Abstract
Until recently, bacterial species that inhabit the human vagina have been primarily studied using organism-centric approaches. Understanding how these bacterial species interact with each other and the host vaginal epithelium is essential for a more complete understanding of vaginal health. Molecular approaches have already led to the identification of uncultivated bacterial taxa associated with bacterial vaginosis. Here, we review recent studies of the vaginal microbiome and discuss how culture-independent approaches, such as applications of next-generation sequencing, are advancing the field and shifting our understanding of how vaginal health is defined. This work may to lead to improved diagnostic tools and treatments for women who suffer from, or are at risk for, vaginal imbalances, pregnancy complications, and sexually acquired infections. These approaches may also transform our understanding of how host genetic factors, physiological conditions (e.g. menopause) and environmental exposures (e.g. smoking, antibiotic usage) influence the vaginal microbiome.
Keywords: microbiome, vaginal microbiome, next-generation sequencing
Introduction: the human microbiome
The human microbiome consists of trillions of microorganisms that colonize the human body. Different microbial communities inhabit vaginal, oral, skin, gastrointestinal, nasal, urethral and other sites of the human body. Currently, there is an international effort underway to describe the human microbiome in relation to health and disease [1–5]. The Human Microbiome Project, which was launched in 2008, is a $157 million, five-year effort funded through the NIH Common Fund's Roadmap for Medical Research. The overall project has set out to determine whether there is a shared core microbiome among individuals, to understand how changes in health are correlated with the microbiome, to develop tools to support this research, and to address associated ethical, legal, and social issues [6]. The development of next-generation sequencing and the decreasing cost of data generation using these technologies permit this ambitious project to investigate the complex microbial communities of the human body at unprecedented resolution [7].
The Vaginal Human Microbiome Project at Virginia Commonwealth University (VCU) [8] has been funded as part of this effort. In this project, we are studying: 1) thousands of women from relevant clinical settings to determine the relationship between the vaginal microbiome and various physiological and infectious conditions; and 2) hundreds of monozygotic and dizygotic twins to assess the influence of genetic and environmental factors on the composition of the human vaginal microbiome. We are targeting thousands of mid-vaginal samples for microbiome analysis using the V1–V3 hypervariable region of the 16S rRNA gene employing a deep-sequencing approach (i.e. ~30,000 reads per sample) that permits detection and quantification of less abundant microbes, which are often important constituents of the vaginal ecosystem. We are also cultivating and sequencing the genomes of hundreds of bacterial clones isolated from relevant vaginal samples, and applying whole metagenome shotgun sequencing (WMGSS) to explore the metabolic potential of the microbiota and characterize abundant bacterial taxa in vaginal samples of particular interest. Here, we review studies of the vaginal microbiome and highlight the advances afforded by next-generation sequencing and other molecular approaches.
The vaginal microbiome of healthy women
The vaginal epithelium and microbiota undergo dramatic shifts that coincide with hormonal changes that occur throughout a woman’s life. As estrogen levels increase during puberty, glycogen is deposited in the stratified, squamous, non-keratinized vaginal epithelium [9] [10]. For many women, this physiological change coincides with and likely mediates a natural increase in the prevalence of species of lactobacilli, which then generally predominate during the reproductive years. The importance of species of the genus Lactobacillus has been appreciated for over a hundred years, dating back to Albert Döderlein’s late-19th-century discovery of long, thick gram-positive rods in the normal vaginal secretions of premenopausal women [11–13]. Lactobacillus species are able to ferment glycogen, thereby producing lactic acid, which is thought to establish the acidic vaginal environment (i.e. pH < 4.5) that has traditionally been considered a hallmark of vaginal health [14]. As estrogen levels gradually decline during menopause, the glycogen content in vaginal epithelial cells also declines, leading to a varying degree of depletion of lactobacilli [15–17]. Also during menopause, the vaginal epithelium thins and loses its elasticity, vaginal blood flow diminishes, and there is a marked decrease in vaginal secretions [18]. Thus, not surprisingly, the representation of lactobacilli in the vagina is diminished.
