Abstract
Objective
To determine the genetic cause of Duane’s retraction syndrome (DRS) in two families segregating DRS as an autosomal dominant trait.
Method
Members of two unrelated pedigrees were enrolled in an ongoing genetic study. Linkage analysis was performed using fluorescent microsatellite markers flanking the CHN1 locus. Probands and family members were screened for CHN1 mutations.
Results
Of the six clinically affected individuals in the two pedigrees, three have bilateral and three have unilateral DRS. Both pedigrees are consistent with linkage to the DURS2 locus, one with complete and one with incomplete penetrance. Sequence analysis revealed the pedigrees segregate novel heterozygous missense CHN1 mutations, c.422C>T and c.754C>T, predicted to result in α2-chimaerin amino acid substitutions P141L and P252S, respectively.
Conclusion
Genetic analysis of two pedigrees segregating nonsyndromic DRS reveals two novel mutations in CHN1, bringing the number of DRS pedigrees know to harbor CHN1 mutations, and the number of unique CHN1 mutations, from seven to nine. Both mutations identified in this study alter residues that participate in intramolecular interactions that stabilize the inactive, closed conformation of α2-chimerin, and thus are predicted to result in its hyper-activation. Moreover, amino acid residue P252 was altered to a different residue in a previously reported DRS pedigree; thus, this is the first report of two CHN1 mutations altering the same residue, further supporting a gain-of-function etiology.
Clinical Relevance
Members of families segregating DRS as an autosomal dominant trait should be screened for mutations in the CHN1 gene, enhancing genetic counseling and permitting earlier diagnosis.
Congenital cranial dysinnervation disorders (CCDDs) are associated with abnormal cranial motor neuron and axon development, causing errors in ocular and facial muscle innervation.1 Duane’s retraction syndrome (DRS) is the most common of the CCDDs.2, 3 Individuals with DRS typically present with limitation or absence of globe abduction, variable limitation of adduction, and palpebral fissure narrowing on attempted adduction secondary to globe retraction. Postmortem studies of individuals with DRS have found absence of abducens motor neurons and nerve, and aberrant innervation of the lateral rectus muscle by a branch of the oculomotor nerve.4, 5
We previously reported that seven families segregating DRS as an autosomal dominant trait each harbor a unique heterozygous missense mutations in the CHN1 gene.6 Affected family members had a higher incidence of bilateral DRS and of vertical movement abnormalities than is typical of sporadic DRS,7–9 and magnetic resonance imaging (MRI) of their orbits revealed small or absent abducens nerves and, in some cases, small oculomotor nerves.9 We have also examined a large cohort of individuals with sporadic DRS, and did not identify any CHN1 mutations among them.10
We have now ascertained two additional pedigrees (ABK and ACL) that co-segregate DRS as an autosomal dominant trait, and performed linkage and mutational analysis to determine whether these families segregate CHN1 mutations.
Methods
Two families that segregate DRS as a dominant trait were enrolled in our ongoing genetic study of the CCDDs. The Children’s Hospital Boston institutional review board approved this study and informed consent was obtained from participants and/or their guardians. The probands, their parents, and the half-sibling of the ACL proband underwent ophthalmological examinations with full ocular motility testing. The affection status of the remaining participants was determined by review of ophthalmologic records and/or reported family history. Each participant donated a blood or saliva sample. High molecular weight DNA was extracted from blood or saliva samples by using the Puregene extraction kit (Qiagen, Germany), or the purifier solution (DNA Genotek, Canada), respectively.
Linkage studies were conducted using four fluorescently labeled microsatellite markers spanning CHN1 (D2S2330, D2S326, D2S2314, D2S364). Fluorescently labeled primers were purchased from Invitrogen (Carlsbad, CA), and amplicons were generated by 30 cycles of PCR amplification containing 10–30 ng of genomic DNA in 5-μl reaction volumes of Qiagen’s Hotstar Taq PCR Master Mix containing 2 pmol of each fluorescent primer pair, 1 nmol each of dATP, dTTP, dGTP, and dCTP, and 0.15 U Taq polymerase. The resulting products were analyzed in an Applied Biosystems 3730 DNA Analyzer (Foster City, CA).
The proband from each family was screened for mutations in the thirteen coding exons and exon-intron boundaries of the CHN1 gene (primer sequences available upon request). The amplicons were analyzed using a combination of denaturing high performance liquid chromatography (dHPLC, Transgenomic, Omaha, NE) and direct sequencing as previously reported.6 Variants that were detected by dHPLC were confirmed by direct sequencing. When a sequence variant was identified in the proband, the participating family members and control samples were also examined for the presence or absence of the variant by either dHPLC or direct sequencing.
