Abstract
The presentation of tumor antigen-derived peptides by human leukocyte antigen (HLA) class I surface antigens on tumor cells is a key prerequisite to trigger effective T-cell responses in cancer patients. Multiple complementary strategies like cDNA and serological expression cloning, reverse immunology and different ‘ome’-based methods have been employed to identify potential T-cell targets. This report focuses on a ligandomic profiling approach leading to the identification of 49 naturally processed HLA class I peptide ligands presented on the cell surface of renal cell carcinoma (RCC) cells. The source proteins of the defined HLA ligands are classified according to their biological function and subcellular localization. Previously established cDNA microarray data of paired tissue specimen of RCC and renal epithelium assessed the transcriptional regulation for 28 source proteins. In addition, HLA-A2-restricted, peptide-specific T cells directed against a HLA ligand derived from sulfiredoxin-1 (SRXN1) were generated, which were able to recognize and lyse ligand-presenting target cells in a HLA class I-restricted manner. Furthermore, tumor-infiltrating T cells isolated from a RCC patient were also able to kill SRXN1 expressing tumor cells. Thus, this experimental strategy might be suited to define potential candidate biomarkers and novel targets for T-cell-based immunotherapies of this disease.
Keywords: Biomarkers, Cell biology, HLA antigens, Peptide ligands, Renal cell carcinoma, T-cell response
1 Introduction
Human leukocyte antigen (HLA) class I molecules display peptides mainly derived from the processing of intracellular generated proteins for their presentation to CD8+ cytotoxic T lymphocytes (CTL) [1, 2]. Whereas cells presenting self-antigens via HLA class I molecules remain unharmed, cells exhibiting peptide ligands unique to diseased or transformed cells become targets for T-cell responses [1, 2]. Thus, the development of immunotherapeutic strategies that target such exclusively presented ligands by specifically inducing CTL responses directed against neoplastic cells requires the identification of tumor-associated antigens (TAA) or epitopes that can distinguish between healthy and malignant cells.
These TAA can be classified into five major categories: (i) cancer testis (CT) antigens, which are specifically expressed in tumors, but not in normal tissue with the exception of testis; (ii) unique antigens derived from mutations; (iii) antigens, which are misfolded or overexpressed, aberrantly degraded or aberrantly post-translationally modified; (iv) differentiation antigens and (v) viral antigens [3–5]. The identification of tumor-specific targets for T-cell-mediated immunity may lead to the definition of novel biomarkers for cancer diagnosis, prognosis and tumor classification as well as for the monitoring of immune responses, of the modulation of immune recognition and its regulation during cancer progression.
So far, a number of different methods have been employed to define potential candidate antigens suitable for mounting immune responses, such as the serological identification of antigens by recombinant expression cloning (SEREX), cDNA cloning and reverse immunology [6–8], cDNA microarrays and proteome-based approaches [9–13]. The latter also include phage display, protein microarrays and in particular quantitative proteome-based approaches, such as DIGE, ICAT, iTRAQ or ICPL as well as PROTEO-MEX, a combination of classical 2-DE-based proteome analysis with serology [14–17]. A more comprehensive method is the systematic identification of peptide ligands by their elution from HLA molecules followed by mass spectrometry, which allows the exploration of the repertoire of self and non-self derived peptides presented by HLA class I molecules [18–25]. The identification of such naturally processed CTL epitopes benefits from the increased sensitivity of mass spectrometry, which now allows the successful identification of single epitopes from highly complex peptide mixtures [26–29]. Despite the requirement of a relatively high number of cells, the peptide elution-based ligandomic approach has a significant advantage over the cDNA and SEREX-based antigen identification strategies, since it even allows the direct identification of unmodified and post-transcriptionally modified T-cell epitopes [30, 31].
In renal cell carcinoma (RCC), various strategies have been employed for the identification of putative candidate biomarkers, which upon their verification and validation might be implemented for early diagnosis, for improving prognosis or as potential therapeutic targets for this disease. A number of differentially expressed proteins have been described, but their implementation as accurate and stable markers allowing early detection or prognosis of disease has failed so far [32–37]. Although novel targeted therapies such as monoclonal antibodies or tyrosine kinase inhibitors have revolutionized the treatment options for RCC patients, the discovery of putative biomarkers in RCC for the determination and monitoring of therapy efficacy and immune response is still urgently required [38–41].
Exploring the HLA ligandome of RCC cells based on a peptide release approach in combination with an ESI MS/ MS-based identification strategy led to the definition of a panel of 49 naturally processed HLA class I ligands. Thus, the current report extends the still limited pool of potential targets for T-cell recognition and provides at least initial evidence that this approach can lead to putative biomarkers and/or functional vaccination strategies in RCC.
2 Materials and methods
2.1 Tissue culture of RCC cell lines
The RCC cell line MZ2733RC was established from a biopsy specimen of a patient with an undifferentiated primary RCC of the clear cell subtype as previously described [42–44]. The MZ2733RC cells were grown in RPMI 1640 supplemented with 10% FCS, 2 mM L-glutamine, 100 IU/mL penicillin, 100 μg/mL streptomycin, 10 mM sodium pyruvate, 100 mM HEPES and 1 × non-essential amino acids (Gibco Life Technology, Karlsruhe, Germany) in cell factory cultures (Nunc, Roskilde, Denmark). The HLA phenotype of this cell line is A*02:01/A*01, B*07:02/B*49 and C*05:01/Cw*07:02 (http://hla.alleles.org/).
A panel of human RCC cell lines (MZ1275RC, MZ2733RC, MZ2858RC, MZ2861RC, MZ2905RC and HAL31RC) established from patients with primary RCC and three corresponding SV40LT-transformed normal kidney epithelium representing cell lines [43] were cultured in complete medium. For SV40LT transformants, the medium was further supplemented with 600 μg/mL hygromycin B (Roche Diagnostics, Mannheim, Germany). T2 cells [45] were grown in RPMI supplemented with 10% FCS, 2 mM L-glutamine and respective antibiotics.
2.2 Tumor specimen
The tissue specimen used for the establishment of the transcriptomic profiling data has been previously described and was collected upon routine surgery and directly frozen until further use. Tissue procurement followed standard ethical procedures according to the institutional policy [44].
2.3 Elution of HLA class I-presented peptides and analysis of cell culture supernatants
5 × 109 MZ2733RC cells were employed for the extraction of naturally processed HLA class I-binding peptides. These HLA class I-presented peptides were obtained by immunoprecipitation of HLA molecules from MZ2733RC cells according to a slightly modified protocol [46, 47] using the pan-specific-anti-HLA-A-, HLA-B-, HLA-C-specific antibody W6/32 [48], protein A affinity chromatography, acid treatment and ultrafiltration. In addition, 4 L of cell culture supernatant of MZ2733RC cells collected during the cell expansion process was analyzed for the presence of HLA class I-binding peptides. The supernatant was ultracentrifuged, filtrated to remove cellular debris and then used for immunoprecipitation as described above.
2.4 Microcapillary LC-MS
The HLA class I-eluted peptide pool was analyzed by a reversed phase ultimate high-performance liquid chromatography (RP-HPLC) system (Dionex, Amsterdam, The Netherlands) coupled to a hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Q-TOF, Micromass, Manchester, UK) equipped with a micro-ESI source [49]. Samples were loaded onto a C18 pre-column for concentration and desalting. After loading, the pre-column was placed in line for the separation by a fused-silica microcapillary column (75 μm id × 250 mm) packed with 5 μm C18 reversed-phase material (Dionex). Solvent A was 4 mM ammonium acetate/water. Solvent B was 2 mM ammonium acetate in 80% acetonitrile/water. Both the solvents were adjusted to pH 3.0 with formic acid. A binary gradient of 15–40% B within 120 min was performed, applying a flow rate of 200 μL/min reduced to approximately 300 nL/min by the Ultimate split system. A gold-coated glass capillary (PicoTip, New Objective, Cambridge, MA, USA) was used for introduction into the micro-ESI source. The integration time for the TOF analyzer was 1 s with an interscan delay of 0.1 s. For online microcapillary HPLC MS/MS experiments, the integration time for the TOF analyzer was 4 s with an interscan delay of 0.1 s. Fragmentation of the parent ion was achieved by automatic MS/MS switching [49].