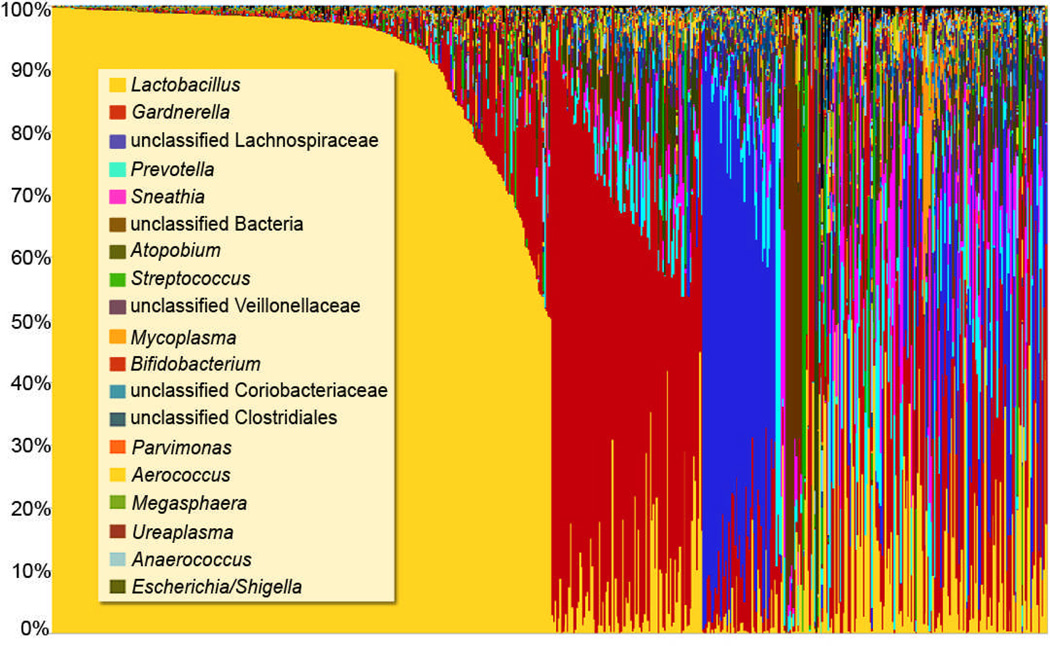
Recent studies of the vaginal microbiome suggest that the traditional paradigm of vaginal health does not apply to all women. Several groups have reported that the average vaginal pH for African American and Hispanic women is higher than that of Caucasian women [19–21], and that in fact, a majority of women in these racial and ethnic groups have high vaginal pH (i.e., pH > 4.5) values, which is outside the range traditionally associated with health. Furthermore, recent studies using molecular techniques have found that Lactobacillus species do not predominate in the vaginal microbiome for a substantial proportion of healthy women of reproductive age [20] [22] [23]. In preliminary analyses of the vaginal microbiomes of a cadre of over 600 women, our results support these findings (Figure 1). Thus, a simple clustering of these microbiomes suggests the existence of multiple ‘vagitypes’, many of which are dominated by a single bacterial taxon. However, other vagitypes exhibit a broad range of taxa. It is not yet clear that these gross microbiome profiles are causal indicators of states of health or disease (Vaginal Microbiome Consortium, unpublished data).
Figure 1. Mid-vaginal microbiome profiles using genus-level taxonomic classification.
A total of 650 microbiome profiles are represented. Each bar represents the genus-level microbiome profile for one mid-vaginal sample and each color represents a distinct genus. Mid-vaginal swab samples were obtained from participants enrolled in the Vaginal Human Microbiome Project at VCU. The V1–V3 hypervariable of the 16S rRNA gene was targeted, and on average, approximately 30,000 reads per sample were generated using the Roche 454 GS FLX Titanium platform.
It has yet to be determined whether apparently healthy women with diverse vagitypes are in a transition state between bacterial vaginosis (BV) and health, but several research groups are pursuing longitudinal studies that will help to address this question [24] [25]. Large, cross-sectional studies may allow for the identification of bacterial taxa or groups of taxa that distinguish vaginal microbiome profiles of healthy, asymptomatic women who have lactobacilli-deficient profiles from those of women with BV or other vaginal conditions.
Novel organisms in the vaginal microbiome
Compared to other body sites, such as those in the gastrointestinal tract, fewer bacterial species have been reported to inhabit the vagina. Nonetheless, many vaginally relevant bacterial species have yet to be characterized. Cultivation-independent approaches allow for the study of organisms that cannot be cultured in the laboratory using current techniques [26] [27]. David Fredricks and colleagues have recently used molecular techniques to identify several uncultivated organisms that are associated with BV [28] [29]. The bacterial vaginosis-associated bacteria in the Clostridiales order (e.g. BVAB1, BVAB2, BVAB3) were identified using broad-range 16S rDNA assays, and they were validated using a combination of fluorescence in situ hybridization (FISH) and bacterium-specific PCR [28]. BVAB3 is the only BVAB that has since been cultivated and sequenced (GenBank accession number CP001850.2). As part of the Vaginal Human Microbiome Project [8], we have recently identified several additional bacterial taxa that apparently have not yet been described (Fettweis et al., in preparation). We are currently validating these taxa, assessing the role of these organisms in vaginal health, and targeting them for cultivation. Additionally, we are employing whole-metagenome shotgun sequencing to learn more about the genetic content of the fastidious organisms that have yet to be cultured. Recently, single-cell genome sequencing technologies have been developed to permit genome assemblies from single uncultivated microbial cells [30]. Thus, the application these emerging technologies to uncultivated vaginal bacterial species would provide cell-specific genetic information.
Environmental and genetic factors
While racial disparities have previously been reported in vaginal microbiome profiles [20] [21] [31], the relative influence of various genetic and environmental factors has yet to be established. Environmental factors, such as smoking [32] [33], douching [32] [34–36], and some sexual practices [32] [35–37], have been associated with lower levels of vaginal lactobacilli. Estrogen has perhaps the most well-established environmental influence on vaginal flora. The deficiency of estrogen that occurs during menopause often leads to a decline in lactobacilli [16] [17] [23], and in some cases, this shift is also associated with urogenital infections [17]. Notably, estrogen replacement generally leads to a reestablishment of lactobacilli [16].
It is unknown how genetic variations affect vaginal microbiome constitution and maintenance, but it has been hypothesized that genetic polymorphisms that disrupt normal signaling of the innate immune system may be associated with less healthy flora [38]. For example, Verstraelen and colleagues [39] found that carriage of Atopobium vaginae and Gardnerella vaginalis during the first half of pregnancy was associated with polymorphisms in genes involved with Toll-like receptor mediated signaling (i.e. CD14 intron 2 1342G>T, TLR1 exon 4 743A>G, CARD15 exon 4 14772A>T; and −1155A>G in the MD2 promoter respectively). Genç et al. [39] [40] demonstrated that an allelic polymorphism in the intron of a gene in the interleukin 1 cytokine family, specifically IL1RN2*, is associated with elevated vaginal pH and, in black women, a decrease in lactobacilli levels. Additionally, several groups have reported maternal single nucleotide polymorphisms (SNPs) that are associated with an increased risk for preterm labor [41–44], several of which affect genes associated with immune modulation. Studies that resolve the specific contributions of genetic and environmental influences on the composition of the vaginal microbiome may lead to better diagnostics and treatments for a variety of conditions affecting women’s health.
