
- 24.Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res. 1997;12:2014–23. doi: 10.1359/jbmr.1997.12.12.2014. [DOI] [PubMed] [Google Scholar]
- 25.Collins TJ. ImageJ for microscopy. BioTechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 26.Acestor N, Masina S, Walker J, Saravia NG, Fasel N, Quadroni M. Establishing two-dimensional gels for the analysis of Leishmania proteomes. Proteomics. 2002;2:877–9. doi: 10.1002/1615-9861(200207)2:7<877::AID-PROT877>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 27.Starkweather R, Barnes CS, Wyckoff GJ, Keightley JA. Virtual polymorphism: finding divergent peptide matches in mass spectrometry data. Analytical chemistry. 2007;79:5030–9. doi: 10.1021/ac0703496. [DOI] [PubMed] [Google Scholar]
- 28.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 30.Southwick FS, DiNubile MJ. Rabbit alveolar macrophages contain a Ca2+-sensitive, 41,000-dalton protein which reversibly blocks the “barbed” ends of actin filaments but does not sever them. The Journal of biological chemistry. 1986;261:14191–5. [PubMed] [Google Scholar]
- 31.Sun HQ, Kwiatkowska K, Wooten DC, Yin HL. Effects of CapG overexpression on agonist-induced motility and second messenger generation. The Journal of cell biology. 1995;129:147–56. doi: 10.1083/jcb.129.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witke W, Li W, Kwiatkowski DJ, Southwick FS. Comparisons of CapG and gelsolin-null macrophages: demonstration of a unique role for CapG in receptor-mediated ruffling, phagocytosis, and vesicle rocketing. The Journal of cell biology. 2001;154:775–84. doi: 10.1083/jcb.200101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saba R, Johnson JE, Saito T. Commissural neuron identity is specified by a homeodomain protein, Mbh1, that is directly downstream of Math1. Development (Cambridge, England) 2005;132:2147–55. doi: 10.1242/dev.01781. [DOI] [PubMed] [Google Scholar]
- 34.Dabiri GA, Young CL, Rosenbloom J, Southwick FS. Molecular cloning of human macrophage capping protein cDNA. A unique member of the gelsolin/villin family expressed primarily in macrophages. The Journal of biological chemistry. 1992;267:16545–52. [PubMed] [Google Scholar]
- 35.Prendergast GC, Ziff EB. Mbh 1: a novel gelsolin/severin-related protein which binds actin in vitro and exhibits nuclear localization in vivo. The EMBO journal. 1991;10:757–66. doi: 10.1002/j.1460-2075.1991.tb08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Corte V, Van Impe K, Bruyneel E, Boucherie C, Mareel M, Vandekerckhove J, et al. Increased importin-beta-dependent nuclear import of the actin modulating protein CapG promotes cell invasion. Journal of cell science. 2004;117:5283–92. doi: 10.1242/jcs.01410. [DOI] [PubMed] [Google Scholar]
- 37.Parikh SS, Litherland SA, Clare-Salzler MJ, Li W, Gulig PA, Southwick FS. CapG(−/−) mice have specific host defense defects that render them more susceptible than CapG(+/+) mice to Listeria monocytogenes infection but not to Salmonella enterica serovar Typhimurium infection. Infection and immunity. 2003;71:6582–90. doi: 10.1128/IAI.71.11.6582-6590.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellieux C, Desgeorges A, Pigeon CH, Chambaz C, Yin H, Hayoz D, et al. Cap G, a gelsolin family protein modulating protective effects of unidirectional shear stress. The Journal of biological chemistry. 2003;278:29136–44. doi: 10.1074/jbc.M300598200. [DOI] [PubMed] [Google Scholar]
- 39.Janmey PA, Chaponnier C, Lind SE, Zaner KS, Stossel TP, Yin HL. Interactions of gelsolin and gelsolin-actin complexes with actin. Effects of calcium on actin nucleation, filament severing, and end blocking. Biochemistry. 1985;24:3714–23. doi: 10.1021/bi00335a046. [DOI] [PubMed] [Google Scholar]
- 40.Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. Journal of neurobiology. 2004;58:103–17. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- 41.Dawe HR, Minamide LS, Bamburg JR, Cramer LP. ADF/cofilin controls cell polarity during fibroblast migration. Curr Biol. 2003;13:252–7. doi: 10.1016/s0960-9822(03)00040-x. [DOI] [PubMed] [Google Scholar]
- 42.Padmanabhan J, Shelanski ML. Process formation in astrocytes: modulation of cytoskeletal proteins. Neurochemical research. 1998;23:377–84. doi: 10.1023/a:1022413718447. [DOI] [PubMed] [Google Scholar]
