Abstract
The development of insulin resistance and type 2 diabetes is determined by various factors, including defects within the insulin signaling pathway. Mediators of insulin resistance operate through activation of various protein kinase C (PKC) isoforms, IκB kinase β (IKKβ) and/or c-Jun N-terminal kinase (JNK), and subsequent inhibition of the proximal insulin signaling pathway via the insulin receptor substrate 1 (IRS1) and Akt. These mechanisms are still largely unresolved because of the complexity of the molecular events. In this study, an expression and activation state profiling of multiple known key signaling biomolecules involved in insulin metabolic and mitogenic signaling pathways was evaluated using a phosphospecific antibody array platform. The results of the arrayed antibodies were verified by the multiplexed bead array assay and conventional western blot analysis, and confirmed the well-known inhibitory effects of phorbol esters on insulin signaling pathway activation. Of interest, the increase in PKC signaling responses with phorbol esters was associated with activation of the lipid phosphatase PTEN and a 27 kDa heat shock protein. Thus, this insulin signaling antibody array provides a powerful and effective way to investigate the mechanism of insulin resistance and likely assist the development of innovative therapeutic drugs for type 2 diabetes.
Keywords: Antibody array, insulin resistance, insulin signaling pathway, multiplexed bead array, phorbol 12-myristate 13-acetate (PMA)
1 Introduction
Insulin is a pleiotropic hormone involved in multiple integrated metabolic and mitogenic signaling pathways [1]. The binding of insulin triggers the activation of the cell surface insulin receptor (IR) and, as a consequence, the receptor becomes phosphorylated at several tyrosine residues located in the cytoplasmic portion of its β-subunit. This autophosphorylation event is accompanied by substantial increase in the receptor intrinsic tyrosine kinase activity. In response to insulin stimulation, a number of adaptor proteins interact at either the Src homology 2 (SH2) or phosphotyrosine binding (PTB) domain of the activated IR. These include insulin receptor substrates (IRS) 1–4, Src and collagen homologous (Shc) molecules, cannabinoid receptor-1 (Cbl), Grb2-associated binder 1 (Gab1), the protein downstream of tyrosine kinases (Doks) and adapter protein with a pleckstrin homology and a Src homology 2 domain (APS). The recuitment of adaptor molecules allows the assembly of multi-protein complexes, generation of second messagers, and activation of enzymes and transcription factors involved in the control of metabolism and gene expression [2, 3, 4, 5].
The ebb and flow of cellular insulin action depends largely on the signaling pathways that are regulated by specific protein-protein interactions and enzymatic activities responsible for posttranslational modifications of proteins, such as the phosphorylation/dephosphorylation processes. Defects within the insulin signaling pathways are often associated with the development of insulin resistance, a condition that is not only a leading metabolic feature of obesity, but also a key factor in the etiology of a number of diseases, including type 2 diabetes [6, 7]. The identification of protein targets that undergo alteration in expression and/or posttranslational modification is essential for understanding their role in the etiology of insulin resistance. Because classical techniques such as western blot analysis do not allow rapid and sensitive identification of many proteins, there is a pressing need to develop fast and reliable methods for the detection of key biomarkers in the insulin signaling pathway.
Protein arrays have become an increasingly powerful tool in the study of protein-protein interactions, enzyme activities, protein profiling, and antibody screening [8, 9, 10]. The advantages of protein arrays include miniaturization, multiplexing and the generation of a large amount of information with relatively small amount of samples. However, there are a number of technical issues for the simultaneous detection of multiple proteins and their modifications, as this type of analysis depends on the availability of specific, high affinity antibodies generated against target molecules, their proper immobilization on the array surface and detection strategies.
Cell treatment with phorbol 12-myristate 13-acetate (PMA) elicits an insulin resistance phenotype by activating protein kinase C (PKC), IκB kinase β (IKKβ) and the c-Jun N-terminal kinase (JNK), which are responsible for serine phosphorylation of the insulin receptor substrate 1 (IRS1) at residue 307 [11, 12]. This posttranslational modification prevents efficient tyrosine phosphorylation of IRS-1, thus abrogating subsequent formation of multiprotein signaling complexes in response to insulin [11, 12]. In order to assess the PMA-induced defects in insulin signaling cascade in a larger scale, an antibody array was developed whereby more than 97 antibodies, many of which recognizing phosphorylated proteins, were covalently immobilized on a glass surface coated with polymeric 3D material to retain their functional integrity. The proteins in cell lysates were labeled with biotin and the targeted proteins that were captured by the immobilized antibodies were detected with Cy3-labeled streptavidin. The results of the antibody array were confirmed with the multiplexed bead array assay and conventional western blot method. This approach represents a versatile method for the capture and rapid detection of known proteins and their posttranslational modifications characteristic of normal and pathological states.
2 Materials and methodsδ
2.1 Antibodies and reagents
PMA (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) to make a 100 µmol/L stock solution and used at a final concentration of 100 nmol/L. Recombinant human insulin (Calbiochem, San Diego, CA) was prepared in 0.01 mol/L HCl as a 100 µmol/L stock solution, then separated into aliquots and stored frozen at −20 °C. Insulin was used at a final concentration of 100 nmol/L. Polyclonal antibodies against phospho-Akt (Ser473), phospho-p44/42 mitogen-activated protein kinase (Thr202/Tyr204) and glycogen synthase kinase 3β were purchased from Cell Signaling Technology, Inc. (Beverly, MA). Monoclonal antibodies against GAPDH (Cat# G8795) and β–actin (Cat# A1978) were obtained from Sigma-Aldrich, and anti-phosphotyrosine antibody, clone 4G10, was from Upstate Biotechnology (San Diego, CA). Unless indicated otherwise, all other polyclonal antibodies were obtained from Full Moon BioSystems, Inc. (Sunnyvale, CA).
2.2 Cell cultures and treatments
HepG2 cells were purchased from the American Type Cell Collection (ATCC, Manassas, VA). Cell culture was carried out in minimal essential medium supplemented with 10 % (mass fraction) fetal bovine serum, 1 mmol/L pyruvate, 50,000 units/L penicillin, 50 mg/L streptomycin, and 2 mmol/L L-glutamine. HepG2 cells were starved in serum-free medium overnight and then treated with or without 100 nmol/L PMA for 30 min followed by a 10 min stimulation with 100 nmol/L human recombinant insulin. Two million cells were subsequently lysed in 200 µL extraction buffer [10 mmol/L sodium biocarbonate, pH 8.3, 134 mmol/L NaCl, 0.1 % (mass fraction) Nonidet P-40, 0.1 % (mass fraction) SDS, phosphatase inhibitor cocktail set I and protease inhibitor cocktail set I (Calbiochem)], and centrifuged at 14,000 g for 20 min at 4 °C. The supernatant was collected and total protein concentration was determined to be ~5 mg/mL by using BCA Protein Assay Kit (Pierce, Rockford, IL). The protein solutions were separated into aliquots and processed immediately or they were stored at −80 °C until analysis.
