Abstract
The endoplasmic reticulum (ER) is a multifunctional intracellular organelle responsible for the synthesis, processing and trafficking of a wide variety of proteins essential for cell growth and survival. Thesefore, comprehensive characterization of the ER proteome is of great importance to the understanding of its functions and has been actively pursued in the past decade by scientists in the proteomics field. This review summarizes major proteomic studies published in the past decade that focused on the ER proteome. We evaluate the data sets obtained from two different organs, liver and pancreas each of which contains a primary cell type (hepatocyte and acinar cell) with specialized functions. We also discuss how the nature of the proteins uncovered is related to the methods of organelle purification, organelle purity and the techniques used for protein separation prior to mass spectrometry. In addition, this review also puts emphasis on the biological insights gained from these studies regarding to the molecular functions of the endoplasmic reticulum including protein synthesis and translocation, protein folding and quality control, ER-associated degradation and ER stress, ER export and membrane trafficking, calcium homeostasis, and detoxification and drug metabolism.
Keywords: endoplasmic reticulum, proteomics, mass spectrometry
Introduction
The endoplasmic reticulum (ER) is an intracellular organelle responsible for the synthesis, processing and trafficking of a wide variety of proteins, accounting for about 1/3 of the proteins in human genome, including hormones, enzymes, receptors, ion channels and transporters. The ER serves many general functions including protein synthesis, translocation, quality control of protein folding, export of proteins from ER to Golgi. In addition, ER is also the site for steroid or xenobiotic metabolism, intracellular calcium homeostasis, and intracellular signaling [1]. Although universally present, some of these functions are more abundant in specialized cell types. Proteins entering the secretory pathway are synthesized on ER bound ribosomes and co-translationally translocated into ER where they are folded with the facilitation of the ER quality control system. The properly folded proteins are allowed to exit the ER by COPII dependent vesicular transport, whereas terminally misfolded proteins are retrotranslocated to the cytosol and degraded by proteasome [2, 3]. Morphologically, ER is comprised of a network of sheet-like flattened saccules or cisternae, tubules and the nuclear envelope that all share a common luminal space and extends throughout the cell [3, 4]. The ER has specialized sub-domains with distinct morphologies and functions. The rough ER (rER) with membrane bound ribosomes is mostly engaged in protein synthesis and folding, and the smooth ER (sER), characterized by the absence of membrane bound ribosomes, is dedicated to calcium storage, lipid synthesis and detoxification of drugs. The transitional ER (tER) is believed the site of newly synthesized proteins to exit ER. COPII (coat protein complex II) coated vesicles are believed to mediate protein export from ER and its anteograde transport to the Golgi whereas COPI (coat protein complex I) coated vesicles mediate retrograde transport to bring back to the ER the recycling proteins as well as escaped ER resident proteins [4].
Because of its importance to cell functions, comprehensive characterization of ER proteome is of great importance to the understanding of its functions and has been actively pursued in the past decade by scientists in the field of proteomics. This review evaluates major proteomic studies of ER proteome published in the past decade. Because of space limits, we will not be able to include all the earlier studies and those focusing on purified ribosomes. Furthermore, the proteomic studies on plant ER proteome are not included in discussion. Recent excellent reviews related to this subject can be found elsewhere [5, 6]. In this review, we evaluate differences between two organs, liver and pancreas each of which contains a primary cell type (hepatocyte and acinar cell) with specialized functions. We also discuss how the nature of the proteins uncovered is related to the methods of organelle purification, organelle purity and the technique of protein separation as well as the mass spectrometry technologies. In addition to summarizing the recent studies and comparing relevant results, this review also puts emphasis on the capabilities and potentials of proteomic approaches to the molecular characterization of the ER.
Purification of the Endoplasmic Reticulum
For organellar proteomic studies of ER, it is critical to consider the procedure by which it is purified prior to the analysis. Early cell fractionation studies used differential centrifugation to isolate nuclear, mitochondrial, microsomal, and cytosol fractions. Microsomes are the membrane vesicles and sheets that remain in suspension after a 10 or 12 thousand g centrifugation for 10–20 min and are then pelleted by centrifugation at 100,000 g for one hour. This fraction was shown to be enriched for RNA (from ribosomes) and enzymes present in the ER with NADPH Cytochrome C Reductase normally used as an ER enzymatic marker [7]. However it also contains contaminating organelles as shown by enzymatic markers and by electron microscopy. Most studies therefore further fractionate the total microsomes by density gradient sedimentation using either a step or continuous gradient of sucrose [7, 8] or other media such as iodixanol (OptiPrep) [9]. Because of its attached ribosomes and therefore higher density, the rER can be readily separated from sER and other membrane fractions and collected at an interface between 1.3 and 1.5 M sucrose [8]. Protocols for purification from both liver and pancreas have been described [8–11]. Such fractions are highly enriched but may still contain some contaminants. In more recent studies, purities has been assessed by electron microscopy and primarily Western blotting for known proteins with calreticulin, BiP and calnexin used as common ER markers [12, 13]. The smooth ER is harder to separate into its components and specialized protocols have been used to isolate plasma membrane and Golgi [8, 14]. The contamination in the ER preparations with even small amounts of other organelles is an issue because the dynamic range of proteins in each organelle may range to 3 or even 4 orders of magnitude. This is not a problem for the major protein components in the ER, however, the highest level proteins from any contaminating organelles will exceed the lowest level proteins authentic to the ER. Even a few percentage of the membrane contamination will lead to detectable protein identifications. This is the impetus behind development of methods such as correlation analysis [15, 16], enrichment studies [17]. It is crucial during sample preparations to monitor the enrichment of well documented ER markers as well as to monitor the diminishment of the contaminating organellar markers. Only high purity ER samples should be used for the subsequent mass spectrometry analysis. Proteins that are found in more than one organelle are particularly challenging and require independent validation (e.g., immunohistochemistry).
Further fractionation of the rER has involved stripping ribosomes with high salt and puromycin [12, 18, 19], extracting with sodium carbonate at basic pH to remove peripheral proteins and enrich for integral membrane proteins [19], or extracting hydrophobic proteins into Triton X-114 [20]. As an alternative to discontinuous gradients, organelles in a postnuclear supernatant have been separated on continuous gradients of sucrose or iodixanol [16, 21]. Either peak fractions for an organelle marker have been evaluated or all fractions used often with application of protein correlation profiling [16] (see below for more details). Not only the method for isolating cellular organelles, but also the downstream sample preparation such as solubilization and subsequent separation of the organellar proteins will affect the outcomes of the proteomic analysis. Techniques applied to ER proteomics studies have included 2D Gel electrophoresis [13, 22–24], 1D gel electrophoresis followed by LC-MS [20, 22] and 2D LC-MS/MS [12, 20] and as will be noted, the LC-MS/MS approaches have generally led to more protein identifications.