Vaginal infections and imbalances
Common vaginal infections and imbalances can be broadly characterized in two classes: 1) those caused by sexually transmitted pathogenic organisms, and 2) those caused by overgrowth of resident flora. Sexually transmitted infections can be caused by bacterial species (e.g., Chlamydia trachomatis, Neisseria gonorrhoeae, Treponema pallidum), parasites (Trichomonas vaginalis) and viruses (e.g., human immunodeficiency virus, human papillomavirus, herpes simplex virus) [45]. Alternatively, dysbioses that have been described include BV [46], Candida (yeast) infections [47], aerobic vaginitis [48] [49], overgrowth of lactobacilli [50], and atrophic vaginitis [51]. Both sexually transmitted infections and vaginal dysbioses must be considered in the vaginal ecosystem.
Bacterial vaginosis is the most common vaginal imbalance, and it has been reported to affect between 10–20% of white, non-Hispanic women, and 30–50% of African American women [52]. Bacterial vaginosis is a disorder characterized by an imbalance in vaginal flora, which often causes malodorous discharge. Notably, women with BV are at an increased risk for acquisition of sexually transmitted infections and for the occurrence of complications during pregnancy [53–55]. Typically, BV is clinically diagnosed using Amsel’s criteria, which requires that three of the following four conditions be met: 1) presence of a thin homogeneous vaginal discharge, 2) amine odor following addition of potassium hydroxide to vaginal secretions, 3) presence of clue cells (i.e. vaginal epithelial cells covered with bacteria), and 4) and vaginal pH greater than 4.5. Alternatively, BV can be diagnosed using the Nugent criteria, which involves scoring Gram-stained vaginal smears to quantify bacterial populations consistent with the disorder. The etiology of BV has yet to be determined; however, it is likely that complex polymicrobial factors are involved. Organisms that have been associated with BV include, but are not limited to, G. vaginalis [56], A. vaginae [57], BV-associated bacteria (BVAB1, BVAB2, and BVAB3) in the order Clostridiales [28], Megasphaera species [58], and Sneathia species [59]. Given that BV is not caused by a single pathogen and that many BV-associated organisms have not been cultivated, applications of next-generation sequencing methods are well suited to the study of the disorder. In addition to broad-range16S rDNA surveys, whole metagenome sequencing of vaginal samples and the subsequent analysis of gene content and metabolic potential will be useful in gaining insights into the etiology of the disorder.
Pregnancy and preterm labor
Estrogen and progesterone levels soar during pregnancy, the vaginal epithelium thickens, and additional glycogen is deposited into the vaginal epithelium [9] [10]. Considering that hormonal changes associated with menarche and menopause are associated with significant changes in vaginal flora, one might expect changes in vaginal microbiome to accompany pregnancy as well, but these effects have not been characterized in detail. However, it has been established that BV is associated with a wide-array of pregnancy complications [60] [61] including preterm premature rupture of membranes (PPROM) [62] and spontaneous preterm labor and preterm birth [60] [61] [63] [64]. Recent studies have identified bacterial species that are routinely found in amniotic fluid [65], and that Mycoplasma [66–68], Ureaplasma [66] [67], and Sneathia [69] (previously grouped with Leptotrichia) species are often identified in the amniotic fluid of women with complications in pregnancy. It remains unclear how bacterial colonizers of the vaginal environments are associated with these infections. Clearly, a better understanding of the dynamics of the vaginal microbiome during pregnancy may lead to better treatments, prognostics and diagnostics for complications associated with the complex processes of pregnancy, labor and birth.
Species- and strain-level analyses of the vaginal microbiome
Species- and strain-level analyses are important for understanding the vaginal ecosystem, and multiple approaches to the study of the vaginal microbiome are often required. These include the following next-generation sequencing applications: 1) metagenomic surveys of single-gene taxonomic markers such as the 16S rRNA gene, 2) multi-gene and whole-genome analyses of bacterial genomes of interest, and 3) whole metagenomic shotgun surveys of vaginal samples. In 16S rDNA metagenomic studies that permit identification and relative quantification of bacterial taxa, typically only a portion of the 16S rRNA gene (e.g. V1–V3 hypervariable region) is sequenced because of the read-length limitations of next-generation sequencing technologies [7]. While not all species can be differentiated using a portion of the 16S rRNA gene, the common vaginal Lactobacillus species can be easily distinguished using this approach [24] [70] [71]. As part of the Vaginal Human Microbiome Project [8], we are developing a protocol to more broadly permit species-level resolution in metagenomic 16S rDNA datasets from vaginal samples. Several groups have recently targeted the chaperonin-60 gene as an alternative to the 16S rRNA gene for the identification of microbes in vaginal samples [72–75]. The vaginal microbiome profiles generated by targeting the chaperonin-60 gene had a similar taxonomic distribution compared to those generated by targeting the 16S rRNA gene, but some differences in species-level resolution were reported [72]. An important advantage to targeting the 16S rRNA gene is that it has been widely adopted in microbiome research, which allows for cross-study comparisons of microbiome profiles [5]. Moreover, existing databases of bacterial 16S rDNA sequences are more comprehensive and cover more strains and species than do similar databases for other potentially useful genes.