2.5 Peptide sequence analysis
Peptide sequence analysis was manually carried out. Therefore, MS/MS spectra were smoothed using MassLynx 4.0 software (Savitzky-Golay, three smooth windows and two smooths). For manual peptide identification, sequence tag searches were done using the MASCOT 2.0 software (peptide and MS/MS tolerance, 0.2 Da; National Center for Biotechnology Information non-redundant (NCBInr) database and relevant hits were manually assessed. Criteria for manual identification were a reasonable interpretation of at least 95% of all fragment peaks, complete sequence coverage with MS/MS fragments and signal intensities of fragment ion peaks that match breakage probabilities of the respective sequence [25]. To assess the false-positive rate of peptide identification, a total number of seven peptides, including STDHIPILY, NTDSPLRY, ALADGVQKV, VLIPKLPQL, RPELVRPAL, RPTLWAAAL and TLSDLRVYL, were chemically synthesized and fragmentation spectra of these synthetic peptides were compared with that recorded from manual analyses. All synthetic peptides showed exactly the same fragmentation pattern as the manually identified peptides demonstrating a false-positive rate in our sequence analysis of <1% [25].
2.6 Transcriptomic profiling
Total RNA was extracted from frozen tissue specimen using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and amplified into anti-sense RNA (aRNA) as previously described [44]. Test and reference RNAs were labeled with Cy5 (red) and Cy3 (green) and co-hybridized to custom-made 17.5K cDNA microarrays. Quality validation as well as unsupervised and supervised analysis were performed as previously outlined [44].
2.7 Data mining
The raw data obtained from the identification of HLA ligands and the transcriptomic profiling of a panel of paired tissue samples representing renal cancer along with tumor adjacent renal epithelium were analyzed with an in-house developed web-based tool, programmed in php and linked to a mySQL database as recently described [37]. For the HLA ligands also, the SYFPEITHI database (www.syfpeithi.de) was employed to obtain information about the amino acid anchor residues of given HLA class I alleles [50]. The heterogeneous data sets of the genes/proteins identified were linked to the Swiss-Prot identities (UniProt Knowledgebase Release 2010_12) serving as major source for information regarding their chromosomal localization, gene ontology (geneontology.org), gene/ protein function and cellular localization.
2.8 Real-time quantitative RT-PCR (qPCR) analyses
qPCR analyses were carried out as previously described [51]. Briefly, the total RNA was extracted from the samples using the RNeasy Mini Kit (Qiagen, Hilden Germany) according to the manufacturer’s instructions. cDNA was synthesized from 2 μg RNA treated with DNase I (Invitrogen) using oligo-(dT)18 primers (Promega, Mannheim, Germany) and Superscript II reverse transcriptase (Invitrogen). Real-time quantitative PCR was performed with the sulfiredoxin-1 (SRXN1)-specific primer set (sense: 5′-CATCGATGTCC-TCTGGATCA-3′; anti-sense: 5′-TGCAAGTCTGGTGTG-GATG-3′) using an annealing temperature of 60°C. Amplification of β-actin served as reference gene (sense: 5′-GAAGCATTTGCGGTGGACGAT-3′; anti-sense: 5′-TCC-TGTGGCATCCACGAAACT-3′). All real-time PCR analyses were performed in a thermo cycler (Rotorgene, Corbett Life Science, Sydney, Australia) using the QuantiTect SYBR-Green PCR Kit (Qiagen). SRXN1 mRNA expression levels were normalized against β-actin.
2.9 In silico target validation
The protein expression pattern of defined HLA ligands was assessed using the immunohistochemical stainings of tissue sections representing normal renal epithelium and renal cancers accessible via the Human Protein Atlas (www.proteinatlas.org, Version 7.0) [52].
2.10 Induction of SRXN1-specific CTL and cytotoxicity assay
CD8+ T cells were purified from peripheral blood mononuclear cells (PBMC) of HLA-A2+ healthy donors by density gradient centrifugation on a Pancoll layer (PAN-Biotech, Aidenbach, Germany) followed by purification using anti-CD8 magnetic beads (Miltenyi Biotech, Cologne, Germany). HLA-A2-restricted CTL specific for the SRXN1-derived peptide ligand p114-122 (TLSDLRVYL (Table 1); purchased from Peptides & Elephants, Potsdam, Germany) were induced by three weekly stimulations with irradiated (30 Gy) peptide-pulsed autologous peripheral blood lymphocytes (PBL) in X-VIVO 15 (Lonza, Basel, Switzerland). IL-2 (150 U/mL; Chiron) and IL-7 (10 ng/mL, Immunotools, Friesoythe, Germany) were added 3 days after the first simulation and 2 days after the two other stimulations.
Table 1.
Characteristics of RCC-presented peptides identified by MS
| Peptide | HLA | Protein | Gene ID | Gene symbol | UniProtKB ID | Position | RT exp. (min) | RT calc. (min) | (M+H)+ calc. | (M+2H)2+ calc. | (M+2H)2+ obs. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LTYEALGLCP | n.d. | Sterol carrier protein 2 | 6342 | SCP2 | P22307 | 56–65 | 54.7 | 43.9 | 1079.537 | 540.2683 | 540.29 |
| STDHIPILY | A*01 | Glutamine-fructose-6-phosphate transaminase 1 | 2673 | GFPT1 | Q06210 | 218–226 | 80.2 | 52.1 | 1058.544 | 529.772 | 529.77 |
| NTDSPLRY | A*01 | Ribosomal protein SA | 3921 | RPSA | P08865 | 149–156 | 31.6 | 21.6 | 965.4611 | 483.2306 | 483.24 |
| ALADGVQKV | A*02 | Apolipoprotein L, 1 | 8542 | APOL1 | O14791 | 160–168 | 27.8 | 13.9 | 900.5073 | 450.7537 | 450.74 |
| SIFEVVWAI | A*02 | Calcium channel, voltage-dependent, α 1E subunit | 777 | CACNA1E | Q15878 | 462–470 | 69.6 | 57.4 | 1063.575 | 532.2874 | 532.29 |
| LLMVLSPRL | A*02 | Clathrin, heavy chain | 1213 | CLTC | Q00610 | 1422–1430 | 122.2 | 56.7 | 1041.641 | 521.3207 | 521.34 |
| RLLDVLAPL | A*02 | Collagen, type XVIII, α 1 | 80781 | COL18A1 | P39060 | 14–22 | 138.1 | 50.4 | 1009.633 | 505.3164 | 505.34 |
| AGLPRPDLSL | A*02 | Hypothetical LOC400614 | 400614 | LOC400614 | XP_378693 | 2–11 | 51.3 | 40.9 | 1038.587 | 519.7933 | 519.78 |
| RIGSGLKALL | A*02 | Chromosome 2 open-reading frame 54 | 79919 | C2orf54 | Q53QU5 | 219–228 | 75.3 | 35.8 | 1027.655 | 514.3273 | 514.28 |
| RLLEVPVML | A*02 | LSM8 homolog, U6 small nuclear RNA associated | 79763 | LSM8 | O95777 | 49–57 | 135.8 | 46.2 | 1069.636 | 535.3182 | 535.35 |
| VLIPKLPQL | A*02 | ORM1-like 3 (S. cerevisiae) | 94103 | ORMDL3 | Q8N138 | 118–126 | 124.3 | 61.9 | 1020.674 | 510.8369 | 510.84 |
| LVLMVLYLI | A*02 | Phosphatidylinositol glycan anchor biosynthesis, class M | 93183 | PIGM | Q9H3S5 | 153–161 | 70.6 | 80.2 | 1076.671 | 538.8356 | 538.79 |
| VMLDVPIRL | A*02 | RAS protein activator like 2 | 9462 | RASAL2 | Q9UJF2 | 855–863 | 129.5 | 52.7 | 1055.621 | 528.3103 | 528.3 |