2.3 Antibody array
Antibody array experiments were performed according to the protocol of Full Moon BioSystems. Briefly, the antibodies were covalently immobilized on glass surface coated with polymeric 3-D materials that contain mainly epoxy, aldehyde, and hydroxyl functional groups allowing covalent attachment of antibodies (Full Moon BioSystems). Each slide consists of an array of antibodies with six replicates per antibody and multiple positive and negative controls to maximize data quality and reproducibility. A FluoReporter Mini-Biotin-XX-Protein Labeling Kit (Invitrogen, Carlsbad, CA) was used for the biotinylation of cell lysate proteins, which was performed according to the manufacturer’s protocol. The procedure used to process the array was as followed: The slide was soaked in a Full Moon BioSystems array blocking solution for 30 to 45 min at room temperature, rinsed extensively with Milli-Q water for 3 to 5 min and then dried with a stream of compressed nitrogen. Approximately 80 µg of biotinylated cell lysate diluted in 6 mL of coupling solution from Full Moon BioSystems was applied to each array, followed by 3 h incubation at room temperature in a chamber with 100 % relative humidity. The slide was washed 4 to 5 times with PBS, rinsed extensively with Milli-Q water, and then incubated with a Cy3-streptavidin solution (0.5 µg/mL) for 45 min at room temperature. This was followed by PBS washes and rinsing steps with Milli-Q water. After drying by centrifugation, the slide was scanned on a GenePix 4000 scanner and the images were analyzed with GenePix Pro 6.0 (Molecular Devices, Sunnyvale, CA). Fluorescence intensity of each array spot was quantified; the mean value and the standard deviation of replicates were calculated. The final fluorescence signal (I) was obtained from the fluorescence intensity of each antibody spot after subtraction of the blank signal (spot in the absence of antibody). For each treatment group, a phosphorylation signal ratio induction (Δ) was calculated based on the following equation,
where Ip and Inp are fluorescence signals of the phosphorylated and non-phosphorylated/total protein from the treated sample, respectively. Parameters, I0p and I0np, are the respective fluorescence signals of the phosphorylated and non-phosphorylated/total protein from the untreated control sample. Hence, the phosphorylation signal ratio induction Δ for the control group is given a value of 1. The array data was obtained from three to four independent arrays generated from at least three different culture dishes (60 mm in diameter) for each control and treatment condition.
2.4 Western blot analysis
The cell lysates were separated by SDS-polyacrylamide gel electrophoresis under reducing conditions. The gels were transferred onto polyvinylidene difluoride membranes and subjected to immunoblotting as previously described [13, 14]. After an overnight incubation with primary affinity-purified antibodies, the membranes were incubated with horseradish peroxidase-labeled secondary antibodies and subsequent chemiluminescence detection using ECL or ECL-plus reagents (Amersham Biosciences, Piscataway, NJ). Western blot experiments were repeated at least three times for each control and treatment conditions.
2.5 Multiplexed bead arrays
Antibody coupling to the carboxylated beads (-COOH Microspheres, Luminex Corp., Austin, TX) was carried out according to the manufacturer’s protocol with minor modifications. The antibody-coupled beads were stored in a blocking buffer (PBS, pH 7.4, 0.1 % (mass fraction) bovine serum albumin, 0.02 % (mass fraction) Tween-20, 0.05 % (mass fraction) NaN3) at 4 °C in the dark. Following a 2-min sonication treatment, each color-coated, antibody-coupled bead population (1000 to 1500 beads, 20 µL) was used to capture the targeted molecules from the biotinylated cell lysate (20 µL), which was prepared as described above. Incubation of the lysates with the immobilized antibody was carried out for 30 min in a well of a 96-well plate (Model P, Corning Inc., Corning, NY), to which phycoerythrin-labeled streptavidin (PE-SA) was added (10 µL, 1.5 µg) as a fluorescent reporter and incubated for 10 min prior to the fluorescence readout. In a single well, up to seven different bead populations, each coated with a different antibody, were used to perform multiplexed protein array assays. The concentration of cell lysate proteins was tested in the range of 250 µg/L to 50 mg/L. A bead assay buffer (PBS, pH 7.4, 1 % (mass fraction) bovine serum albumin, 0.05 % (mass fraction) NaN3) was used to make stock solutions and dilutions. The multiplexed assays were performed on a Luminex 100 instrument (Luminex Corp.) by counting 120 events per bead population. The blank signals obtained from beads processed in the absence of cell lysate proteins were subtracted from the signals generated by the fluorescent complexes from beads processed in the presence of cell lysates. Multiplexed bead array assays were repeated at least three times for each control and treatment conditions.
3 Results and discussion
3.1 Antibody selection for the array production
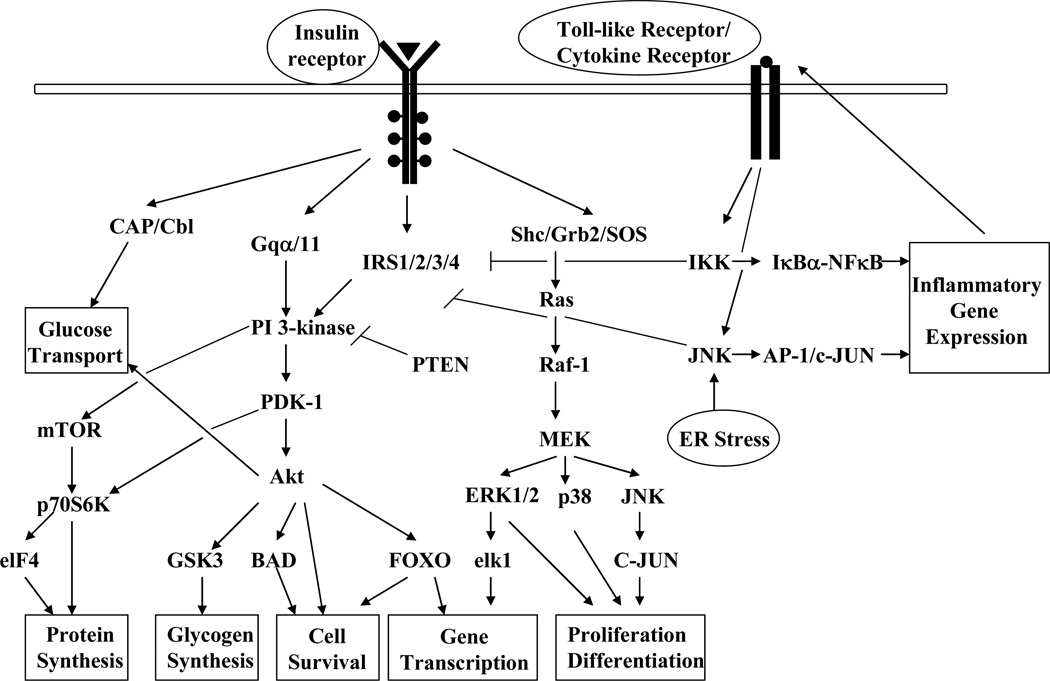
Insulin signal transduction includes a complex activation cascade downstream of the insulin receptor, and any defects within this network lead to the development of insulin resistance (Fig. 1). The major signaling events responsible for the metabolic and mitogenic functions of insulin include the activation of the phosphatidylinositol 3-kinase (PI 3-kinase)-Akt complex and the Ras-mitogen activated protein kinase (MAPK) cascade. Stress and inflammatory cytokines activate serine kinases, e.g. IKKβ and JNK1, responsible for IRS-1 phosphorylation at serine 307 and concomitant attenuation in IR signaling [15, 16, 17]. To monitor the expression pattern of several of these key molecules, 97 antibodies were chosen, including 48 antibodies developed against specific phosphorylation sites of target proteins (Table 1). Detailed information regarding the isotope, source and species specificity of these antibodies can be found in the supporting information table S-1.