Several efforts are underway to sequence bacterial genomes of organisms that colonize the vaginal epithelium [2] [8] [56] [76]. This collection of genomes will provide a valuable resource for future studies. These genomes will aid in the identification of biologically significant single-gene markers that are specific to bacterial species of interest. One such example is the sialidase gene, which has been found to be absent in one of three identified genotypes of G. vaginalis [77]. Given that vaginal sialidase levels have been associated with BV [78] [79] and preterm labor [80] [81], this difference in gene content is of particular biological interest. Genome-wide strain-level comparisons of bacterial species of interest and comprehensive studies of mobile elements, such as bacteriophages and transposons, promise additional insights into how common vaginal bacterial species and strains impact women’s health.
Lactobacilli and exclusion of other bacterial species
While over twenty species of lactobacilli have been isolated from vaginal samples, only one or two Lactobacillus species typically predominate in any one sample, and the most common of these include L. crispatus, L. iners, L jensenii, and L. gasseri [20] [31] [82]. Lactobacillus species have been reported to differ in their ability to exclude other bacteria, and L. iners is the species that most often co-exists with other ‘less-healthy’ microorganisms [76]. Understanding of the interactions between bacterial species and the host is also essential to a full understanding of the vaginal microbiome, thus here we review the proposed mechanisms of exclusion by lactobacilli.
In premenopausal women, two general strategies have been proposed to explain how lactobacilli inhibit the growth of other microorganisms in the vaginal niche: 1) competitive exclusion through adherence to vaginal epithelial cells; and 2) production of antimicrobials, such as bacteriocins, hydrogen peroxide (H2O2), and lactic acid [83]. Lactobacillus species are thought to compete for binding to receptors on host epithelial cells, and numerous studies have demonstrated that they can inhibit adherence of various urogenital pathogens including group B Streptococcus species [84], Staphylococcus aureus [84], G. vaginalis [85], N. gonorrhoeae [86], Pseudomonas aeruginosa [87], and Klebsiella pneumonia [87]. Notably, Lactobacillus species and strains vary in their ability to adhere to host cells [88] [89], which is consistent with the fact that the mechanism of competitive exclusion is only one of several defense mechanisms.
The production of antimicrobials by lactobacilli also contributes to the maintenance of the vaginal flora. At least two bacteriocins have been identified in vaginal Lactobacillus strains [46] [90–93]. One of the identified bacteriocins, lactocin 160, has been shown to target the cytoplasmic membrane of G. vaginalis, suggesting that it is likely to be a biologically relevant defense mechanism [46]. The association between H2O2-producing lactobacilli and vaginal health has also been well established [36] [94], but it is controversial whether the production of H2O2 is directly responsible for the exclusion of other species. The absence of H2O2-producing lactobacilli has been associated with several unhealthy vaginal conditions and states including BV [36] [94], vaginal colonization of E. coli [95], preterm labor [96], and endometriosis [97]. Despite the consistent finding that H2O2-producing lactobacilli are associated with vaginal health, H2O2 production was not found to be required for the lactic acid-induced inhibition of BV-associated bacteria [98], or with lactobacillus-induced inhibition of N. gonorrhoeae growth [89] [99]. O’Hanlon and colleagues [100] also reported that H2O2 antimicrobial activities can be blocked by both semen and cervicovaginal fluid, which would suggest the mechanism may be ineffective in vivo. While the rationale for why H2O2-producing lactobacilli seem to be associated with health has yet to be conclusively determined, one interesting hypothesis suggests that the role of H2O2 as a mediator of the SOS response and prophage induction may be important. Lactobacillus species are commonly lysogenized with temperate bacteriophages, and it has been hypothesized that induction of the lytic cycle may trigger a transition to a less-healthy microbiome profile [101]. Moreover, Martin et al. [102] suggested that production of H2O2 may select for lactobacilli strains harboring defective prophages. These strains would thus be more stable reducing the likelihood of a transition to the abnormal vaginal microbiome state.
There is a mounting body of evidence that suggests that lactic acid production is important defense mechanism that prevents the growth of acid-sensitive bacterial species such as those associated with BV [103]. O’Hanlon and colleagues [98] recently found that lactic acid reduces the viability of a panel of bacteria associated with BV (e.g. G. vaginalis, A. vaginae) at concentrations typically found in women with a lactobacillus-dominated vaginal microbiome, and Graver et al. [99] found that in cocultivation experiments, lactic acid-producing Lactobacillus species inhibited the growth of N. gonorrhoeae. Notably, O’Hanlon et al. [98] reported that a change in pH from 7 to 4.5 reduced the viability of BV-associated bacteria between two-fold and 106-fold, depending on the species. However, physiological concentrations of lactic acid, but not acetic acid, dramatically reduced the viability of BV-associated organisms (106-fold to 108-fold), which suggests that the antimicrobial activity of lactic acid is not solely based on vaginal acidification.
While most studies of vaginal acidification have focused on the role of lactobacilli, host epithelial cells may make a significant contribution to vaginal acidification as well [83] [104]. Two potential host roles have been suggested: 1) an upregulation of proton secretion at the surface of the cervicovaginal epithelium in response to estrogen [104], and 2) host production of lactic acid and fatty acids [83] [105]. Thus, it is likely that host epithelial cells, in concert with lactic acid producing bacteria, contribute to vaginal acidification.
