Abstract
In this study, we investigated the effects of eccentric cleavage products of β-carotene, i.e. β-apocarotenoids (BACs), on retinoid X receptor alpha (RXRα) signaling. Transactivation assays were performed to test whether BACs activate or antagonize RXRα. Reporter gene constructs (RXRE-Luc, pRL-tk) and RXRα were transfected into Cos-1 cells and used to perform these assays. None of the BACs tested activated RXRα. Among the compounds tested, β-apo-13-carotenone was found to antagonize the activation of RXRα by 9-cis-retinoic acid and was effective at concentrations as low as 1nM. Molecular modeling studies revealed that β-apo-13-carotenone makes molecular interactions like an antagonist of RXRα. The results suggest a possible function of BACs on RXRα signaling.
Keywords: β-apocarotenoids, retinoids, retinoid receptors, carotenoids
INTRODUCTION
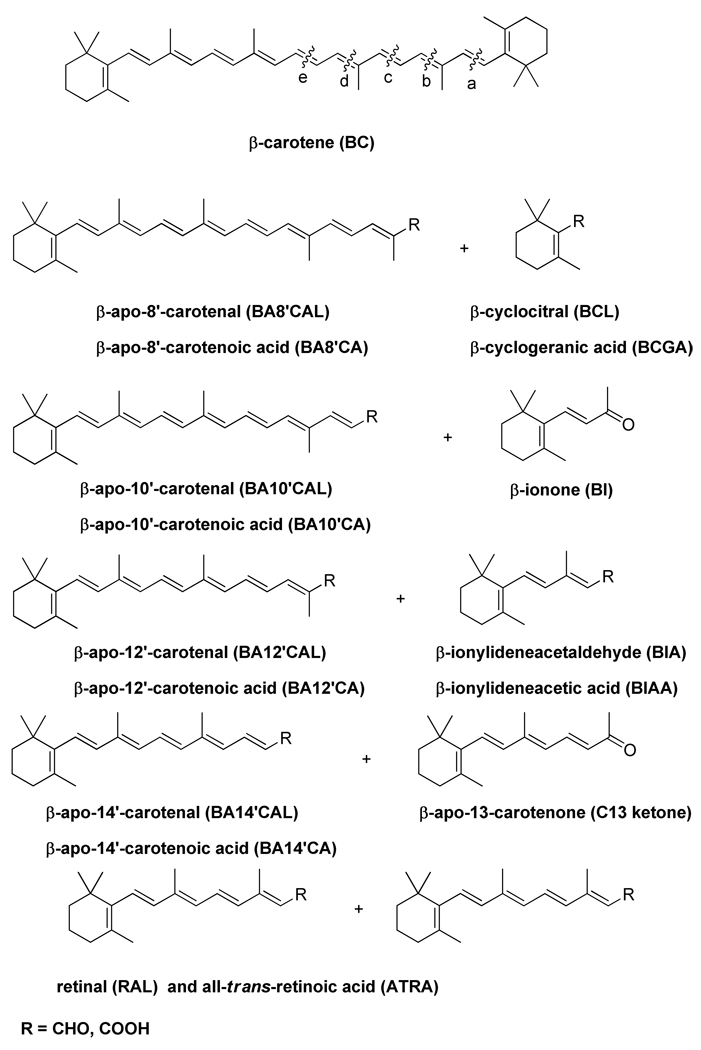
Carotenoids are C40 polyisoprenoids that are biosynthesized from eight isoprene units followed by cyclization, isomerization, and oxidation [1]. Approximately 600 carotenoids have been characterized in nature [2]. Among them, 50–60 display provitamin A activity [3–4]. In order to exhibit a provitamin A activity, the carotenoid molecule must have at least one unsubstituted β-ionone ring and the correct number and position of methyl groups in the polyene chain [5]. Two pathways have been described for the cleavage of BC in mammals [6]. β,β-Carotene-15,15′-oxygenase (BCO1) catalyzes the central cleavage of BC to yield retinal (the initial product of symmetric or central cleavage of BC). BCO1 enzymes from fruit fly [7], chicken [8], mouse [9] and human [10] have been cloned and biochemically characterized. The second pathway of BC metabolism is the eccentric cleavage which occurs at double bonds other than the central 15,15’ double bond of the polyene chain of BC to produce β-apocarotenals and β-apocarotenones. Little is known about the biological function of β-apocarotenoids (BACs) in higher animals.
The retinoid X receptor (RXR) is a member of the nuclear receptor superfamily of ligand dependent transcription factors and consists of three distinct subtypes (α, β, and γ) [11–13]. RXRs are nuclear receptor proteins which can modulate the transcriptional activity of target genes by binding as RXR heterodimeric complexes or RXR homodimers to gene promoters. The first identified RXR, referred to as RXRα, was initially described as an orphan receptor [14]. 9-cis-Retinoic acid, a stereoisomer of all-trans retinoic acid (ATRA) is a high-affinity ligand for RXRα, as well as for the two additional related subtypes, RXRβ and RXRγ, that were later identified [14–20]. RXRs form heterodimers with a number of orphan and nuclear hormone receptors including retinoic acid receptors (RARs), thyroid receptor (TR), vitamin D receptors (VDRs), peroxisome proliferator activator receptors (PPARs), liver X receptors (LXRs), farnesoid X receptors (FXRs), and pregnane X receptors (PXRs) [16, 21–23]. RXRs can also form homodimers in vitro, indicating the existence of a RXR-specific signaling [24–25]. It was observed that liver-specific inactivation of RXR in mice was associated with abnormalities in major metabolic pathways which supports the pleiotropic role of this receptor [26]. RXR-selective synthetic retinoids, i.e. rexinoids, are valuable in elucidating the role of RXRs. One particular rexinoid, LGD 1069, is currently used for the treatment of refractory advanced-stage cutaneous T-cell lymphoma [27–32].