Figure 1.
Graphic illustration of the perturbation of the insulin signal transduction pathways by cellular stressors. The binding of insulin to its receptor triggers the activation of downstream signaling cascades important for the metabolic and mitogenic actions of insulin. The phosphorylation of insulin receptor substrates on select tyrosine residues leads to the activation of PI-3 kinase with subsequent increase in the activity of serine/threonine kinase Akt. Insulin also activates the Ras/MAPK signaling pathway and gene transcription. Under stressful conditions, the inflammatory pathways are activated, leading to the phosphorylation of IRS-1 at serine 307 by the kinases IKKβ and JNK1 and subsequent attenuation in insulin signal transduction. Activation of the lipid phosphatase PTEN decreases PI-3 kinase activity and, therefore, reduces Akt signaling and activation of downstream target proteins involved in multiple aspects of cell physiology.
Table 1.
List of the proteins from which antibodies were produced and spotted on the present array.
| Accession Numbera | Name b | Description |
|---|---|---|
| NP_004086 | 4E-BP1(Ab-36) | eukaryotic translation initiation factor 4E binding protein 1 |
| 4E-BP1(Ab-45) | ||
| 4E-BP1(Phospho- Thr36) | ||
| 4E-BP1(Phospho- Thr45) | ||
| NP_001014432 | Akt(Ab-308) | v-akt murine thymoma viral oncogene homolog 1 |
| Akt(Ab-473) | ||
| Akt(Phospho- Thr308) | ||
| Akt(Phospho- Ser473) | ||
| NP_001617 | Akt2(Ab-474) | v-akt murine thymoma viral oncogene homolog 2 |
| Akt2(Phospho- Ser474) | ||
| Q13131 | AMPK1/AMPK2(Phospho- Ser485/Ser491) | 5'-AMP-activated protein kinase catalytic subunit alpha-1/2 |
| NP_116784 | BAD(Ab-112) | BCL2-associated agonist of cell death |
| BAD(Ab-136) | ||
| BAD(Ab-155) | ||
| BAD(Phospho- Ser112) | ||
| BAD(Phospho- Ser136) | ||
| BAD(Phospho- Ser155) | ||
| NP_001092 | β-actin | beta actin |
| NP_001744 | Caveolin-1(Ab-14) | Cavelin-1 |
| Caveolin-1(Phospho- Tyr14) | ||
| NP_002219 | c-Jun(Ab-63) | jun oncogene |
| c-Jun (Phospho- Ser63) | ||
| NP_004085 | eIF2α(Ab-51) | eukaryotic translation initiation factor 2, subunit 1 alpha |
| eIF2α (Phospho- Ser51) | ||
| P06730 | elF4E(Ab-209) | Eukaryotic translation initiation factor 4E |
| elF4E (Phospho- Ser209) | ||
| NP_002006 | FOXO1a(Ab-256) | Forkhead box O1, FKHR |
| FOXO1a(Ab-319) | ||
| FOXO1a (Phospho- Ser256) | ||
| FOXO1a (Phospho- Ser319) | ||
| NP_963853 | FOXO3(Ab-253) | Forkhead box O3, FKHRL1 |
| FOXO3 (Phospho- Ser253) | ||
| NP_005929 | FOXO4 (Ab-197) | Forkhead box O4, AFX |
| FOXO4 (Phospho- Ser197) | ||
| NP_997006 | Gab1(Ab-627) | GRB2-associated binding protein 1 |
| Gab1 (Phospho- Tyr627) | ||
| NP_002037 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| NP_063937 | GSK3α (Ab-21) | Glycogen synthase kinase 3 alpha |
| GSK3α (Phospho- Ser21) | ||
| NP_002084 | GSK3β (Ab-9) | Glycogen synthase kinase 3 beta |
| GSK3β (Phospho- Ser9) | ||
| NP_001531 | Hsp27(Ab-82) | heat shock 27kDa protein |
| Hsp27 (Phospho- Ser82) | ||
| O15111 | IKKα (Ab-23) | Inhibitor of nuclear factor kappa-B kinase subunit alpha |
| IKKα (Phospho- Thr23) | ||
| O14920 | IKKβ | Inhibitor of nuclear factor kappa-B kinase subunit beta |
| P35568 | IRS-1(Ab-307) | Insulin receptor substrate 1 |
| IRS-1(Ab-312) | ||
| IRS-1(Ab-636) | ||
| IRS-1(Ab-639) | ||
| IRS-1 (Phospho- Ser307) | ||
| IRS-1 (Phospho- Ser312) | ||
| IRS-1 (Phospho- Ser636) | ||
| IRS-1 (Phospho- Ser639) | ||
| NP_065390 | IκB-α(Ab-32/36) | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| IκB-α (Ab-42) | ||
| IκB-α (Phospho- Ser32/Ser36) | ||
| IκB-α (Phospho- Tyr42) | ||
| P45983 | JNK(Ab-183) | Mitogen-activated protein kinase 8, c-Jun N-terminal kinase 1 |
| JNK (Phospho- Thr183) | ||
| Q02750 | MEK1(Ab-217) | Dual specificity mitogen-activated protein kinase kinase 1 |
| MEK1(Ab-221) | ||
| MEK1(Ab-291) | ||
| MEK1 (Phospho- Ser217) | ||
| MEK1 (Phospho- Ser221) | ||
| MEK1 (Phospho- Thr291) | ||
| P36507 | MEK-2(Ab-394) | Dual specificity mitogen-activated protein kinase kinase 2 |
| MEK-2 (Phospho- Thr394) | ||
| NP_033071 | NFκB-p65(Ab-536) | v-rel reticuloendotheliosis viral oncogene homolog A |
| NFκB-p6 (Phospho- Ser536) | ||
| NP_003945 | p38 MAPK (Phospho- Thr180) | mitogen-activated protein kinase kinase kinase 14 |
| NP_620407 | p44/42 MAP Kinase(Ab-202) | mitogen-activated protein kinase 1 |
| p44/42 MAP Kinase(Ab-204) | ||
| p44/42 MAP Kinase (Phospho- Thr202) | ||
| p44/42 MAP Kinase (Phospho- Tyr204) | ||
| NP_003152 | p70 S6 Kinase (Ab-424) | ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| p70 S6 Kinase (Phospho- Ser424) | ||
| p70 S6 Kinase(Ab-411) | ||
| p70 S6 Kinase (Phospho- Ser411) | ||
| O15530 | PDK1(Ab-241) | 3-phosphoinositide-dependent protein kinase 1 |
| PDK1 (Phospho- Ser241) | ||
| P19525 | PKR(Ab-446) | Interferon-induced, double-stranded RNA-activated protein kinase |
| PKR(Ab-451) | ||
| PKR (Phospho- Thr446) | ||
| PKR (Phospho- Thr451) | ||
| P60484 | PTEN(Ab-370) | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN |
| PTEN(Ab-380) | ||
| PTEN(Ab-380/382/383) | ||
| PTEN (Phospho- Ser370) | ||
| PTEN (Phospho- Ser380) | ||
| PTEN (Phospho- Ser380/Thr382/Thr383) | ||
| P04049 | Raf1(Ab-259) | RAF proto-oncogene serine/threonine-protein kinase |
| Raf1(Ab-338) | ||
| Raf1 (Phospho- Ser259) | ||
| Raf1 (Phospho- Ser338) | ||
| NP_001001 | S6 Ribosomal Protein(Ab-235) | ribosomal protein S6 |
| S6 Ribosomal Protein (Phospho- Ser235) |
Accession Numbers were obtained from PubMed.