Concluding remarks
The application of next-generation sequencing and other molecular techniques to the study of the vaginal microbiome is transforming our understanding of vaginal health through the detection of uncultivated organisms and other fastidious organisms that have gone undetected using culture-based techniques. Molecular approaches allow for species- and strain-level resolution of bacteria that have important implications for vaginal health. Microbiome profiles, which elucidate the complex patterns of bacterial taxa present in a sample, provide insight into the polymicrobial nature of the bacterial populations in the healthy and unhealthy vagina. The application of whole metagenome shotgun sequencing to vaginal samples will permit study of the gene content and metabolic capabilities of uncultivated organisms. Studies using molecular techniques in combination with cultured-based studies may help to elucidate how bacterial species interact with each other and the host vaginal epithelium and how these interactions are associated with host genetic factors, and various physiological and infectious conditions.
Acknowledgement
This work was supported by Grant 1UH2AI083263/4UH3AI083263, The Vaginal Microbiome: Disease, Genetics and the Environment, from the National Institutes of Health. Additional members of the Vaginal Microbiome Consortium who have contributed to the preliminary findings from the Vaginal Human Microbiome Project at VCU include (in alphabetical order): João P. Alves, Joseph F. Borzelleca, James P. Brooks, Cynthia N. Cornelissen, Lindon J. Eaves, Christopher J. Friedline, Abigail L. Glascock, Michael D. Harwich, Stephanie L. Hendricks, Vladimir Lee, Michael C. Neale, Melissa A. Prestosa, Federico A. Puma, Mark A. Reimers, Maria C. Rivera, Nihar U. Sheth, Judy L. Silberg, Jerome F. Strauss III, Logan J. Voegtly, and Timothy P. York.
References
- 1.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M’rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, Feldgarden M, Gevers D, Haas BJ, Madupu R, Ward DV, Birren BW, Gibbs RA, Methe B, Petrosino JF, Strausberg RL, Sutton GG, White OR, Wilson RK, Durkin S, Giglio MG, Gujja S, Howarth C, Kodira CD, Kyrpides N, Mehta T, Muzny DM, Pearson M, Pepin K, Pati A, Qin X, Yandava C, Zeng Q, Zhang L, Berlin AM, Chen L, Hepburn TA, Johnson J, McCorrison J, Miller J, Minx P, Nusbaum C, Russ C, Sykes SM, Tomlinson CM, Young S, Warren WC, Badger J, Crabtree J, Markowitz VM, Orvis J, Cree A, Ferriera S, Fulton LL, Fulton RS, Gillis M, Hemphill LD, Joshi V, Kovar C, Torralba M, Wetterstrand KA, Abouellleil A, Wollam AM, Buhay CJ, Ding Y, Dugan S, FitzGerald MG, Holder M, Hostetler J, Clifton SW, Allen-Vercoe E, Earl AM, Farmer CN, Liolios K, Surette MG, Xu Q, Pohl C, Wilczek-Boney K, Zhu D. Science. 2010;328:994–999. [Google Scholar]
- 3.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Clin. Chem. 2009;55:856–866. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fettweis JM, Alves JP, Borzelleca JF, Brooks JP, Friedline CJ, Gao Y, Gao X, Girerd P, Harwich MD, Hendricks SL, Jefferson KK, Lee V, Mo H, Neale MC, Puma FA, Reimers MA, Rivera MC, Roberts S, Serrano MG, Sheth N, Judy L, Voegtly Silberg, L, Prom-Wormley EC, Xie B, York TP, Cornelissen CN, Strauss JF, III, Eaves LJ, Buck GA. Nature Precedings. 2011 [Google Scholar]
- 9.Bourne A. The British Medical Journal. 1947;1:79–83. doi: 10.1136/bmj.1.4489.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The British Medical Journal. 1943;2:753. [Google Scholar]
- 11.Cruickshank R. The Journal of Hygiene. 1931;31:375–381. doi: 10.1017/s0022172400010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lash AF, Kaplan B. The Journal of Infectious Diseases. 1926;38:333–340. [Google Scholar]
- 13.Thomas S. The Journal of Infectious Diseases. 1928;43:218–227. [Google Scholar]
- 14.Redondo-Lopez V, Cook RL, Sobel JD. Reviews of Infectious Diseases. 1990;12:856–872. doi: 10.1093/clinids/12.5.856. [DOI] [PubMed] [Google Scholar]
- 15.Hillier SL, Lau RJ. Clin. Infect. Dis. 1997;25(Suppl 2):S123–S126. doi: 10.1086/516221. [DOI] [PubMed] [Google Scholar]
- 16.Heinemann C, Reid G. Can. J. Microbiol. 2005;51:777–781. doi: 10.1139/w05-070. [DOI] [PubMed] [Google Scholar]
- 17.Pabich WL, Fihn SD, Stamm WE, Scholes D, Boyko EJ, Gupta K. J. Infect. Dis. 2003;188:1054–1058. doi: 10.1086/378203. [DOI] [PubMed] [Google Scholar]
- 18.Nyirjesy P. Curr Infect Dis Rep. 2007;9:480–484. doi: 10.1007/s11908-007-0073-5. [DOI] [PubMed] [Google Scholar]