Proteomic analyses of the ER
Proteomics studies on isolated rough and smooth ER
Because of the importance of the ER in cell function a number of studies with increasing sophistication have analyzed the ER proteome. The separation methods, mass spectrometry approaches as well as the numbers of proteins identified in these studies are summarized in Table 1. In an early study, Galeva and Altermann [22] analyzed the protein composition of hepatic microsomes prepared by differential centrifugation using one-dimensional and two-dimensional gel electrophoresis followed by tryptic peptide mapping using MALDI-TOF instrument. The proteomic analysis was carried out on control rats as well as rats treated with phenobarbital to induce xenobiotic metabolizing enzymes such as cytochrome(s) P450. A total of 60 and 41 proteins including 22 and 3 membrane proteins were identified from 1-DE and 2-DE respectively. Both approaches showed that phenobarbital induced not only cytochromes P450 but also stress related ER proteins. This study represented an early effort to compare ER components in control vs. treatment using non-quantitative proteomic approach.
Table 1.
Summary of proteomics studies of ER proteome
| Species | Tissue | Organelle | ER purification | Protein/peptide separation | Mass spectrometry | Proteins detected | Reference |
|---|---|---|---|---|---|---|---|
| Rat | Liver | ER | Differential centrifugation; sodium cholate wash | Proteins, 1D and 2D gels | MALDI-TOF MS | 60 (1-DE) 41 (2-DE) |
[22] |
| Mouse | Liver | RER | Step sucrose gradient | Proteins, 2D gels | MALDI-TOF MS, Tandem MS | 141 | [13] |
| Hamster | Liver | ER | Continuous iodixanol gradient | Proteins, 2D gels | MALDI-Q-TOF MS | 34a | [32] |
| Mouse | Liver | ER fraction | Rate-zonal centrifugation on continuous sucrose gradient | Peptides, LC | linear ion-trap Fourier transform MS, Tandem MS | 229 | [16] |
| Rat | Liver | RER | Step sucrose gradient | Proteins, 1D gels, LC | QTOF-2 MS, Tandem MS | 787 | [20] |
| Rat | Liver | SER | Step sucrose gradient | Proteins, 1D gels, LC | QTOF-2 MS, Tandem MS | 998 | [20] |
| Bovine | Mammary Gland | Microsomes | Differential centrifugation | Proteins, 1D gels, LC | Tandam MS, LTQ | 703 | [36] |
| Mouse | Brain | Microsomes | Differential centrifugation | Peptides, 2D-LC separation (SCX, rpHPLC) | Tandam MS, LTQ-FT | 1914 | [37] |
| Rat | Pancreas | RER | Step sucrose gradient | Peptides, iTRAQ, 2D-LC separation (SCX, rpHPLC) | Tandam MS, MALDI-TOF/TOF | 469 | [12] |
| Canine | Pancreas | RER | Step sucrose gradient; carbonate wash | Proteins, SDS-PAGE, 2D BAC/SDS-PAGEb | nanoLC-MS/MS, | 258 | [19] |
| Chicken | Pre-B-cell | ER fraction | Continuous iodixanol density gradient | Peptides, iTRAQ, 2D-LC separation (SCX, rpHPLC) | Tandam MS, QSTAR | 79 | [21] |
| Canine | Pancreas | RER | Step sucrose gradient; fractionation of RER membrane | Proteins, 2D gels, | Tandam MS, LCQdecaXP | 32c | [24] |
only differentially expressed proteins were identified;
BAC:16-benzyldimethyl-n-hexadecylammonium chloride;
most of these are integral membrane proteins
In a later study, Knoblach et al [13] conducted the first systematic identification of ER proteins in mouse liver with a special emphasis on the identification of new ER luminal proteins. In this study, the ER vesicles were fractionated using step sucrose gradient and the quality of the fractionation was carefully assessed by ultrastructural and immunological analysis using ER, Golgi, mitochondria, nuclear and plasma membrane markers. 141 proteins were identified using 2D gel electrophoresis combined with peptide mass fingerprinting and tandem mass spectrometry. Among these proteins, 6 had not been described previously. Two newly discovered ER luminal proteins, ERp19 and ERp46, are members of the PDI (protein disulphide isomerase) family of proteins containing one and three thioredoxin motifs, respectively. Functional analyses of ERp19 and ERp46 using yeast complementation studies showed that ERp46 but not ERp19 could substitute for protein disulphide isomerase function in vivo. After its identification as a novel ER protein in this study, the function of ERp46 has been investigated in more detail in very recent studies [25, 26]. It was suggested that ERp46 plays important role in insulin [26] and adiponectin signaling [25].
Cells that are specialized in protein secretion, such as pancreatic cells, are particularly rich in rER. In the process of cell homogenization, the rER is converted into ribosome-studded vesicles, the so-called rough microsomes. Zahedi et al [19] reported a membrane proteomic analysis of canine pancreatic rough microsomes which led to the identification of a novel ER Hsp40 family member, ERj7. Using both SDS-PAGE and 2D-BAC/SDS-PAGE with tandem MS analysis, 258 non-redundant proteins were identified. Because the rough microsomes were subjected to carbonate extraction to enrich membrane proteins, a high percentage, 52%, of these proteins contained at least one transmembrane domain. A number of the proteins identified were potential candidates for novel ER proteins including two membrane resident Hsp40s (DnaJB12 and ERj7). The authors then characterized and confirmed that ERj7 was indeed an ER localized type I membrane protein containing a lumenal J-domain which interacts with Hsp70 protein Bip and stimulates its ATPase activity.
In a recent study, to comprehensively characterize not only the normal but also the diseased pancreatic rER proteome, Chen et al [12] quantitatively compared the protein compositions of pancreatic rER between normal and acute pancreatitis rats using isobaric tags (iTRAQ) and 2D LC-MALDI-MS/MS. Pancreatic rER was purified by ultracentrifugation on a discontinuous sucrose gradient. To detect lower-abundance ER proteins, ribosomes were then drastically reduced from rER preparations by high salt and puromycin treatment. The high purity of the isolated rER was carefully confirmed by electron microscopy and Western blotting using the known ER marker proteins as well as markers for potential contaminating organelles. A total of 469 unique proteins were revealed from four independent experiments including all major functional categories such as protein translation and translocation, protein folding/chaperone, membrane trafficking, cytoskeleton, transporter proteins/calcium store, oligosaccharide biosynthesis, lipid and sterol biosynthesis, secretory and zymogen granule membrane proteins [12]. In addition to providing a comprehensive catalog of pancreatic ER proteome, this study represented the first effort to illustrate molecular mechanism of exocrine pancreatic disease in the ER using state-of-the-art quantitative proteomics. This will be further discussed in the following section.