The polyclonal antibody against the “phosphorylated protein” was generated from a short peptide which contains phosphorylated amino acid residue at specific site of the protein of human origin (noted as “Phospho-xxxxxx”, “xxxxxx” is the specific phosphorylation amino acid residue and site); the polyclonal antibody against the “whole protein” was generated either from the same short peptide sequence but without the phosphorylated residue (noted as “Ab-xxx”, “xxx” is the specific phosphorylation site) or from the full-length protein except that anti-GAPDH, β-actin and IKKβ antibody are monoclonal antibodies.
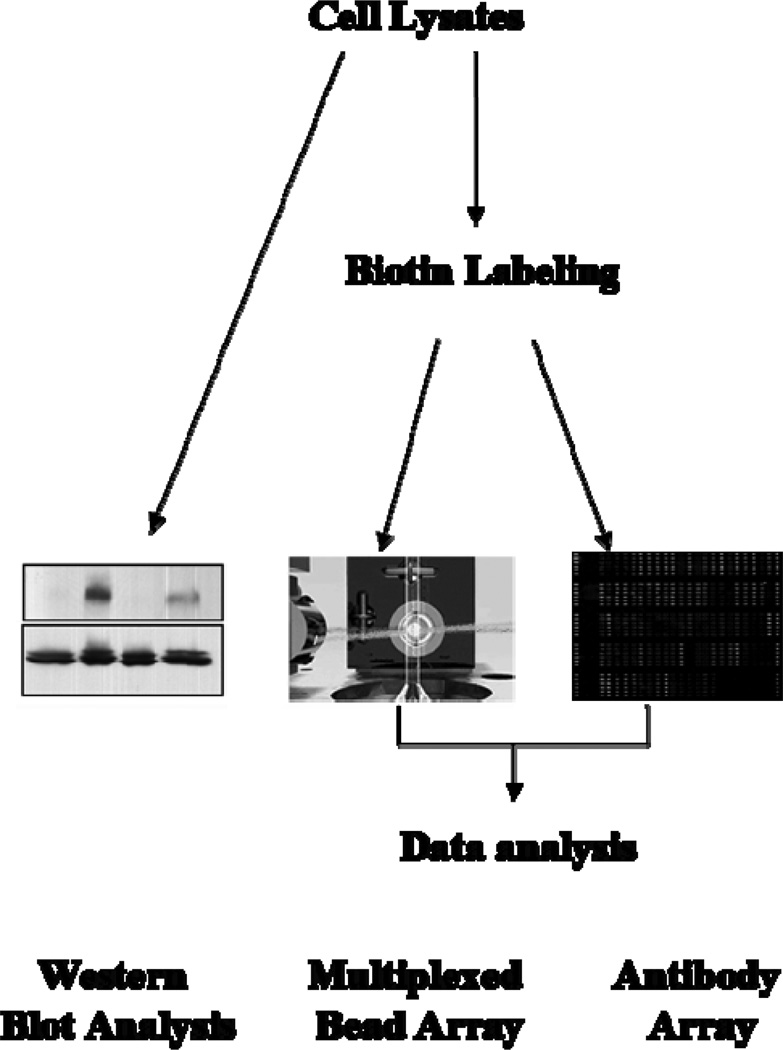
One of the technical challenges in the development of an antibody array is the retention of function and specificity of antibodies after their immobilization on glass slides and downstream array processing [10]. The antibodies chosen for this study were found suitable for this strategy by showing specificity to bind a target protein in a competitive manner in the presence of peptide antigens (See the supporting information table S-1 for more information). Figure 2 gives an overview of the study design. Immobilization of the antibodies was carried out as outlined in “Materials and methods”. Cell lysate samples were labeled with biotin, incubated with the antibody array and the captured target molecules were detected with Cy3-labeled streptavidin. Of the 97 antibodies used, 18 displayed very weak signals or signals equivalent to the blank signal (See the supporting information S-2 for representative image and spot morphology of insulin array). These 18 antibodies include antibodies specific for the phosphorylated forms of caveolin-1 (pTyr14), elF4E (pSer209), and PTEN (pSer380), as well as 15 antibodies against non-phosphorylated proteins (data not given in the supporting information table S-3 showing the raw antibody array data generated only for the performing antibodies). These results could be potentially due to biotin modification of proteins in cell lysates affecting epitope recognition and/or binding affinity between antibody and analyte protein. The poor detection could also be due to the low abundance of protein targets.
Figure 2.
Schematic overview of the experimental design and procedure. Two sets of experiments were carried out in different time periods, and each set of experiments contained 4 independent replicates. Each independent replicate included one cell culture dish (60 mm in diameter) serving as the control, one for insulin treatment, one for PMA treatment, and one for treatment with PMA followed by insulin stimulation (4 samples per one replicate). After treatment with PMA and insulin, HepG2 cells were lysed in the extraction buffer described in the “Materials and methods” section. Four out of 8 dish lystaes were analyzed by western blot, and the others were labeled with biotin and analyzed with either the antibody array or the multiplexed bead array.
3.2 Identification of insulin-induced effectors in HepG2 cells using the antibody array
For the majority of proteins targeted by insulin for which phosphorylation has been reported [1, 2, 18], we found similar increase in the inducible phosphorylated form for key effectors involved in the pleiotropic actions of insulin. These include 3-phosphoinositide-dependent protein kinase 1 (PDK1), Akt, glycogen synthase kinase 3 (GSK3), Forkhead box O (FOXO) proteins, p70S6 Kinase, eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1), BCL2-associated agonist of cell death (BAD), Raf1, MEK1, and MAPK (Table 2). Moreover, additional downstream IR targets from other signaling pathways were identified, such as the 5'-AMP-activated protein kinase catalytic subunit alpha-1/2 (AMPK) which plays a role in cellular energy homeostasis, and interferon-induced, double-stranded RNA-activated protein kinase (PKR) involved in stress pathway (Table 2). The phosphorylation of these new targets provides evidence of the usefullness of antibody arrays for the identification of new components in the insulin signaling network.
