Abstract
Resonance Raman spectroscopy is used to document, for the first time, a 6 cm−1 decrease of the Fe-S stretch by introducing an H-bond donor into the proximal pocket of a cytochrome P450, which interacts with the key cysteine thiolate axial ligand. The anticipated trans-effect on bound exogenous ligands is also confirmed and evidence is obtained supporting intimate interaction of the new histidyl-imidazole fragment with the heme periphery.
Keywords: biophysics, heme proteins, oxidoreductases, Raman spectroscopy
Members of the widely distributed cytochrome P450 class of monoxygenases, or CYPs, generate highly reactive oxygen-derived intermediates that effect a diverse set of reactions, including hydroxylation and epoxidation of relatively inert substrates,[1–3] thereby facilitating such important physiological functions as steroid biosynthesis and metabolism of pharmaceuticals.[4,5] The active site heme b is bound by a proximal cysteine thiolate, while on the distal side the heme is presented with an intricately arranged set of residues facilitating proton transfer to ferric peroxo- and ferric hydroperoxo-species.[6] Substrate binding to the six coordinate, low-spin (6cLS) ferric resting state converts it to a five-coordinate, high-spin state (5cHS) that is reduced by natural redox partners, with the resulting ferrous form then binding dioxygen. Normally, the enzymatic cycle continues on to completion (oxidation of substrate) by accepting one more electron from a reductase to generate a peroxo-intermediate. Sequential delivery of two protons generates an initial hydroperoxo-intermediate and then the so-called Compound I, a potent oxidizing species generally formulated as a ferryl heme π-cation radical.[1–3,7] Understandably, there is considerable interest in gaining insight into the structural factors that control the reactivity of the intermediates encountered within this cycle.[8–10] One obvious point of interest is that the strongly electron donating axial thiolate of all CYPs and structurally related enzymes, such as Nitric Oxide Synthases (NOSs), can promote O-O bond cleavage or impact the behavior of subsequent intermediates.[11–15] The effective negative charge on this proximal cysteine-thiolate could be fine-tuned by the nature and number of suitably positioned H-bond donors known to be present in the proximal pocket[14–23] A direct measure of the immediate consequences of such H-bonding is documentation of the strength of the Fe-S linkage, a task most readily accomplished by resonance Raman (rR) spectroscopy, using excitation within the S→ Fe charge transfer transition envelope that occurs near 360 nm in the ferric HS form.[14,16–18,24–26] In the present work this direct spectroscopic method is used to interrogate active site structural changes that accompany a single site mutation of a truncated form of CYP2B4; i.e., the F429H variant, where the highly conserved proximal-side phenylalanine is replaced with the potentially H-bonding histidyl residue. The essential results obtained provide new definitive experimental evidence for the influence of proximal-side H-bonding on the key Fe-S linkage and its effects on the cytochrome P450 cycle.