The purpose of the present study was to test BACs as potential ligands for RXRα. We utilized a transient cotransfection (receptor/reporter) assay to screen these compounds as agonists or antagonists of RXRα. Our data indicate that β-apo-13-carotenone acts as a potent antagonist of activated (liganded) RXRα. Molecular modeling studies suggest that β-apo-13-carotenone makes molecular interactions which are similar to those made by ATRA bound as an antagonist to RXRα.
MATERIALS & METHODS
The preparation of β-apocarotenoids
β-Cyclocitral and β-ionone were purchased (Sigma-Aldrich; Milwaukee, WI, USA) and purified (preparative TLC) prior to use. β-apo-8’-carotenal (Sigma-Aldrich) and β-apo-12’-carotenal (CaroteNature; Lupsingen, Switzerland) were purchased and used as obtained. β-Cyclogeranic acid was prepared by air oxidation of β-cyclocitral and crystallization of the product. In the other instances where a β-apocarotenal precursor was purchased, the aldehyde group was converted to its methyl ester (KCN/acetic acid/MnO2/methanol) according to the procedure of Corey and co-workers [33], followed by saponification to the acid (β-apo-12’- and β-apo-8’carotenoic acid). For preparation of the remaining β-apocarotenoids, the ethyl ester of β-ionylideneacetic acid was prepared (as an 80:20 trans/cis isomer mixture about the newly formed double bond) by the Wadsworth-Emmons reaction of β-ionone with triethylphosphonoacetate. In addition to saponifying this ester to β-ionylideneacetic acid, reduction to the alcohol (DIBAL-H) followed by allylic oxidation (MnO2/dichloromethane) produced β-ionylideneacetaldehyde (BIA). BIA was converted to apo-13-carotenone (C13 ketone) by Wittig reaction with 2-oxopropyltriphenylphoshonium chloride [34]. Prior to any conversions of BIA, its cis/trans isomers were separated by column chromatography. Wadsworth-Emmons reaction of retinaldehyde with triethylphosphonoacetate provided essentially isomerically pure β-apo-14’-carotenoic acid after saponification. Two stage reduction-oxidation (DIBAL-H; MnO2/dichloromethane) provided β-apo-14’-carotenal. Wadsworth-Emmons reaction as for the conversion of retinal to ethyl-β-apo-14’-carotenoic acid converted β-apo-12’-carotenal to β-apo-10’-carotenoic acid after saponification. All reactions and compound handling were performed under gold fluorescent lights in oven dried glassware under a dry argon atmosphere. All products were purified by appropriate column or preparative thin layer chromatography and analyzed by HPLC for purity (Model 127 pump and 166 detector, Beckman Instruments, San Ramon, CA, USA; Metachem Polaris 5 um C18, 250 × 4.6 mm column, Varian Inc., Palo Alto, CA, USA). Chemical and structural characterization was carried out using ultraviolet spectrophotometry (Beckman DU-40 spectrophotometer), NMR spectroscopy (Bruker DRX400; Billerica, MA, USA) and electrospray mass spectrometry (Micromass QTOF; Milford, MA, USA).
Cell Culture
Cos-1 cells (ATCC CRL-1650 from the American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle medium (Sigma), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 mg/ml streptomycin, and 0.2 ml /100 ml fungizone (antifungal reagent). The cells were maintained in an incubator at a high humidity, 5% CO2, and 37°C.
Plasmid Constructs
The control reporter plasmid pRL-TK was obtained from Promega (Madison, WI, USA). It contains the herpes simplex virus thymidine kinase (HSV-TK) promoter region upstream of cDNA encoding the native Renilla (Renilla reniformis) luciferase enzyme [36]. This plasmid was used as an internal control in cotransfection experiments to normalize for transfection efficiency. The pRXRE-tk-Luc and pSG5-RXRα expression vectors were kind gifts from Dr. Noa Noy (Case Western Reserve University School of Medicine, Cleveland, OH, USA). The experimental reporter plasmid pRXRE-tk-Luc contains five tandem repeats of a 35-bp sequence (DR-1) from the promoter of the mouse CRBP-II gene [24].
