Abstract
The shoot apical meristem (SAM) is a small population of stem cells that continuously generates organs and tissues. This review covers our current understanding of organ initiation by the SAM in Arabidopsis thaliana. Meristem function and maintenance involves two major hormones, cytokinins and auxins. Cytokinins appear to play a major role in meristem maintenance and in controlling meristematic properties, such as cell proliferation. Self-organizing transport processes, which are still only partially understood, lead to the patterned accumulation of auxin at particular positions, where organs will grow out. A major downstream target of auxin-mediated growth regulation is the cell wall, which is a determinant for both growth rates and growth distribution, but feedbacks with metabolism and the synthetic capacity of the cytoplasm are crucial as well. Recent work has also pointed at a potential role of mechanical signals in growth coordination, but the precise mechanisms at work remain to be elucidated.
INTRODUCTION: THE SHOOT APICAL MERISTEM, PHYLLOTAXIS, AND PATTERNING
Plants grow and generate new organs throughout their lifespan. This is possible through the activity of small populations of cells, called meristems, which continuously generate organs and tissues. This review deals with the aerial parts of the plant, where so-called shoot apical meristems (SAMs) play a central role. Positioned at the tips of shoots, they produce different types of organs (leaves, flowers, and floral organs), depending on the physiological state of the plant. A second type of aerial meristem, the secondary meristem, is responsible for the secondary growth in the girth (thickening) of stems. These have a number of characteristics and molecular mechanisms in common with SAMs Schrader et al., 2004), but will not be discussed further here. Instead, we will focus on the SAM itself and describe our current knowledge on how this meristem and adjacent tissues contribute to creating plant architecture.
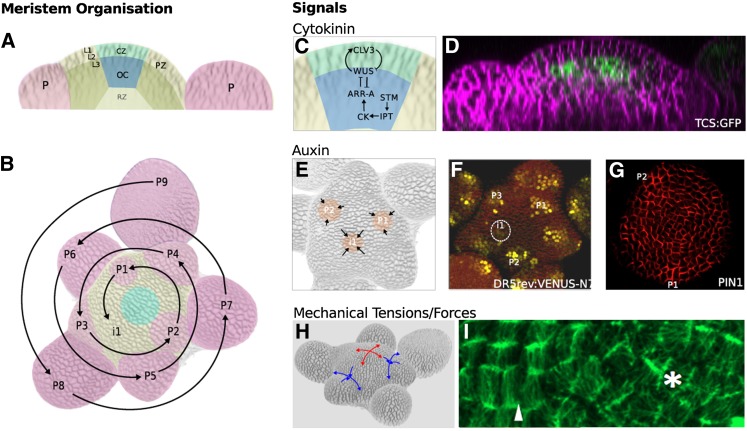
The SAM is a highly organized structure, and a simple histological analysis is sufficient to distinguish different zones and layers (Figure 1). At the summit of the meristematic dome lies the so-called central zone, where a small population of stem cells assures the maintenance of the meristem. This central zone is surrounded by a peripheral zone, where organ primordia are initiated. Superimposed on this zonation is an organization in layers, with a so-called tunica layer at the meristem surface, containing cells that divide in anticlinal (perpendicular to the surface) directions only. A striking feature of the SAM in most species is its capacity to generate organs in very regular arrangements (Figure 1). Although subsequent growth of the stem can affect patterning, these arrangements of the primordia, also called phyllotaxis, largely contribute to the final architecture of the plant. Different types of phyllotaxis exist: from simple alternate arrangements, where organs are produced in alternate, opposite positions, to the more complex whorled phyllotactic patterns where organs are arranged in multiple spirals. The type of phyllotaxis is species dependent, but it is not necessarily constant throughout development (reviewed in Steeves and Sussex, 1989; for a historical perspective, see Adler et al., 1997).
Figure 1.
The Organized Structure of the Arabidopsis SAM Is Generated through a Combination of Hormonal and Mechanical Signals and the Cellular Responses to These Signals.
(A) The SAM consists of the central zone (CZ), containing the stem cells and the surrounding peripheral zone (PZ). Primordia (P), the progenitors of new lateral organs, are initiated in the PZ. The CZ is maintained by an underlying organizing center (OC). Below the OC is the rib zone (RZ), which is responsible for the elongation of the stem. In dicotyledonous species, the two outermost cell layers (L1 and L2) divide anticlinally and make up the so-called tunica layer. From the third cell layer (L3) inwards, cell divisions take place in nonuniform orientations, resulting in the less organized tissue structure known as the corpus. Organ initiation is largely controlled by auxin transport and distribution within the L1 of the tunica (see [E]), whereas maintenance of the CZ requires communication between the tunica and the corpus (see [C]).
(B) Primordia are spaced according to a regular pattern or phyllotaxis. In Arabidopsis, spacing follows a spiral phyllotaxy. P9 indicates the oldest primordium and P1 the youngest. Successive organs are separated by an angle of ∼137.5. The position at which the next primordium (i1) will be initiated can be predicted based on this rule.
(C) and (D) Cells in the OC express the transcription factor WUS, which promotes the expression of CLV3, a small peptide that moves into the surrounding tissue. In the central zone, CLV3 interacts with the receptor-like kinase CLAVATA1, inhibiting WUS expression and promoting stem cell fate. The hormone cytokinin (CK) is also required to establish and maintain the central zone. The transcription factor STM upregulates the expression of IPT genes that are rate limiting for cytokinin biosynthesis. Cytokinin activates A-type ARR transcriptional regulators via a phosphorelay system. A-type ARRs stimulate downstream cytokinin responses but also downregulate the expression of WUS. WUS inhibits the expression of A-type ARRs, creating a negative feedback loop that regulates size and position of the organizing center and, thus, of the central zone. Consistent with this role for cytokinin, the cytokinin response reporter Two-Component-Output-Sensor: Green Fluorescent Protein (TCS:GFP) (Müller and Sheen, 2008) (D) is detected at high levels in the organizing center, as previously shown by Yoshida et al. (2011).