- 19.Stevens-Simon C, Jamison J, McGregor JA, Douglas JM. Sex Transm Dis. 1994;21:168–172. doi: 10.1097/00007435-199405000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiscella K, Klebanoff MA. American Journal of Obstetrics and Gynecology. 2004;191:747–750. doi: 10.1016/j.ajog.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 22.Neggers YH, Nansel TR, Andrews WW, Schwebke JR, Yu K-fun, Goldenberg RL, Klebanoff MA. J. Nutr. 2007;137:2128–2133. doi: 10.1093/jn/137.9.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson JD, Lee RA, Balen AH, Rutherford AJ. Int J STD AIDS. 2007;18:308–311. doi: 10.1258/095646207780749583. [DOI] [PubMed] [Google Scholar]
- 24.Fredricks DN. Anaerobe. 2011;17:191–195. doi: 10.1016/j.anaerobe.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCook A. Nat Med. 2011;17:765–767. doi: 10.1038/nm0711-765. [DOI] [PubMed] [Google Scholar]
- 26.Handelsman J. Microbiol. Mol. Biol. Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pace NR. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 28.Fredricks DN, Fiedler TL, Marrazzo JM. N. Engl. J. Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 29.Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. Appl. Environ. Microbiol. 2008;74:4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lasken RS. Biochem. Soc. Trans. 2009;37:450–453. doi: 10.1042/BST0370450. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, Foster JA, Forney LJ. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 32.Cherpes TL, Hillier SL, Meyn LA, Busch JL, Krohn MA. Sex. Transm. Dis. 2008;35:78–83. doi: 10.1097/OLQ.0b013e318156a5d0. [DOI] [PubMed] [Google Scholar]
- 33.Ryckman KK, Simhan HN, Krohn MA, Williams SM. Mol. Hum. Reprod. 2009;15:131–137. doi: 10.1093/molehr/gan081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ness RB, Hillier SL, Richter HE, Soper DE, Stamm C, McGregor J, Bass DC, Sweet RL, Rice P. Obstet. Gynecol. 2002;100:765. doi: 10.1016/s0029-7844(02)02184-1. [DOI] [PubMed] [Google Scholar]
- 35.Beigi RH, Wiesenfeld HC, Hillier SL, Straw T, Krohn MA. J. Infect. Dis. 2005;191:924–929. doi: 10.1086/428288. [DOI] [PubMed] [Google Scholar]
- 36.Hawes SE, Hillier SL, Benedetti J, Stevens CE, Koutsky LA, Wolner-Hanssen P, Holmes KK. J. Infect. Dis. 1996;174:1058–1063. doi: 10.1093/infdis/174.5.1058. [DOI] [PubMed] [Google Scholar]
- 37.Marrazzo JM, Koutsky LA, Eschenbach DA, Agnew K, Stine K, Hillier SL. J. Infect. Dis. 2002;185:1307–1313. doi: 10.1086/339884. [DOI] [PubMed] [Google Scholar]
- 38.Genç MR, Onderdonk A. BJOG. 2011;118:154–163. doi: 10.1111/j.1471-0528.2010.02772.x. [DOI] [PubMed] [Google Scholar]
- 39.Verstraelen H, Verhelst R, Nuytinck L, Roelens K, De Meester E, De Vos D, Van Thielen M, Rossau R, Delva W, De Backer E, Vaneechoutte M, Temmerman M. J. Reprod. Immunol. 2009;79:163–173. doi: 10.1016/j.jri.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Genc MR, Vardhana S, Delaney ML, Onderdonk A, Tuomala R, Norwitz E, Witkin SS. Eur. J. Obstet. Gynecol. Reprod. Biol. 2004;116:152–156. doi: 10.1016/j.ejogrb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Romero R, Friel LA, Velez Edwards DR, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Erez O, Chaiworapongsa T, Pearce BD, Bartlett J, Salisbury BA, Anant MK, Vovis GF, Lee MS, Gomez R, Behnke E, Oyarzun E, Tromp G, Williams SM, Menon R. Am. J. Obstet. Gynecol. 2010;203:361.e1–361.e30. doi: 10.1016/j.ajog.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gómez LM, Sammel MD, Appleby DH, Elovitz MA, Baldwin DA, Jeffcoat MK, Macones GA, Parry S. American Journal of Obstetrics and Gynecology. 2010;202:386.e1–386.e6. doi: 10.1016/j.ajog.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 43.Plunkett J, Doniger S, Orabona G, Morgan T, Haataja R, Hallman M, Puttonen H, Menon R, Kuczynski E, Norwitz E, Snegovskikh V, Palotie A, Peltonen L, Fellman V, DeFranco EA, Chaudhari BP, McGregor TL, McElroy JJ, Oetjens MT, Teramo K, Borecki I, Fay J, Muglia L. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001365. e1001365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harper M, Zheng SL, Thom E, Klebanoff MA, Thorp J, Jr, Sorokin Y, Varner MW, Iams JD, Dinsmoor M, Mercer BM, Rouse DJ, Ramin SM, Anderson GD. Obstet Gynecol. 2011;117:125–130. doi: 10.1097/AOG.0b013e318202b2ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biggs WS, Williams RM. Prim. Care. 2009;36:33–51. viii. doi: 10.1016/j.pop.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Turovskiy Y, Ludescher RD, Aroutcheva AA, Faro S, Chikindas ML. Probiotics Antimicrob Proteins. 2009;1:67–74. doi: 10.1007/s12602-008-9003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soong D, Einarson A. Can Fam Physician. 2009;55:255–256. [PMC free article] [PubMed] [Google Scholar]