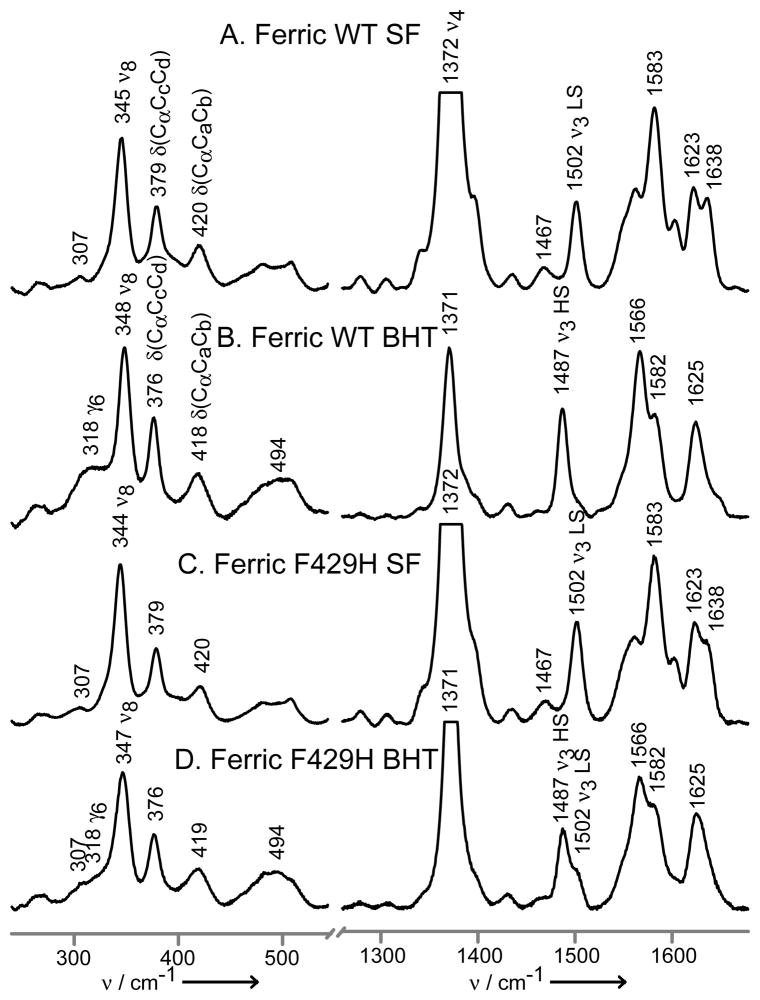
This F429H mutant is of great interest, because the presence of this additional H-bond drastically diminished the enzyme’s ability to promote O-O bond cleavage.[9] Additionally, computational studies have suggested that the mutant’s heme oxygenase activity could arise as a result of a predicted weakening of the Fe-S bond strength.[15] Figure 1 shows the Soret band-excited rR spectra obtained for the substrate-free (SF) and butylated hydroxytoluene (BHT)-bound “wild type” ferric CYP2B4 and its F429H mutant. In the case of the WT enzyme (traces A and B), substrate binding causes an almost complete conversion from a 6cLS to 5cHS state; i.e. the ν3 “spin-state marker” shifts from 1502 to 1487 cm−1.[27–29]
Figure 1.
The low (left) and high (right) frequency rR spectra of ferric CYP2B4, wild-type substrate-free (A), wild-type BHT-bound (B), F429H mutant substrate-free (C) and F429H mutant BHT-bound. Spectra measured with 406 nm excitation line and normalized to the ν3 mode.
Similar behavior is documented for the F429H mutant, though the conversion is not as complete. This residual 6cLS fraction seen for the mutant might arise from decreased affinity for the substrate or from a conformational heterogeneity that increases the stability of the aquo-bound form. However, the absence of the SF form for the CO adduct (vide infra) argues against the former case, unless one invokes a significantly different substrate affinity between the ferric and ferrous forms. In the low frequency region the bending modes associated with the propionate and vinyl substituents are observed near ~370 cm−1 and 400–440 cm−1, respectively. In contrast to behavior seen for bacterial P450s,[16,30] only small shifts are typically seen for microsomal mammalian CYPs such as those of interest here,[26,31] whose distal pockets are larger and more flexible.[1,2] It is noted, on the other hand, that BHT binding to both enzymes studied here does activate out-of-plane (oop) modes at 318 and ~500 cm−1.
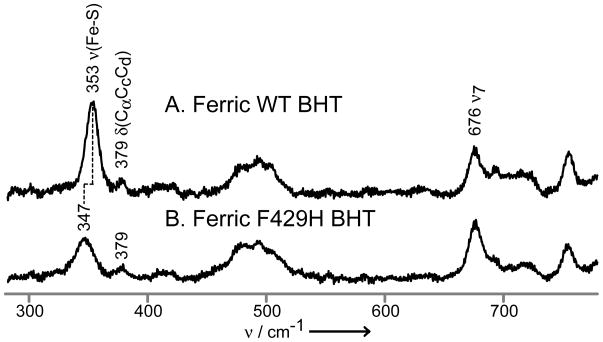
The key ν(Fe-S) mode is enhanced only via excitation within the Fe-S charge transfer transition that occurs near 360 nm for the (substrate-bound) ferric HS state.[24] Thus, in Figure 2 this mode is observed as a relatively strong feature appearing at 353 cm−1 in the BHT-bound WT enzyme and at 347 cm−1 for the BHT-bound CYP2B4 F429H. While the 347 cm−1 feature observed in the spectrum of the F429H mutant is accidently degenerate with the 347 cm−1 feature seen in with Soret excitation (Figure 1, Trace D left), the two features correspond to two different modes; i.e., the ν(Fe-S) and the ν8 modes, respectively. Convincing evidence for this is presented in the Supporting Information, where it is shown that the observed bandwidths are substantially broader for these 353/347 cm−1 features than those observed for the heme modes (Figure S2).