In another study utilizing highly enriched organelles isolated from rat liver homogenates, Gilchrist et al. [20] reported a quantitative organelle map of the secretory pathway including the rER, sER, and Golgi apparatus. In this study, isolated organelles were solubilized and their protein content was separated by 1D SDS-PAGE. Each protein lane was cut into slices and subjected to trypsin digestion followed by tandem mass spectrometry. The redundant peptide counting approach was used as an index to quantify relative protein abundance in different samples. The quantitative data were then analyzed and visualized as for microarray data using principal component analysis (PCA) and hierachical clustering. A set of protein were selected as markers for the ER and Golgi and other proteins co-clustered with the markers were assigned to the corresponding organelles. For example, redundant peptides assigned to the translocon constituent sec61α, a rER marker, were highly enriched in the three biological preparations of rER samples, less were observed in the three sER, and only a few peptides were seen in the three Golgi samples. In another example, a cluster with proteins mainly concentrated in smooth microsomes included the AAA family member p97 as well as the glycoprotein folding sensor UGGT.
In the same study [20], in order to detect lower abundant proteins, organellar membranes from rER and sER fractions were further subfractionated and analyzed. Taken all these analyses together, 1430 proteins were assigned to ER and Golgi after removing biosynthetic cargo and contaminating proteins from other organelles, among which 832 proteins were assigned as unique constituents of the ER with no peptides detected in Golgi/COPI vesicles. These largely corresponded to ribosomal proteins, translocon constituents, molecular chaperones, and proteins involved in lipid oxidation and drug detoxification, as well as proteasome subunits and ubiquitin ligases involved in ER associated degradation (ERAD). In addition, 405 proteins were found to be shared between the ER and the Golgi. This study currently represents the most comprehensive label-free quantitative proteomics analysis of the secretory pathway including the ER.
Proteomics studies of ER using protein correlation profiling from sucrose gradient
Isolated organelles are well suited to proteomics because of their reduced complexities and enriched protein concentrations which overcome the dynamic range problem in current MS-based proteomics efforts. They can also contribute functional and spatial information about the identified proteins. However, due to the high sensitivity of mass spectrometers and the difficulties to purify organelles to 100% homogeneity by biochemical subcellular fractionations, it has been challenging to distinguish between bona fide organellar proteins and proteins from contaminating organelles. Recent developments in proteomics have made it possible to analyze the protein composition of organelles using a variety of quantitative approaches. To address the problems of co-purified organelles, several groups have utilized quantitative proteomics in the high throughput assignment of proteins to subcellular compartments partially separated by centrifugations on a continuous density gradient.
In one approach, protein correlation profiling (PCP), was first introduced to study the human centrosome [15]. The PCP approach used peptide ion intensity profiles from given organellar marker proteins to define a consensus profile through a density centrifugation gradient, in direct analogy to Western blotting profiling of gradient fractions or specific activity in enzyme purification by chromatography. Distribution curves generated from the intensities of tens of thousands of peptides from consecutive fractions could then establish organelle-specific proteins by their similarity to the consensus profile of the corresponding organelle using mean squared deviation (χ2 value). In other words, peptides that have similar profiles share similar organelle associations. In a more recent study, Foster et al. [16] extended the PCP approach to localize 1400 proteins from mouse liver homogenates to 10 subellular compartments resolved by rate zonal centrifugation using continuous sucrose gradient. Among these proteins, 229 proteins were assigned to ER. The localizations of some previously unassigned proteins were then confirmed with biochemical and cell biological methods. Furthermore, proteomic and genomic data were integrated to identify networks of coexpressed genes, cis-regulatory motifs, and putative transcriptional regulators involved in organelle biogenesis using expression neighborhood analysis. This integrated approach can potentially be powerful on the basis of accurate subcellular assignments of the identified proteins.
Different from the above label-free PCP technique, LOPIT (localization of organelle proteins by isotope tagging) represents a quantitative approach employing isotope labeling originally using ICAT [27] and more recently iTRAQ reagents [28]. In the LOPIT approach, rather than processing each sample separately, isotopically labeled subcellular organellar fractions were pooled before the LC-MS/MS analysis. This approach has the advantage of reducing the number of steps at which the samples are processed separately and therefore reducing experimental variations. As for the PCP approach, LOPIT also begins with the partial separation of organelles by continuous density gradient centrifugation. Protein profiles along the gradient are quantified by isotopically coded tags in conjunction with 2D LC-MS/MS. Multivariate statistical techniques are then used to assign localizations to proteins by comparing their gradient profiles to those of established organelle markers in an unbiased manner. As the first application of LOPIT to a vertebrate system, Hall et al [21] conducted a proteomic analysis of the major organelles of DT40 chicken lymphocyte cell line. They used the distributions of 102 known organelle resident proteins as a basis to assign a further 223 proteins to five organelles including 79 to the ER. An important finding in this study was that a high proportion of the identified proteins are not localized to a single organelle rather a substantial fraction of proteins are in transit between compartments at steady state reflecting the dynamic nature of intracellular organelles in eukaryotic cells.
Although the PCP or LOPIT approach has been successfully applied to multiple studies and in theory has the strength to resolve constituents of different subcellular compartments even if their gradient distributions overlap, it is worth noting that the advantage of the profiling correlation based organelle mapping approach over that using highly enriched organelle samples has been questioned [20]. When the Golgi proteins were compared in two studies, one using the PCP approach [16] and the other using highly enriched samples [20], the authors in the later study concluded that the highly enriched samples may be a preferred strategy for organellar proteomics [20]. It is clear, however, that with the PCP approach, the better resolved the organelles are, the more certain the protein assignments will be.
Quantitative analysis of diseased ER proteome
To assure the production of fully functional protein molecules, ER possesses a very stringent quality control system achieved through the association of ER chaperones with unfolded or misfolded polypeptide chains [2, 3]. If some proteins fail to assume their native structure after a prolonged period of time, they are extruded from ER and degraded by the proteasome in the cytosol [3, 29]. The disruption of the ER homeostasis between protein synthesis, translocation, folding, export and degradation can lead to numerous human diseases [30]. For example, Cystic fibrosis (CF), an inherited childhood human disease, is primarily triggered by defective folding and export of transmembrane conductance regulator (CFTR), a multidomain cAMP-regulated chloride channel, from the ER [31]. A recent proteomic analysis revealed that the ΔF508 CFTR, the most common cystic fibrosis–associated mutant, was trapped as a folding intermediate in the ER, sequestering more chaperones and co-chaperones than wild-type CFTR [29].
In the past few years, quantitative proteomics technologies have been utilized not only for characterizing the ER proteome in normal animal tissues but also for unveiling protein alterations in ER under disease conditions. Morand et al [32] reported a systematic analysis of protein expression in the hepatic ER fractions to identify new candidate proteins involved in hepatic complications of insulin resistance and lipoprotein dysregulation. They compared hepatic ER-associated proteins from chow-fed (control) and fructose-fed (insulin-resistant) hamsters using two-dimensional gel electrophoresis and identified 34 differentially expressed protein spots by tandem mass spectrometry. Among these proteins, ER60, ERp46, ERp29, glutamate dehydrogenase, and TAP1 were more than 2-fold down-regulated, whereas α-glucosidase, P-glycoprotein, fibrinogen, protein disulfide isomerase, GRP94, and apolipoprotein E were up-regulated in the hepatic ER of the fructose-fed hamster. This study yielded new targets in the hepatic ER for further investigation whose expression correlated with the onset of insulin resistance in the liver.