(E) to (G) The positions of primordia are determined by auxin maxima (orange) that are created by self-organizing patterns of auxin transport (arrows) (E). The auxin response reporter DR5:3xVENUS-N7 is detected in primordia before outgrowth begins (see circled i1 in [F]). Directional movement of auxin is produced by the activity of PIN1 proteins, which transport auxin out of cells and are polarly localized. Immunolocalization of PIN1 in the L1 of the SAM shows that PIN1 proteins are oriented toward the auxin maxima (G).
(H) and (I) Mechanical principle directions of stresses experienced by the tissue varies across the SAM as a result of the geometry of the structure and as a result of local differences in growth rates. Stresses at the dome of the meristem are isotropic (red arrows), whereas in the fold between the outgrowing primordia and the SAM, the direction of stresses is highly anisotropic (blue arrows). Microtubules orient according to the principle direction of the mechanical stress that they experience. [I] shows a detail of a meristem expressing Microtubule Associated Protein4: Green Fluorescent Protein (as previously shown in Hamant et al., 2008). Note that microtubules at the meristem dome do not show a clear orientation (asterisk), but in organ boundaries a marked coordination of tubules is observed (arrowhead).
([D], F. Besnard and T. Vernoux, unpublished data, construct from Müller and Sheen [2008]; [G], reprinted from Barbier de Reuille et al. [2006], Figure 3F; [F] A. Jones, unpublished data, construct from Laskowski et al., 2008, [I], J. Traas, unpublished data, live imaging as described in Fernandez et al. [2010].)
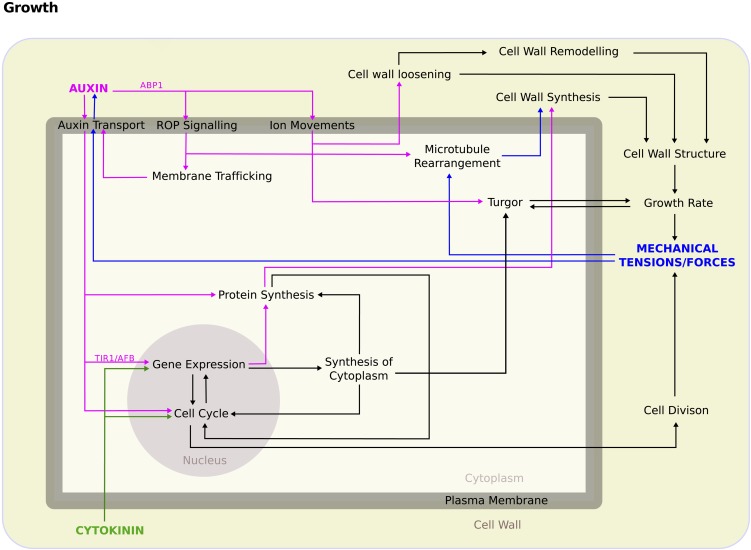
Here, we give a brief overview of our current understanding of organ initiation by the SAM in Arabidopsis thaliana, including positioning and outgrowth at the shoot apex. In this context, an important concept is that multicellular organisms are complex systems (i.e., are composed of elements that interact according to local rules). These interactions lead to the emergence of collective properties at a higher level of organization. Thus, macromolecules self-organize into well-defined, cellular structures and molecular networks to form cells, while at a higher level of organization cells interact to assemble into tissues and organs with particular shapes. Importantly, these emergent properties cannot be deduced from adding up local behavior (i.e., the whole is not simply a sum of the parts). This is partially linked to the presence of multiple feedbacks between the different levels of organization (Figure 2). Considering living systems as complex systems has important implications. First, since emergent properties cannot be understood by studying local properties alone, complex systems must be analyzed at multiple scales. Second, since novel properties at higher scales depend on the interactions between the individual elements, it is essential to consider the properties of these interactions, which can be both chemical and mechanical in nature.
Figure 2.
Cytokinin, Auxin, and Mechanical Forces Provide Inputs That the Cell Responds to by Regulating a Range of Cellular Processes That Affect Growth.
Auxin-induced pathways are indicated by magenta arrows, cytokinin-induced pathways are indicated by green arrows, and pathways induced by changes in the mechanical forces experienced by the cell are indicated by blue arrows. Black arrows indicate pathways that may be induced by more than one input. Note that many processes can be regulated by more than one pathway and that some pathways feedback in such a way as to change the input, for example, auxin concentration in the cell wall leads to changes in auxin transport and therefore affects auxin concentration. It is also important to note that growth of the cell, and those around it, will affect both mechanical forces and hormone gradients. Therefore, the system must not only be considered over multiple levels of organization but also as a dynamic system over time.
PATTERNING REQUIRES SIGNALING: A FOCUS ON CHEMICAL SIGNALS
We first discuss the main signaling molecules involved in setting up and maintaining patterning at the SAM. This includes two major hormones, cytokinins and auxins. Other classes of plant hormones, including brassinosteroids, gibberellins, and strigolactones, have been associated with SAM function (reviewed in Alabadí et al., 2009), but their role there is less well defined. Therefore, they will not be discussed in more detail.
Cytokinins and Patterning of the Meristem Proper
Cytokinins have been associated with the SAM proper (i.e., the peripheral and central zones). First, there is a strong link between cytokinins and the undifferentiated state of the meristematic cells in the central zone. The central regions of the SAM have high cytokinin levels and activated cytokinin responses, which appear to be important to sustain the stem cell population (Shani et al., 2006). This has been studied in detail in Arabidopsis. In this species, SHOOT MERISTEMLESS (STM), a homeodomain transcription factor of the Class 1 KNOX gene family, plays a major role in SAM function (Long et al., 1996). STM is expressed throughout the nonorganogenic regions of the SAM but excluded from regions of primordia formation. It is essential both for SAM establishment during embryogenesis and its subsequent maintenance, since it maintains the environment in which the stem cell niche can be sustained (Scofield et al., 2008). STM acts in part by promoting cytokinin synthesis by increasing ISOPENTENYL TRANSFERASE (IPT) gene expression (Jasinski et al., 2005; Yanai et al., 2005). Conversely, expressing IPT under control of the STM promoter can partially rescue some aspects of stm mutant phenotypes, as can external applications of cytokinin (Yanai et al., 2005).