Figure 2.
The low frequency rR spectra of ferric CYP2B4, wild-type BHT-bound (A) and F429H mutant BHT-bound (B). Spectra measured with 356 nm excitation line and normalized to the ν7 mode at 676 cm−1.
The observed 6 cm−1 shift of ν(Fe-S) to lower frequency for F429H is qualitatively consistent with expectations based on the reduction of effective negative charge on the thiolate, in agreement with recent computations.[14,15] Similar earlier rR work with CYP101[17] and CYP102,[16] led to unexpected results; i.e., removal of existing H-bond donors led either to no effect on the observed ν(Fe-S) frequency or an unexpected decrease,[17] while an attempt to introduce a new H-bond donor residue to the proximal pocket of CYP102 (i.e., F393H) failed to induce any observable shift of its ν(Fe-S) mode.[16] The former result was later suggested to be attributable to vibrational mode reorganization.[14] The absence of any changes on the ν(Fe-S) mode for the F393H mutant of CYP102 was interpreted as evidence that the histidyl residue was not in a proper orientation to H-bond to the proximal cysteine thiolate fragment.[16,32] Given these complications encountered in these previous rR studies, it is satisfying that the present results agree with the predicted influence on the Fe-S bond strength.[14,15]
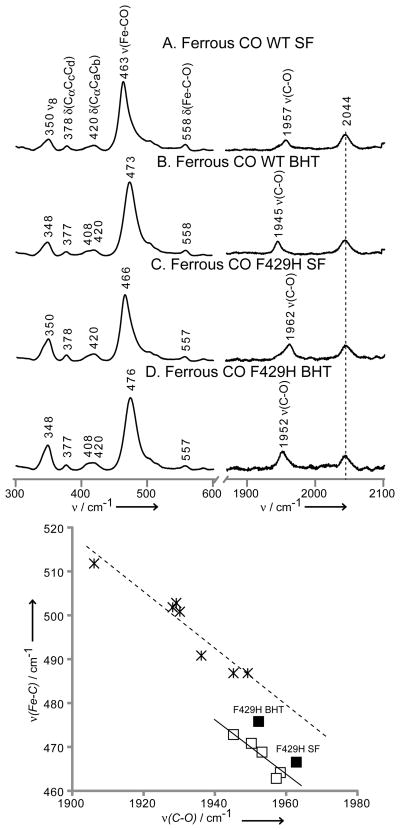
The new H-bonding modulation of the Fe-S bond strength is also reflected in the altered electronic structure of the trans-axial Fe-C-O linkage of the ferrous CO adduct. Thus, as shown in Figure 3 (top), the ν(Fe-C) shifts up by 3 cm−1 for the mutant (i.e., 463 → 466 and 473 →476 cm−1 for the SF and BHT-bound comparisons), with the ν(C-O) modes also shifting up by 5–7cm−1 for the same comparisons (i.e., 1957 →1962 and 1945→1952 cm−1). These directly correlated shifts of the ν(Fe-C) and ν(C-O) modes, both to higher frequency, are indicative of a diminished σ-donation of the trans-axial proximal thiolate ligand resulting from the newly introduced H-bonding histidine residue, such behavior being in agreement with predictions made from several high-level computational studies.[14,15,33] It is noted that earlier computational work, supported with modest experimental data for models, has previously been used to argue this point for the isoelectronic ferric NO adducts.[34]
Figure 3.