RXRα Transactivation Assay
Cos-1 cells were plated at 1.5 × 105 cells/35-mm2 tissue-culture plate in DMEM with 10% FBS. The cells were grown overnight at 37°C with 5% CO2. The next day, the cells were transfected in serum-free medium with three plasmids mixed in the following amounts per well, 0.05 µg of pRL-TK, 2 µg of pRXRE-luciferase, 2.5 µg of pSG5-RXRα in triplicates using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Following transfection, the plates were incubated at 37°C in 5 % CO2 for 4 h. The medium was then changed to complete DMEM. Note that complete DMEM contains 10% charcoal stripped FBS instead of 10% FBS. Charcoal stripped FBS has been treated with activated carbon to adsorb lipophilic compounds including retinoids. Twenty hours after transfection, cells were treated with test compounds that were dissolved in ethanol or 0.1% ethanol alone for an additional 24 h. Cells were washed once with PBS and lysed by incubation with 500 µl passive lysis buffer (Promega) for 15 min at room temperature. A 20-µl aliquot of cell lysate was then assayed for luciferase activities using a GloMax 96 Microplate Luminometer (Promega) and the Dual Luciferase Reporter (DLR) assay system (Promega), according to the supplier's recommendations. For each experiment, the firefly luciferase activity (experimental reporter) was normalized to Renilla luciferase (control reporter) activity. The change in normalized firefly luciferase activity was calculated relative to that for cells that were transfected with vehicle (ethanol), which was set as 1.
Fold Activation = Average (Firefly/Renilla) from test compound / Average (Firefly/Renilla) from vehicle (Ethanol treated cells)
Molecular Modeling and Docking
Structures of proteins and their co-crystallized ligands were recalled from the protein data bank and were displayed, along with test ligands that were built and manipulated, using Sybyl 7.1 (Tripos, Inc.; St. Louis, MO). The RXRα which binds all-trans retinoic acid in an antagonist tetrameric form [36] is available as PDB entry: 1G5Y, while the dimeric RXRα which binds 9-cis-retinoic acid in the agonist form [37] is recalled from PDB entry 1FBY. Computational docking of the C13 ketone (β-apo-13-carotenone) to these proteins was conducted using Surflex-Dock v. 2.11 [38] which is available from Tripos, Inc. Surflex-Dock allows ring and acyclic structure flexibility and all-atom optimization during determination of the docked ligand poses and uses specific information about molecular interactions between known ligands and the target protein to guide the search process for docked poses. Thus, in the case of both protein structures, ligand was extracted from the protein crystal structure and re-docked to the protein to determine if the program successfully finds the original docked pose of the crystallized ligand. Subsequently, the C13 ketone, built to resemble the native bound ligand was then docked to the protein structure and inspected for the quality of the match relative to the ligand in the crystal structure.
RESULTS & DISCUSSION
The eccentric cleavage products of β-carotene: β-apocarotenoids
β-Carotene, the most potent dietary provitamin A carotenoid, can be metabolized in mammals via two enzymatic pathways. BCO1 catalyzes the cleavage of the 15,15’ double bond resulting in two retinaldehyde molecules, and the eccentric cleavage takes place at double bonds other than the central 15,15’ double bond of the polyene chain of β-carotene to produce β-apocarotenals and β-apocarotenones, i.e. β-apocarotenoids (BACs) with different chain lengths (Figure 1). Presumably the aldehydes can be oxidized to the corresponding carboxylic acids or reduced to the alcohols. β-apo-10’-Carotenal and β-ionone were shown to be products of β,β-carotene-9′,10′-oxygenase (BCO2) [39]. β-apo-13-Carotenone and β-apo-14'-carotenal were identified as enzymatic cleavage products of β-carotene in homogenates of intestinal mucosa of rat [40] but the enzyme responsible was not determined. Another study using tissue homogenates from human, monkey, ferret and rat which were incubated with β-carotene found β-apo-8'-,10'-, and 12'-carotenals, retinal, and all-trans retinoic acid [41]. More recently, β-apo-8'-carotenal was detected in plasma after digestion of β-carotene in a healthy human subject [42]. There is no doubt that the formation of BACs occurs; however, it is not known whether the products of this pathway are biologically active. We have tested all possible eccentric cleavage products of β-carotene (except the 10’-aldehyde) for their ability to transactivate RXRα.
Figure 1. The cleavage products of β-carotene.
β-carotene can be cleaved either symmetrically (denoted as “e” cleavage) by β,β-carotene-15,15′-oxygenase (BCO1) yielding all-trans retinaldehyde that can be further oxidized to all-trans retinoic acid (ATRA) by retinal dehydrogenases. The second pathway of β-carotene cleavage is called the eccentric cleavage (denoted as “a”, “b”, “c”, “d” cleavages) and it occurs at double bonds other than the central 15,15’ double bond of the polyene chain of β-carotene to produce β-carotenals and β-apo-carotenones with different chain lengths.
Effects of β-apocarotenoids on transactivation of RXRα
The biological properties of the BACs were characterized using a cell-based receptor/reporter cotransfection (transactivation) assay [24, 43–44] in which the full length RXRα in a eukaryotic expression vector was co-transfected with two reporter plasmids, a firefly luciferase reporter (experimental reporter) containing the RXRE from CRBPII and renilla luciferase (control reporter) which serves an internal control to normalize for transfection efficiency. Initially, assays using 9-cis-RA for transactivation of RXRα were used to ensure that the assay was reproducible. Dose response experiments employed a range of 10−5 M (10 µM) to 10−10 M (0.1 nM) 9-cis-RA. As expected, the receptor was activated with increasing concentrations of 9-cis-RA (data not shown) with half maximal activation observed at 10−8 M. Maximal fold induction at 10−5 M 9-cis-RA varied from 2.2 – 5.1 in individual experiments. We next screened all BACs at 1 µM+ 10 µM for activation of RXRα. None of the BACs activated RXRα (data not shown).