Results
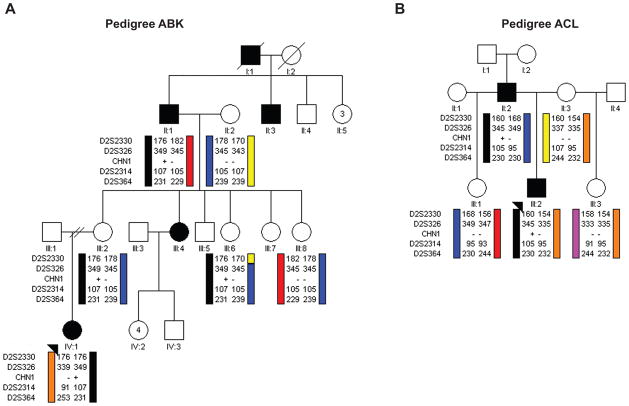
Pedigree ABK is Caucasian of Northern European ancestry, and segregates DRS as an autosomal dominant trait with reduced penetrance in four generations, as described previously by Zhu-Tam et al.11 (Figure 1A). The proband has congenital bilateral limitation of abduction, and globe retraction and narrowing of the palpebral fissure on adduction. Her mother was examined and has no defect in ocular motility, but reported the family history of unilateral left-sided DRS as depicted in Figure 1A. Five family members (2 affected and 3 clinically unaffected) and one spouse participated in the genetic study.
Figure 1. Schematic pedigrees and haplotype analysis of families segregating autosomal dominant DRS at the CHN1 locus.
Pedigree members are denoted by circles (females) and squares (males) and by their generation and position. Black circles/squares indicate clinically affected individuals, and black arrowheads denote the probands. Haplotype analysis of (A) pedigree ABK and (B) pedigree ACL are shown schematically for markers surrounding the CHN1 gene. Genotyping data and schematic segregating haplotype bars for four chromosome 2 markers and CHN1 mutation status are shown below the symbol for each individual who participated in the study. Black bars indicate the affected haplotype that was passed from the affected grandfather in pedigree ABK or the affected father in pedigree ACL. For CHN1, (+) indicates presence of the mutation while (−) indicates its absence. For Pedigree ABK, the affected haplotype and CHN1 mutation are also inherited by two unaffected family members, III:2 and III:6.
Pedigree ACL is a previously unpublished family of African-American ancestry who segregates DRS as an autosomal dominant trait in the proband (II:2) and his father (Figure 1B). The proband has bilateral DRS with esotropia and absent abduction. Globe retraction is present on attempted adduction bilaterally; adduction is full in the right eye and moderately limited with mild upshoot in the left eye. He also demonstrates a chin down posture when attempting fine focusing, with fusion on upgaze. The proband’s father also has bilateral DRS, and had undergone surgery as a child. Both affected and three unaffected family members participated in the study.
Analysis of the four genetic markers flanking CHN1 in each pedigree revealed co-segregation of the DRS phenotype with the CHN1 locus. Pedigree ACL demonstrated complete penetrance. In pedigree ABK both III:2 and III:6 carry the disease-associated haplotype but do not, or are not reported to manifest DRS, respectively.
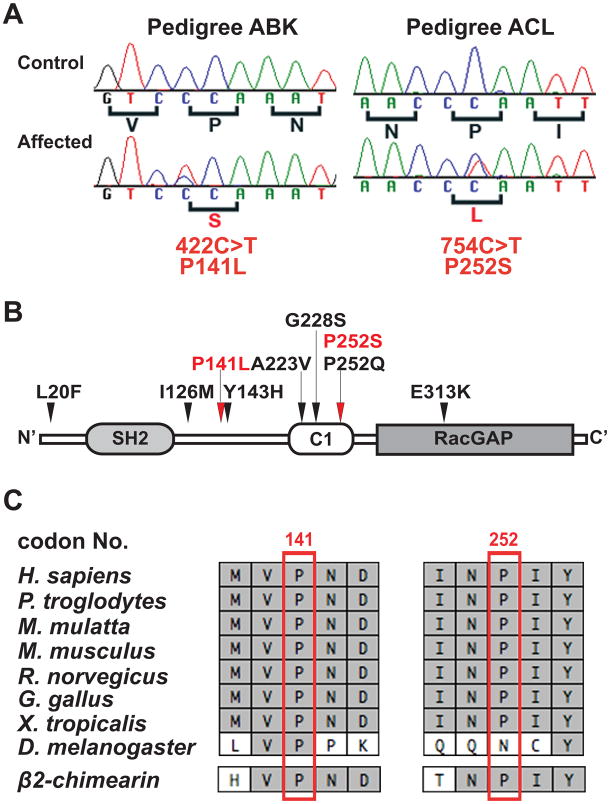
A heterozygous missense mutation (c.422C>T) in CHN1 exon 6 was identified in the ABK proband IV:1 (Figure 2A). This mutation was also present in the affected grandfather, and in the unaffected mother and maternal aunt who harbor the affected haplotype, and absent in the remaining participants from the family. This missense mutation is predicted to result in a conservative amino acid substitution of a non-polar proline to a non-polar leucine at amino acid residue 141 (p.P141L), located in the SH2-C1 linker region of α-chimaerin (Figure 2B).