Alteration of ER functions in pancreatic acinar cells has been implicated in the pathogenesis of acute pancreatitis (AP) [33]. To comprehensively characterize the normal and diseased rER subproteome, Chen et al [12] quantitatively compared the protein compositions of pancreatic rER between normal and AP animals using isobaric tags (iTRAQ) and 2D LC-MALDI-MS/MS. A total of 469 unique proteins were identified and quantified using two different AP models, arginine-induced [34] and caerulein-induced AP [35]. By quantitatively comparing pancreatitis RER samples with controls, 37 proteins, 25 in arginine-induced AP, 6 in caerulein-induced AP and 6 common in both models, were found significantly increase or decrease. In arginine AP, almost all the digestive enzymes showed dramatic reduction (decreased 50% to over 80%) and this was fully reproduced in duplicate experiments. Compared to the large decrease of digestive enzymes, several ER chaperone proteins including 78 kDa glucose regulated protein precursor (GRP78 also known as BiP), peptidylprolyl cistrans isomerase and ERp27 showed a relatively mild decrease. The proteins showing increases in AP were those involved in protein translocation and translation and fibrinogen alpha, beta and gamma chains. Compared to the significant changes in arginine AP, caerulein-induced AP showed similarities as well as differences. Common features between these two different animal models included the mild decreases of several ER chaperone proteins and the substantial increases of fibrinogen alpha, beta and gamma chains. In contrast to arginine-induced AP, only a few digestive enzymes showed mild decrease in caerulein-induced AP and several proteins functioning in protein translocation showed slight decreases rather than increase as detected in arginine AP. In spite of their common features, the different protein changes in these two animal models reflected different molecular mechanisms in AP pathogenesis which will inspire future investigations. These results suggested that the early stages of AP involve changes of multiple RER proteins that may affect the synthesis and processing of digestive enzymes.
Combined database of ER proteins
The above reviewed studies analyzed the ER mainly from two organs, liver and pancreas each of which contains a primary cell type (hepatocyte and acinar cell) with specialized functions. We did not include data sets from crude microsomes listed in Table 1 [36, 37] because a significant portion of the identified proteins are from co-purifying organelles. In order to assess the protein compositions of liver and pancreas ER, we generated a master list from four studies which both carried out extensive purifications and identified a large number of proteins [12, 13, 19, 20]. This table which is present in Supplementary Table 1 included two data sets of rat and mouse liver and two from rat and canine pancreas. We put the ER data sets in the Gilchrist et al, study in two separate columns, one for proteins unique for ER, the other one for proteins common between ER and COPI. Despite recent progress in proteomics and data annotation tools, performing a direct comparison of several datasets (often from different organisms) published by different groups remains difficult and time consuming. Due to the database redundancy it is not always possible to match sequences by accession numbers. In addition, protein accession numbers reported in the literature may become obsolete which complicates sequence retrieval. To do the comparison, proteins were retrieved from the EBI International Protein Index and the NCBI databases. In the cases where the accession numbers became obsolete, we attempted to retrieve the original sequence. Sequences were correlated to their accessions using BLAST (Basic Local Alignment Search Tool). Since we compared proteins from different organisms we did not expect to find exact matches, therefore sequences had to be at least 65% identical over at least 50% of the sequence length. This approach does not guarantee identification of the orthologous sequences, rather we expected to find the closest homolog. We then proceeded to identify the closest human homolog which is also listed in the table along with the categorization by GeneOntology as to Molecular Function, Cellular component, and Biological Process.
Together 1536 proteins were identified in one or more data sets. Although some were identified in multiple sets others were present in only one, most likely due to the specialized nature of the purification and sample preparation. We performed no further curation of the data sets beyond that performed by the authors. Some of the 1536 proteins are likely from contaminating organells but not all known proteins have appeared in the proteomic data sets, thus the numbers of ER proteins is likely to change with further study. A total of 1003 proteins were found in liver (736 of these where specific to liver ER), 202 in pancreas ER only and 331 in both tissues. A previous compilation of human ER proteins from the literature contained about 500 proteins [38]. Further research is needed to determine ER proteins common to all cell types and those are tissue-specific.
A major future need is to confirm the localization as true ER components for these 1500 proteins. Some are known to be localized in the ER by prior immunohistochemistry (IHC) or functional studies, but this information is not available for most. The first confirmatory approach when specific antibodies are available is to confirm the presence and enrichment by Western blotting of the organelle fraction used for proteomic analyses. This confirms the presence of the proteins identified by MS/MS but does not establish whether the protein is present in the organelle of interest or is from co-purified contaminants. To resolve this requires immunolocalization in the intact cell or for some organelles on isolated organelles stuck to a glass slide. Unfortunately, not all antibodies used for WB work for IHC. The alternative is to express a tagged protein in the cell type in question and visualize its location by IHC or fluorescence of fluorescent protein chimeras. Here attention has to be paid to the fact that overexpression may alter cellular localization. It also need to be determined whether specific proteins are uniquely localized in an organelle or are present in multiple cellular compartments.
Molecular architecture of the ER unveiled by proteomic studies
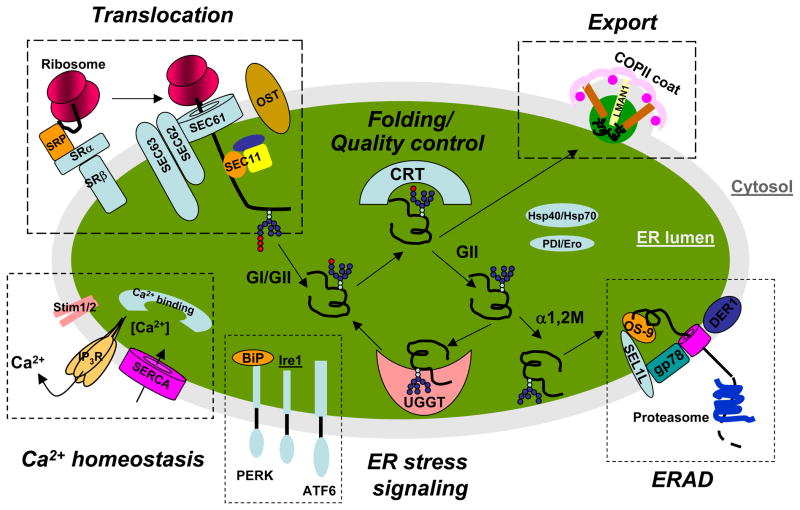
In this section, we will discuss the biological insights gained from the above reviewed proteomic studies regarding to the molecular functions of the endoplasmic reticulum with the emphasis on protein synthesis and translocation, protein folding and quality control, ER export, ER-associated degradation and calcium homeostasis (Fig. 1). Mass spectrometry-based proteomic studies have made significant contributions to the understanding of the molecular compositions of the ER. As will be revealed in the following sections, modern proteomics technologies have the capability to identify and more importantly quantify almost all the players in ER machinery. Mass spectrometry-based proteomic analysis has the potential and is expected to play an increasingly significant role in comprehensively understanding the molecular architecture of ER, quantifying dynamic changes of ER proteome under different physiological and pathophysiological conditions.