Cytokinin signaling has also been associated with maintaining the spatial boundaries of the central zone of the meristem. In this region, a local signaling loop operates in the maintenance of the stem cell population at the meristem summit. This maintenance is directed by an organizing center in the internal layers of the meristem, comprised of cells expressing the transcription factor WUSCHEL (WUS) (Mayer et al., 1998; Figure 1C), which promotes stem cell identity in the overlying cells. It also induces them to express the peptide ligand CLAVATA3 (CLV3), which via interaction with the receptor-like kinase CLV1, feeds back to inhibit WUS expression and maintain a stem cell pool of a constant size (Schoof et al., 2000). This feedback loop interacts with the downstream signaling network of cytokinin, comprised of membrane-localized receptors, which upon cytokinin perception activate both positive (B-type) and negative (A-type) transcriptional regulators (ARRs) via a phosphorelay system (Hwang et al., 2012) and positions the WUS expression domain relative to the L1 layer (Gordon et al., 2009). Ectopic expression of WUS represses the negative A-type ARRs, which stimulate downstream cytokinin responses (Leibfried et al., 2005). Conversely, overexpression of an A-type ARR inhibits WUS expression. Cytokinins, therefore, not only contribute to maintaining SAM cells in the meristematic state, but are also involved in regulating the size of the stem cell population at the meristem summit. Computational modeling demonstrates that while the feedback between WUS and CLV3 is sufficient to produce a stem cell niche of a determined size, additional feedback created by the cytokinin signaling pathway must also be included to produce accurate positioning of the stem cell niche that is maintained when cell divisions are included (Chickarmane et al., 2012).
Cytokinin also mediates light responses of the SAM. When shoot apices are grown in the dark, organ primordial are not formed. Application of cytokinin alone is sufficient to increase apical growth and lead to initiation of primordia (Yoshida et al., 2011). This suggests that cytokinin stimulates growth, which can then be redirected to organ initiation by auxin.
It is notable that auxin responses are excluded from the central region, even when high levels of auxin are directly placed on the meristem tip (Reinhardt et al., 2003). Since combined modeling and experimental approaches have shown that auxin levels are high in the SAM center (Barbier de Reuille et al., 2006; Vernoux et al., 2011), there must be active repression of auxin responses alongside the high levels of cytokinin signaling, but the mechanism by which this is achieved is unknown. Conversely, at least in Arabidopsis, cytokinin responses are apparently repressed when cells leave the meristem and are incorporated in the young primordia, since young primordia express ARABIDOPSIS PHOSPHOTRANSFER PROTEIN6 (Bartrina et al., 2011). This encodes a nonfunctional phosphotransfer protein that negatively interferes with the cytokinin-activated phosphorelay chain mentioned above (Mähönen et al., 2006).
Patterning at the Meristem Periphery: A Central Role for Auxin and Auxin Transport
Many lines of evidence, including experiments involving the precise placement of auxin on the SAM surface, show that auxin is the major signal associated with organ initiation and positioning at the SAM. The most abundant form, indole-3-acetic acid (IAA), is not able to diffuse freely from cell to cell, and its transport throughout tissues is facilitated by auxin importers and exporters at the plasma membranes (recently reviewed in Grunewald and Friml, 2010). In particular, the auxin exporters (transmembrane proteins of the so-called PIN-FORMED or PIN family) show a polar localization within individual cells. Since the localization between neighboring cells is often coherent, it has been assumed that these transporters create fluxes of auxin through the tissues, causing the formation of auxin maxima and minima.
The first auxin exporter to be identified was PIN-FORMED1 (PIN1) (Okada et al., 1991; Gälweiler et al., 1998) in Arabidopsis. The pin1 mutant has a needle-like inflorescence stem, due to its inability to initiate organs from the SAM. PIN1 is strongly expressed in the SAM, where it is mainly found in the outermost cell layer and in vascular tissues. The pin1 phenotype can be complemented by local external auxin applications, which are able to induce the formation of flower buds (Reinhardt et al., 2000). These initial observations suggested that high local auxin concentrations are required for the initiation of a new organ and that PIN transporters are required for the creation of such auxin maxima. Direct evidence demonstrating the existence of auxin maxima at the periphery of the meristem is still lacking due to the absence of a true marker for auxin concentrations. However, this scenario is supported by three different lines of evidence. First, the patterns of polar localization of PIN1 (Wisniewska et al., 2006) suggest auxin fluxes directed toward the young primordia (Benková et al., 2003; Reinhardt et al., 2003; Barbier de Reuille et al., 2006). Second, the synthetic DR5 promoter, which is activated by auxin-responsive transcription factors, is strongly expressed in very young organs, indicating the activation of auxin-induced genes during the early stages of organ initiation (Benková et al., 2003; Barbier de Reuille et al., 2006). Third, a recently developed perception sensor that is degraded in the presence of high auxin concentrations confirms the presence of auxin maxima at the sites of organ formation (Vernoux et al., 2011). However, it should be noted that in the vegetative SAM of pin1 mutants, leaf primordia are formed whose positioning is nonrandom, suggesting that further PIN1-independent mechanisms exist (Guenot et al., 2012).