Table 2.
Antibody array data for proteins activated by insulin stimulation and attenuated by pre-treatment of HepG2 cells with PMA.
| Ratio a | ||
|---|---|---|
| Name | Insulin | PMA+Insulin |
| 4E-BP1(Thr36) | 1.51 | 1.19 |
| 4E-BP1(Thr45) | 7.68 | 4.78 |
| Akt(Ser473) | 2.03 | 1.24 |
| Akt(Thr308) | 1.51 | 0.97 |
| Akt2(Ser474) | 1.28 | 0.86 |
| AMPK1/AMPK2(Ser485/Ser491) | 1.28 | 0.84 |
| BAD(Ser112) | 1.52 | 0.91 |
| BAD(Ser136) | 1.13 | 0.63 |
| BAD(Ser155) | 2.02 | 0.97 |
| c-Jun(Ser63) | 0.85 | 0.60 |
| eIF2α(Ser51) | 1.46 | 0.77 |
| FOXO1a(Ser256) | 1.35 | 0.87 |
| FOXO1a(Ser319) | 2.21 | 0.81 |
| FOXO3(Ser253) | 0.89 | 0.47 |
| FOXO4(Ser197) | 2.01 | 1.70 |
| GSK3β(Ser9) | 2.21 | 0.82 |
| IκB-α(Ser32/Ser36) | 1.05 | 0.85 |
| MEK1(Ser217) | 1.60 | 1.10 |
| MEK1(Ser221) | 1.59 | 0.96 |
| MEK1(Thr291) | 1.29 | 1.12 |
| MEK-2(Thr394) | 1.80 | 1.29 |
| p38 MAPK(Thr180) | 1.24 | 0.68 |
| p44/42 MAP Kinase(Thr202) | 1.51 | 1.03 |
| p44/42 MAP Kinase(Tyr204) | 1.61 | 1.15 |
| p70 S6 Kinase (Ser424) | 1.50 | 0.96 |
| p70 S6 Kinase(Ser411) | 1.65 | 1.04 |
| PDK1(Ser241) | 1.65 | 0.82 |
| PKR(Thr446) | 0.94 | 0.77 |
| PKR(Thr451) | 1.97 | 1.04 |
| PTEN(Ser370) | 2.37 | 1.27 |
| Raf1(Ser259) | 2.09 | 1.48 |
| Raf1(Ser338) | 1.23 | 1.00 |
| S6 Ribosomal Protein(Ser235) | 1.51 | 1.06 |
For each treatment group, the phosphorylation signal ratio induction was calculated and further normalized by the ratio obtained from the untreated, control group, which was given a value of 1. The coefficients of variation for the fluorescence intensity are ≤ 6% from replicate spots and arrays; the CVs of phosphorylation signal ratio induction are therefore ≤ 12 %. The statistical analysis shows that the ratio change between insulin and PMA pretreatment followed by insulin treatment is significant with p < 0.05.
3.3 PMA attenuates insulin-mediated phosphorylation of effector proteins
Table 2 shows the phosphorylation pattern of 22 known signaling molecules representing a total of 33 specific phosphorylation sites in response to insulin alone or following addition of insulin to PMA-treated cells. Using our phospho-antibody array, a reasonably consistent profiling was obtained, such that the key components of the insulin signal transduction cascade were attenuated under PMA treatment, including Akt-GSK3β and Raf-1-p42/44 MAPK activation. Moreover, the suppression by PMA of insulin-dependent increase in phosphorylation of several serine/threonine residues on transcription factors, such as FOXO1a, FOXO4 and eIF2α, and the translational repressor protein 4E-BP1 were detected as well. In spite of inhibition of insulin stimulated activation of insulin signaling, pretreatment of PMA activates the proteins involved in insulin resistance pathways, i.e. phosphoryalation and activation of NFκB-p65 at Ser536 with a 2.29 fold increase, IKK α at Thr23 with an increase of 1.73 fold and JNK at Thr183 with a small 1.15 fold increase with respect to insulin treatment alone (See supporting information S-4).
3.4 Protein phosphorylation profile in response to PMA
Serine/threonine phosphorylation on IRS proteins results in inhibition of the insulin signal cascade [12, 19], and the phosphorylation of IRS-1 on serine-307 correlates with impairment in insulin signaling in animal models with type 2 diabetes [20, 21, 22]. It is believed that this phosphorylation step impedes tyrosine phosphorylation and/or induces intracellular degradation of IRS-1. In the current study, our antibody array data showed that PMA elicited phosphorylation of IRS-1 on serine-307 by 2.73-fold and on serine-639, although to a much lower extent (Table 3). The serine/threonine kinases JNK and IKKα, which are responsible for Ser-307 phosphorylation of IRS-1, were also activated by phosphorylation upon addition of PMA. There was a 1.5-fold increase in the phosphorylation of the chaperone protein Hsp27, presumably through the Raf-1/MAPK pathway [23, 24]. Moreover, the lipid phosphatase PTEN, a negative insulin signal regulator [25, 26], was phosphorylated nearly two-fold in response to PMA treatment, while phosphorylation of Akt at Ser-473 and FOXO4 at Ser-197 was increased 1.4- and 1.5-fold, respectively. We can surmise that the induction of the stress pathway by PMA leads to the activation of Ser/Thr kinases responsible for inactivating phosphorylation of IRS-1 and subsequent attenuation of insulin responsiveness. PMA by itself also inhibits mitogensis signaling as well, for instance, p44/42 MAPK phosphorylation at Thr202/ Tyr204 and p38 MAPK at Thr180 (See supporting information S-5). The results from our antibody array are fully consistent with the results of other studies [11, 12]. However, additional experiments will be required to explore the role of PTEN and Hsp27 in PMA-induced insulin resistance.
Table 3.
Activation of the signal effectors by PMA treatment only
| Ratio a | |
|---|---|
| 4E-BP1(Thr36) | 1.15 |
| 4E-BP1(Thr45) | 6.65 |
| Akt(Ser473) | 1.37 |
| FOXO4(Ser197) | 1.52 |
| Gab1(Tyr627) | 1.59 |
| HSP27(Ser82) | 1.49 |
| IKK α(Thr23) | 1.29 |
| IRS-1(Ser307) | 2.73 |
| IRS-1(Ser639) | 1.11 |
| JNK(Thr183) | 1.56 |
| PTEN(Ser380/Thr382/Thr383) | 1.97 |
| Raf1(Ser259) | 1.18 |
For each treatment group, the phosphorylation signal ratio induction was calculated and further normalized by the ratio obtained from the untreated, control group, which was given a value of 1. The coefficients of variation for the fluorescence intensity are ≤ 6% from replicate spots and arrays; the CVs of phosphorylation signal ratio induction are therefore ≤ 12 %. The statistical analysis shows that the ratio change between PMA treatment and the control is significant with p < 0.05.