- 48.Tempera G, Furneri PM. Gynecol. Obstet. Invest. 2010;70:244–249. doi: 10.1159/000314013. [DOI] [PubMed] [Google Scholar]
- 49.Donders G, Bellen G, Rezeberga D. BJOG. 2011;118:1163–1170. doi: 10.1111/j.1471-0528.2011.03020.x. [DOI] [PubMed] [Google Scholar]
- 50.Horowitz BJ, Mårdh PA, Nagy E, Rank EL. Am. J. Obstet. Gynecol. 1994;170:857–861. doi: 10.1016/s0002-9378(94)70298-5. [DOI] [PubMed] [Google Scholar]
- 51.Stika CS. Dermatol. Ther. 2010;23:514–522. doi: 10.1111/j.1529-8019.2010.01354.x. [DOI] [PubMed] [Google Scholar]
- 52.Sobel JD. Infect. Dis. Clin. North Am. 2005;19:387–406. doi: 10.1016/j.idc.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. AIDS. 2008;22:1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carey JC, Klebanoff MA, Hauth JC, Hillier SL, Thom EA, Ernest JM, Heine RP, Nugent RP, Fischer ML, Leveno KJ, Wapner R, Varner M. N. Engl. J. Med. 2000;342:534–540. doi: 10.1056/NEJM200002243420802. [DOI] [PubMed] [Google Scholar]
- 55.Rathod SD, Krupp K, Klausner JD, Arun A, Reingold AL, Madhivanan P. Sex. Transm. Dis. 2011;38:882–886. doi: 10.1097/OLQ.0b013e31821f91a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harwich MD, Jr, Alves JM, Buck GA, Strauss JF, 3rd, Patterson JL, Oki AT, Girerd PH, Jefferson KK. BMC Genomics. 2010;11:375. doi: 10.1186/1471-2164-11-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradshaw CS, Tabrizi SN, Fairley CK, Morton AN, Rudland E, Garland SM. J. Infect. Dis. 2006;194:828–836. doi: 10.1086/506621. [DOI] [PubMed] [Google Scholar]
- 58.Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, Li L, Nelson KE, Xia Y, Xiang C. BMC Genomics. 2010;11:488. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. J. Clin. Microbiol. 2007;45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, Romero R. BJOG. 2011;118:533–549. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou X, Brotman RM, Gajer P, Abdo Z, Schüette U, Ma S, Ravel J, Forney LJ. Infect. Dis. Obstet. Gynecol. 2010;2010 doi: 10.1155/2010/737425. 737425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silva MG, Peraçoli JC, Sadatsune T, Abreu ES, Peraçoli MTS. Int. J. Gynaecol. Obstet. 2003;81:175–182. doi: 10.1016/s0020-7292(03)00043-2. [DOI] [PubMed] [Google Scholar]
- 63.Hay PE, Lamont RF, Taylor-Robinson D, Morgan DJ, Ison C, Pearson J. BMJ. 1994;308:295–298. doi: 10.1136/bmj.308.6924.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gratacós E, Figueras F, Barranco M, Vila J, Cararach V, Alonso PL, Fortuny A. Acta. Obstet. Gynecol. Scand. 1998;77:37–40. [PubMed] [Google Scholar]
- 65.Hitti J, Riley DE, Krohn MA, Hillier SL, Agnew KJ, Krieger JN, Eschenbach DA. Clin. Infect. Dis. 1997;24:1228–1232. doi: 10.1086/513669. [DOI] [PubMed] [Google Scholar]
- 66.Perni SC, Vardhana S, Korneeva I, Tuttle SL, Paraskevas L-R, Chasen ST, Kalish RB, Witkin SS. Am. J. Obstet. Gynecol. 2004;191:1382–1386. doi: 10.1016/j.ajog.2004.05.070. [DOI] [PubMed] [Google Scholar]
- 67.Figueroa R, Garry D, Elimian A, Patel K, Sehgal PB, Tejani N. J. Matern. Fetal. Neonatal. Med. 2005;18:241–247. doi: 10.1080/13506120500223241. [DOI] [PubMed] [Google Scholar]
- 68.Marconi C, de Andrade Ramos BR, Peraçoli JC, Donders GGG, da Silva MG. Am. J. Reprod. Immunol. 2011;65:549–556. doi: 10.1111/j.1600-0897.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 69.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. PLoS ONE. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR, Temmerman M, Vaneechoutte M. BMC Microbiol. 2007;7:115. doi: 10.1186/1471-2180-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schellenberg J, Links MG, Hill JE, Dumonceaux TJ, Peters GA, Tyler S, Ball TB, Severini A, Plummer FA. Appl. Environ. Microbiol. 2009;75:2889–2898. doi: 10.1128/AEM.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Kimani J, Jaoko W, Wachihi C, Mungai JN, Peters GA, Tyler S, Graham M, Severini A, Fowke KR, Ball TB, Plummer FA. Appl. Environ. Microbiol. 2011;77:4066–4074. doi: 10.1128/AEM.02943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill JE, Goh SH, Money DM, Doyle M, Li A, Crosby WL, Links M, Leung A, Chan D, Hemmingsen SM. Am. J. Obstet. Gynecol. 2005;193:682–692. doi: 10.1016/j.ajog.2005.02.094. [DOI] [PubMed] [Google Scholar]