The low (left) and high (right) frequency rR spectra of ferrous CO adducts of CYP2B4, wild-type substrate-free (A), wild-type BHT-bound (B), F429H mutant substrate-free (C) and F429H mutant BHT-bound (D). Spectra measured with 442 nm excitation line and normalized to the ν7 and ν4 modes for low and high frequency region, respectively. The bottom graph shows linear correlation between ν(Fe-C) and ν(C-O) frequencies, the open squares represent wild type truncated CYP2B4,[40] the solid squares indicate points for F429H mutants and the stars show mammalian NOSs.[37–39]
Furthermore, the behavior seen here for the vibrational modes of the Fe-C-O fragment is satisfyingly consistent with data reported for CO adducts of NOS.[23] Thus, the plots shown at the bottom of Figure 3 present data acquired for CO adducts of mammalian NOS (stars), the CO adducts of WT CYP2B4 under various conditions (open squares) and the two points obtained here for the F429H mutant of CYP2B4 (solid squares); additional data reported for bacterial NOS proteins, as well as the isoelectronic Fe(III)NO adducts of cytochromes P450 and model compounds, are given in Figure S3 of Supporting Information. The inverse correlations of the ν(Fe-C) and ν(C-O) modes are generally accepted to reliably reflect variations in polarity of the distal heme pocket,[33,35] with displacements from one another along the vertical direction being the result of differences in the strength of the proximal ligand. Thus, the displacement of the NOS line from the CYP2B4 line arises because the Fe-S bonds of the NOSs are weakened by the presence of an H-bond from a conserved tryptophan residue in the proximal pocket; i.e., their ν(Fe-S) stretching frequencies generally occur near 338 cm−1,[36] approximately 12–14 cm−1 lower than for cytochromes P450. As can be seen in Figure 3, the points derived for the F429H mutant of CYP2B4 indeed are shifted towards the line for the NOS proteins, a shift entirely consistent with the observed 6 cm−1 lowering of the ν(Fe-S) for this mutant.[37–39]
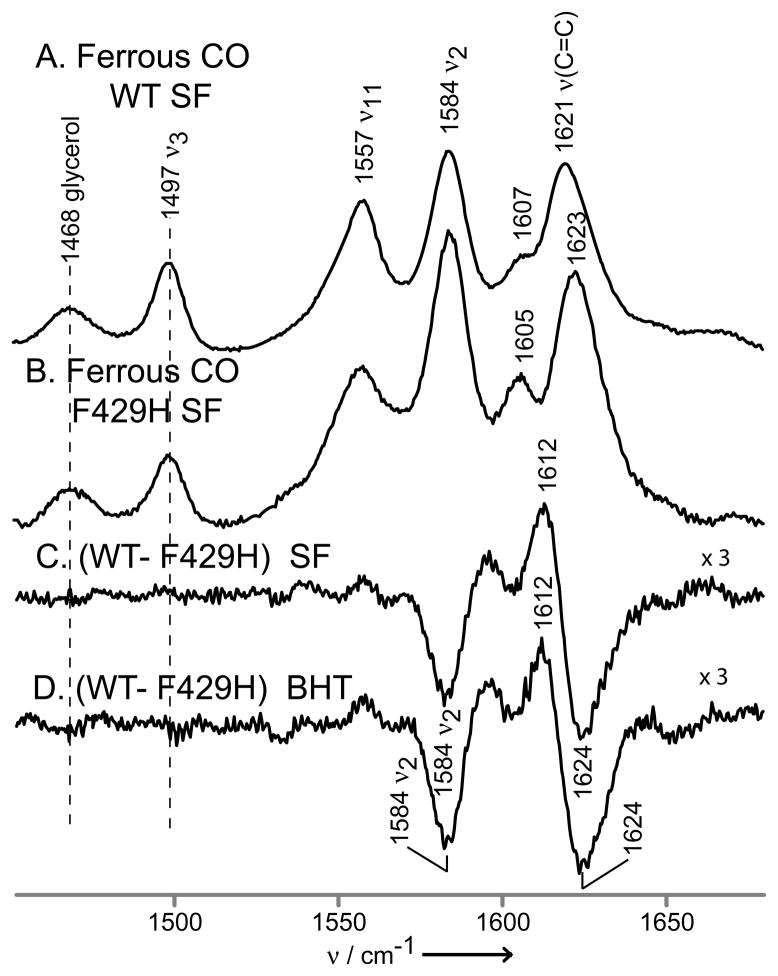
Finally, it is noteworthy that the remarkable structural sensitivity of the rR technique reveals small, but clearly detectable, mutation-induced changes for particular heme modes of the F429H mutant. A complex difference pattern (Figure 4, trace C) is obtained when comparing the rR spectra for the SF forms of ferrous CO adducts of the WT and F429H mutant. A similar difference plot obtained for the pair of BHT-bound forms is given in trace D. These data show that, while the mutation does not significantly change the basic heme structure, as reflected in the constancy of the core-sensitive ν3 and ν11 modes, changes are evident in the spectral region associated with the ν(C=C) modes of the peripheral vinyl groups. The most reasonable interpretation of these difference patterns is that the vinyl mode envelope is shifted to slightly higher frequency, behavior which is consistent with out-of-plane displacement of the vinyl groups.