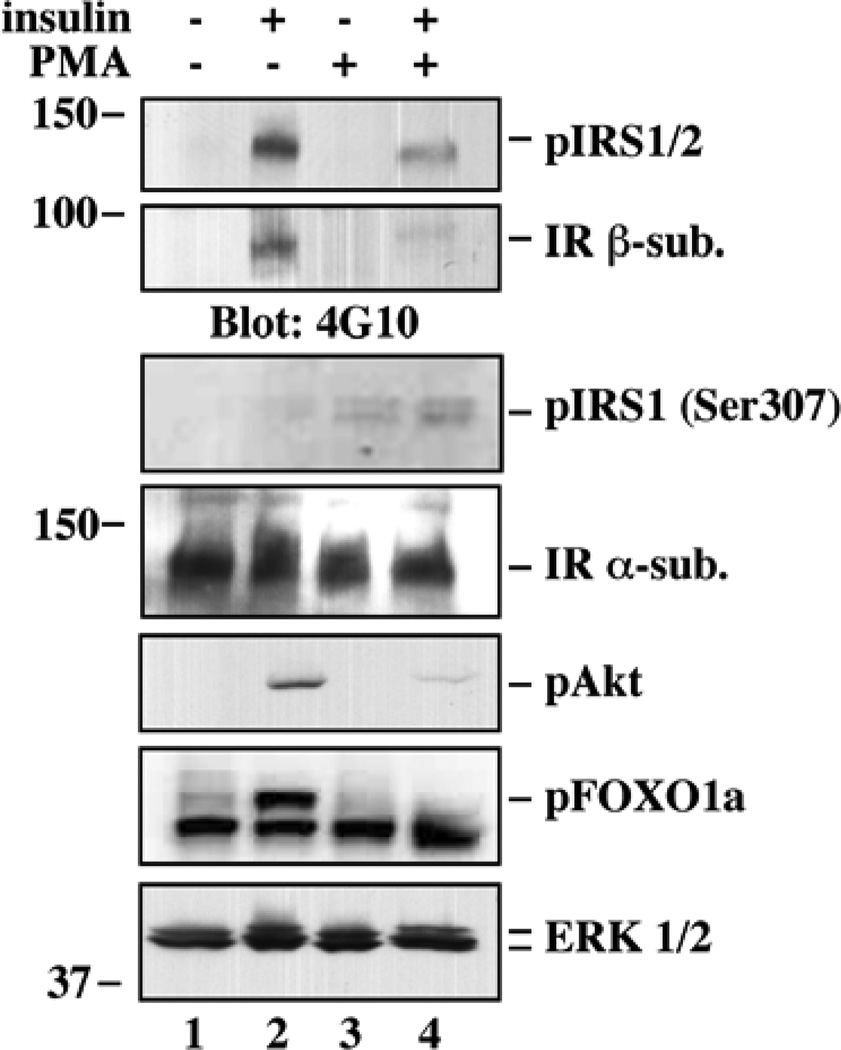
3.5 Western blot analysis of selected insulin-dependent effectors
To validate the antibody array results, conventional Western blot analysis was carried out using lysates from control and treated HepG2 cells (Fig. 3). In response to cell pretreatment with PMA, there was a marked reduction in insulin-stimulated tyrosine phosphorylation of the insulin receptor and IRS1, which was accompanied by subsequent attenuation in the phosphorylation of Akt and its downstream target, FOXO1a. Furthermore, phosphorylation of IRS-1 on serine-307 was detected after PMA stimulation. The fold increase in phosphorylation for some of these proteins was somewhat different as compared to that obtained from the antibody array. Such differences are likely due to the detection sensitivity of each method, the use of immobilized (array platform) versus soluble forms of the antibodies, and the fact that the cell lysates were maintained either in native condition (array assay) or denatured (Western blot).
Figure 3.
Western blot analysis of selected insulin-dependent effectors. HepG2 cells were treated with or without PMA for 30 min, followed by insulin stimulation for 5 min. Cells were lysed, and proteins were resolved by SDS-polyacrylamide gel electrophoresis. Western blot analysis was performed with antibodies against the indicated phosphorylated proteins (a lower case ‘p’ in front of the protein name) and/or total proteins. Blots were reprobed with anti-IR α-subunit or p44/42 MAPK (ERK1/2) to ensure equal protein load in each lane.
3.6 Multiplexed bead array analysis
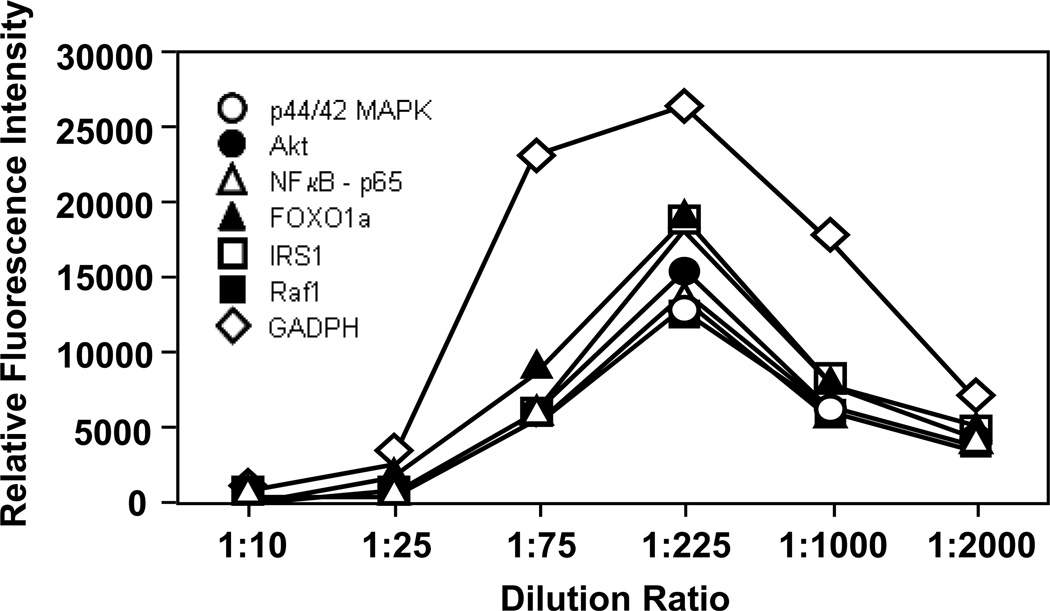
Multiplexed bead arrays have been proven as a sensitive and reliable measurement tool for quantifying proteins and bacteria in our laboratory [27, 28, 29]. Because the bead arrays were assayed in a similar fashion as the antibody array in terms of sample processing, assay format and fluorescence detection (Fig. 2), the results of the two measurement platforms can be readily compared. In this case, seven different antibodies were immobilized to the surface of seven differently color-coated carboxylated bead populations. Antibodies against six phosphorylated proteins and an anti-GAPDH antibody were used in this assay. The antibody-conjugated beads were incubated with biotin-labeled proteins prepared from cell lysate, and then probed with PE-labeled streptavidin. Because the analyte concentration could significantly affect the assay results, serial dilution of the cell lysate was performed to attain reliable multiplexed bead array results. A control experiment was performed whereby the fluorescence readout of a single bead as a function of the cell extract dilution was carried out (Figure 4). It is apparent that a dilution factor of 1 to 225 showed the most sensitive bead array detection. At low dilution ratio, a marked reduction in fluorescence signal was noted likely due to the interference by high protein concentration used in the bead array assay, consistent with the observations of Selby [30]. On the other hand, at high dilution ratio low fluorescence signal was observed, which could be due to the low protein concentration used in the assay. From these results, lysates from control and treated HepG2 cells were prepared at a 1:225 dilution, which corresponded to a protein concentration of ~ 2.22 mg/L. The results of the bead array assay, summarized in Table 4, correlated very well with both the antibody array and western blot analysis.