Figure 2. Nucleotide sequence, amino acid position, and conservation of the CHN1 mutations.
(A) Heterozygous CHN1 mutations in probands of pedigree ABK and ACL. Sequence chromatographs of the control individuals are normal (top row), while sequence chromatographs of the affected individual with DRS (bottom row) each reveals a heterozygous CHN1 mutation. The normal sequence and corresponding amino acid residues are indicated under each control sequence chromatograph (black), while the mutation and resulting amino acid substitution are denoted under each affected sequence (red). (B) Predicted α2-chimaerin protein structure. The amino acid residues altered by the two novel heterozygous mutations are depicted in red, with red arrows above the protein pointing to their predicted positions. The seven previously reported CHN1 mutations are indicated in black and their locations are indicated by black arrows above the protein. (C) Portions of the human CHN1 amino acid sequence that surround each substitution are aligned with homologous sequence in seven different species, followed by alignment with the paralogous β2-chimerin sequence at the bottom. Identical amino acid residues are highlighted in light gray. The residues altered by the two new mutations are boxed in red.
A heterozygous missense mutation (c.754C>T) in CHN1 exon 9 was identified in the ACL proband, III:2 (Figure 2A). The affected father, but none of the unaffected members of the pedigree, also harbored this mutation. This mutation is predicted to result in a non-conservative amino acid substitution of a non-polar proline to a polar serine at amino residue 252 (P252S).
These two missense changes have not been previously reported and are not in SNP databases from UCSC Genome Browser (http://genome.ucsc.edu) or NCBI (www.ncbi.nih.gov/SNP). Using PolyPhen12 (http://genetics.bwh.harvard.edu/pph), P141L is predicted to have a probably damaging impact and P252S is predicted to have a benign impact on the structure and function of α2-chimaerin. Neither change was present on 400 chromosomes of European-derived Caucasian ethnicity and 388 chromosomes of mixed ethnicity. In addition, 754C>T was also absent from 200 chromosomes of African-American ethnicity. α2-chimaerin p.P141 and p.P252 are evolutionally conserved in multiple species and in α2-chimaerin’s close human paralog,β2-chimaerin (Figure 2C).
Comment
We have identified two novel heterozygous missense CHN1 mutations in two dominant DRS pedigrees. Clinical examinations reveal that the probands from both families have isolated bilateral DRS with limited or no abduction and with retraction of the globe and narrowing of the palpebral fissure on attempted adduction. Interestingly, while the affected father of proband in pedigree ACL, II:2, also has bilateral DRS, all three affected relatives of the ABK proband IV:1 have unilateral DRS. In addition, none of the affected family members in either pedigree were noted to have significant errors in vertical motility. Thus, while these DRS phenotypes fall within the spectrum of clinical finding from previously described DRS families carrying CHN1 mutations, they are less atypical than most.6 Similar to AKB III:2 and III:3, we previously reported mutation-positive individuals in whom the DRS phenotype is not penetrant.6 Such clinically asymptomatic patients have not yet undergone detailed MR imaging to determine if they might harbor an endophenotype similar to what we have reported for CFEOM3.13
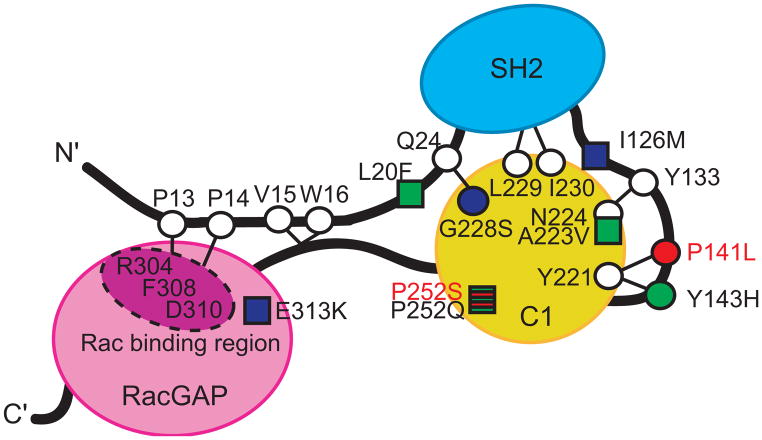
CHN1 encodes the Rac guanosine triphosphatase-activating (RacGAP) signaling molecule α2-chimaerin (Figure 3). When inactive,α2-chimaerin is found in the cytoplasm in a closed conformation. In response to DAG signaling, it unfolds and translocates to the membrane, exposing its RacGAP domain and inactivating Rac. Crystallization of its close relative,β2-chimaerin, and studies of mutant α2- and β2-chimaerin revealed that the inactive closed conformation is maintained by intramolecular interactions that impede access to the Rac and DAG binding sites (Figure 3).6, 14–16 The protein modeling and functional studies of the seven CHN1 mutations we previously reported in DRS pedigrees revealed that each hyperactivatesα2-chimaerin and lowers Rac-GTP levels in the cell, and a subset do so by destabilizing the inactive closed conformation of α2-chimaerin and, thus, increasing its translocation to the cell membrane and its signaling activity.6
Figure 3. Schematic of α2-chimaerin structure in its closed conformation showing the predicted α2-chimaerin intramolecular interactions.