| IMLEALERV | A*02 | Small nuclear ribonucleoprotein polypeptide G | 6637 | SNRPG | P62308 | 68–76 | 103 | 29.1 | 1073.595 | 537.2975 | 537.31 |
| TLSDLRVYL | A*02 | Sulfiredoxin 1 | 140809 | SRXN1 | Q9BYN0 | 114–122 | 114.7 | 38.5 | 1079.602 | 540.301 | 540.32 |
| RVMAPRALL | A*02/ B*07 | Major histocompatibility complex, class I, C | 3107 | HLA-C | P10321 | 2–10 | 45.7 | 35.7 | 1026.617 | 513.8083 | 513.77 |
| TAHLMVVVL | A*02/ B*07 | Olfactory receptor, family 13, subfamily F, member 1 | 138805 | OR13F1 | Q8NGS4 | 242–250 | 53.7 | 45.7 | 982.5679 | 491.784 | 491.75 |
| APRAVFPSI | B*07 | Actin, α 1, skeletal muscle | 58 | ACTA1 | P68133 | 28–36 | 71.9 | 43.1 | 957.5441 | 479.2721 | 479.26 |
| RPELVRPAL | B*07 | Aldo-keto reductase family 1, member C3 | 8644 | AKR1C3 | P42330 | 91–99 | 39.3 | 29.2 | 1050.634 | 525.8172 | 525.79 |
| FPNIPGKSL | B*07 | Degenerative spermatocyte homolog 1, lipid desaturase | 8560 | DEGS1 | O15121 | 265–273 | 59.4 | 49.2 | 972.5436 | 486.7718 | 468.75 |
| VPATDRNAL | B*07 | Eukaryotic translation initiation factor 3, subunit 6 48 kDa | 3646 | EIF3S6 | P60228 | 109–117 | 23.1 | 20.2 | 956.5084 | 478.7542 | 478.77 |
| PSRDSLPLPV | B*07 | G-protein signaling modulator 1 (AGS3-like, C. elegans) | 26086 | GPSM1 | Q86YR5 | 418–427 | 123.1 | 39.1 | 1080.597 | 540.7986 | 540.84 |
| RPQKVCSFL | B*07 | Guanine nucleotide binding protein (G protein), γ 5 subunit | 79218 | Gng5 | P63218 | 60–68 | 46.5 | 25.6 | 1077.58 | 539.2899 | 539.31 |
| MPRGVVVTL | B*07 | HECT domain containing 1 | 25831 | HECTD1 | Q9ULT8 | 851–859 | 69.7 | 38 | 971.5631 | 486.2816 | 486.28 |
| RPTLWAAAL | B*07 | Insulin-like growth factor binding protein 3 | 3486 | IGFBP3 | P17936 | 5–13 | 95.6 | 49.3 | 998.5706 | 499.7853 | 499.79 |
| APRTLVLLL | B*07 | Major histocompatibility complex, class I, A | 3105 | HLA-A | P16189 | 5–13 | 130.8 | 44.7 | 995.6535 | 498.3268 | 498.34 |
| IPRSITVLV | B*07 | Minichromosome maintenance complex component 7 | 4176 | MCM7 | P33993 | 73–81 | 82.6 | 48 | 997.6328 | 499.3164 | 499.31 |
| IPRAALLPL | B*07 | HtrA serine peptidase 1 | 5654 | HTRA1 | Q92743 | 3–11 | 94.6 | 59.6 | 963.6273 | 482.3137 | 482.31 |
| LPKQPPLAL | B*07 | RAN binding protein 9 | 10048 | RANBP9 | Q96S59 | 691–699 | 64.1 | 54.7 | 976.6112 | 488.8056 | 488.8 |
| GPRAVFVLL | B*07 | Transcription factor binding to IGHM enhancer 3 | 7030 | TFE3 | P19532 | 18–26 | 125.5 | 49.9 | 971.5961 | 486.2981 | 486.28 |
| APRPGLLSL | B*07 | Widely interspaced zinc finger motifs | 58525 | WIZ | Q96IG5 | 80–88 | 76.3 | 43.7 | 923.5596 | 462.2798 | 462.3 |
| ISVGISLLLL | B*07? | GRB2-binding adaptor protein, transmembrane | 202309 | GAPT | Q8N292 | 12–21 | 101.3 | 65.5 | 1027.668 | 514.3342 | 514.31 |
| LSLENLEKI | B*07? | Inositol polyphosphate-5-phosphatase F | 22876 | INPP5F | Q5W135 | 667–675 | 127.6 | 24.8 | 1058.602 | 529.8008 | 529.82 |
| REAPSPLMI | B*49 | Bromodomain-containing 4 | 23476 | BRD4 | O60885 | 1060–1068 | 66.5 | 35.6 | 1013.537 | 507.2687 | 507.25 |
| LEVIPRTLI | B*49 | Chaperonin-containing TCP1, subunit 3 γ | 7203 | CCT3 | P49368 | 406–414 | 104.4 | 50.9 | 1053.659 | 527.3295 | 527.33 |
| GEYPKLLRL | B*49 | Component of oligomeric golgi complex 5 | 10466 | COG5 | Q9UP83 | 411–419 | 68.6 | 38.3 | 1088.639 | 544.8193 | 544.83 |
| TEITHAVVI | B*49 | Eukaryotic translation initiation factor 3, subunit 8, 110 kDa | 8663 | EIF3S8 | Q99613 | 322–330 | 49 | 32.9 | 982.5492 | 491.7746 | 491.75 |
| AEFIKFTVI | B*49 | GTF2IRD2 β | 389524 | GTF2IRD2B | Q6EKJ0 | 397–405 | 129.6 | 51.9 | 1067.606 | 534.303 | 534.31 |
| KEFDGKSLV | B*49 | Heat shock protein 90 kDa α, class B member 1 | 3326 | HSP90AB1 | P08238 | 526–534 | 31.6 | 12.8 | 1022.544 | 511.7721 | 511.77 |
| RESFSLVQV | B*49 | HLA-B-associated transcript 3 | 7917 | BAT3 | P46379 | 820–828 | 76.3 | 15.1 | 1064.566 | 532.783 | 532.81 |
| GEFIIGRVI | B*49 | LIM and senescent cell antigen-like domain 2 | 55679 | LIMS2 | Q7Z4I7 | 80–88 | 107.2 | 46.1 | 1003.586 | 502.293 | 502.29 |
| RESFPMILV | B*49 | Muscle RAS oncogene homolog | 22808 | MRAS | O14807 | 116–124 | 134.6 | 44.9 | 1091.584 | 546.2922 | 546.31 |
| IEIERILSV | B*49 | Myosin IE | 4643 | MYO1E | Q12965 | 816–824 | 114.7 | 32 | 1071.633 | 536.3167 | 536.33 |
| REFLFNAI | B*49 | Ribonucleotide reductase M2 B (TP53 inducible) | 50484 | RRM2B | Q7LG56 | 152–159 | 113.8 | 42.4 | 1009.539 | 505.2696 | 505.28 |
| VEVGRVAYV | B*49 | Ribosomal protein L14 | 9045 | RPL14 | P50914 | 7–15 | 29.5 | 15.3 | 991.5497 | 496.2749 | 496.26 |
| REMIPFAVV | B*49 | Septin 9 | 10801 | Sep 09 | Q9UHD8 | 485–493 | 114.7 | 41.8 | 1061.574 | 531.2869 | 531.29 |
| GEHTLLVTV | B*49 | Similar to CG14977-PA | 392758 | LOC392758 | XP_374498 | 77–85 | 63 | 28.9 | 968.5335 | 484.7668 | 484.77 |
| REANLQALI | B*49 | Ubiquitin 1 | 29979 | UBQLN1 | Q9UMX0 | 562–570 | 73.2 | 22 | 1027.582 | 514.291 | 514.25 |
| IYAHGALPII | HLA-C? | Progestin and adipo Q receptor family member IV | 124222 | PAQR4 | Q8N4S7 | 52–61 | 70.7 | 68.2 | 1067.617 | 534.3086 | 534.31 |
The table lists the identified peptide sequence along with the respective HLA class I restriction element, the protein name, the gene ID, gene symbol, the UniProtKB ID, the relative position of the identified peptide within the open-reading frame, the experimental (RT exp.) and calculated (RT calc.) retention time of the peptide within the total ion current as well as its calculated/observed ion masses (M+H); n.a., not assigned.
Subcloning of antigen-specific T cells was performed by seeding 5 cells/well in 96-well U-bottom plates together with 1 × 105 HLA-A2+, peptide-pulsed, irradiated PBMC in the presence of 150 U/mL IL-2. After 2 wk, growing clones were further expanded with IL-2 and pulsed PBMC before their cytotoxic activity was determined via a 4-h standard 51Cr release assay.
Briefly, bulk CTL cultures and expanded clones from healthy donors were subjected to a standard 51Cr release assay using T2 cells pulsed with an irrelevant or the SRXN1-derived p114–122 peptide, respectively. In addition, tumor infiltrating lymphocytes (TIL), characterized at the time of their asservation in regard to the relative frequency of distinct lymphocyte subsets represented in the given TIL culture, were used as effector cells and either co-incubated with HLA-A2+ SRXN1-expressing RCC cell line MZ2905RC or the HLA-A2− RCC cell line HAL31RC (negative control) as target cells at different effector to target (E/T) ratios. Specific lysis was calculated as 100 × (experimental lysis–-spontaneous release)/(maximum release–spontaneous release), where spontaneous and maximum release were obtained by incubating target cells in medium alone or supplemented with 1% Triton X-100, respectively.
