Plants are complex, multicellular organisms whose growth and development responds to a plethora of intrinsic and environmental signals. Over the last several decades, molecular genetics has provided a highly effective means to dissect this complexity and pinpoint key regulatory genes and signals that control plant growth and development. However, the advent of genomic technologies has revealed that plant cells and tissues contain many thousands of molecular components that interact to form gene regulatory networks and biochemical complexes, which control organ growth and development. Researchers are currently challenged with making sense of the avalanche of -omics information about these components and their relationships. Systems biology provides a means to analyze these complex data sets and reveal new biological insights.
In this issue, four invited reviews describe how systems biology approaches are being used by plant researchers to generate mechanistic insights that span from genomic to cellular scales. Bassel et al. (pages 3859–3875) describe how high-throughput -omics approaches are being used to construct genome-scale plant regulatory networks. The authors review the generation of coexpression networks, mapping, and analysis of large-scale gene regulatory and metabolic networks and the protein interactome.
The availability of large-scale data sets has not eliminated the need for complementary smaller scale analyses. Indeed, many of the success stories in plant systems biology to date derive from modeling smaller scale networks inferred from genetic evidence. Pertinent examples include the gene regulatory networks that control circadian rhythms, hormone signaling, and tissue patterning. Middleton et al. (pages 3876–3891) review such examples, while introducing readers to the relevant mathematical and computational approaches needed to model regulatory networks.
Plants, as multicellular organisms, often require researchers to consider more than just the network and cellular scales and employ multiscale modeling techniques to predict emergent dynamics at the tissue and organ levels. Multiscale models consider behaviors on two or more scales, ranging from the subcellular up to whole-organism scales. This subdivision has led to discussions of bottom-up versus top-down approaches, respectively. Although it may appear that bottom-up approaches (building networks from information about individual components and their interactions) have seen explosive growth in recent years due to the development of new genomic technologies (reviewed in Bassel et al.), the distinction between these approaches is often blurred, and using -omics data (i.e., captured on a whole-genome or whole-organism level) to infer network properties may also be viewed as top-down. The most natural and appropriate approach arguably might rather be thought of as middle-out, in which models are constructed starting with the level at which we have the most information. For example, cell-based models are an attractive approach, where individual cells can be endowed with internal machinery (representing the subcellular networks) and the interactions of numerous individual cells lead to the (emergent) tissue-scale properties (see figure). Band et al. (pages 3892–3906) and Murray et al. (pages 3907–3919) review recent developments in multiscale modeling, illustrating how they are generating valuable new mechanistic insights into the regulation of root and shoot growth and development, respectively. These reviews also introduce readers to the relevant mathematical and computational approaches and describe the elements needed to develop dynamic multiscale models, such as information about the key gene regulatory networks, cell and tissue mechanics, and hydraulics. While this sounds challenging, it is clear that multiscale models are set to become increasingly important to plant researchers if we are finally to bridge the gap from genotype to phenotype.
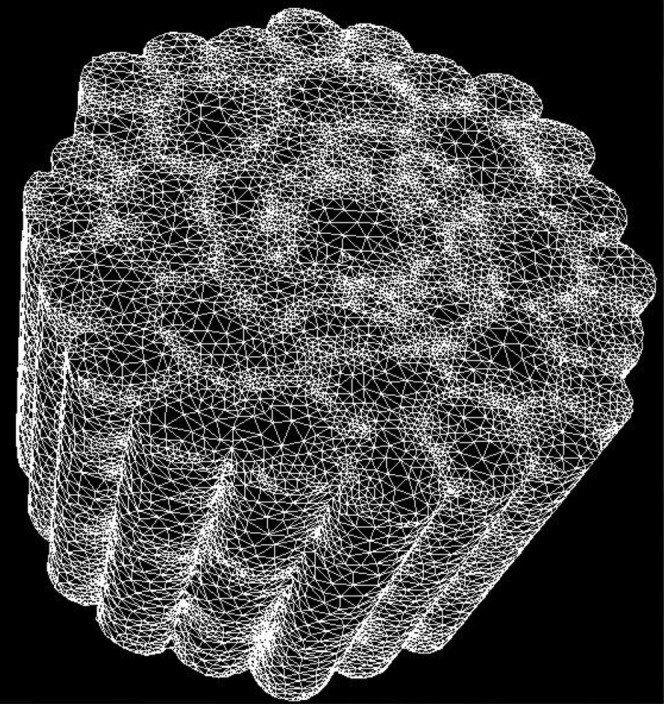
Individual cell reconstruction by a three-dimensional surface mesh, generated from an Arabidopsis thaliana root cross section. The triangular mesh allows for an accurate surface geometry representation and will host mathematical models of biological processes, such as hormone transport and/or signal transduction pathways. (Image courtesy of Andrew French, Alistair Middleton, and Benjamin Peret, Centre for Plant Integrative Biology, University of Nottingham, UK.)
Acknowledgments
The motivation for this set of Review articles came out the Plant Growth and Biology Modeling workshop held in Elche, Spain, in September of 2011. Funding for the workshop was provided by the Agron-Omics Consortium (http://www.agron-omics.eu/), the Centre for Plant Integrative Biology, Nottingham, U.K. (http://www.cpib.ac.uk/), and Universidad Miguel Hernández (http://www.umh.es/). Thanks to workshop organizers Jose Luis Micol and Héctor Candela and to Universidad Miguel Hernández for providing additional funding for N.A.E. to attend the workshop.
References
- Band L.R., Fozard J.A., Godin C., Jensen O.E., Pridmore T., Bennett M.J., King J.R. (2012). Multiscale systems analysis of root growth and development: Modeling beyond the network and cellular scales. Plant Cell 24: 3892–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel G.W., Gaudinier A., Brady S.M., Hennig L., Rhee S.Y., De Smet I. Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton A.M., Farcot E., Owen M.R., Vernoux T. (2012). Modeling regulatory networks to understand plant development: Small is beautiful. Plant Cell 24: 3876–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.A.H., Jones A., Godin C., Jan Traas J. (2012). Systems analysis of shoot apical meristem growth and development: Integrating hormonal and mechanical signaling. Plant Cell 24: 3907–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]