Figure 4.
Use of the multiplexed bead array system for the determination of the relative fluorescence intensity as a function of the dilution ratio of a cell lysate sample. Seven antibodies, including 6 antibodies against phosphorylated proteins and one against GAPDH, were immobilized to the surface of seven differently color-coated carboxylated beads. The solubilized proteins from untreated HepG2 cells were labeled with biotin, incubated with antibody-conjugated beads, and the resultant immuno-complexes were detected using PE-labeled streptavidin. The initial concentration of cell lysates was 500 mg/L, and a series of dilutions (1:10, 1:25, 1:75, 1:225, 1:1000, and 1:2000) of the lysate were applied to the bead array. Data represent the means ± standard deviations from multiple experimental repeats (N ≥ 4).
Table 4.
Multiplex Beads Array assays for selected antibodies.
| Ratio a | |||
|---|---|---|---|
| Insulin | PMA | PMA+Insulin | |
| Akt(Ser473) | 1.21 | 1.17 | 1.07 |
| FOXO1a(Ser256) | 1.23 | 1.17 | 1.17 |
| IRS-1(Ser307) | 0.47 | 1.47 | 1.10 |
| p44/42 MAP Kinase(Thr202) | 1.23 | 1.13 | 1.13 |
| NFκB-p65(Ser536) | 0.48 | 1.50 | 1.08 |
| Raf1(Ser259) | 1.67 | 1.11 | 1.21 |
For each treatment group, the ratio induction of fluorescence intensity from phosphorylated protein to GADPH protein was calculated and further normalized by the intensity ratio obtained from the untreated, control group, which was given a value of 1. The coefficient of variation for the fluorescence intensity values is ≤ 5.5%, when analyzing replicates within an experiment and from different experiments.
4 Concluding remarks
Reversible protein phosphorylation is essential in the regulation of their subcellular localization, degradation and complex formation, and leads to cellular signaling transduction and biological processes. The insulin signal transduction is controlled through the fine-tuning of protein modifications, including serine/threonine phosphorylation as well as tyrosine phosphorylation. Mapping phosphorylation events of proteins enables the understanding of the cellular signaling process and mechanisms. The majority of current high throughput phosphoproteomic methods rely on mass spectrometry (MS) and microarray technology [31, 32].
Although several MS advancements, such as MS/MS/MS (MS3), multistage activation (MSA), electron capture dissociation (ECD) and electron transfer dissociation (ETD), have been made for improving the fragmentation efficiency and facilitating the analysis of phosphorylated peptides [32, 34], the enrichment of phosphopeptides and phosphoproteins poses an additional challenge. Moreover, knowing the modifications of protein isoforms rather than of peptides derived from these protein isoforms is crucial for understanding the underlying fundamental of individual protein phosphorylations. Top-down proteomics, i.e. the analyses of entire proteins, to decipher the concerted action of phosphorylations at a single protein level, will be a significant challenge [33, 34]. Nonetheless, Kruger et al. identified seven new insulin-induced effectors out of 40 effectors in differentiated brown adipocytes by using MS in combination with phosphotyrosine immunoprecipitation and stable isotope labeling of amino acids in cell culture (SILAC) [35, 36]. A limitation of this technique is that it only allowed the detection of abundant tyrosine phosphorylated proteins and displayed low sensitivity. Hence, there is an urgent need for a more versatile method with higher sensitivity and throughput capability, i.e. the present antibody array. The antibody array enables the identification of a variety of protein expressions associated with the activation of the insulin signal transduction pathway. An essential question the study was designed to answer is whether there are significant differences between the degrees of phosphorylation on specific phosphosites under three conditions, e.g. insulin vs. no treatment control, PMA vs. no treatment control, insulin vs. pretreatment with PMA followed by insulin treatment. By cross validations with multiplexed bead arrays and conventional western blot analyses, this antibody array will be a powerful alternative to MS-based methods for phosphoproteome.
In the present study, we applied 6 replicates in one array to minimize the variation, and at the same time employed GAPDH as the housekeeping protein for the normalization of different arrays. For each protein we applied both antibody against phosphorylated form and antibody against non-phosphorylated protein in the same array; the ratio of phosphorylated protein to non-phosphorylated protein in essence carries a similar meaning as Cy3/Cy5 ratio in a 2-color DNA array [37, 38] while it has no dye related, fluorescence-dependent biases associated with the 2-color DNA array. The signal ratio induction (Δ) introduced in this study is independent of housekeeping protein to avoid a potential error that GAPDH may be subjective to insulin stimulation [39].
Our data obtained from three independent methods (antibody array, multiplexed bead arrays and western blot analysis) clearly show that the present antibody array assay is well suited for the analysis and comparison of signaling events triggered by insulin. This array approach has enabled us to uncover protein targets acting as new candidate effectors in the mediation of insulin resistance by PMA. An emerging concept from this study is that the level of PTEN and Hsp27 phosphorylation may be linked to insulin resistance. New antibody arrays with additional phospho-specific antibodies are being developed, which will allow us further to explore in greater details the complex nature of insulin signal transduction and its regulation through cross-talk between insulin receptor and pro-inflammatory signaling pathway. However, to provide more comparable ratio data with the western blot analysis, it is possible to use denatured cell lysates in the antibody array, which could extend the sources of antibodies for protein array research. Finally, antibody array assays are versatile and provide useful tools for the screening of new pharmacological agents and improve the functional characterization of the molecular basis for various forms of insulin resistance. This knowledge may facilitate identification of new therapeutic targets for the treatment of diseases, such as type 2 diabetes.
Supplementary Material
Acknowledgements
We thank Sutapa Kole for her expert assistance with cell culture and Western blot analyses. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. A portion of that support was through an R&D contract with MedStar Research Institute.
Abbreviations
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IKK
IκB kinase
- IR
insulin receptor
- IRS
insulin receptor substrates
- JNK
c-Jun N-terminal kinase
- PMA
phorbol 12-myristate 13-acetate
Footnotes
Certain commercial equipment, instruments, and materials are identified in this paper to specify adequately the experimental procedure. In no case does such identification imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment are necessarily the best available for the purpose.
The authors have decleared no conflicts of interest.