3 Results
3.1 Identification of HLA-A, -B and -C presented peptides from the ccRCC cell line MZ2733RC by MS
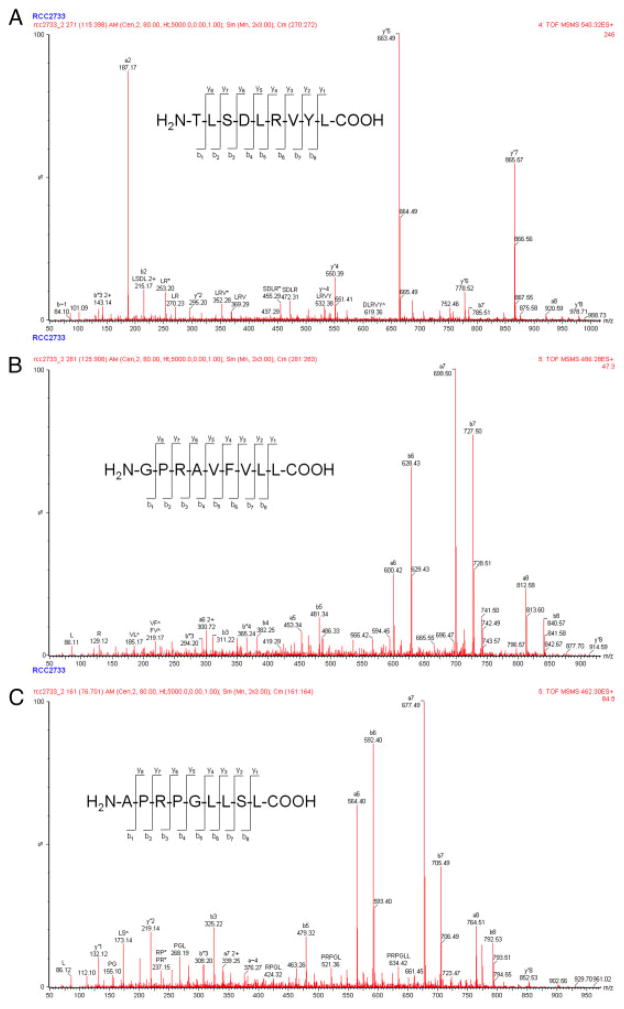
By applying a combination of immunoprecipitation of detergent-solubilized HLA-peptide complexes, followed by their affinity chromatography purification, acid elution of peptides, separation of the resulting peptide pool via microcapillary RP-HPLC chromatography and subsequent identification via ESI MS/MS (Supporting Information Fig. 1) 49 HLA class I-presented peptides representing members of the ligandome of MZ2733RC cells were successfully identified (Table 1). These include the HLA-A*02-presented ligand derived from SRXN1 and the HLA-B*07-presented ligands derived from TFE3 and WIZ, for which representative annotated MS/MS spectra are shown in Fig. 1. The total ion current profile for the pool of peptide ligands isolated from MZ2733RC cells along with one of four MS/MS chromatograms to detect doubly charged ions at m/z 300–550 is provided as Supporting Information Fig. 2.
Figure 1.
Representative MS/MS spectra of peptides eluted from HLA class I molecules. MS/MS spectra annotated with the fragment assignments of peptides. Resulting peptide sequences leading to the identification of SRXN1 (A), TFE3 (B) and WIZ (C), respectively.
Overall, the pool of peptide ligands identified was presented by various HLA class I alleles with all assignments in line with the HLA phenotype of MZ2733RC cells (HLA-A1, -A2, -B7 and -B49). Supporting Information Table 1 summarizes information about the source proteins for the panel of identified HLA ligands such as their name, UniProtKB and gene IDs and their chromosomal localization. For approximately 64% of the source proteins defined as naturally presented on MZ2733RC cells independently defined HLA ligands have already been previously described (Supporting Information Table 1) [9, 13, 24, 28, 47, 50, 53, 54]. In addition, some of the source proteins identified yield antigenic peptides presented by various HLA class I alleles (Supporting Information Table 1). Bioinformatic analysis based on gene ontology information demonstrated that the most frequent characteristic of the source proteins from the identified HLA ligands is protein binding, followed by catalytic activity, nucleic acid/nucleotide, metal ion and structural molecule binding (Supporting Information Table 2A). At the functional level, the source proteins represent components of the antigen processing machinery, of the cytoskeleton and of signal transduction pathways, metabolic enzymes, proteins involved in cell proliferation, protein synthesis and folding, membrane proteins, stress proteins, proteins associated with DNA/RNA processing and transcription factors as well as proteins of yet unknown function. As expected, the majority of the presented peptides are derived from intracellular source proteins (Supporting Information Table 2B). In terms of their cellular localization, the panel of source proteins for the identified HLA ligands is distributed across a number of cellular compartments. The most frequent cellular compartment for the source proteins is the cytoplasm followed by membranes, the nucleus, the cytoskeleton, the ER, the extracellular space and the mitochondria, whereas for a few of the identified source proteins the exact cellular localization is as yet not known. Therefore, even this relatively small panel of source proteins indicates that HLA ligands can not only be derived from proteins of quite different cellular functions but also from almost every cellular compartment.
In order to determine whether the HLA ligands defined on MZ2733RC cells represent naturally processed peptides generated via the classical MHC class I antigen processing pathway, the peptide content of the MZ2733RC cell culture supernatant was also analyzed. Only one single peptide with the sequence KEIFLRELI was recovered in the supernatant of MZ2733RC cells. Employing BLAST alignment, this HLA-B49-presented peptide is either derived from a heat shock protein (HSP) 90 family member, such as HSP90 α (P07900; aa 41–49), HSP90 β (P08238, aa 36–44) or grp94 also known as endoplasmin (P14625, aa 97–105). These data confirm that the HLA class I ligands identified in MZ2733RCC cells represent naturally processed antigens and not peptides derived from necrotic cells or proteolytic degradation processes of medium components. Thus, there is very little evidence for the presence of free peptides within the cell culture supernatant, which might compete for binding to HLA molecules on the cell surface of MZ2733RC cells.
3.2 Analysis of the transcriptional regulation for source proteins defined by HLA ligandomics
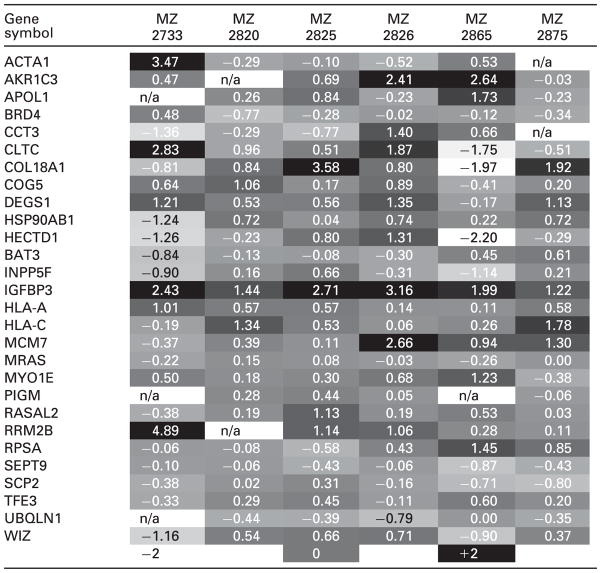
In order to classify the identified source proteins as oncoantigens and/or potential targets for immunotherapy, their transcriptional regulation was evaluated. Based on the availability of previously performed transcriptomic data sets of paired RCC tissue samples including the MZ2733 system [44], the transcriptional regulation for those genes encoding source proteins of HLA class I ligands was reassessed. Overall, 28/49 genes encoding for source proteins were represented on the cDNA microarray, whereas for the remaining 22 potential target structures corresponding probes were missing on these custom-made arrays [44]. The heat map (Table 2) summarizes their transcriptional regulation in the MZ2733 system as well as in five additional members of undifferentiated RCC lesions (Supporting Information Table 3). Interestingly, 2/28 of the genes encoded for proteins represented as HLA ligands: the minichromosome maintenance complex component 7 (MCM7) and the insulin-like growth factor binding protein 3 (IGFBP3) are members of a previously defined clusterogram comprising 48 genes, which are differentially expressed in undifferentiated RCC [44]. Moreover, two other genes encoding source proteins of HLA ligands, namely aldo-keto reductase family 1 member C3 (AKR1C3) and chaperonin-containing TCP1 subunit 3 γ (CCT3) are at least functionally closely related to members of this clusterogram specified as AKR1A1 and CCT5. HLA-A- and HLA-C-derived HLA ligands are also in line with the previously established cDNA array data being classified as genes frequently upregulated in RCC [44]. Moreover, the degenerative spermatocyte homolog 1 (DEGS1), initially identified as predominantly upregulated in tumors other than RCC, is also upregulated in undifferentiated RCC lesions [44, 55]. In the MZ2733 system, CTLC, ACTA1, DEGS1, IGFBP3, HLA-A and RRM2B are upregulated and CCT3, HSP90AB1, HECTD1 and WIZ are downregulated, whereas MCM7, HLA-C and AKR1C3 are not regulated.