Antagonist effects of β-apocarotenoids on activated (liganded) RXRα
Next, we tested whether any of the BACs could act as antagonists of 9-cis-RA activation in RXRα. We screened the BACs in our assays to test antagonistic behavior by incubating 9-cis-RA alone or equimolar mixtures of 9-cis-RA and the test compound. The results shown in Table 1 are expressed as a percent of the activation observed with 9-cis-RA alone that is set to 100%. The maximal, normalized fold induction for 9-cis-RA alone varied from 2.2 to 3.8 (2.9 average). The data shown in Table 1 are a compilation of multiple experiments in each of which the compounds used were tested in triplicate wells. β-apo-13-Carotenone was the most potent antagonist of the compounds tested under the conditions of these experiments.
Table 1. Inhibition of 9-cis-Retinoic Acid Induced Transactivation of RXRα by β-Apocarotenoids.
Transactivation assays were carried out in the presence of 10 µM 9-cis RA and 10 µM of the indicated β-apocarotenoids in triplicate wells. Results are expressed as the percent activation observed with 9-cis RA alone that is defined as 100%. For most compounds two or more independent experiments were conducted.
| COMPOUND | %ACTIVATION of 9-cis-RA ALONE |
|---|---|
| β-cyclocitral (“a” cleavage) | 83 |
| β-cyclogeranic acid (“a” cleavage) | 93 |
| β-ionone (“b” cleavage) | 83, 106 |
| β-ionylideneacetaldehyde (“c” cleavage) | 87, 98 |
| β-ionylideneacetic acid (“c” cleavage) | 99 |
| β-apo-13-carotenone (“d” cleavage) | 7, 9 |
| retinaldehyde (“e” cleavage) | 31, 78 |
| β-apo-14’-carotenal (“d” cleavage) | 75, 75 |
| β-apo-14’-carotenoic acid (“d” cleavage) | 87 |
| β-apo-12’-carotenal (“c” cleavage) | 69, 114 |
| β-apo-12’-carotenoic acid (“c” cleavage) | 96 |
| β-apo-8’-carotenal (“a” cleavage) | 73, 78 |
| β-apo-8’-cartenoic acid (“a” cleavage) | 73, 84, 86 |
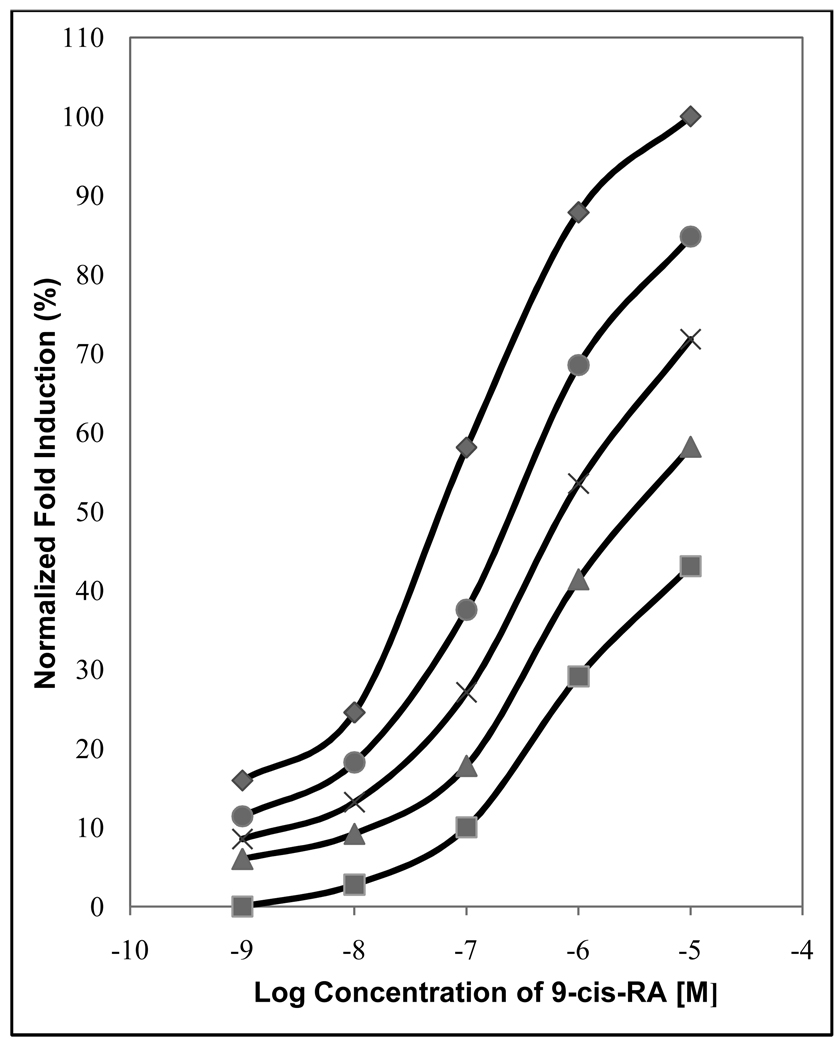
Following the screening experiments shown in Table 1, we tested more completely the effects of β-apo-13-carotenone on 9-cis-RA-induced gene expression. A classic antagonist is defined as a compound that shifts the concentration-response curve of an agonist to the right [45–46]. To characterize the interaction between both 9-cis-RA and β-apo-13-carotenone, a cumulative concentration-response curve of the agonist was established in the absence or presence of increasing fixed concentrations of the antagonist. As shown in Figure 2, increasing concentrations of β-apo-13-carotenone shifted the curves to the right in a concentration-dependent manner. Importantly, concentrations as low as 1 nM shifted the 9-cis-RA dose response curve.