Figure 1. Overview of molecular functions of the ER.
Major ER functions are illustrated in this diagram. These include protein synthesis and translocation machinery, protein folding and quality control, ER export, ER-associated degradation (ERAD), ER stress signaling, and calcium homeostasis. The names of some of the key molecules in each functional module are provided. Most of these proteins have been detected and some are quantified in proteomic studies reviewed here. More details on the indicated molecules can be found in the text or in the Supplemental Table 1. CRT: calreticulin; DER1: derlin 1; OST: oligosaccharyltransferase; Stim 1 or 2: stromal interaction molecule 1 or 2; UGGT: UDP-glucose:glycoprotein glucosyltransferase.
Protein synthesis and translocation
Secretory proteins and many transmembrane proteins are synthesized on ribosomes bound to the ER membrane. Their synthesis or translation involves a number of initiation and elongation factors that participate often transiently in the process [39] and are the primary sites for controlling the rate of translation [40]. The emergence of the amino terminal of the nascent proteins, the signal peptides, from the ribosome leads to interaction with the translocon and co-traslationally translocated across the ER membrane [37, 38]. The emerging signal peptide binds the signal recognition particle (SRP) and docks onto the translocon through the SRP receptor (SR) [41, 42]. As a result, the nascent polypeptides directly enter the translocon pore [43] which consists of the SEC61α, β and γ subunits. In addition, signal peptidase, oligosaccharyltransferase (OST), TRAM, TRAP, Sec62, Sec63, BiP and GRP170 (also called oxygen regulated protein 150) comprise associated protein complexes that assist in the signal sequence-mediated targeting, co-translational translocation, and processing of nascent polypeptide chains [43–45].
Proteins involved in protein synthesis and translocation comprised one of the largest functional groups among all identified ER proteins (Supplemental Table 1). For the protein translational machinery, essentially all of the 86 known ribosomal proteins from both subunits have been observed in pancreatic and 78 in liver RER. In addition, twenty two known initiation, elongation and termination factors such as eukaryotic initiation factors (eIFs) 2, 3, 4A, 4B, 4G, 4H, 5A, and the eukaryotic elongation factors (eEFs) 1α, β, γ and 2 and other associated proteins such as the polyA binding proteins have been observed in the proteomic analyses. Because not all initiation factors remain associated with ribosomes during translation, a lower fraction of known initiation factors were observed in ER proteomic studies than for ribosomal proteins. In terms of the protein translocation, ER proteomic studies unveiled a large number of proteins in this category including components of signal recognition particle (SRP) and both α and β subunits of its receptor, the heterotrimeric SEC61 complex, SEC62 and SEC63 as the translocase, signal sequence receptor subunits and SEC11 signal peptidases which bind and remove signal peptides in ER lumen. In addition, several subunits of OST complex have been identified including STT3-A, ribophorin I, ribophorin II, OST48, and DAD1. Shibatani et al. [23] carried out proteomics analysis of isolated ribosome-associated membrane protein from canine rough microsomes using native gel electrophoresis to resolve protein complexes and they reported the identification of the known subunits of OST as well as two previously uncharacterized proteins, DC2 and KCP2 that co-purified with these subunits. It is the newly identified proteins that can be associated with specific ER structures or functions through their experimental context that has been one of the early payoffs of ER proteomics studies.
Protein folding and quality control
The ER provides an optimized environment for protein folding and maturation: the ER lumen has unique oxidizing potential that supports disulphide bond formation during protein folding, and a very high protein concentration to form a gel-like protein matrix of chaperones and folding enzymes [2, 46–48]. As a versatile protein folding factory, ER contains a specialized set of chaperones and folding enzymes including members of Hsp (heat shock protein) proteins from Hsp40, Hsp70 and Hsp90 families, peptidyl-prolyl cis-trans isomerases (PPIases), oxidoreductases, glycan trimming enzymes and lectins [2, 3, 47, 49, 50]. Many such proteins have been detected as abundant proteins by multiple proteomics analyses of the ER proteome discussed in this review (Supplemental Table 1). In purified rough microsomes from mouse liver, Knoblach et al. [17] detected BiP, GRP94, lectin-like chaperones (calnexin, calreticulin), peptidylprolyl isomerases, thiol disulphide oxidoreductases (PDI, P5 (CaBP1), ERp72, ERp57, ERp44, ERp29, and ERp46). Among these, a number of proteins including ERp19, ERp46 and Erj7 were identified as novel ER proteins by mass spectrometry-based proteomics studies [13, 19]. Quantitative proteomics revealed these same chaperones to be in similar concentrations in rough and smooth microsomes from rat liver [20].
Besides providing a unique folding environment, the ER has a crucial quality control role to permit only correctly folded proteins to leave ER and reach their final destinations. ER chaperones especially BiP and calnexin/calreticulin are active in ER quality control to proof-read newly synthesized proteins [2, 3, 48]. If persistently misfolded, these proteins are ultimately degraded. In addition, UDP-glucose:glycoprotein glucosyltransferase (UGGT) acts as folding sensor [51] to monitor local deviations from the native state and allow the incompletely folded substrate to reassociate with calnexin and/or calreticulin for an additional folding cycle. Polypeptides fulfilling the quality control requirements are released from this sensor system and can exit ER to their final destination. Essentially all the above mentioned critical components of the quality control machinery have been identified as abundant proteins in multiple ER proteomics studies (Supplemental Table 1).
ER stress, the Unfolded Protein Response and ER -associated degradation
The accumulation of unfolded or misfolded proteins in the ER lumen induces a coordinated adaptive program called the unfolded protein response (UPR) [52]. The UPR is activated by ER stress, an imbalance between the demand of client proteins on the ER and the organelle’s folding capacity [53]. The UPR alleviates stress by (i) inhibiting protein synthesis, (ii) upregulating protein folding capacity by enhancing synthesis of ER chaperones, and (iii) activating degradation pathways associated with the ER [54]. The three ER-resident transmembrane proteins sensors of ER stress are: the kinase and endoribonuclease inositol requiring element 1 (IRE1) [55], the PKR-like ER kinase (PERK) [56] and the basic leucine-zipper activating transcription factor 6 (ATF6) [57]. The activation of the UPR may lead either to cell survival, by triggering the synthesis of ER chaperone proteins along with a decrease in general protein translation, or to programmed cell death, apoptosis. A number of ER stress response proteins have been identified in the reviewed proteomic studies including ERS-1, ERS-25, stress-induced phosphoprotein 1, ER associated dnaJ protein 3 (ERdj3), protein chaperones (Hsc70/Hsp90, Hsp5, Hsp8, Hsp10, Hsp40, Hsp60, Hsp72, Hsp105, HSJ2, stress 70 protein mitochondrial or 75 kDa glucose regulated protein (GRP75), GRP78 or BiP, GRP58 or ERp57, GRP94), UPR (UBX domain protein 4, X-box binding-like-1) (Supplemental table 1). However, the ER resident transmembrane sensors PERK, IRE and ATF6 were not reported in published proteomic analyses likely because of low copy number, insolubility, or poorly ionizing fragments, although the first two are the most likely because the GPMdb reports for these proteins indicates relatively few or no identifications (thegpmdb.org).