A second set of transporters associated with auxin distribution at the SAM is the family of AUX/LIKE AUX (LAX) influx carriers (for a thorough functional analysis, see Péret et al., 2012). The founding member of the gene family, AUX1, is expressed at the outer cell layer (also called L1) of the SAM (Bennett et al., 1996; Reinhardt et al., 2003). The protein seems to be evenly localized over all membranes of the L1, indicating that it is not involved in the creation of hormone fluxes. Instead, it might concentrate auxin at the meristem surface. The AUX1 sequence shows a high degree of similarity with three other sequences in the Arabidopsis genome, named LAX1, LAX2, and LAX3 (Swarup and Bennett, 2003), but even the quadruple mutant is still able to produce a viable, moderately fertile plant, albeit with significant changes in architecture. It is therefore conceivable that other proteins, such as certain P-Glycoprotein transporters, which have also been associated with auxin import, share a redundant function with the members of the LAX family in the meristem (Boutté et al., 2007). Another clue on the precise role of AUX1 comes from a double pin1 aux1 mutant. Auxin applications do not induce single flowers on the apex of such a double mutant, but rather very large, fused organs, suggesting that somehow AUX1 is required for the restriction of organ boundaries (Reinhardt et al., 2003). Altogether, the available data indicate that the formation of local auxin maxima mainly depends on the action of PIN exporters at the meristem surface. AUX and LAX proteins act to facilitate organ positioning, probably by guaranteeing a sufficient supply of auxin in the L1 layer.
Models for Polar Auxin Transport Regulation: Canalization or Up-the-Gradient?
What coordinates auxin fluxes in the meristem? The principles underlying the observed transport patterns could be very simple. Jönsson et al. (2006) and Smith et al. (2006) proposed models where cells check the auxin concentrations in their direct environment and subsequently pump auxin toward neighbors with a higher concentration (i.e., they move auxin against the concentration gradient). Because these patterning processes require the interaction of hundreds of cells, it is impossible to estimate on a purely intuitive basis if a particular scenario is plausible or not. Therefore, computational modeling was used as a powerful means to test this hypothesis. Interestingly, these models showed that transport against the gradient is sufficient to reproduce realistic PIN1 patterns and to generate different types of phyllotactic patterns (Jönsson et al., 2006; Smith et al., 2006). It should be noted that these models show that the mechanism is plausible, but by no means do they represent absolute, mathematical proof. Indeed, other models are able to explain the distribution of auxin transporters. Another hypothesis, for instance, was based on the pioneering work of Sachs (1969), who proposed the existence of a positive feedback between flux and transport. It was subsequently shown that, in principle, this mechanism is able to amplify small fluxes and can potentially create canals of auxin between hormone sources and sinks (Mitchison, 1981). A range of experiments supports the canalization hypothesis, at least in the inner tissues of the plant, where it can account for the formation of veination patterns (Scarpella et al., 2004, 2006; Sauer et al., 2006). Note that this would imply the coexistence of two radically different mechanisms for PIN allocation in the membrane, one based on flux sensing (in the inner tissues) and the other on local concentration sensing (at the meristem surface). Therefore, Stoma et al. (2008) tested whether canalization could potentially also account for the behavior of auxin transporters at the SAM surface. Using a dedicated computer simulation tool, they showed that this was indeed the case, thus providing a unifying concept for the control of auxin distribution in the plant (Stoma et al., 2008). More recently, Bayer et al. (2009) tested a third scenario where both modes of auxin transport (i.e., up-the-gradient and canalization) coexist. One or the other mechanism would prevail, depending on the absolute auxin concentrations (Bayer et al., 2009). Further experiments are now required to distinguish between flux-based and other hypotheses. Testing of the models will first of all require identification of the molecular mechanism controlling PIN1 polarity. It will then be possible to determine if these mechanisms are compatible with any of the proposed hypotheses. Some attempts to construct more mechanistic models (including cellular processes) have already been made. Wabnik et al. (2010), for example, proposed a flux-based model wherein auxin feedbacks on PIN endocytosis and production were combined with extracellular auxin signaling. This model was able to reproduce several patterning processes. In parallel, Heisler et al. (2010) proposed a model involving wall mechanics to explain transport against the gradient (see also below).
Beyond Auxin Maxima
What happens once a localized auxin maximum is generated? The hormone appears to act via two distinct pathways. (1) First, it interacts with AUXIN BINDING PROTEIN1 (ABP1), an auxin binding protein localized at the plasma membrane and endoplasmic reticulum (Robert et al., 2010; Xu et al., 2010). The precise function and regulation of ABP1 is still poorly understood, but it appears to mediate rapid cellular responses involving membrane traffic and the cytoskeleton as discussed below. (2) The second response pathway involves a complex network of transcriptional regulators (reviewed in Leyser, 2006; Guilfoyle and Hagen, 2007; Chapman and Estelle, 2009). This network is composed of the so-called auxin response factors (ARFs), transcription factors that activate or repress auxin-regulated genes. In the absence of auxin, ARFs themselves are inactivated by dimerization with repressors of the Aux/IAA family (Guilfoyle and Hagen, 2007; Szemenyei et al., 2008). In the presence of auxin, the Aux/IAAs are targeted to the proteasome by an ubiquitin ligase complex (Leyser, 2006; Chapman and Estelle, 2009) that functions as an auxin receptor. In this process, auxin promotes the interaction between Aux/IAA proteins and the TIR1 F-box protein (or homologs) of this complex (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Tan et al., 2007). The auxin-induced degradation of Aux/IAAs then releases ARFs to regulate transcription of their target genes.
Vernoux et al. (2011) found that a subset of 13 ARFs and 12 AUX/IAAs were coexpressed at the SAM. Interestingly, most of these genes were more strongly expressed at the periphery of the meristem where the organs are initiated. Expression was mostly weak at the meristem center. A second important observation concerned a large-scale two-hybrid interaction analysis, which suggested a very simple network structure, where the 25 ARFs and Aux/IAAs function as a double-brake system. In the absence of auxin, ARF transcriptional activators are predicted to be inhibited both by the interaction with Aux/IAAs and competition with a second group of ARFs that function as transcriptional repressors. By contrast, in the presence of auxin, the ARF activators are released from the inhibition by Aux/IAA, but still must compete with ARF repressors for specific sites. The very simple structure of this network provides robustness to the system. In addition, the strong expression levels at the meristem periphery suggest high auxin sensitivity there.
FROM SIGNALS TO GROWTH PATTERNS: FROM BIOCHEMISTRY TO PHYSICS
Hormones first interact with perception and downstream signaling pathways involving a network of receptors and transcriptional regulators. Next, these regulators must somehow control local growth rates, but how is this achieved? How do local biochemical activities translate into particular changes in geometry, and what feedbacks exist between growth and geometry? These questions are further complicated by the fact that a growing biological system is not simply a geometrical structure in which chemical gradients and molecular networks operate. It is also a physical structure governed and constrained by mechanical cues. Therefore, if we want to understand how organs are initiated, we also need to take into account the physical properties of the system. In plants, this means understanding the nature of the plant cell wall.