Figure 2. The antagonistic effect of β-apo-13-carotenone on RXRα, dose response curve of 9-cis-RA alone (♦), and increasing concentrations of β-apo-13-carotenone: 10−9 M (●), 10−8 M (✖), 10−7 M (▲), and 10−6 M (■).
The fold induction of 10−5 M 9-cis-RA (the absolute fold induction of this point was 2.29 over the vehicle treated cells) was set to 100% and the other experimental points were calculated relative to this.
Molecular Modeling Experiments to identify interaction between β-apo-13-carotenone and RXRα
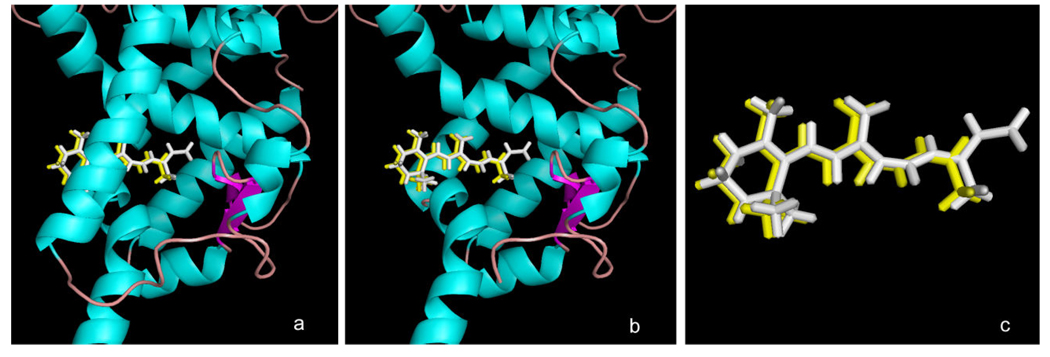
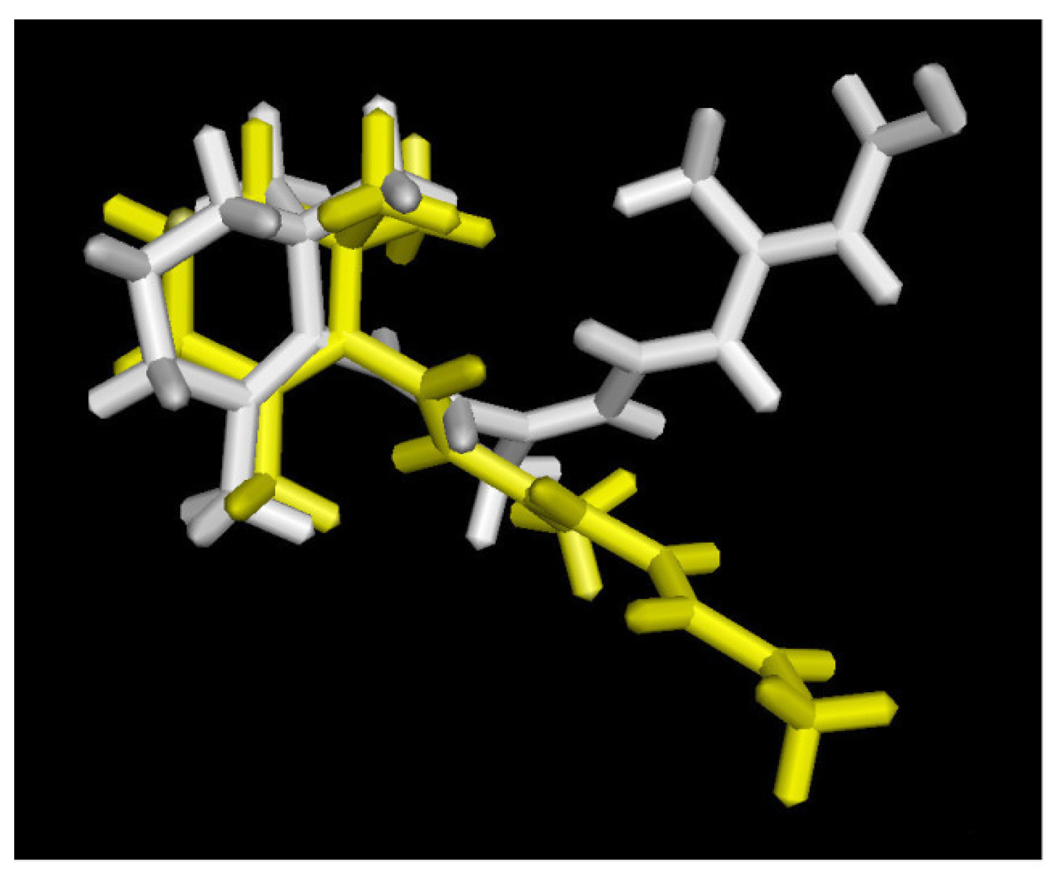
Computational docking experiments were performed to determine if the antagonistic activity observed for β-apo-13-carotenone might be due to direct association with the ligand binding site of RXRα. ATRA has been crystallized with a tetrameric form of RXRα that antagonizes RXR signaling [36]. Shown in Figure 3 is the ligand binding domain of this tetramer (in green) with bound ATRA (in white). When ATRA is removed from the protein and redocked using Surflex-Dock, the rebound ATRA assumes the same position as in the crystal structure (not shown). More importantly, when β-apo-13-carotenone is constructed using the bound ATRA as template, its docked structure with RXRα (shown in yellow) is virtually coincident with the bound ATRA. Alternatively, 9-cis-RA binds as an agonist to a dimeric form of RXRα but in a twisted, high energy conformation [37] we find to be ca. 70 kcal/mol above a calculated low energy ligand conformation. Shown in Figure 4 is a simplified view of efforts to dock β-apo-13-carotenone with this agonist-bound form of RXRα. In white is the structure of the bound 9-cis-RA crystallized in the RXRα ligand binding site. In yellow is the best pose found by Surflex-Dock in attempts to dock β-apo-13-carotenone into this binding site when it was built in a conformation resembling that of the 9-cis-RA. The RXR protein structure has been removed to facilitate observing the very different position β-apo-13-carotenone assumes in the docking experiment. It should be noted that a similar result was obtained if the β-apo-13-carotenone conformation from Figure 3 was docked (not shown) or even when redocking of the extracted 9-cis-RA was attempted (not shown), perhaps reflecting the effects of the highly strained structure of the bound 9-cis-RA. Regardless, the results of these modeling experiments suggest that β-apo-13-carotenone should be capable of as an antagonist to the RXRα protein.
Figure 3. Docking of β-apo-13-carotenone with the RXRα tetramer.