The three domains of α2-chimaerin are depicted as follows: the N-terminal Src homology-2 (SH2) domain is depicted in blue, the C1 domain that binds to the second message signaling lipid diacylglycerol (DAG) is depicted in yellow; and the RacGAP domain that interacts with Rac and down-regulates its activity is depicted in pink. Linker regions are depicted as black lines. Specific amino acid residues are highlighted as circles or squares, with circles representing the positions of amino acids predicted to be involved in intramolecular interactions that stabilize the closed conformation of α2-chimaerin based on homology with β2-chimaerin.14 The seven previously reported mutations alter amino acid residues that are represented by circles or squares filled with green or blue; those filled with green were previously demonstrated to enhance translocation of α2-chimaerin to the membrane when mutated, while those filled with blue did not.6 The circle filled with red and the square striped red and green represent the residues altered by the new novel mutations: P252S alters the same residue as P252Q (thus the residue is striped), while P14lL alters a residue predicted to interact with Y221. Thus, both residues are anticipated to destablize the closed conformation of α2-chimaerin and result in its pathological hyperactivation. Figure adapted with permission from Miyake et al.6
Based on the positions of residues altered by mutations in pedigrees ABK and ACL, we predict that these mutations will behave in a similar fashion to those we reported previously.6 The mutation that segregates in pedigree ACL alters amino acid residue P252, which was also altered by one of the seven original DURS2 mutations6 (Figure 3). The previous pedigree harbored CHN1 755C>A (P252Q), while ACL harbors 754C>T (P252S); both mutations alter the polar uncharged proline in a conserved fashion. We previously established that P252Q enhances the translocation of α2-chimearin to the membrane and lowers Rac-GTP levels in vitro.6 Thus, this is the first report of two DRS mutations altering the same amino acid residue and, because the ACL mutation alters P252 in a similar fashion as the previous mutation, we predict that it will behave in a similar fashion, despite its benign prediction by PolyPhen. Of note, this program also predicts the previous reported I126M amino acid substitution to be benign, despite our finding that it lowers RacGAP levels and causes DRS.6
The mutation that segregates in pedigree ABK alters residue P141. Based on the crystal structure of β2-chimerin,14 we previously predicted that α2-chimerin residues P141 and Y143, both in the SH2-C1 linker region, form intramolecular interactions with residue Y221 in the C1 domain, thus stabilizing the closed conformation of α2-chimaerin6, 14 (Figure 3). Moreover, one of the original seven DURS2 pedigrees harbored the heterozygous CHN1 mutation 427C>T resulting in Y143H, and this mutation enhanced the translocation of α2-chimearin to the membrane and lowered Rac-GTP levels in vitro.6 Thus, we predict that P141L will behave in a similar fashion.
In conclusion, we have identified two novel heterozygous missense CHN1 mutations that cause autosomal dominant bilateral DRS, bringing the total number of known CHN1 mutations to nine. These two new mutations alter residues previously shown to stabilize, or implicated in the stabilization of, the closed conformation of α2-chimearin, and provide further support that the DRS phenotype results from specific CHN1 mutations that hyperactivate the α2-chimearin signaling molecule. While we have previously demonstrated that overexpression of mutant or wildtype α2-chimaerin in the embryonic chick oculomotor nerve results in axon stalling with aberrant branching and/or defasciculation,6 the molecular pathway by which hyperactivation of α2-chimearin in developing abducens and oculomotor axons results in the DRS phenotype has yet to be elucidated.