[16] This displacement diminishes the conjugation of the vinyl groups with the heme macrocyle π-system, with the result that the ν2 core mode, associated with stretching of the Cβ-Cβ bonds of the pyrrole ring to which the vinyls are attached,[28] is also altered, experiencing an increased resonance enhancement. Significantly, all of these changes are observed for modes of molecular fragments localized at or near the 2- and/or 4-vinyl groups and are therefore entirely consistent with the proposed interaction of the newly introduced histidyl imidazole with the pyrrole A-ring, based on inspection of recently the acquired x-ray crystal structure of the mutant.[LAW, unpublished]
Figure 4.
The high frequency rR spectra of ferrous CO adducts of substrate-free wild-type CYP2B4 (A) and F429H mutant (B). Spectra were normalized to the glycerol mode at 1468 cm−1. The traces (C) and (D) are difference traces of ferrous CO adducts of CYP2B4 wild-type and 429H mutant in substrate-free form (C) and in BHT-bound state (D).
In summary, the present results show that the newly introduced H-bond does indeed lead to a shift of the ν(Fe-S) stretching mode to lower frequency, documenting a weakening of this bond, as predicted by theory.[14,15] Furthermore, the impact of this modulation of Fe-S bond strength is transmitted to the heme iron linkage with a trans-axial diatomic ligand (CO), as reflected in the observed behavior of the ν(Fe-C) and ν(C-O ) modes of the Fe-C-O fragment,[23,41] such electronic effects holding potential implications for functional properties. Finally, changes in certain heme vibrational modes provide convincing evidence for the presence of an intimate interaction between the replacement residue, 429H, and the heme pyrrole A ring, a structural feature that is suggested from inspection of preliminary X-ray crystallographic data.[LAW, unpublished]
Experimental Section
Plasmid for the truncated mutant was constructed from pLW01-wild type full length P4502B4 plasmid.[42] The cDNA coding for the 19 amino acids at the N-terminus (amino acids 3-21 of WT CYP2B4) were removed using NcoI restriction sites which had been inserted at position 3 and 21 by site directed mutagenesis. After removing the cDNA between the two NcoI sites, the remainder of the plasmid was religated by T4 DNA ligase (Invitrogen, NY). The following mutations, E2A, G22K, H23K, P24T, K25S, A26S, H27K, R29K, H226Y and F429H were then introduced into the Δ3–21 WT CYP2B4, using a QuikChange Site-Directed Mutagenesis Kit (Stratagene) to generate the F429H mutant of the truncated form of CYP2B4. The truncated form of CYP2B4 (all of the mutations indicated above, minus the F429H mutation), considered WT” above, is the form which had been previously engineered to allow successful crystallization when bound to different compounds.[43]
The plasmid pLW01 CYP450 2B4 F429H with cDNA for the mutant or WT CYP2B4 was transformed into C41 (DE3) cells. The cells were grown in TB medium at 23°C and induced with IPTG, supplemented with delta-aminolevulinic acid at induction and ethanol thirty minutes after induction. The induced cell culture was harvested ~120 hr after induction.