Shown is the structure of the ligand-binding domain of the tetrameric RXRα (in green) from PDB entry 1G5Y with: (a) the bound crystallized all-trans retinoic acid (ATRA) isomer (in white) and the Surfle-Dock docked structure of β-apo-13-carotenone (yellow) when built in a conformation resembling that of the bound ATRA; (b) same as in (a) with the protein helix 3 removed to facilitate visualization of the quality of docking in the binding site; (c) as in (a) with the RXR protein removed to show details of the similarity of the crystallized ATRA ligand and the docked pose of β-apo-13-carotenone.
Figure 4. Docking of β-apo-13-carotenone with the RXRα dimer.
Shown is the structure of 9-cis-retinoic acid (9-cis-RA) (in white) crystallized in the binding site of the RXRα from PDB entry 1FBY, along with the docked structure of β-apo-13-carotenone (in yellow) in the ligand binding site after its construction in a conformation that resembles that of the bound 9-cis-RA (the protein structure has been removed to facilitate appreciation of the significant difference between the bound and docked structures).
Implications
The biological activities of BACs are well understood in plant biology but there is limited information regarding their role in mammals. β-Ionone is a pollinator attractant and β-cyclocitral and β-ionone contribute to fruit or vegetable flavor [47–48]. β-apo-13-Carotenone blocks the growth of root hairs by interfering with PIN2-mediated auxin transport [49]. Also, this compound is believed to be the precursor of the well-known fungal pheromone trisporic acid [50]. Previous work by others has demonstrated that β-apo-14’-carotenal can function to inhibit RXRα at 10 µM concentrations. The results reported here suggest that β-apo-13-carotenone, the other product of the oxidative cleavage of β-carotene at the 13,14 double bond may affect gene expression mediated by RXRs in mammals. The pathways for the production of these cleavage products are not fully known and may involve enzymatic or non-enzymatic processes. These compounds are also likely present in the plant-derived foods consumed by mammals.
Acknowledgments
We thank Vanessa Reed for expert technical assistance and Dr. Rohit Tiwari for assistance in working with Surflex-Dock. We also thank Drs. Dianne R. Soprano (Temple University) and Noa Noy (Case Western Reserve University) for their advice on conducting transactivation assays, and Dr. Noy for the vectors used in this work. This work was supported by grants from the NIH (R01-DK044498 and R01-HL-049879) and the Ohio Agricultural Research and Development Center.