Table 2.
Overlap of targets shared between the ligandomics and microarray analyses
|
The table lists the gene symbols along with the log fold changes defined in a subset of six previously analyzed tissue systems comprising renal cancer versus tumor adjacent normal renal epithelium [44]. The bottom line defines the coding of the regulation mode. Data that were not interpretable are marked by n/a.
3.3 Target verification via the Human Protein Atlas and by qPCR
Next to the microarray profiling data covering 28 of the 49 genes encoding for source proteins defined via the HLA ligandomics approach, the protein expression pattern in normal renal epithelium and renal cancers was assessed by screening target protein-specific immunohistochemical stainings provided by the Human Protein Atlas. For 30 of the 49 source proteins identified by the HLA ligandomics approach (Table 1), target-specific antibodies along with representative immunohistochemistry (IHC) are available (Supporting Information Table 4) and classified according to distinct validation criteria. Antibodies targeting SCP2, RPSA, COL18A1, RASAL2, AKR1C3, DEGS1, GNG5, HECTD1, IGFBP3, WIZ, GAPT, BRD4, CCT3, HSP90AB1 or MYO1E received either a very low or low validation score and are therefore classified as uncertain. In contrast, antibodies targeting APOL1, CLTC, LSM8, ACTA1, EIF3E, MCM7, HTRA1, RANBP9, TFE3, COG5, BAT3, RRM2B or RPL14 and UBQLN1 had a medium or high validation score. For the source proteins CLTC and MCM7, even several informative antibodies are listed in the depository of the Human Protein Atlas. IHC stainings classified by the Human Protein Atlas by a medium and high validation score are considered as supportive. Two representative IHC patterns for the latter subgroups either targeting BAT3 (medium validation score) or MCM7 (high validation score) are provided in Supporting Information Figs. 3 and 4.
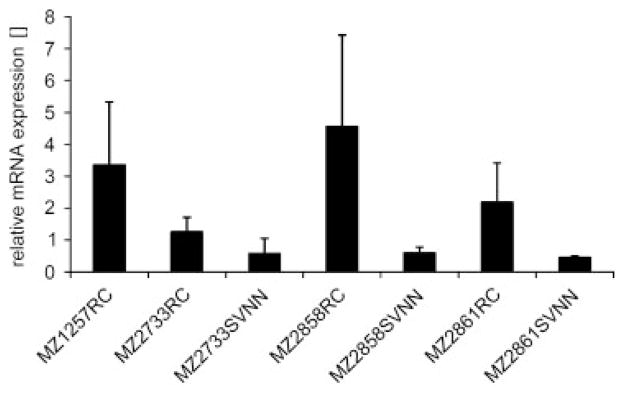
So far, SRXN1 was neither represented on the previously performed cDNA microarrays nor in the antibody depository of the Human Protein Atlas. Since metabolic changes frequently occur in tumor lesions either mediated by the tumor itself or by infiltrating immunosuppressive monocytes, such as myeloid-derived suppressor cells or tumor-associated macrophages, the transcriptional regulation of the target SRXN1, which might protect tumor cells from oxidative stress was analyzed by comparative qPCR analyses of RCC cell lines and corresponding tumor adjacent renal epithelial representing cells. As shown in Fig. 2, SRXN1 transcription was increased in RCC cells when compared with the corresponding tumor adjacent renal epithelium cultures, which might contribute to its presentation by RCC cells as an overexpressed protein.
Figure 2.
Representative qPCR-based expression analyses for sulfoxiredoxin 1 (SRXN1) in paired RCC and corresponding tumor adjacent renal epithelium representing cell lines. q-PCR analyses of three paired RCC (MZ2733RC, MZ2858RC and MZ2861RC) and corresponding SV40TL transformed tumor adjacent renal epithelium representing cell lines (MZ2733SVNN, MZ2858SVNN and MZ2861SVNN) along with the RCC cell line MZ1257RC, here serving as positive control, were performed as described in Section 2. The graph represents means and standard deviations of at least two independent biological replicates.
3.4 Functional relevance of HLA class I-presented peptides
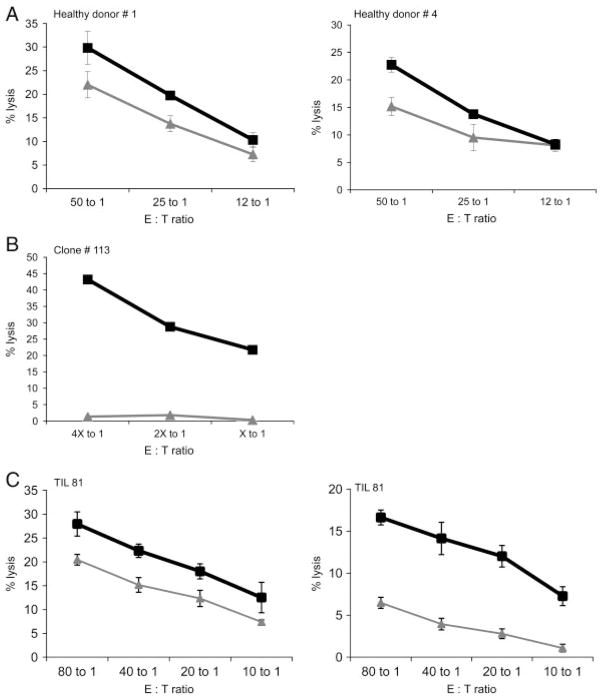
The proof of principle that the CTL-independent MS-based approach might lead to the identification of T-cell epitopes is the induction of T-cell responses against HLA ligands identified by mass spectrometry. Therefore, as a pilot study CTL recognizing the HLA-A2-restricted SRXN1-specific epitope (peptide p114–122) were generated by three weekly stimulations with irradiated peptide-pulsed autologous PBL as targets. HLA-A2+ T2 cells loaded with the SRXN1-specific peptide were recognized by SRXN1-peptide-specific CTL obtained from several healthy donors (Fig. 3A), whereas T2 cells pulsed with non-targeting control peptide (here gp100) could not be effectively recognized by these target-specific CTL. Subcloning of T cells in the presence of HLA-A2+, peptide-pulsed, irradiated PBMC resulted in the isolation of a clone with low background activity against T2 cells and high specificity for SRXN1-derived p114–122 peptide-loaded T2 cells (Fig. 3B). Furthermore, as representatively shown in Fig. 3C T cells with cytotoxic activity against the SRXN1-derived epitope were not only detectable in peripheral blood isolated from healthy donors, but also in TIL obtained from a RCC patient. By targeting peptide-loaded T2 cells, T cells represented in the TIL 81 population showed a higher cytotoxic activity against T2 cells pulsed with the SRXN1-derived peptide than with the control peptide (Fig. 3C, left graph), indicating that the SRXN1-derived peptide is indeed recognized. There is also a significant difference in the recognition of RCC cell lines by TIL 81. Whereas the HLA-A2+ RCC cell line MZ2905RC is recognized by TIL 81, the HLA-A2− RCC cell line HAL31RC is less efficiently killed (Fig. 3C, right graph). Based on the results obtained with the peptide pulsed T2 cells, it can be assumed that the SRXN1-derived peptide contributes at least in part to the recognition of the HLA-A2-matched RCC cell line MZ2905RC.
Figure 3.
Induction of SRXN1-specific CTL. Cytotoxic activity of bulk cultures obtained from two different healthy donors (A) and a representative clone derived from a healthy donor (B) as well as from tumor-infiltrating lymphocytes (TIL) of a RCC patient (C). The gray lines in (A), (B) and the left panel in (C) indicate T2 cells pulsed with an irrelevant peptide (gp100), whereas the black lines represent T2 cells pulsed with the SRXN1-derived peptide (aa 114–122). However, in the right panel (C), the gray line represents the TIL 81-mediated killing of the HLA-A2− RCC cell line HAL31RC, whereas the black line represents the killing of the SRXN1-expressing HLA-A2+ RCC cell line MZ2905RC. The percentage of CD3+, CD8+ cells among the TIL was 46%. Each graph (except graph B) represents mean values and standard deviations of three replicates.