The Cell Wall as a Central Element in Growth Control
How do the individual cells of a tissue grow? Mechanistically, plant cells are under high internal turgor pressure. They are prevented from bursting by the dense network of cellulose microfibrils that are cross-linked to each other by other polysaccharides within the cell wall (Keegstra, 2010). Cell expansion can only take place as long as the cells are under pressure, and there is strong evidence that the irreversible yielding of the cell wall to this pressure is inextricably linked with cell growth. A widely accepted hypothesis proposes that, for a given turgor pressure, cell expansion is driven initially by cell wall loosening and subsequent remodeling and synthesis. In growing leaves, this has been linked to acidification of the cell wall (Dale, 1986) and the activity of cell wall–loosening enzymes such as expansins (Cosgrove, 1996) or pectin-modifying enzymes. Expansins cause cell wall relaxation, which appears to involve the disruption of hydrogen bonds between cellulose microfibrils and cross-linking glycans in the wall (reviewed in Li et al., 2003). Importantly, classical experiments by Fleming et al. (1997) showed that local, external applications of expansins could also induce outgrowths on tomato (Solanum lycopersicum) meristems, and several expansin genes are indeed expressed at high levels in young organ primordia. More recently, Peaucelle et al. (2008, 2011) also showed a link with pectin-modifying enzymes. Pectins, which form the amorphous matrix surrounding the rigid cellulose microfibrils, are believed to play a major role in the definition of cell wall stiffness, depending on the degree of methylation of the pectins. Indeed, Peaucelle et al. (2008) found that pectin methylesterase (PME) activity in the wall is correlated with organ initiation. Inhibiting PMEs prevented organ outgrowth causing the formation of pin-like apices.
In summary, the data suggest that a combination of wall-modifying activities causes organs to bulge out as a consequence of changes they produce in the physical properties of the wall (Figures 1H to 1J). To demonstrate effectively that this is the case, several groups have started directly to determine these physical parameters, in particular wall stiffness. This work is still in an early phase and has so far only provided a partial view, which nevertheless tends to confirm that patterning at the meristem is correlated with changes in wall stiffness. In particular, the slow-growing cells at the tip of the meristematic dome have walls that are probably stiffer than those at the periphery (Milani et al., 2011; Kierzkowski et al., 2012), whereas organ outgrowth is correlated with a reduced elasticity, in particular in the inner layers of the meristem (Peaucelle et al., 2011). However, it should be cautioned that experiments showing that cell wall loosening can induce cell growth (e.g., in transgenic lines) are not proof of causation in normal growth. This will require elucidation of the interaction mechanisms between cell wall growth and other aspects of cellular growth.
Shape changes depend not only on growth rates, but also on growth directions, for example, growth anisotropy, which in turn depends on the orientation of the cellulose microfibrils in the extracellular matrix (i.e., the cell wall texture). Walls with highly oriented cellulose fibers tend to grow perpendicular to the main fibril direction. Cellulose is synthesized by membrane-bound enzyme complexes, and there is strong evidence that the microtubular cytoskeleton guides these enzymes and thus controls cell wall texture. How precisely this occurs is still not well understood, but it seems that microtubules, through their tight association with the plasma membrane, somehow restrict the movements of cellulose synthase complexes (Bringmann et al., 2012). What regulates the structure of the microtubule cytoskeleton is also unclear, but recent evidence indicates that microtubule dynamics and stability could play a role. Studies where microtubules were depolymerized showed that organs still tend to grow out but that organ boundaries are no longer defined physically, showing that cell anisotropy is required for tissue folding (buckling) between floral primordia and the rest of the meristem. A second role for wall anisotropy is in the stem of the growing inflorescence, which loses its cylindrical shape when microtubules are removed (Hamant et al., 2008). These data show that while the orientation of cell growth is not required for organ level patterning, it is necessary for its correct elaboration through morphogenesis. This exemplifies the principle that instructions (patterns) can be established at a higher level but can only be implemented through the correct behavior of lower level elements.
Hormonal Regulation of Cell Wall Properties
The primary hormone linked with wall modifications is auxin. Evidence suggests that auxin can affect cell wall structure through both transcriptional and nontranscriptional pathways, although it is important to appreciate that much of the data on cell growth in plants is derived primarily from expanding cells outside of the meristematic regions, and the general applicability of these mechanisms to meristem cells is not certain.
Auxin-mediated growth has classically been described by the acid growth hypothesis, which proposes a nontranscriptional mechanism for the acidification-linked loosening of the wall. In expanding nonmeristematic cells, where cell growth is largely associated with fluid uptake and vacuole expansion, application of exogenous auxins rapidly induces H+ extrusion from the cytosol into the cell wall due to auxin-induced activation of plasma membrane H+-ATPases through phosphorylation (Takahashi et al., 2012) and reduced endocytic recycling (Paciorek et al., 2005). Cell wall–modifying expansin proteins show a measurable increase in activity within the pH range created in the cell wall by this movement of H+ (McQueen-Mason et al., 1992), so the electrophysiological response to auxin can be linked to a known mechanism of cell wall loosening. The movement of H+ also stimulates uptake of K+ from the cell wall into the cytosol through the activation of voltage-gated ion channels (Philippar et al., 1999; Bauer et al., 2000). This inward movement both rectifies the hyperpolarization of the membrane caused by the movement of H+ and serves to lower the osmotic potential of the cytosol such that water is drawn into the cell and turgor pressure increased.