Abbreviations used
- BC
β-carotene
- BAC
β-apocarotenoid
- BCO1
β,β-carotene-15,15′-oxygenase
- BCO2
β,β-carotene-9’,10’-oxygenase
- ATRA
all-trans-Retinoic Acid
- RXR
retinoid-X receptor
- TR
thyroid receptor
- VDR
vitamin D receptor
- PPAR
peroxisome proliferator activator receptor
- LXR
liver X receptor
- FXR
farnesoid X receptor
- PXR
pregnane X receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Römer S, Fraser PD. Recent advances in carotenoid biosynthesis, regulation, and manipulation. Planta. 2005;221:1533–1535. doi: 10.1007/s00425-005-1533-5. [DOI] [PubMed] [Google Scholar]
- 2.Spurgeon SL, Porter JW. Biosynthesis of carotenoids, Biosynthesis of lsoprenoid Compounds. vol. 2. New York: Wiley & Sons; 1983. pp. 1–122. [Google Scholar]
- 3.Olson JA, Krinsky NI. Introduction: the colorful, fascinating world of the carotenoids: important physiologic modulators. FASEB J. 1995;9:1547–1550. doi: 10.1096/fasebj.9.15.8529833. [DOI] [PubMed] [Google Scholar]
- 4.Parker RS. Absorption, metabolism, and transport of carotenoids. FASEB J. 1996;10:542–551. [PubMed] [Google Scholar]
- 5.Wirtz GM, Bornemann C, Giger A, Muller RK, Schneider H, Schlotterbeck G, Schiefer G, Woggon WD. Helv. Chim. Acta. 2001;84:2315–3201. [Google Scholar]
- 6.Harrison EH. Mechanism of Digestion and Absorption of Dietary Vitamin A. Ann. Rev. Nutr. 2005;35:87–103. doi: 10.1146/annurev.nutr.25.050304.092614. [DOI] [PubMed] [Google Scholar]
- 7.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving b-carotene to retinal. J. Biol. Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 8.Wyss A, Wirtz G, Woggon W, Brugger R, Wyss M, Friedlein A, Bachmann H, Hunziker W. Cloning and expression of β,β-carotene 15,15′-dioxygenase. Biochem. Biophys. Res. Commun. 2000;271:334–336. doi: 10.1006/bbrc.2000.2619. [DOI] [PubMed] [Google Scholar]
- 9.Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, X F., Jr Cunningham, Identification, expression, and substrate specificity of a mammalian β-carotene 15,15′-dioxygenase. J. Biol. Chem. 2001;276:6560–6565. doi: 10.1074/jbc.M009030200. [DOI] [PubMed] [Google Scholar]
- 10.Yan W, Jang GF, Haeseleer F, Esumi N, Chang J, Kerrigan M, Campochiaro M, Campochiaro P, Palczewski K, Zack DJ. Cloning and characterization of a human β,β-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics. 2001;72:193–202. doi: 10.1006/geno.2000.6476. [DOI] [PubMed] [Google Scholar]
- 11.Allegretto EZ, McClurg MR, Lazarchik SB, Clemm DL, Kerner SA, Elgort MG, Boehm MF, White SK, Pike JW, Heyman RA. Transactivation properties of retinoic acid and retinoid X receptors in mammalian cells and yeast. J. Biol. Chem. 1993;26:625–633. [PubMed] [Google Scholar]
- 12.Keidel S, LeMotte P, Apfel C. Different agonist- and antagonist-induced conformational changes in retinoic acid receptors analyzed by proteases mapping. Mol. Cell Biol. 1994;14:287–298. doi: 10.1128/mcb.14.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostrowski J, Hammer L, Roalsvig T, Pokornowski K, Reczek PR. The N-terminal portion of domain E of retinoic acid receptors and is essential for the recognition of retinoic acid and various analogs. Proc. Natl. Acad. Sci. 1995;92:1812–1816. doi: 10.1073/pnas.92.6.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 15.Rowe A, Eager NS, Brickell PM. A member of the RXR nuclear receptor family is expressed in neural-crest-derived cells of the developing chick peripheral nervous system. Development. 1991;111:771–778. doi: 10.1242/dev.111.3.771. [DOI] [PubMed] [Google Scholar]
- 16.Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Naar AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. RXRβ: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- 17.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis Retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 18.Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, Zacharewski T, Chen JY, Staub A, Garnier JM, Mader S. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- 19.Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A. 9-cis Retinoic acid stereoisomer binds and activates the nuclear receptor RXRα. Nature. 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 20.Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, Oro AE, Kakizuka A, Evans RM. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 21.Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signaling. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 24.Mangelsdorf DJ, Umesono K, Kliewer SA, Borgmeyer U, Ong ES, Evans RM. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell. 1991;66:555–561. doi: 10.1016/0092-8674(81)90018-0. [DOI] [PubMed] [Google Scholar]