Some misfolded proteins are beyond rescue and will never be able to pass the ER quality control standards. These terminally misfolded proteins are cleared from ER by the process called ER associated degradation (ERAD). In ERAD, the misfolded proteins are retro-translocated to the cytosol and become subject to ubiquitination and subsequent degradation by the multi-subunit 26S proteasome complex [58, 59]. Multiple components of the ERAD machinery have been identified in the ER proteomics studies. These include SEL1L, OS-9, Derlin-1 and 2, and E3 ubiquitin-ligase gp78 as well as AAA family member p97 and multiple subunits of the proteasomal endopeptidase complexes (Supplemental Table 1).
ER export and membrane trafficking
Once the newly synthesized proteins are fully folded, they are exported from ER, generally believed via COPII coated vesicles [4, 60]. A number of proteins involved in vesicular transport between ER to Golgi have been detected in several of the proteomics studies reviewed here (Supplemental Table 1). Components of the COPII protein complexes such as Sec23A, Sec24A and Sec24D were also detected in the same fraction. This is consistent with the fact that the smooth microsomes used in this study are derivatives of the tER which contains the ER exit sites. In addition to the coat proteins, several proteins that recycle between the ER and the cis-Golgi to mediate the vesicular transport between these compartments were reported in several proteomics studies (Supplemental Table 1). These proteins included ERGIC-53, also called LMAN1, a cargo receptor of several soluble glycoproteins, the P24 family members 2, 5, 6, 9 and 10, Rab1A, Sec22b, the SNARE protein mediating transport between ER and Golgi, and the KDEL receptor. In addition to the proteins involved in COPII transport, several components of the COPI coat including subunit alpha, beta′ and delta were detected in smooth microsomes fraction in the study by Gilchrist et al. [20].
Calcium homeostasis
The ER plays a major role in Ca2+ homeostasis and signaling. The lumen of the ER contains a total Ca2+ concentration of 1–3 mM with a free Ca2+ concentration of 60–400 uM [61]. This is brought about by the presence of Ca2+ transporting ATPases of the Serca and Spca families although the latter is primarily localized to the Golgi [62] and a number of low to medium affinity Ca2+ binding proteins including calreticulin, calnexin, Bip, Grp94, calumenin and the reticulocalbins [61, 63]. The Serca 2 isoform, which is broadly expressed, was found in both pancreas and liver and examples of the other Ca2+ binding proteins were also observed. Stim 1 and 2 (stromal interaction molecule 1 and 2) are recently discovered ER Ca2+ sensor which upon depletion of Ca2+ stores signal to the plasma membrane to increase Ca2+ influx [64]. Both forms of Stim were observed in pancreas which may indicate its abundant expression in this tissue type. The presence of Ca2+ in the ER lumen serves two functions [65]. First, it facilitates the chaperone action of a number of foldase proteins as shown by the fact that Ca2+ depletion will initiate an unfolded protein response. It has been speculated that the higher Ca2+ in the lumen mimics the extracellular Ca2+ and helps proteins assume a stable conformation for secretion. Second, the luminal Ca2+ provides a reservoir for release of Ca2+ into the cytoplasm by gated ion channels, the IP3 and Ryanodine receptors (IP3R and RyR). Essentially all of the Ca2+ binding proteins have been identified in multiple studies while the ion channels and pumps have been identified only to a limited extent. This may be due to the tissue specific expression of some proteins such as the IP3Rs (only Type 2 was found) and RyRs (none found) but is most likely due to their being large hydrophobic proteins expressed at low abundance as more targeted studies using immunoblotting have shown their presence in liver and pancreas.
Detoxification and Drug Metabolism Proteins
The ER is the site of a group of enzymes that metabolize endogenous hydrophobic molecules and exogenous foreign molecules termed xenobiotics that include molecules in food as well as drugs. The proteins involved include the cytochrome p450 family (CYPs), UDP-glucuronyl transferase, glutathione S-transferase (GSTs) and carboxyl esterases. These proteins show organ, cell type and species specificities [66]. The human genome contains 57 and the mouse genome 102 CYPs. They are present primarily in the ER or the inner mitochondrial membrane and 15 CYPs all from the CYP1, CYP2, and CYP3 families metabolize xenobiotics. Because of the localization of this function most studies have been carried out in liver and a number have utilized mass spectrometry. When rat liver microsomes were fractionated on a 1D gel twenty four drug metabolizing proteins were identified while only three were found by evaluation of 2D gels [22]. Lane et al identified 16 cytochrome P450s from mouse liver microsomes and showed the effect of a toxin to alter their expression [67]. In a more recent study, the yield of CYPs was improved by better membrane solubilization and 26 mouse liver CYPs were identified [68]. Rodlich et al identified 18 CYPs in microsomes prepared from human liver biopsies and demonstrated eight phosphorylation sites [69]. In another recent study of human liver, the coverage of CYPs was improved by analysis of sequential thin slices (0.2 mm) from 1D gels and this allowed demonstration of single amino acid polymorphisms [70]. The five comprehensive ER proteomic studies in Supplemental Table 1 show a clear tissue specific pattern. The two liver studies identified a large number of proteins in this category. Gilchrist et al [20] identified 54 proteins classified as detoxification although some are actually involved in biosynthetic reactions. By contrast, the pancreas studies identified only a few detoxification enzymes.