It has long been suggested that ABP1 functions as an auxin receptor in the acid growth pathway. Electrophysiological responses are altered in cells that under- or overexpress ABP1 (Ruck et al., 1993; Leblanc et al., 1999) and an ABP1 knockout mutant appears to have defects in cell growth during embryogenesis (Chen et al., 2001). The exact function of ABP1 has remained enigmatic, but significant progress has recently been made in demonstrating that cell wall–localized ABP1 is required for auxin to regulate clathrin coat–mediated endocytosis of plasma membrane–localized proteins, including H+ ATPase (Paciorek et al., 2005; Robert et al., 2010).
The acid growth hypothesis offers an explanation for rapid cellular expansion. However, it is accepted that a transcriptional response is required to sustain growth over longer periods of time since a supply of cell wall proteins will be needed. Many mutants associated with the Aux/IAA pathway have dwarfed phenotypes, indicating reduced auxin-induced growth, including the axr3-1 mutant, a gain-of-function mutation of IAA17 that prevents its degradation in response to auxin (Leyser et al., 1996; Worley et al., 2000). Transcriptomic analysis of the axr3-1 mutant identified 108 genes that were repressed in the mutant compared with the wild type, of which 28 were associated with roles in cell wall biosynthesis and remodeling. Normally, in response to auxin, IAA17 would be degraded and the repression of these genes relieved. These regulated genes included both structural cell wall proteins and proteins with cell wall–modifying activities, such as expansins and PMEs (Overvoorde et al., 2005), which, as mentioned earlier, have been demonstrated to be associated with organ initiation in the SAM (Pien et al., 2001; Peaucelle et al., 2008, 2011).
Understanding the different pathways through which auxin modifies the cell wall highlights an important point, namely, that the structure of the cell wall can be regulated by cytoplasmic processes, such as vesicular trafficking and protein synthesis. Coordination of processes occurring in the cytoplasm and in the cell wall is therefore important for cell growth, and we will return later to the role of the cytoplasm in growth.
Growth Coordination: A Role for Mechanical Signals?
So far, we have mainly considered the physical properties and resulting growth patterns as an output of chemical patterning. However, evidence has started to accumulate indicating that physical properties themselves could feed back on the chemical gradients and cellular structures.
Different parts of the meristem grow at different rates, and even neighboring cells can have very different expansion rates. This is typically the case at the organ boundaries, where cells grow much more slowly than their direct neighbors in the young forming organ (Kwiatkowska and Dumais, 2003). As a result, there needs to be some mechanical compromise between these neighbors. In other words, the overall tensions generated in growing tissues have the potential to feed back on local expansion rates (reviewed in Hamant and Traas, 2010). Hamant et al. (2008) proposed a hypothesis where patterned fields of forces at the meristem surface influence cell wall and microtubule organization. To this end, they had to consider the overall mechanical properties of the meristem. Although these are not well characterized, a number of properties of the meristem have been proposed in the literature: (1) the meristematic tissue is elastic; (2) the outer epidermal wall is under a uniform turgor pressure generated by the internal tissues; and (3) this, in turn, limits meristem growth (Hamant et al., 2008, and references therein). If these assumptions are correct, the meristem can be considered as a pressure vessel, a shell inflated by an inner pressure. Since the physical properties of these types of objects are well known, a theoretical distribution of the principal stress directions can be calculated that largely depends on the geometry of the system. Interestingly, these predicted patterns at the meristem surface matched those of the microtubules next to the outer epidermal wall: for example, (1) randomly oriented at the dome of the SAM and young primordia, (2) oriented along the organ boundaries, and (3) perpendicular to the long axis of the growing stem. This led to the hypothesis that microtubules somehow align along the main force directions. Since they orient the cellulose microfibrils, they would reinforce the wall anisotropically to resist the forces. This would tend to reinforce or maintain the tissue stresses and, as a result, cause tissue folding at organ boundaries and guarantee the formation of a cylindrical stem. In this context, by providing directional, positional information to the cells, the physical forces would function as a signal able to coordinate growth over long distances. This hypothesis was further supported by several experiments. First, applying external mechanical forces to the meristem caused changes in the orientation of the microtubules over a period of 4 to 6 h. Second, similar changes of the microtubules were observed along predicted stress patterns around ablated cells (Hamant et al., 2008; Heisler et al., 2010; Uyttewaal et al., 2012).
This behavior of the microtubules has some interesting implications for auxin signaling. Heisler et al. (2010) observed that the main orientation of the microtubular arrays is frequently parallel to the membranes that bear PIN proteins. In addition, PIN proteins move to membranes that are parallel to microtubule reorientation in response to cell ablations with similar dynamics. Therefore, there seems to be some type of coupling between the organization of the cytoskeleton and auxin transport. In this context, it is interesting to note that in leaf epidermal pavement cells, actin-rich domains and domains where microtubules are attached are well separated (Xu et al., 2010). Interestingly, the actin-rich domains are also labeled by PIN proteins in these cells. It was proposed that this separation of microtubule domains and actin/PIN-rich domains was regulated by small Rho GTPases. If such a system indeed operates in the SAM, it is tempting to propose that the effect of mechanics on cytoskeleton organization also feeds back on cell polarity and auxin transport, although this remains to be proven.
Although the mechanical feedback hypothesis is attractive, it is also facing some major challenges. In particular, it is not obvious how the forces are sensed and translated by the cells. A glimpse of what a sensing mechanism might look like was obtained by analyzing the botero mutant in Arabidopsis (Uyttewaal et al., 2012). BOTERO encodes a katanin (i.e., a microtubule-severing protein involved in microtubule bundling). Knocking out the gene reduces considerably the capacity of microtubules to organize into ordered arrays and simultaneously reduces growth coordination at the meristem. As a result, the surface of the SAM becomes highly irregular and the mutant is no longer able to form well-defined organ boundaries. Part of the force-sensing mechanism could therefore be based on microtubule dynamics. Another element involved in such a mechanism could be the cell wall itself. Obviously even minute changes in stress patterns will immediately cause changes in wall structure, including changes in the relative positions of the cellulose fibers. This information, stored, as it were, in the cell wall, could potentially be transmitted to the plasma membrane and thence to the cytoplasm.