4 Discussion
Under physiological conditions, the peptide repertoire presented by HLA class I molecules on the cell surface of cells also termed HLA class I ligandome is mainly composed of peptides with 8–12 amino acids in length, which are predominantly derived from endogenously synthesized cellular proteins. Although the number of successful identifications of such naturally processed HLA class I and/or HLA class II ligands has been steadily growing over the last decade, the collection of known HLA ligands still represents only a small fraction of their theoretically expected repertoire size. Moreover, even phosphopeptides can be presented by both classes of HLA molecules [30]. The key technologies to define the ligandome are comprehensively summarized by Klug et al. [29]. However, in addition to technical challenges and limitations, cellular parameters such as the extent of protein misfolding, the cellular localization or the linkage to given signal transduction pathways are relevant for some, but not for all HLA class I-represented peptides [56–59]. Thus, the HLA ligandome is hardly determined by a single factor and might also differ between normal and diseased cells. Experimental evidence for this assumption was provided by employing stable isotope labeling coupled with MS for the comparison of the HLA class I peptide repertoires of colon carcinoma and RCC tissues to that of adjacent control tissue [18, 22, 53]. This approach led to the identification of HLA class I-represented peptides either overexpressed or exclusively restricted to cancerous tissue.
Since primary renal parenchyma cells cannot be expanded in vitro to adequate numbers, this study focused on the analysis of the ccRCC-specific ligandome of MZ2733RC cells. Using the peptide elution technology, 49 naturally processed peptides were identified, mostly nonamers presented by the HLA class I alleles HLA-A*01, -A*02, -B*07, -B*49 and -C*05:01/Cw*07:02. The pool of identified ligands extends the list of previously defined naturally processed peptide ligands isolated from RCC tissues or cell lines and presented by a variety of HLA class I molecules [9, 22, 24, 30, 60]. Some of these HLA ligands are known to be differentially presented between primary tumor lesions, metastases and normal tissues [22]. In addition, a number of the defined HLA ligands presented in this report such as APOL1 [9, 22], IGFBP3 [9], LSM8, DEGS1, CLTC [22], SRXN1 [25] and HLA-B [60] have also been described independently. For the source protein BRD4, even phosphorylated HLA class I ligands have been identified [30]. Others were at least functionally related to already known HLA ligands, such as AKR1C3, MCM7 or RRM2B [24]. The partial overlap and/or close relationship of HLA ligands identified support(s) the assumption that currently predominantly high abundant ligands and/or ligands with high binding affinity are accessible with this approach. This is strengthened by the conditional peptide exchange technology, which not only allows monitoring the peptide loading or exchange process for HLA ligands but also requires the use of high affinity binders [61].
The most prominent protein families identified in this study represent proteins involved in the synthesis, modification and degradation of proteins, followed by metabolic enzymes, in particular enzymes involved in the lipid and redox metabolisms and components of signal transduction pathways, which have been demonstrated to be frequently modified during neoplastic transformation (Supporting Information Table 2A [9, 22, 25, 30]).
Some of the HLA class I-eluted peptides derived from RCC cells represent house-keeping genes ubiquitously expressed in both normal and malignant tissues. Based on currently available gene ontology information and due to the fact that some proteins can be in more than one cluster, 20 of the 49 identified source proteins for RCC-derived HLA ligands presented in this report were of cytosolic origin, while 16 were membrane-associated, 14 of nuclear origin, 5 linked to the cytoskeleton whereas 4 each were representing proteins of the ER or extracellular space and 3 were of mitochondrial origin (Supporting Information Table 2B). This compartment heterogeneity further supports the finding that a significant number of epitope-containing peptides (15–25% of the total proteasome products) are extended at the N-terminus and are initially incapable of HLA binding unless further processed. A large number of cytosolic, ER-resident or nuclear amino-and endopeptidases like, e.g. the tripeptidyl peptidase II, the bleomycin hydrolase, the ER-resident aminopeptides 1 and 2 as well as nuclear proteasomes, have the potential to create HLA-binding peptides from the proteasome-generated precursors [62–70]. In addition, the source proteins of the HLA class I ligands identified in MZ2733RC cells were encoded by genes randomly distributed over >70% of the chromosomes. These results suggest that the immune surveillance mechanisms can monitor HLA class I-presented peptides independent of their chromosomal and cellular localization thereby leading to the activation of innate and adaptive responses against qualitatively (PTM) or quantitatively (newly expressed/over expressed) altered proteins upon neoplastic transformation. However, alterations of proteins in multiple cellular compartments may be necessary to report complex malignant processes to the immune system, which then might have an impact on the shaping of the resulting NK and T-cell responses.
Based on transcriptomic profiling data previously established with paired biopsy systems comprising RCC tumor lesions and corresponding tumor adjacent renal epithelium, it was possible to reassess the transcriptional regulation for those 28 members of the 49 genes encoding for source proteins which were represented on the custom-made cDNA array and in part classified as differentially expressed (Table 2). In addition, the published literature on some of these potential candidate markers, including APOL1 [9, 22], IGFBP3 [9], LSM8, DEGS1 and CLTC [22], SRXN1 [25], HLA-B [59] and BRD4 [30] provide further evidence that the HLA ligands defined in this report are not only valid but also likely of functional relevance in RCC. However, the profiling results also demonstrate the lack of a strict correlation between the mRNA and protein expression levels, which is in line with the report by Weinzierl et al. [25].
The screening of the Human Protein Atlas for antibodies specifically recognizing source proteins contributing to the ligandome of the cell line MZ2733RC provided comparative IHC on RCC and normal renal kidney tissue sections for a total number of 29 source proteins (Supporting Information Table 4). The validation score for 14 of the representative staining pattern such as for APOL1, CLTC, LSM8, ACTA1, EIF3E, MCM7 (Supporting Information Fig. 4), HTRA1, RANBP9, TFE3, COG5, BAT3 (Supporting Information Fig. 3), RRM2B, RPL14 and UBQLN1 are classified as supportive, indicating that the respective staining pattern are in line with the corresponding data from the literature, whereas the remaining staining pattern are as yet classified as uncertain, due to either none informative or at least to some extent controversial staining pattern.
In regard to the current verification status, there exist both comparative cDNA microarray profiling (Supporting Information Table 3) as well as IHC staining data (Supporting Information Table 4) for 23 of the 49 identified source proteins (Supporting Information Table 4, dark gray shading), whereas for another 13 source proteins at least either transcriptional profiling or IHC staining data (Supporting Information Table 4, light gray shading) are available. Thus, 36 of the 49 identified ligands are at least verified via an independent experimental strategy. The functional relevance of altered metabolic profiles frequently detected in tumors has recently rekindled interest and seems to play an important role in the development of tumors. Peptide epitopes derived from metabolic enzymes might represent suitable and powerful targets for T-cell-based therapeutic strategies. However, despite their known natural processing and presentation on RCC cells, there exists no information about their immunogenicity. Thus, a peptide derived from SRXN1, one of the source proteins for a HLA-A*02-restricted ligand defined in the current report, was chosen for the induction of T-cell responses against this protein. The characterization of SRXN1 as potential oncoantigen was first achieved by demonstrating an increased SRXN1-transcript level in different RCC cell lines compared with their corresponding cell cultures derived from normal kidney epithelium (Fig. 2). The selection of this SRXN1-derived peptide was further supported by the report of Weinzierl et al. [25], who not only described the same peptide sequence as a HLA ligand in the RCC099 tissues but also demonstrated higher SRXN1-specific mRNA levels in tumor tissues when compared with renal epithelium. SRXN1 is an enzyme regulating oxygen species signaling by reducing hyperoxidized peroxiredoxins [71]. Moreover, SRXN1 expression is often elevated in tumors and might contribute to tumorigenesis [72], thus being a potential target for treatment. The present report extends these studies and demonstrates for the first time that (i) an immune response can be mounted against an SRXN1-derived HLA-presented peptide, (ii) HLA-A2-restricted SRXN1-specific T cells are able to lyse SRXN1-peptide presenting cells and (iii) SRNX1-specific T cells can be detected in TILs isolated from a RCC lesion. However, it is noteworthy that even though the total number of samples is still rather low, the overall response rate of T cells derived from PBMC of healthy donors (3/7) and RCC patients (4/9) is quite similar. For T cells derived from TIL preparations, the number of tested samples is still too low to draw any conclusions. But taking into account that TIL preparations differ from patient to patient both in regard of the overall lymphocyte subset composition and in terms of their cytotoxic capacity, further studies will be needed to define the overall frequency of SRXN1-specific CTLs in healthy controls and RCC patients as well as their cytotoxic potential.