Future perspectives
Proteomic studies of the ER have lead to insights into the normal functions as well as disease-related changes of this versatile organelle. With continuing improvements in mass spectrometry and informatics technologies, we are approaching the goal of obtaining the complete, accurate and permanent proteome for ER. However, several issues need to be taken into account and some currently are still challenging. First, because subcellular fractions can not reach 100% purity, it is inevitable to detect proteins from co-purified organelles by increasingly sensitive mass spectrometry approaches. Therefore, rigorous confirmation, with independent techniques, of the subcellular localization of newly identified ER proteins is required for definitive conclusions. The correlation profiling strategy provides a high throughput way for organelle proteomic analysis, however, it does not completely overcome the problem and still require the independent validations. The combination of correlation profiling with high resolution organelle isolation might yield more authentic protein inventories. Currently the most powerful validation is by immunohistochemistry. Second, molecular dynamics at the cytosolic and luminal surfaces of the ER have to be taken into consideration. The composition of the ER is subject to change based on molecular interactions occurring on both cytosolic and luminal sides of the ER membrane. These are particularly challenging to analyze. Dynamics of protein interactions in organelles are often controlled by posttranslational modifications including phosphorylation, acetylation, and ubiquitination. Characterizing such modifications in a quantitative and dynamic manner is the key to a thorough understanding of the regulation of the ER functions. Third, in addition to the ubiquitously expressed ER proteins, we and others have found the presence of tissue specific proteins which correlate with specific functions of the tissue. Analysis of more tissues and cell types beyond liver and pancreas is essential to understand which ER proteins are general and which are tissue specific. Identification and confirmation of more such tissue specific ER proteins will shed light on the adaptation of ER function to tissue-specific demands and regulations. Last, but not least, it cannot be emphasized enough that the most useful proteomics data on the ER has been quantitative in nature. As the complete complement of ER proteins in different tissues is established, attention will shift to understanding alterations of the organellar proteome during diseases, in response to physiological perturbations and genetic manipulations.
Supplementary Material
Acknowledgments
We acknowledge the supported by NIH grant R37 DK41122 to J.A.W., the National Resource for Proteomics and Pathways (P41 RR018627) to P.C.A. and the University of Michigan Gastrointestinal Peptide Center (P30 DK34933). Support was also received as a pilot grant from the Center for Computational Medicine and Biology at the University of Michigan. We also thank Dr. Laszlo Prokai at the University of North Texas Health Science Center for providing us his data set for our bioinformatics analysis.
Footnotes
Note Added during Revisions
We thank a reviewer for calling our attention to a recent study of ER proteins from mouse liver [71]. Using 1D gel electrophoresis and LC-MS/MS, this study identified 903 and 1042 proteins in rough and smooth ER respectively. Among these proteins, 662 proteins were common to both rat [20] and mouse liver ER proteome. The authors proposed that these common proteins were essential for the maintenance of ER function [71].
Conflict of interest statement The authors declare no conflict of interest.
References
- 1.Lavoie C, Paiement J. Topology of molecular machines of the endoplasmic reticulum: a compilation of proteomics and cytological data. Histochem Cell Biol. 2008;129:117–128. doi: 10.1007/s00418-007-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 3.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 4.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 5.Groen AJ, de Vries SC, Lilley KS. A proteomics approach to membrane trafficking. Plant Physiol. 2008;147:1584–1589. doi: 10.1104/pp.108.123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lilley KS, Dupree P. Plant organelle proteomics. Curr Opin Plant Biol. 2007;10:594–599. doi: 10.1016/j.pbi.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Tartakoff AM, Jamieson JD. Subcellular fractionation of the pancreas. Methods Enzymol. 1974;31:41–59. doi: 10.1016/0076-6879(74)31006-3. [DOI] [PubMed] [Google Scholar]
- 8.Croze EM, Morre DJ. Isolation of plasma membrane, golgi apparatus, and endoplasmic reticulum fractions from single homogenates of mouse liver. J Cell Physiol. 1984;119:46–57. doi: 10.1002/jcp.1041190109. [DOI] [PubMed] [Google Scholar]
- 9.Plonne D, Cartwright I, Linss W, Dargel R, et al. Separation of the intracellular secretory compartment of rat liver and isolated rat hepatocytes in a single step using self-generating gradients of iodixanol. Anal Biochem. 1999;276:88–96. doi: 10.1006/abio.1999.4311. [DOI] [PubMed] [Google Scholar]
- 10.Preissler M, Williams JA. Localization of ATP-dependent calcium transport activity in mouse pancreatic microsomes. J Membr Biol. 1983;73:137–144. doi: 10.1007/BF01870437. [DOI] [PubMed] [Google Scholar]
- 11.Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Sans MD, Strahler JR, Karnovsky A, et al. Quantitative organellar proteomics analysis of rough endoplasmic reticulum from normal and acute pancreatitis rat pancreas. J Proteome Res. 9:885–896. doi: 10.1021/pr900784c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoblach B, Keller BO, Groenendyk J, Aldred S, et al. ERp19 and ERp46, new members of the thioredoxin family of endoplasmic reticulum proteins. Mol Cell Proteomics. 2003;2:1104–1119. doi: 10.1074/mcp.M300053-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Simon ES, Xiang Y, Kachman M, et al. Quantitative proteomics analysis of cell cycle-regulated Golgi disassembly and reassembly. J Biol Chem. 285:7197–7207. doi: 10.1074/jbc.M109.047084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 16.Foster LJ, de Hoog CL, Zhang Y, Xie X, et al. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Walker AK, Strahler JR, Simon ES, et al. Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics. 2006;5:306–312. doi: 10.1074/mcp.M500172-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Adelman MR, Sabatini DD, Blobel G. Ribosome-membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol. 1973;56:206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahedi RP, Volzing C, Schmitt A, Frien M, et al. Analysis of the membrane proteome of canine pancreatic rough microsomes identifies a novel Hsp40, termed ERj7. Proteomics. 2009;9:3463–3473. doi: 10.1002/pmic.200800722. [DOI] [PubMed] [Google Scholar]
- 20.Gilchrist A, Au CE, Hiding J, Bell AW, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 21.Hall SL, Hester S, Griffin JL, Lilley KS, Jackson AP. The organelle proteome of the DT40 lymphocyte cell line. Mol Cell Proteomics. 2009;8:1295–1305. doi: 10.1074/mcp.M800394-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galeva N, Altermann M. Comparison of one-dimensional and two-dimensional gel electrophoresis as a separation tool for proteomic analysis of rat liver microsomes: cytochromes P450 and other membrane proteins. Proteomics. 2002;2:713–722. doi: 10.1002/1615-9861(200206)2:6<713::AID-PROT713>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- 24.Sakai K, Hamanaka R, Yuki H, Watanabe M. A novel fractionation method of the rough ER integral membrane proteins; resident proteins versus exported proteins? Proteomics. 2009;9:3036–3046. doi: 10.1002/pmic.200800803. [DOI] [PubMed] [Google Scholar]