SIGNALING AND GROWTH: BEYOND THE WALL
Growth and Cytoplasmic Synthesis
The previous paragraphs might give the impression of a relatively straightforward scenario where growth patterns depend on wall loosening, synthesis, and cellulose microfibril orientation, largely in response to auxin signals. The situation is more complex, however, as all cells are filled with cytoplasm. Therefore, growth is also associated with an increase in cytoplasm requiring the production of macromolecules, primarily proteins, itself dependent on ribosome number and activity. It is unclear whether this could be triggered by cell wall–derived processes or whether alternative pathways coordinate overall metabolic activities, including wall synthesis. In other words, does cell wall expansion drive the internal processes or vice versa? One direct line of evidence comes from the highly localized induction of expansin gene expression on tobacco (Nicotiana tabacum) SAMs, which led to the induction of normal leaf primordia (Pien et al., 2001). Induction on the flank of an existing primordium also led to an outgrowth of the developing leaf blade. This might suggest that cell wall loosening is driving growth and that the synthesis of cytoplasm simply follows. However, there are multiple lines of evidence suggesting that this is not necessarily the case. Inhibition of DNA synthesis, for example, also arrests growth (Grandjean et al., 2004) and mutants perturbed in diverse metabolic activities also show reduced size. Therefore, there is no doubt that growth requires multiple feedbacks between the cell wall and cytoplasmic synthesis.
The coordination between metabolic activity and growth has been intensively studied in animals because of its close relationship with cell transformation leading to tumorigenesis (Jorgensen and Tyers, 2004). The TOR kinase plays a key role in stimulating growth in response to nutrient signals (yeast) or growth factors (metazoans) acting by multiple routes. These include increasing the production of ribosomes through multiple mechanisms, such as stimulating rRNA and ribosomal protein synthesis, as well as by activating ribosomal protein S6 kinase (S6K). The phosphorylation of S6 enhances the translation of a subset of RNAs, and S6K also activates eukaryotic initiation factor 4E to stimulate translation. Loss of function of TOR or its downstream effectors, including S6K and ribosomal protein genes, reduces cell growth. These effects are clearly seen in minute flies with crippled ribosomal protein production as well as s6k mutants. In these examples, the effects on cell size are different, with minute flies having normal sized cells and s6k very small cells, suggesting S6K also affects the coupling of cell growth and division (Jorgensen and Tyers, 2004). Signaling to TOR in animals occurs through receptors, such as the IGF receptor via PI3 kinase, PDK1, and Akt. An alternative pathway in animals involves Ras and myc, which promotes ribosome synthesis as well as directly regulating genes encoding metabolic enzymes and other proteins required for cell growth.
Evidence is beginning to suggest that mechanisms analogous to those found in animals are likely also to be relevant to plant cells. Indeed, plants have both the TOR pathway (Garrocho-Villegas and Sanchez de Jiménez, 2012; Robaglia et al., 2012; Xiong and Sheen, 2012) and its important target S6K (Henriques et al., 2010), and TOR has been shown to have a growth-limiting function in plant cells as in animals (Henriques et al., 2010). Phosphorylation of S6K has been used as an assay of TOR activity to identify regulators of the pathway. In Arabidopsis cell culture, S6K activity and ribosome biogenesis are upregulated upon removal of starvation (Turck et al., 2004). This is consistent with the proposed ancestral role for the TOR pathway in nutrient sensing and response.
Increasing evidence is also linking auxin to changes in these cytoplasmic aspects of cell growth. Auxin increases the overall protein synthetic capacity of the cell due to multiple effects, including increasing the rate of translation, the accumulation of rRNA and of ribosomal proteins, as well as their recruitment to ribosomal complexes (Garcia-Hernandez et al., 1996). Auxin also regulates the phosphorylation of ribosomal proteins by S6K (Turck et al., 2004), and this auxin-dependent activation of S6K is dependent on PI3 kinase just as the IGF pathway is in animals (Turck et al., 2004). Downstream targets of the TOR pathway, including EBP1 and translationally controlled tumor protein (TCTP), are also conserved in plants and have functions in stimulating protein synthesis and growth. TCTP acts as the guanine nucleotide exchange factor of the Ras GTPase that controls TOR activity in Drosophila melanogaster, and its reduced level in Arabidopsis causes reduced growth (Berkowitz et al., 2008). EBP1 expression in plants is correlated with the expression of genes with functions in ribosome biogenesis, and it has been shown to affect growth in a dose-dependent manner. Degradation of EBP1 protein has been shown to be regulated by auxin; when auxin is present, degradation of EBP1 is inhibited (Horváth et al., 2006).
Growth and Cell Division: Another Level of Complexity
The meristem is distinguished by another important characteristic, which is cell division. The link between growth and the cell cycle has been subject to debate. On the one hand, cell division is often used as synonymous to growth; on the other hand, it has been suggested that cell division is rather unimportant to plant growth, serving simply to subdivide tissue space into conveniently sized units (Kaplan and Hagemann, 1991; Kaplan, 2001). Nevertheless there appears to be a close interplay between growth and division in the SAM, since meristematic cell size is maintained within quite tight boundaries as a consequence of cell division activity. The precise coordination of cell division and growth in the SAM suggests that it may play significant roles. This could be linked to coupling between growth and the cell cycle or could be connected with the significance of the new cell walls and membranes generated during division. These create new mechanical elements that may be important in morphogenesis or overall structure but also allow auxin gradients to be established, since these are built up through the polar concentration of PIN proteins in specific areas of the plasma membrane. In addition, while cell division may not be important in morphogenesis processes in meristems with the exception of specific formative divisions of initial cells (Sozzani et al., 2010), overall growth is strongly dependent on cell production rate as exemplified in plants dwarfed by bonsai pruning (Korner et al., 1989) or by a jasmonate-dependent response to herbivores (Zhang and Turner, 2008).