In conclusion, metabolic alterations of tumor cells could be monitored by immune cells, suggesting that proteins involved in metabolic processes might be developed into potential targets for immunomonitoring and/or therapies.
Supplementary Material
Acknowledgments
Part of the work in this report was supported by the BMBF, Bonn, project grant 031U101H (B.S.), the Mildred Scheel Foundation, grant 10-1712-Se5 (B.S.) and the Wilhelm Sander Foundation, grant 2007.001.1 (C.M.). In addition, the authors thank Professor. Dr. Paolo Fornara (Clinic for Urology and Kidney Transplantation Center, University Hospital Halle) for providing us with clinical samples following standard ethical procedures according to the institutional policy, Dr. Dagmar Riemann for asservating tumor and control tissue specimen along with TIL, Barbara Malenica for her excellent technical assistance and Sylvi Magdeburg and Nicole Ott for secretarial help in preparing the manuscript.
Abbreviations
- AKR1C3
aldo-keto reductase family 1 member C3
- CCT3
chaperonin-containing TCP1 subunit 3 γ
- CTL
cytotoxic T lymphocytes
- HLA
human leukocyte antigen
- IHC
immunohistochemistry
- MCM7
minichromosome maintenance complex component 7
- PBMC
peripheral blood mononuclear cells
- qPCR
real-time quantitative RT-PCR
- RCC
renal cell carcinoma
- SRXN1
sulfiredoxin-1
- TIL
tumor-infiltrating lymphocytes
Footnotes
The authors have declared no conflict of interest.
References
- 1.Scholz C, Tampe R. The peptide-loading complex – antigen translocation and MHC class I loading. Biol Chem. 2009;390:783–794. doi: 10.1515/BC.2009.069. [DOI] [PubMed] [Google Scholar]
- 2.Storkus WJ, Herrem C, Kawabe M, Cohen PA, et al. Improving immunotherapy by conditionally enhancing MHC class I presentation of tumor antigen-derived peptide epitopes. Crit Rev Immunol. 2007;27:485–493. doi: 10.1615/critrevimmunol.v27.i5.60. [DOI] [PubMed] [Google Scholar]
- 3.Gordan JD, Vonderheide RH. Universal tumor antigens as targets for immunotherapy. Cytotherapy. 2002;4:317–327. doi: 10.1080/146532402760271091. [DOI] [PubMed] [Google Scholar]
- 4.Jager D, Jager E, Knuth A. Vaccination for malignant melanoma: recent developments. Oncology. 2001;60:1–7. doi: 10.1159/000055289. [DOI] [PubMed] [Google Scholar]
- 5.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 6.Jager D, Taverna C, Zippelius A, Knuth A. Identification of tumor antigens as potential target antigens for immunotherapy by serological expression cloning. Cancer Immunol Immunother. 2004;53:144–147. doi: 10.1007/s00262-003-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawakami Y, Robbins PF, Rosenberg SA. Human melanoma antigens recognized by T lymphocytes. Keio J Med. 1996;45:100–108. doi: 10.2302/kjm.45.100. [DOI] [PubMed] [Google Scholar]
- 8.Maecker B, von BB, Anderson KS, Vonderheide RH, Schultze JL. Linking genomics to immunotherapy by reverse immunology – “immunomics” in the new millennium. Curr Mol Med. 2001;1:609–619. doi: 10.2174/1566524013363447. [DOI] [PubMed] [Google Scholar]
- 9.Kruger T, Schoor O, Lemmel C, Kraemer B, et al. Lessons to be learned from primary renal cell carcinomas: novel tumor antigens and HLA ligands for immunotherapy. Cancer Immunol Immunother. 2005;54:826–836. doi: 10.1007/s00262-004-0650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otsuka M, Kato M, Yoshikawa T, Chen H, et al. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem Biophys Res Commun. 2001;289:876–881. doi: 10.1006/bbrc.2001.6047. [DOI] [PubMed] [Google Scholar]
- 11.Seliger B, Dressler SP, Lichtenfels R, Kellner R. Candidate biomarkers in renal cell carcinoma. Proteomics. 2007;7:4601–4612. doi: 10.1002/pmic.200700415. [DOI] [PubMed] [Google Scholar]
- 12.Seliger B, Dressler SP, Wang E, Kellner R, et al. Combined analysis of transcriptome and proteome data as a tool for the identification of candidate biomarkers in renal cell carcinoma. Proteomics. 2009;9:1567–1581. doi: 10.1002/pmic.200700288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinschenk T, Gouttefangeas C, Schirle M, Obermayr F, et al. Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 2002;62:5818–5827. [PubMed] [Google Scholar]
- 14.Rauch J, Gires O. SEREX, Proteomex, AMIDA, and beyond: serological screening technologies for target identification. Proteomics Clin Appl. 2008;2:355–371. doi: 10.1002/prca.200780064. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- 16.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 17.Rajcevic U, Petersen K, Knol JC, Loos M, et al. iTRAQ-based proteomics profiling reveals increased metabolic activity and cellular cross-talk in angiogenic compared with invasive glioblastoma phenotype. Mol Cell Proteomics. 2009;8:2595–2612. doi: 10.1074/mcp.M900124-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flad T, Spengler B, Kalbacher H, Brossart P, et al. Direct identification of major histocompatibility complex class I-bound tumor-associated peptide antigens of a renal carcinoma cell line by a novel mass spectrometric method. Cancer Res. 1998;58:5803–5811. [PubMed] [Google Scholar]
- 19.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2. 1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 20.Purcell AW, Gorman JJ. Immunoproteomics: mass spectrometry-based methods to study the targets of the immune response. Mol Cell Proteomics. 2004;3:193–208. doi: 10.1074/mcp.R300013-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Boss CN, Grunebach F, Brauer K, Hantschel M, et al. Identification and characterization of T-cell epitopes deduced from RGS5, a novel broadly expressed tumor antigen. Clin Cancer Res. 2007;13:3347–3355. doi: 10.1158/1078-0432.CCR-06-2156. [DOI] [PubMed] [Google Scholar]
- 22.Stickel JS, Weinzierl AO, Hillen N, Drews O, et al. HLA ligand profiles of primary renal cell carcinoma maintained in metastases. Cancer Immunol Immunother. 2009;58:1407–1417. doi: 10.1007/s00262-008-0655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer BF, Schoor O, Kruger T, Reichle C, et al. MAGED4-expression in renal cell carcinoma and identification of an HLA-A*25-restricted MHC class I ligand from solid tumor tissue. Cancer Biol Ther. 2005;4:943–948. doi: 10.4161/cbt.4.9.1907. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann S, Gluckmann M, Kausche S, Schmidt A, et al. Rapid and sensitive identification of major histocompatibility complex class I-associated tumor peptides by nano-LC MALDI MS/MS. Mol Cell Proteomics. 2005;4:1888–1897. doi: 10.1074/mcp.M500076-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Weinzierl AO, Lemmel C, Schoor O, Muller M, et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol Cell Proteomics. 2007;6:102–113. doi: 10.1074/mcp.M600310-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Woods AS, Huang AY, Cotter RJ, Pasternack GR, et al. Simplified high-sensitivity sequencing of a major histocompatibility complex class I-associated immunoreactive peptide using matrix-assisted laser desorption/ ionization mass spectrometry. Anal Biochem. 1995;226:15–25. doi: 10.1006/abio.1995.1185. [DOI] [PubMed] [Google Scholar]
- 27.Zhen Y, Xu N, Richardson B, Becklin R, et al. Development of an LC-MALDI method for the analysis of protein complexes. J Am Soc Mass Spectrom. 2004;15:803–822. doi: 10.1016/j.jasms.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Hillen N, Mester G, Lemmel C, Weinzierl AO, et al. Essential differences in ligand presentation and T cell epitope recognition among HLA molecules of the HLA-B44 supertype. Eur J Immunol. 2008;38:2993–3003. doi: 10.1002/eji.200838632. [DOI] [PubMed] [Google Scholar]
- 29.Klug F, Miller M, Schmidt HH, Stevanovic S. Characterization of MHC ligands for peptide based tumor vaccination. Curr Pharm Des. 2009;15:3221–3236. doi: 10.2174/138161209789105180. [DOI] [PubMed] [Google Scholar]
- 30.Meyer VS, Drews O, Gunder M, Hennenlotter J, et al. Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J Proteome Res. 2009;8:3666–3674. doi: 10.1021/pr800937k. [DOI] [PubMed] [Google Scholar]