The protein samples were isolated and purified as previously described.[44] Butylated hydroxytoluene (BHT) was added to all buffers to prevent conversion to cytochrome P420 during purification. The protein was eluted from hydroxyapatite column using buffer containing 0.4 M K2HPO4/KH2PO4 pH 7.4 and 20% glycerol. The purified protein was diluted into a final buffer containing 20% glycerol and 0.1 M K2HPO4/KH2PO4 pH 7.4 and concentrated to ~1 mM using 50,000 MWCO VivaSpin 20.
The rR measurements
All samples are ~ 0.4mM in heme. All further details, including solution conditions and spectrometer parameters are given in Supporting Information.
Supplementary Material
Footnotes
This work was supported in part by National Institutes of Health grants R01GM096117 (to JRK) from the National Institutes of General Medical Sciences and National Institutes of Health grants R01GM035533 and R01GM094209 and VA Merit Review Grant (to LAW).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Piotr J. Mak, Department of Chemistry, Marquette University, Milwaukee, WI 53233 (USA), Fax: 414 288 7066
Dr. Yuting Yang, Department of Anesthesiology, University of Michigan and VA Medical Center, Ann Arbor, MI 48105 (USA), Fax: 734 845 3096
Dr. Sangchoul Im, Department of Anesthesiology, University of Michigan and VA Medical Center, Ann Arbor, MI 48105 (USA), Fax: 734 845 3096
Prof. Lucy A. Waskell, Email: waskell@med.umich.edu, Department of Anesthesiology, University of Michigan and VA Medical Center, Ann Arbor, MI 48105 (USA), Fax: 734 845 3096.
Prof. James R. Kincaid, Email: james.kincaid@marquette.edu, Department of Chemistry, Marquette University, Milwaukee, WI 53233 (USA), Fax: 414 288 7066.
References
- 1.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. 3. Kluwer/Plenum; New York: 2004. [Google Scholar]
- 2.Sigel A, Sigel H, Sigel RKO, editors. Metal Ions in Life Sciences. Vol. 3. John Wiley & Sons, Ltd; Chichester: 2007. [Google Scholar]
- 3.Coon MJ. Ann Rev Pharmacol Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich FP. Chem Res Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 5.Hollenberg PF, Kent UM, Bumpus NN. Chem Res Toxicol. 2008;21:189–205. doi: 10.1021/tx7002504. [DOI] [PubMed] [Google Scholar]
- 6.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 7.Terner J, Palaniappan V, Gold A, Weiss R, Fitzgerald MM, Sullivan AM, Hosten CM. J Inorg Biochem. 2006;100:480–501. doi: 10.1016/j.jinorgbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Davydov R, Macdonald IDG, Makris TM, Sligar SG, Hoffman BM. J Am Chem Soc. 1999;121:10654–10655. [Google Scholar]
- 9.Davydov R, Razeghifard R, Im SC, Waskell L, Hoffman BM. Biochemistry. 2008;47:9661–9666. doi: 10.1021/bi800926x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denisov IG, Mak PJ, Makris TM, Sligar SG, Kincaid JR. J Phys Chem A. 2008;112:13172–13179. doi: 10.1021/jp8017875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rittle J, Green mT. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 12.Shaik S, Cohen S, Wang Y, Chen H, Kumar D, Thiel W. Chem Rev. 2010;110:949–1017. doi: 10.1021/cr900121s. [DOI] [PubMed] [Google Scholar]
- 13.Stone KL, Behan RK, Green MT. Proc Natl Acad Sci USA. 2006;103:12307–12310. doi: 10.1073/pnas.0603159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galinato MGI, Spolitak T, Ballou DP, Lehnert N. Biochemistry. 2011;50:1053–1069. doi: 10.1021/bi101911y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Usharani D, Zazza C, Lai W, Chourasia M, Waskell L, Shaik S. J Am Chem Soc. 2012;134:4053–4056. doi: 10.1021/ja211905e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Ost TWB, Schelvis JPM. Biochemistry. 2004;43:1798–1808. doi: 10.1021/bi034920g. [DOI] [PubMed] [Google Scholar]