- 43.Ikeda S, Cunningham LA, Boggess D, Hawes N, Hobson CD, Sundberg JP, et al. Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor) Human molecular genetics. 2003;12:1029–37. doi: 10.1093/hmg/ddg112. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi H, Koshimizu U, Miyazaki J, Nakamura T. Impaired spermatogenic ability of testicular germ cells in mice deficient in the LIM-kinase 2 gene. Developmental biology. 2002;241:259–72. doi: 10.1006/dbio.2001.0512. [DOI] [PubMed] [Google Scholar]
- 45.Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, et al. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–33. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 46.Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell motility and the cytoskeleton. 2005;60:153–65. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- 47.Omata N, Murata T, Takamatsu S, Maruoka N, Yonekura Y, Fujibayashi Y, et al. Region-specific induction of hypoxic tolerance by expression of stress proteins and antioxidant enzymes. Neurol Sci. 2006;27:74–7. doi: 10.1007/s10072-006-0570-6. [DOI] [PubMed] [Google Scholar]
- 48.Ikematsu K, Tsuda R, Kondo T, Kondo H, Ozawa K, Ogawa S, et al. The expression of ‘150-kDa oxygen regulated protein (ORP-150)’ in human brain and its relationship with duration time until death. Legal medicine (Tokyo, Japan) 2004;6:97–101. doi: 10.1016/j.legalmed.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Kitano H, Nishimura H, Tachibana H, Yoshikawa H, Matsuyama T. ORP150 ameliorates ischemia/reperfusion injury from middle cerebral artery occlusion in mouse brain. Brain research. 2004;1015:122–8. doi: 10.1016/j.brainres.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 50.Ogawa S. ORP150 (150 kDa oxygen regulated protein) suppressed neuronal cell death. Nippon yakurigaku zasshi. 2003;121:43–8. doi: 10.1254/fpj.121.43. [DOI] [PubMed] [Google Scholar]
- 51.Bando Y, Tsukamoto Y, Katayama T, Ozawa K, Kitao Y, Hori O, et al. ORP150/HSP12A protects renal tubular epithelium from ischemia-induced cell death. Faseb J. 2004;18:1401–3. doi: 10.1096/fj.03-1161fje. [DOI] [PubMed] [Google Scholar]
- 52.Aleshin AN, Sawa Y, Kitagawa-Sakakida S, Bando Y, Ono M, Memon IA, et al. 150-kDa oxygen-regulated protein attenuates myocardial ischemia-reperfusion injury in rat heart. Journal of molecular and cellular cardiology. 2005;38:517–25. doi: 10.1016/j.yjmcc.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Miyagi T, Hori O, Koshida K, Egawa M, Kato H, Kitagawa Y, et al. Antitumor effect of reduction of 150-kDa oxygen-regulated protein expression on human prostate cancer cells. Int J Urol. 2002;9:577–85. doi: 10.1046/j.1442-2042.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- 54.Asahi H, Koshida K, Hori O, Ogawa S, Namiki M. Immunohistochemical detection of the 150-kDa oxygen-regulated protein in bladder cancer. BJU international. 2002;90:462–6. doi: 10.1046/j.1464-410x.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- 55.Cechowska-Pasko M, Bankowski E, Chene P. The effect of hypoxia on the expression of 150 kDa oxygen-regulated protein (ORP 150) in HeLa cells. Cell Physiol Biochem. 2006;17:89–96. doi: 10.1159/000091467. [DOI] [PubMed] [Google Scholar]
- 56.Ozawa K, Kondo T, Hori O, Kitao Y, Stern DM, Eisenmenger W, et al. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. The Journal of clinical investigation. 2001;108:41–50. doi: 10.1172/JCI11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato M, Sugano N, Ohzono K, Nomura S, Kitamura Y, Tsukamoto Y, et al. Apoptosis and expression of stress protein (ORP150, HO1) during development of ischaemic osteonecrosis in the rat. The Journal of bone and joint surgery. 2001;83:751–9. doi: 10.1302/0301-620x.83b5.10801. [DOI] [PubMed] [Google Scholar]
- 58.Hirao M, Hashimoto J, Yamasaki N, Ando W, Tsuboi H, Myoui A, et al. Oxygen tension is an important mediator of the transformation of osteoblasts to osteocytes. Journal of bone and mineral metabolism. 2007;25:266–76. doi: 10.1007/s00774-007-0765-9. [DOI] [PubMed] [Google Scholar]
- 59.Park JE, Lee DH, Lee JA, Park SG, Kim NS, Park BC, et al. Annexin A3 is a potential angiogenic mediator. Biochemical and biophysical research communications. 2005;337:1283–7. doi: 10.1016/j.bbrc.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Bivi N, Bereszczak JZ, Romanello M, Zeef LA, Delneri D, Quadrifoglio F, et al. Transcriptome and Proteome Analysis of Osteocytes Treated with Nitrogen-Containing Bisphosphonates. Journal of proteome research. 2009 doi: 10.1021/pr8005606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.