- 31.Dengjel J, Nastke MD, Gouttefangeas C, Gitsioudis G, et al. Unexpected abundance of HLA class II presented peptides in primary renal cell carcinomas. Clin Cancer Res. 2006;12:4163–4170. doi: 10.1158/1078-0432.CCR-05-2470. [DOI] [PubMed] [Google Scholar]
- 32.Craven RA, Banks RE. Understanding and managing renal cell carcinoma: can proteomic studies contribute to clinical practice? Contrib Nephrol. 2008;160:88–106. doi: 10.1159/000125936. [DOI] [PubMed] [Google Scholar]
- 33.Kim DS, Choi YP, Kang S, Gao MQ, et al. Panel of candidate biomarkers for renal cell carcinoma. J Proteome Res. 2010;9:3710–3719. doi: 10.1021/pr100236r. [DOI] [PubMed] [Google Scholar]
- 34.Johann DJ, Jr, Wei BR, Prieto DA, Chan KC, et al. Combined blood/tissue analysis for cancer biomarker discovery: application to renal cell carcinoma. Anal Chem. 2010;82:1584–1588. doi: 10.1021/ac902204k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamura N, Masuda T, Gotoh A, Shirakawa T, et al. Quantitative proteomic analysis to discover potential diagnostic markers and therapeutic targets in human renal cell carcinoma. Proteomics. 2008;8:3194–3203. doi: 10.1002/pmic.200700619. [DOI] [PubMed] [Google Scholar]
- 36.Perroud B, Ishimaru T, Borowsky AD, Weiss RH. Grade-dependent proteomics characterization of kidney cancer. Mol Cell Proteomics. 2009;8:971–985. doi: 10.1074/mcp.M800252-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lichtenfels R, Dressler SP, Zobawa M, Recktenwald CV, et al. Systematic comparative protein expression profiling of clear cell renal cell carcinoma: a pilot study based on the separation of tissue specimens by two-dimensional gel electrophoresis. Mol Cell Proteomics. 2009;8:2827–2842. doi: 10.1074/mcp.M900168-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linehan WM, Bratslavsky G, Pinto PA, Schmidt LS, et al. Molecular diagnosis and therapy of kidney cancer. Annu Rev Med. 2010;61:329–343. doi: 10.1146/annurev.med.042808.171650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Reilly T, McSheehy PM. Biomarker development for the clinical activity of the mTOR inhibitor everolimus (RAD001): processes, limitations, and further proposals. Transl Oncol. 2010;3:65–79. doi: 10.1593/tlo.09277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rini BI. New strategies in kidney cancer: therapeutic advances through understanding the molecular basis of response and resistance. Clin Cancer Res. 2010;16:1348–1354. doi: 10.1158/1078-0432.CCR-09-2273. [DOI] [PubMed] [Google Scholar]
- 41.McGuire BB, Fitzpatrick JM. Biomarkers in renal cell carcinoma. Curr Opin Urol. 2009;19:441–446. doi: 10.1097/MOU.0b013e32832f0c68. [DOI] [PubMed] [Google Scholar]
- 42.Bukur J, Rebmann V, Grosse-Wilde H, Luboldt H, et al. Functional role of human leukocyte antigen-G up-regulation in renal cell carcinoma. Cancer Res. 2003;63:4107–4111. [PubMed] [Google Scholar]
- 43.Lichtenfels R, Kellner R, Bukur J, Beck J, et al. Heat shock protein expression and anti-heat shock protein reactivity in renal cell carcinoma. Proteomics. 2002;2:561–570. doi: 10.1002/1615-9861(200205)2:5<561::AID-PROT561>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.Wang E, Lichtenfels R, Bukur J, Ngalame Y, et al. Ontogeny and oncogenesis balance the transcriptional profile of renal cell cancer. Cancer Res. 2004;64:7279–7287. doi: 10.1158/0008-5472.CAN-04-1597. [DOI] [PubMed] [Google Scholar]
- 45.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T–B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 46.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 47.Seeger FH, Schirle M, Gatfield J, Arnold D, et al. The HLA-A*6601 peptide motif: prediction by pocket structure and verification by peptide analysis. Immunogenetics. 1999;49:571–576. doi: 10.1007/s002510050539. [DOI] [PubMed] [Google Scholar]
- 48.Barnstable CJ, Bodmer WF, Brown G, Galfre G, et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 49.Morris HR, Paxton T, Dell A, Langhorne J, et al. High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1996;10:889–896. doi: 10.1002/(SICI)1097-0231(19960610)10:8<889::AID-RCM615>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 50.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 51.Recktenwald CV, Kellner R, Lichtenfels R, Seliger B. Altered detoxification status and increased resistance to oxidative stress by K-ras transformation. Cancer Res. 2008;68:10086–10093. doi: 10.1158/0008-5472.CAN-08-0360. [DOI] [PubMed] [Google Scholar]
- 52.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, et al. A Human Protein Atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Lemmel C, Weik S, Eberle U, Dengjel J, et al. Differential quantitative analysis of MHC ligands by mass spectrometry using stable isotope labeling. Nat Biotechnol. 2004;22:450–454. doi: 10.1038/nbt947. [DOI] [PubMed] [Google Scholar]
- 54.Seeger FH, Arnold D, Dumrese T, de la Salle H, et al. The HLA-B* 1516 motif demonstrates HLA-B-specific P2 pocket characteristics. Immunogenetics. 1998;48:156–160. doi: 10.1007/s002510050418. [DOI] [PubMed] [Google Scholar]
- 55.Zhou W, Ye XL, Sun ZJ, Ji XD, et al. Overexpression of degenerative spermatocyte homolog 1 up-regulates the expression of cyclin D1 and enhances metastatic efficiency in esophageal carcinoma Eca109 cells. Mol Carcinog. 2009;48:886–894. doi: 10.1002/mc.20533. [DOI] [PubMed] [Google Scholar]
- 56.Golovina TN, Morrison SE, Eisenlohr LC. The impact of misfolding versus targeted degradation on the efficiency of the MHC class I-restricted antigen processing. J Immunol. 2005;174:2763–2769. doi: 10.4049/jimmunol.174.5.2763. [DOI] [PubMed] [Google Scholar]
- 57.Ostankovitch M, Robila V, Engelhard VH. Regulated folding of tyrosinase in the endoplasmic reticulum demonstrates that misfolded full-length proteins are efficient substrates for class I processing and presentation. J Immunol. 2005;174:2544–2551. doi: 10.4049/jimmunol.174.5.2544. [DOI] [PubMed] [Google Scholar]
- 58.Princiotta MF, Finzi D, Qian SB, Gibbs J, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 59.Shastri N, Schwab S, Serwold T. Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 60.Flad T, Mueller L, Dihazi H, Grigorova V, et al. T cell epitope definition by differential mass spectrometry: identification of a novel, immunogenic HLA-B8 ligand directly from renal cancer tissue. Proteomics. 2006;6:364–374. doi: 10.1002/pmic.200500099. [DOI] [PubMed] [Google Scholar]
- 61.Bakker AH, Hoppes R, Linnemann C, Toebes M, et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc Natl Acad Sci USA. 2008;105:3825–3830. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 63.Saveanu L, Fruci D, van Endert P. Beyond the proteasome: trimming, degradation and generation of MHC class I ligands by auxiliary proteases. Mol Immunol. 2002;39:203–215. doi: 10.1016/s0161-5890(02)00102-5. [DOI] [PubMed] [Google Scholar]
- 64.Seifert U, Maranon C, Shmueli A, Desoutter JF, et al. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol. 2003;4:375–379. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 65.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci USA. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hearn A, York IA, Rock KL. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J Immunol. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 68.York IA, Mo AX, Lemerise K, Zeng W, et al. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18:429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 69.Towne CF, York IA, Watkin LB, Lazo JS, Rock KL. Analysis of the role of bleomycin hydrolase in antigen presentation and the generation of CD8 T cell responses. J Immunol. 2007;178:6923–6930. doi: 10.4049/jimmunol.178.11.6923. [DOI] [PubMed] [Google Scholar]
- 70.Stoltze L, Schirle M, Schwarz G, Schroter C, et al. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 71.Chang TS, Jeong W, Woo HA, Lee SM, et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem. 2004;279:50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 72.Wei Q, Jiang H, Matthews CP, Colburn NH. Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc Natl Acad Sci USA. 2008;105:19738–19743. doi: 10.1073/pnas.0810676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.