- 17.Yoshioka S, Tosha T, Takahashi S, Ishimori K, Hori H, Morishima I. J Am Chem Soc. 2002;124:14571–14579. doi: 10.1021/ja0265409. [DOI] [PubMed] [Google Scholar]
- 18.Lang J, Santolini J, Couture M. Biochemistry. 2011;50:10069–10081. doi: 10.1021/bi200965e. [DOI] [PubMed] [Google Scholar]
- 19.Ogliaro F, Cohen S, de Visser SP, Shaik S. J Am Chem Soc. 2000;122:12892–12893. [Google Scholar]
- 20.Ogliaro F, de Visser SP, Shaik S. J Inorg Biochem. 2002;91:554–567. doi: 10.1016/s0162-0134(02)00437-3. [DOI] [PubMed] [Google Scholar]
- 21.Poulos TL, Finzel BC, Howard AJ. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 22.Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson J, Deisenhofer J. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 23.Hasemann CA, Ravichandran KG, Boddupalli SS, Peterson J, Deisenhofer J. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 24.Champion PM, Stallard BR, Wagner GC, Gunsalus IC. J Am Chem Soc. 1982;104:5469–5472. [Google Scholar]
- 25.Sabat J, Stuehr DJ, Yeh SR, Rousseau DL. J Am Chem Soc. 2009;131:12186–12192. doi: 10.1021/ja901016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak PJ, Im SC, Zhang H, Waskell LA, Kincaid JR. Biochemistry. 2008;47:3950–3963. doi: 10.1021/bi800034b. [DOI] [PubMed] [Google Scholar]
- 27.Kitagawa T, Mizutani Y. Coord Chem Rev. 1994;135/136:685–735. [Google Scholar]
- 28.Spiro TG, editor. Biological Applications of Raman Spectroscopy. III. John Wiley and Sons; New York: 1988. [Google Scholar]
- 29.Kincaid JR. In: Porphyrin Handbook. Kadish KM, Smith KM, Guilard R, editors. Vol. 7. Academic Press; San Diego: 2000. pp. 225–291. [Google Scholar]
- 30.Mak PJ, Kaluka D, Manyumwa EM, Zhang H, Deng T, Kincaid JR. Biopolymers. 2008;89:1045–1053. doi: 10.1002/bip.21058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mak PJ, Denisov IG, Grinkova YV, Sligar SG, Kincaid JR. J Am Chem Soc. 2011;133:1357–1366. doi: 10.1021/ja105869p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ost TWB, Munro AW, Mowat CG, Taylor PR, Pesseguiero A, Fulco AJ, Cho AK, Cheesman MA, Walkinshaw MD, Chapman SK. Biochemistry. 2001;40:13430–13438. doi: 10.1021/bi010717e. [DOI] [PubMed] [Google Scholar]
- 33.Xu C, Ibrahim M, Spiro TG. Biochemistry. 2008;47:2379–2387. doi: 10.1021/bi702254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulat F, Lehnert N. Inorg Chem. 2007;46:1547–1549. doi: 10.1021/ic070023f. [DOI] [PubMed] [Google Scholar]
- 35.Li XY, Spiro TG. J Am Chem Soc. 1988;110:6024–6033. doi: 10.1021/ja00226a017. [DOI] [PubMed] [Google Scholar]
- 36.Schelvis JPM, Berka V, Babcock GT, Tsai A. Biochemistry. 2002;41:5695–5701. doi: 10.1021/bi0118456. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Stuehr DJ, Ikeda-Saito M, Rousseau DL. J Biol Chem. 1993;268:22255–22258. [PubMed] [Google Scholar]
- 38.Fan B, Wang J, Stuehr DJ, Rousseau DL. Biochemistry. 1997;36:12660–12665. doi: 10.1021/bi9715369. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Stuehr DJ, Rousseau DL. Biochemistry. 1997;36:4595–4606. doi: 10.1021/bi962309u. [DOI] [PubMed] [Google Scholar]
- 40.Mak PJ, Zhang H, Hollenberg PF, Kincaid JR. J Am Chem Soc. 2010;132:1494–1495. doi: 10.1021/ja910276s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiro TG, Wasbotten IH. J Inorg Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Bridges A, Gruenke L, Chang YT, Vakser IA, Lowe G, Waskell L. J Biol Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 43.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 44.Saribas AS, Gruenke L, Waskell L. Protein Express Purif. 2001;21:303–309. doi: 10.1006/prep.2000.1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.