- 25.Mader S, Chen JY, Chen Z, White J, Chambon P, Gronemeyer H. The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificities of the DNA binding domains. EMBO J. 1993;12:5029–5041. doi: 10.1002/j.1460-2075.1993.tb06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan YJ, An D, Cai Y, Repa JJ, Hung-Po Chen T, Flores M, Postic C, Magnuson MA, Chen J, Chien KR. Hepatocyte-specific mutation establishes retinoid X receptor α as a heterodimeric integrator of multiple physiological processes in the liver. Mol. Cell. Biol. 2000;20:4436–4444. doi: 10.1128/mcb.20.12.4436-4444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehm MF, Zhang L, Badea BA, White SK, Mais DE, Berger E, Suto CM, Goldman ME, Heyman RA. Synthesis and structure-activity relationships of novel retinoid X receptor-selective retinoids. J. Med. Chem. 1994;37:2930–2941. doi: 10.1021/jm00044a014. [DOI] [PubMed] [Google Scholar]
- 28.Boehm MF, Zhang L, Zhi L, McClurg MR, Berger E, Wagoner M, Mais DE, Suto CM, Davies JA, Heyman RA. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J. Med. Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 29.Heald P. The treatment of cutaneous T-cell lymphoma with a novel retinoid. Clin. Lymphoma. 2000;S1:S45–S49. doi: 10.3816/clm.2000.s.009. [DOI] [PubMed] [Google Scholar]
- 30.Hurst RE. Bexarotene ligand pharmaceuticals. Curr. Opin. Investig. Drugs. 2000;4:514–523. [PubMed] [Google Scholar]
- 31.Kempf W, Kettelhack N, Duvic M, Burg G. Topical and systemic retinoid therapy for cutaneous T-cell lymphoma. Hematol. Oncol. Clin. North Am. 2003;17:1405–1419. doi: 10.1016/s0889-8588(03)00107-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhang C, Duvic M. Retinoids: therapeutic applications and mechanisms of action in cutaneous T-cell lymphoma. Dermatol. Ther. 2003;16:322–330. doi: 10.1111/j.1396-0296.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- 33.Corey EJ, Gilman NW, Ganem BE. New method for the oxidation of aldehydes to carboxylic ccids and esters. J. Am. Chem. Soc. 1968;90:5616–5617. [Google Scholar]
- 34.Kithsiri Wijeratne EM, Liu MX, Kantipudi NB, Brochini CB, Leslie Gunatilaka AA, Canfield LM. Synthesis and preliminary biological evaluations of β-carotene and retinoic acid oxidation products. Bioorg. Med. Chem. 2006;14:7875–7879. doi: 10.1016/j.bmc.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 35.Promega Corporation. Madison, WI, USA: 2001. pRL-TK Vector, Technical Bulletin No. TB240. [Google Scholar]
- 36.Gampe RT, Jr, Montana VG, Lambert MH, Wisely GB, Milburn MV, Xu HE. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000;14:2229–2241. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXRα ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain AJ. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 39.Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001;276:14110–14116. doi: 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- 40.Tang G, Wang XD, Russell RM, Krinsky NI. Characterization of β-apo-13-carotenone and β-apo-14'-carotenal as enzymatic products of the excentric cleavage of β-carotene. Biochemistry. 1991;30:9829–9834. doi: 10.1021/bi00105a003. [DOI] [PubMed] [Google Scholar]
- 41.Wang XD, Tang G, Fox JG, Krinsky NI, Russell RM. Enzymatic conversion of β-carotene into β-apo-carotenals and retinoids by human, monkey, ferret and rat tissues. Arch. Biochem. Biophys. 1991;258:8–16. doi: 10.1016/0003-9861(91)90322-a. [DOI] [PubMed] [Google Scholar]
- 42.Ho CC, de Moura FF, Kim SH, Clifford AJ. Excentral cleavage of β-carotene in vivo in a healthy man. Am. J. Clin. Nutr. 2007;85(3):770–777. doi: 10.1093/ajcn/85.3.770. [DOI] [PubMed] [Google Scholar]
- 43.Boehm MF, McClurg MR, Pathirana C, Mangelsdorf DJ, White SK, Hebert J, Winn D, Goldman ME, Heyman RA. Synthesis of high specific activity [3H]-9-cis-retinoic acid and its application for identifying retinoids with unusual binding properties. J. Med. Chem. 1994;37(3):408–414. doi: 10.1021/jm00029a013. [DOI] [PubMed] [Google Scholar]
- 44.Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 45.Goldstein A, Aronow L, Kalman SM. Principles of Drug Action: The Basis of Pharmacology. 2nd edition. New York: Wiley; 1987. [Google Scholar]
- 46.Kenakin TP. Pharmacologic Analysis of Drug-Receptor Interaction. New York: Raven Press; 1974. [Google Scholar]
- 47.Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004;14:1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 48.Winterhalter P, Rouseff RL. Carotenoid-Derived Aroma Compounds. American Chemical Society; 2002. [Google Scholar]
- 49.Schlicht M, Samajova O, Schachtschabel D, Mancuso S, Menzel D, Boland W. D'orenone blocks polarized tip growth of root hairs by interfering with the PIN2-mediated auxin transport network in the root apex. Plant J. 2008;55:709–717. doi: 10.1111/j.1365-313X.2008.03543.x. [DOI] [PubMed] [Google Scholar]
- 50.Schachtschabel D, Schimek C, Wöstemeyer J, Boland W. Biological activity of trisporoids and trisporoid analogues in Mucor mucedo (−) Phytochemistry. 2005;66:1358–1136. doi: 10.1016/j.phytochem.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 51.Ziouzenkova O, Orasana G, Sukhova G, Lau E, Berger JP, Tang G, Krinsky NI, Dolnikowski GG, Plutzky J. Asymmetric cleavage of β-carotene yields a transcriptional repressor of retinoid X receptor and peroxisome proliferator-activated receptor responses. Mol. Endocrinol. 2007;21:77–88. doi: 10.1210/me.2006-0225. [DOI] [PubMed] [Google Scholar]