- 75.Hill JE, Penny SL, Crowell KG, Goh SH, Hemmingsen SM. Genome Res. 2004;14:1669–1675. doi: 10.1101/gr.2649204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Macklaim JM, Gloor GB, Anukam KC, Cribby S, Reid G. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl 1):4688–4695. doi: 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.dos GL, Santiago S, Deschaght P, El Aila N, Kiama TN, Verstraelen H, Jefferson KK, Temmerman M, Vaneechoutte M. Am. J. Obstet. Gynecol. 2011;204:450.e1–450.e7. doi: 10.1016/j.ajog.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 78.Smayevsky J, Canigia LF, Lanza A, Bianchini H. Infect. Dis. Obstet. Gynecol. 2001;9:17–22. doi: 10.1155/S1064744901000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howe L, Wiggins R, Soothill PW, Millar MR, Horner PJ, Corfield AP. Int. J. STD. AIDS. 1999;10:442–447. doi: 10.1258/0956462991914438. [DOI] [PubMed] [Google Scholar]
- 80.Cauci S, McGregor J, Thorsen P, Grove J, Guaschino S. Am. J. Obstet. Gynecol. 2005;192:489–496. doi: 10.1016/j.ajog.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 81.Cauci S, Culhane JF. Am. J. Obstet. Gynecol. 2011;204:142.e1–142.e9. doi: 10.1016/j.ajog.2010.08.061. [DOI] [PubMed] [Google Scholar]
- 82.Antonio MAD, Hawes SE, Hillier Sharon L. The Journal of Infectious Diseases. 1999;180:1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- 83.Boris S, Barbés C. Microbes Infect. 2000;2:543–546. doi: 10.1016/s1286-4579(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 84.Zárate G, Nader-Macias ME. Lett. Appl. Microbiol. 2006;43:174–180. doi: 10.1111/j.1472-765X.2006.01934.x. [DOI] [PubMed] [Google Scholar]
- 85.Boris S, Suárez JE, Vázquez F, Barbés C. Infect. Immun. 1998;66:1985–1989. doi: 10.1128/iai.66.5.1985-1989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vielfort K, Sjölinder H, Roos S, Jonsson H, Aro H. Microbes and Infection. 2008;10:1325–1334. doi: 10.1016/j.micinf.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 87.Osset J, Bartolomé RM, García E, Andreu A. Journal of Infectious Diseases. 2001;183:485–491. doi: 10.1086/318070. [DOI] [PubMed] [Google Scholar]
- 88.Chavière G, Cocnnier MH, Kernèis S, Fourniat J, Servin AL. Journal of General Microbiology. 1992;138:1689–1696. doi: 10.1099/00221287-138-8-1689. [DOI] [PubMed] [Google Scholar]
- 89.Spurbeck RR, Arvidson CG. Infect. Immun. 2008;76:3124–3130. doi: 10.1128/IAI.00101-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ocaña VS, Pesce de Ruiz Holgado AA, Nader-Macías ME. Appl. Environ. Microbiol. 1999;65:5631–5635. doi: 10.1128/aem.65.12.5631-5635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li J, Aroutcheva AA, Faro S, Chikindas ML. Infect. Dis. Obstet. Gynecol. 2005;13:135–140. doi: 10.1080/10647440500148156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tao L, Pavlova SI, Mou SM, Ma W.-ge, Kiliç AO. Infect. Dis. Obstet. Gynecol. 1997;5:244–251. doi: 10.1155/S1064744997000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vera Pingitore E, Hébert EM, Nader-Macías ME, Sesma F. Res. Microbiol. 2009;160:401–408. doi: 10.1016/j.resmic.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 94.Wasiela M, Misiak G, Pieczara A, Kalinka J. Archives of Perinatal Medicine. 2008;14:23–27. [Google Scholar]
- 95.Gupta K, Stapleton AE, Hooton TM, Roberts PL, Fennell CL, Stamm WE. J. Infect. Dis. 1998;178:446–450. doi: 10.1086/515635. [DOI] [PubMed] [Google Scholar]
- 96.Wilks M, Wiggins R, Whiley A, Hennessy E, Warwick S, Porter H, Corfield A, Millar M. J. Clin. Microbiol. 2004;42:713–717. doi: 10.1128/JCM.42.2.713-717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haggerty CL, Hillier SL, Bass DC, Ness RB PID Evaluation and Clinical Health (PEACH) Study Investigators. Clinical Infectious Diseases. 2004;39:990–995. doi: 10.1086/423963. [DOI] [PubMed] [Google Scholar]
- 98.O’Hanlon D, Moench T, Cone R. BMC Infectious Diseases. 2011;11:200. doi: 10.1186/1471-2334-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Graver MA, Wade JJ. Ann. Clin. Microbiol. Antimicrob. 2011;10:8. doi: 10.1186/1476-0711-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Hanlon DE, Lanier BR, Moench TR, Cone RA. BMC Infect. Dis. 2010;10:120. doi: 10.1186/1471-2334-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pavlova SI, Kiliç AO, Mou SM, Tao L. Infect. Dis. Obstet. Gynecol. 1997;5:36–44. doi: 10.1155/S1064744997000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martín R, Soberón N, Escobedo S, Suárez JE. Int. Microbiol. 2009;12:131–136. [PubMed] [Google Scholar]
- 103.Boskey ER, Telsch KM, Whaley KJ, Moench TR, Cone RA. Infect. Immun. 1999;67:5170–5175. doi: 10.1128/iai.67.10.5170-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gorodeski GI, Hopfer U, Liu CC, Margles E. Endocrinology. 2005;146:816–824. doi: 10.1210/en.2004-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Preti G, Huggins GR. J. Chem. Ecol. 1975;1:361–376. doi: 10.1007/BF01020154. [DOI] [PubMed] [Google Scholar]