Acknowledgments
The authors thank the family members for their participation, and Jonathan Horton MD PhD for referring one of the families for this study. This work was supported by NIH R01EY15298 and HD18655 Intellectual and Developmental Disability Research Centers. ECE is an investigator of the Howard Hughes Medical Institute.
References
- 1.Engle EC. The genetic basis of complex strabismus. Pediatr Res. 2006 Mar;59(3):343–348. doi: 10.1203/01.pdr.0000200797.91630.08. [DOI] [PubMed] [Google Scholar]
- 2.Kirkham T. Inheritance of Duane’s syndrome. Br J Ophthalmol. 1970;54:323–329. doi: 10.1136/bjo.54.5.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeRespinis PA, Caputo AR, Wagner RS, Guo S. Duane’s retraction syndrome. Surv Ophthalmol. 1993;38(3):258–288. doi: 10.1016/0039-6257(93)90077-k. [DOI] [PubMed] [Google Scholar]
- 4.Hotchkiss MG, Miller NR, Clark AW, Green WG. Bilateral Duane’s retraction syndrome: A clinical-pathological case report. Arch Ophthalmol. 1980 May;98:870–874. doi: 10.1001/archopht.1980.01020030864013. [DOI] [PubMed] [Google Scholar]
- 5.Miller NR, Kiel SM, Green WR, Clark AW. Unilateral Duane’s retraction syndrome (type 1) Arch Ophthalmol. 1982 Sep;100(9):1468–1472. doi: 10.1001/archopht.1982.01030040446016. [DOI] [PubMed] [Google Scholar]
- 6.Miyake N, Chilton J, Psatha M, et al. Human CHN1 mutations hyperactivate alpha2-chimaerin and cause Duane’s retraction syndrome. Science. 2008 Aug 8;321(5890):839–843. doi: 10.1126/science.1156121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engle EC, Andrews C, Law K, Demer JL. Two Pedigrees Segregating Duane’s Retraction Syndrome as a Dominant Trait Map to the DURS2 Genetic Locus. Invest Ophthalmol Vis Sci. 2007 Jan;48(1):189–193. doi: 10.1167/iovs.06-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung M, Stout JT, Borchert MS. Clinical diversity of hereditary Duane’s retraction syndrome. Ophthalmology. 2000;107(3):500–503. doi: 10.1016/s0161-6420(99)00090-1. [DOI] [PubMed] [Google Scholar]
- 9.Demer JL, Clark RA, Lim KH, Engle EC. Magnetic Resonance Imaging Evidence for Widespread Orbital Dysinnervation in Dominant Duane’s Retraction Syndrome Linked to the DURS2 Locus. Invest Ophthalmol Vis Sci. 2007 Jan;48(1):194–202. doi: 10.1167/iovs.06-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyake N, Andrews C, Fan W, He W, Chan WM, Engle EC. CHN1 mutations are not a common cause of sporadic Duane’s retraction syndrome. Am J Med Genet A. 2010 Jan;152A(1):215–217. doi: 10.1002/ajmg.a.33168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu-Tam LY, Gurwood AS. Bilateral familial Duane’s retraction syndrome. Optometry. 2007 Sep;78(9):465–468. doi: 10.1016/j.optm.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002 Sep 1;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demer JL, Clark R, Tischfield MA, Engle EC. Magnetic Resonance Imaging Evidence Of an Asymmetrical Endophenotype in Congenital Fibrosis of Extraocular Muscles Type 3 Resulting from TUBB3 Mutations. Invest Ophthalmol Vis Sci. 2010 Apr 14; doi: 10.1167/iovs.10-5438. Apr 14 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canagarajah B, Leskow FC, Ho JY, et al. Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin. Cell. 2004 Oct 29;119(3):407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Brown M, Jacobs T, Eickholt B, et al. Alpha2-chimaerin, cyclin-dependent Kinase 5/p35, and its target collapsin response mediator protein-2 are essential components in semaphorin 3A-induced growth-cone collapse. J Neurosci. 2004 Oct 13;24(41):8994–9004. doi: 10.1523/JNEUROSCI.3184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colon-Gonzalez F, Leskow FC, Kazanietz MG. Identification of an autoinhibitory mechanism that restricts C1 domain-mediated activation of the Rac-GAP alpha2-chimaerin. J Biol Chem. 2008 Dec 12;283(50):35247–35257. doi: 10.1074/jbc.M806264200. [DOI] [PMC free article] [PubMed] [Google Scholar]