- 25.Charlton HK, Webster J, Kruger S, Simpson F, et al. ERp46 binds to AdipoR1, but not AdipoR2, and modulates adiponectin signalling. Biochem Biophys Res Commun. 2010;392:234–239. doi: 10.1016/j.bbrc.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Alberti A, Karamessinis P, Peroulis M, Kypreou K, et al. ERp46 is reduced by high glucose and regulates insulin content in pancreatic beta-cells. Am J Physiol Endocrinol Metab. 2009;297:E812–821. doi: 10.1152/ajpendo.00053.2009. [DOI] [PubMed] [Google Scholar]
- 27.Dunkley TP, Watson R, Griffin JL, Dupree P, Lilley KS. Localization of organelle proteins by isotope tagging (LOPIT) Mol Cell Proteomics. 2004;3:1128–1134. doi: 10.1074/mcp.T400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Dunkley TP, Hester S, Shadforth IP, Runions J, et al. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci U S A. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hebert DN, Bernasconi R, Molinari M. ERAD substrates: Which way out? Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 31.Riordan JR. Assembly of functional CFTR chloride channels. Annu Rev Physiol. 2005;67:701–718. doi: 10.1146/annurev.physiol.67.032003.154107. [DOI] [PubMed] [Google Scholar]
- 32.Morand JP, Macri J, Adeli K. Proteomic profiling of hepatic endoplasmic reticulum-associated proteins in an animal model of insulin resistance and metabolic dyslipidemia. J Biol Chem. 2005;280:17626–17633. doi: 10.1074/jbc.M413343200. [DOI] [PubMed] [Google Scholar]
- 33.Kubisch CH, Sans MD, Arumugam T, Ernst SA, et al. Early activation of endoplasmic reticulum stress is associated with arginine-induced acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2006;291:G238–245. doi: 10.1152/ajpgi.00471.2005. [DOI] [PubMed] [Google Scholar]
- 34.Tani S, Itoh H, Okabayashi Y, Nakamura T, et al. New model of acute necrotizing pancreatitis induced by excessive doses of arginine in rats. Dig Dis Sci. 1990;35:367–374. doi: 10.1007/BF01537416. [DOI] [PubMed] [Google Scholar]
- 35.Lampel M, Kern HF. Acute interstitial pancreatitis in the rat induced by excessive doses of a pancreatic secretagogue. Virchows Arch A Pathol Anat Histol. 1977;373:97–117. doi: 10.1007/BF00432156. [DOI] [PubMed] [Google Scholar]
- 36.Peng L, Rawson P, McLauchlan D, Lehnert K, et al. Proteomic analysis of microsomes from lactating bovine mammary gland. J Proteome Res. 2008;7:1427–1432. doi: 10.1021/pr700819b. [DOI] [PubMed] [Google Scholar]
- 37.Stevens SM, Jr, Duncan RS, Koulen P, Prokai L. Proteomic analysis of mouse brain microsomes: identification bioinformatic characterization of endoplasmic reticulum proteins in the mammalian central nervous system. J Proteome Res. 2008;7:1046–1054. doi: 10.1021/pr7006279. [DOI] [PubMed] [Google Scholar]
- 38.Scott M, Lu G, Hallett M, Thomas DY. The Hera database and its use in the characterization of endoplasmic reticulum proteins. Bioinformatics. 2004;20:937–944. doi: 10.1093/bioinformatics/bth010. [DOI] [PubMed] [Google Scholar]
- 39.Preiss T, MWH Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 40.Sans MD, Williams JA. Translational control of protein synthesis in pancreatic acinar cells. Int J Gastrointest Cancer. 2002;31:107–115. doi: 10.1385/IJGC:31:1-3:107. [DOI] [PubMed] [Google Scholar]
- 41.Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson I, Kelleher DJ, Miao Y, Shao Y, et al. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol. 2003;161:715–725. doi: 10.1083/jcb.200301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 46.Ellgaard L. Catalysis of disulphide bond formation in the endoplasmic reticulum. Biochem Soc Trans. 2004;32:663–667. doi: 10.1042/BST0320663. [DOI] [PubMed] [Google Scholar]
- 47.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudek J, Benedix J, Cappel S, Greiner M, et al. Functions and pathologies of BiP and its interaction partners. Cell Mol Life Sci. 2009;66:1556–1569. doi: 10.1007/s00018-009-8745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 50.Zimmermann R, Muller L, Wullich B. Protein transport into the endoplasmic reticulum: mechanisms and pathologies. Trends Mol Med. 2006;12:567–573. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Ruddock LW, Molinari M. N-glycan processing in ER quality control. J Cell Sci. 2006;119:4373–4380. doi: 10.1242/jcs.03225. [DOI] [PubMed] [Google Scholar]
- 52.Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 53.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 54.Kim R, Emi M, Tanabe K, Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006;11:5–13. doi: 10.1007/s10495-005-3088-0. [DOI] [PubMed] [Google Scholar]
- 55.Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. Embo J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida H, Okada T, Haze K, Yanagi H, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 59.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 60.Fromme JC, Orci L, Schekman R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008;18:330–336. doi: 10.1016/j.tcb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 61.Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32:269–278. doi: 10.1016/s0143416002001884. [DOI] [PubMed] [Google Scholar]
- 62.Missiaen L, Dode L, Vanoevelen J, Raeymaekers L, Wuytack F. Calcium in the Golgi apparatus. Cell Calcium. 2007;41:405–416. doi: 10.1016/j.ceca.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Honore B. The rapidly expanding CREC protein family: members, localization, function, and role in disease. Bioessays. 2009;31:262–277. doi: 10.1002/bies.200800186. [DOI] [PubMed] [Google Scholar]
- 64.Cahalan MD. STIMulating store-operated Ca(2+) entry. Nat Cell Biol. 2009;11:669–677. doi: 10.1038/ncb0609-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 66.Seliskar M, Rozman D. Mammalian cytochromes P450--importance of tissue specificity. Biochim Biophys Acta. 2007;1770:458–466. doi: 10.1016/j.bbagen.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 67.Lane CS, Wang Y, Betts R, Griffiths WJ, Patterson LH. Comparative cytochrome P450 proteomics in the livers of immunodeficient mice using 18O stable isotope labeling. Mol Cell Proteomics. 2007;6:953–962. doi: 10.1074/mcp.M600296-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sutton CW, Sutherland M, Shnyder S, Patterson LH. Improved preparation and detection of cytochrome P450 isoforms using MS methods. Proteomics. 2010;10:327–331. doi: 10.1002/pmic.200900489. [DOI] [PubMed] [Google Scholar]
- 69.Redlich G, Zanger UM, Riedmaier S, Bache N, et al. Distinction between human cytochrome P450 (CYP) isoforms and identification of new phosphorylation sites by mass spectrometry. J Proteome Res. 2008;7:4678–4688. doi: 10.1021/pr800231w. [DOI] [PubMed] [Google Scholar]
- 70.Lisitsa AV, Petushkova NA, Thiele H, Moshkovskii SA, et al. Application of slicing of one-dimensional gels with subsequent slice-by-slice mass spectrometry for the proteomic profiling of human liver cytochromes P450. J Proteome Res. 2010;9:95–103. doi: 10.1021/pr900262z. [DOI] [PubMed] [Google Scholar]
- 71.Song Y, Jiang Y, Ying W, Gong Y, et al. Quantitative proteomic survey of endoplasmic reticulum in mouse liver. J Proteome Res. 2010;9:1195–1202. doi: 10.1021/pr900146t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.