References
- 1.Virkamäki A, Ueki K, Kahn CR. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Invest. 1999;103:931–943. doi: 10.1172/JCI6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell. Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 3.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 4.White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell. Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 5.de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett. 2008;582:97–105. doi: 10.1016/j.febslet.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nandi A, Kitamura Y, Kahn CR, Accili D. Mouse models of insulin resistance. Physiol. Rev. 2004;84:623–647. doi: 10.1152/physrev.00032.2003. [DOI] [PubMed] [Google Scholar]
- 7.Olefsky JM. Fat talks, liver and muscle listen. Cell. 2008;134:914–916. doi: 10.1016/j.cell.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Smith MG, Jona G, Ptacek J, Devgan G, et al. Global analysis of protein function using protein microarrays. Mech. Ageing Dev. 2005;126:171–175. doi: 10.1016/j.mad.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 9.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 10.Nath N, Hurst R, Hook B, Meisenheimer P, et al. Improving protein array performance: Focus on washing and storage conditions. J. Proteome Res. 2008;7:4475–4482. doi: 10.1021/pr800323j. [DOI] [PubMed] [Google Scholar]
- 11.Jiang G, Dallas-Yang Q, Liu F, Moller DE, Zhang BB. Salicylic acid reverses phorbol 12-myristate-13-acetate (PMA)- and tumor necrosis factor alpha (TNFalpha)-induced insulin receptor substrate 1 (IRS1) serine 307 phosphorylation and insulin resistance in human embryonic kidney 293 (HEK293) cells. J. Biol. Chem. 2003;278:180–186. doi: 10.1074/jbc.M205565200. [DOI] [PubMed] [Google Scholar]
- 12.Aguirre V, Werner ED, Giraud J, Lee YH, et al. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J. Biol. Chem. 2002;277:1531–1537. doi: 10.1074/jbc.M101521200. [DOI] [PubMed] [Google Scholar]
- 13.He HJ, Kole S, Kwon YK, Crow MT, Bernier M. Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J. Biol. Chem. 2003;278:27096–27104. doi: 10.1074/jbc.M301003200. [DOI] [PubMed] [Google Scholar]
- 14.He HJ, Zhu TN, Xie Y, Fan J, et al. Pyrrolidine dithiocarbamate inhibits interleukin-6 signaling through impaired STAT3 activation and association with transcriptional coactivators in hepatocytes. J. Biol. Chem. 2006;281:31369–31379. doi: 10.1074/jbc.M603762200. [DOI] [PubMed] [Google Scholar]
- 15.Hirosumi J, Tuncman G, Chang L, Görgün CZ, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 16.Cai D, Yuan M, Frantz DF, Melendez PA, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-β and NF-κB. Nat. Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arkan MC, Hevener AL, Greten FR, Maeda S, et al. IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 18.Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J. Clin. Invest. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87:99–109. doi: 10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Um SH, Frigerio F, Watanabe M, Picard F, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 21.Werner ED, Lee J, Hansen L, Yuan M, Shoelson SE. Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. J. Biol. Chem. 2004;279:35298–35305. doi: 10.1074/jbc.M405203200. [DOI] [PubMed] [Google Scholar]
- 22.Vallerie SN, Furuhashi M, Fucho R, Hotamisligil GS. A predominant role for parenchymal c-Jun amino terminal kinase (JNK) in the regulation of systemic insulin sensitivity. PLoS ONE. 2008;3:e3151. doi: 10.1371/journal.pone.0003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alford KA, Glennie S, Turrell BR, Rawlinson L, et al. Heat shock protein 27 functions in inflammatory gene expression and transforming growth factor-beta-activated kinase-1 (TAK1)-mediated signaling. J. Biol. Chem. 2007;282:6232–6241. doi: 10.1074/jbc.M610987200. [DOI] [PubMed] [Google Scholar]
- 24.Nomura N, Nomura M, Sugiyama K, Hamada J. Phorbol 12-myristate 13-acetate (PMA)-induced migration of glioblastoma cells is mediated via p38MAPK/Hsp27 pathway. Biochem. Pharmacol. 2007;74:690–701. doi: 10.1016/j.bcp.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Vinciguerra M, Foti M. PTEN and SHIP2 phosphoinositide phosphatases as negative regulators of insulin signalling. Arch. Physiol. Biochem. 2006;112:89–104. doi: 10.1080/13813450600711359. [DOI] [PubMed] [Google Scholar]
- 26.Sasaoka T, Wada T, Tsuneki H. Lipid phosphatases as a possible therapeutic target in cases of type 2 diabetes and obesity. Pharmacol. Ther. 2006;112:799–809. doi: 10.1016/j.pharmthera.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Cole KD, Gaigalas AK, Zhang YZ. Fluorescent nanometer microspheres as a reporter for sensitive detection of simulants of biological threats using multiplexed suspension arrays. Bioconjug. Chem. 2005;16:194–199. doi: 10.1021/bc0498020. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Cole KD, He HJ, Hancock DK, et al. Comparison of ovalbumin quantification using forward-phase protein microarrays and suspension arrays. J. Proteome Res. 2006;5:1770–1775. doi: 10.1021/pr060074v. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Cole KD, He HJ, Peterson A, et al. Epitope selection of monoclonal antibodies for interleukin-4 quantification using suspension arrays and forward-phase protein microarrays. J. Proteome Res. 2007;6:4720–4727. doi: 10.1021/pr070535s. [DOI] [PubMed] [Google Scholar]
- 30.Selby C. Interference in immunoassay. Ann. Clin. Biochem. 1999;36:704–721. doi: 10.1177/000456329903600603. [DOI] [PubMed] [Google Scholar]
- 31.Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 32.Piggee C. Phosphoproteomics: miles to go before it's routine. Anal. Chem. 2009;81:2418–2120. doi: 10.1021/ac802740t. [DOI] [PubMed] [Google Scholar]
- 33.de la Fuente van Bentem S, Mentzen WI, de la Fuente A, Hirt H. Towards functional phosphoproteomics by mapping differential phosphorylation events in signaling networks. Proteomics. 2008;8:4453–4465. doi: 10.1002/pmic.200800175. [DOI] [PubMed] [Google Scholar]
- 34.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass. Spectrom. 2009;44:861–878. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 35.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 36.Krüger M, Kratchmarova I, Blagoev B, Tseng YH, et al. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2451–2456. doi: 10.1073/pnas.0711713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckel-Passow JE, Hoering A, Therneau TM, Ghobrial I. Experimental design and analysis of antibody microarrays: applying methods from cDNA arrays. Cancer Res. 2005;65:2985–2989. doi: 10.1158/0008-5472.CAN-04-3213. [DOI] [PubMed] [Google Scholar]
- 38.Zou S, He HJ, Zong Y, Shi L, Wang L. In: Springer Series on Fluorescence, Volume 6. Standardization and Quality Assurance in Fluorescence Measurements II. Ute Resch-Genger, editor. Berlin Heidelberg: Springer-Verlag; 2008. pp. 215–237. [Google Scholar]
- 39.Alexander MC, Lomanto M, Nasrin N, Ramaika C. Insulin stimulates glyceraldehyde-3-phosphate dehydrogenase gene expression through cis-acting DNA sequences. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5092–5096. doi: 10.1073/pnas.85.14.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.