The inter-relationship between cell growth and the cell cycle is complex and much discussed. In other systems, three basic relationships have been described, involving variously (1) cell growth triggering division at a threshold cell size, (2) independent controls of growth and cell division, or (3) common upstream pathways (Jorgensen and Tyers, 2004). In animals, examples can be found of all three, whereas in yeast, the primary mechanism of triggering division is cell size and nutrient sensing (reviewed in Jorgensen and Tyers, 2004). Upstream metabolic activity is measured by overall protein translation rates through the synthesis of unstable regulatory components, such as cyclins. In animals, transcriptional control of D-type cyclins (CYCD) and the subsequent activation of their partner kinase CDK4 promotes division by initiating phosphorylation of the retinoblastoma (RB) protein, which in turn activates E2F transcription factors (Coqueret, 2002). The same CYCD control in Drosophila appears to act mainly by promoting cell growth (Meyer et al., 2000).
Heterodimeric complexes of E2F proteins with their dimerization partners (DP proteins) are important in regulating the expression of genes required for the entry into the S-phase of the cell cycle as well as other cell cycle–related processes and cell growth. In Arabidopsis, E2FA and E2FB (with DPA) promote entry into S-phase, while E2FC and its partner DPB repress cell cycle progression and are associated with differentiation and expansion-driven growth outside the meristems (Inzé and De Veylder, 2006). Overexpression of E2FB promotes premature entry into the S-phase of the cell cycle and thus reduces both the duration of the cell cycle and cell size, indicating that it may be involved in the coupling of cell size to cell division (He et al., 2004; Magyar et al., 2005; Sozzani et al., 2006). Similarly, overexpression in Arabidopsis of the D-type cyclin CYCD3;1, the upstream regulator of the RB-E2F pathway, reduces cell size, whereas mutation has the converse phenotype (Dewitte et al., 2003, 2007). Transcript profiling of induced expression of E2Fs show that genes involved in cell wall biogenesis are well represented among E2FA/DPA regulated genes (Vlieghe et al., 2003; de Jager et al., 2009), including a number of expansins, pectinesterase, and pectin lyases, and some are modulated within 6 h, suggesting that they may be direct targets (de Jager et al., 2009).
Auxin is absolutely required not only for cell growth but also for cell division in both cell cultures and in planta, but the mechanism by which it acts on the cell cycle has proved surprisingly elusive. Could auxin act on division through an indirect effect of its role in promoting cell growth? Indeed, although a number of cell cycle regulators and cyclin-dependent kinase activity have been reported to respond to auxin (John et al., 1993; Oakenfull et al., 2002), in most cases this is on a relatively long timescale. However, auxin does seem to have a direct effect on several key cell cycle regulators in the CYCD-RB-E2F pathway. Kip-Related Proteins (KRP), also known as Inhibitors of CDK (ICKs), are inhibitors of CYCD activity, and expression of KRP2 is rapidly downregulated by auxin (Himanen et al., 2002). However, transcriptomic analysis has not identified a general transcriptional link between auxin and cell cycle regulators. Protein degradation is an important regulatory mechanism within the cell cycle, and cell responses to auxin also involve proteolysis. In this regard, it is notable that auxin has been linked to changes in the stability of cell cycle regulators, promoting turnover of the negative regulator KRP2 (Sanz et al., 2011) and, conversely, the stability of cell cycle–promoting E2FB. This provides a mechanism for auxin to stimulate progression of the cell cycle into S-phase; indeed, ectopic E2FB expression can remove the auxin dependence of cultured tobacco BY-2 cells (Magyar et al., 2005). Auxin also promotes degradation of differentiation-enhancing E2FC, as well as the F-box protein SKP2A involved in E2FC turnover, in the latter case by direct binding of auxin in a mechanism analagous to TIR1 activation. Hence, auxin simultaneously promotes degradation of E2FC and of its degradation machinery (Jurado et al., 2010). It seems clear that auxin plays important regulatory roles in the cell cycle by regulating the degradation of proteins other than Aux/IAAs and that auxin both interacts with the control of cell growth and directly impinges on aspects of cell cycle control.
Classically, cytokinin was identified as a plant hormone required to sustain cell division and inhibit differentiation in plant cell cultures. The primary mechanism of interaction of cytokinins with the cell cycle is through transcriptional regulation of the D-type cyclins of the CYCD3 class (Soni et al., 1995; Riou-Khamlichi et al., 1999), whose ectopic expression is capable of driving cells into the cell cycle (Dewitte et al., 2003; Menges et al., 2006). CYCD3 activity is required to sustain normal response of both division and differentiation to cytokinin (Riou-Khamlichi et al., 1999; Dewitte et al., 2007). CYCD3 activates cyclin-dependent kinase activity of CDKA (Healy et al., 2001), whose inhibition causes differentiation of SAM cells (Gaamouche et al., 2010). Unlike CDKA, CYCD3 is an unstable protein subject to rapid turnover (Planchais et al., 2004), providing a rapid response sensor to the presence of both cytokinin and Suc (Riou-Khamlichi et al., 2000). There are three CYCD3 genes in Arabidopsis, and all are strongly expressed in different regions of the SAM (Dewitte et al., 2007).
Both auxin and cytokinin therefore appear to control the mitotic cell cycle through the conserved CYCD-RB-E2F pathway that regulates the progression of cells from G1-phase into the mitotic cell cycle in S-phase, using different mechanisms acting at different points: cytokinin through transcriptional regulation of the upstream component CYCD3 and auxin primarily through the promoting the stability of the downstream E2FB transcription factor.
CONCLUSIONS AND PERSPECTIVE
From Signaling to Shape
A scenario emerges whereby, through a process of self-organization, plant hormones adopt robust and dynamic distribution patterns at the SAM. This distribution then leads to patterns of growth. Major downstream targets of growth regulation are the structural elements of the cell, and in particular the cell wall, which is a determinant for both growth rates and growth distribution, but feedbacks with cytoplasmic synthesis and metabolic networks are crucial as well. How this coordination works is not well understood.
Auxin appears to be the common linking factor, promoting cell wall synthesis, metabolic activity, and cell growth, as well as cell cycle activity. It represents the best candidate for linking and coordinating biochemical and morphogenetic aspects of growth, since it is intimately involved with organ initiation as well as having cellular effects on both growth and division and cell wall synthesis. It is clear that these processes are highly interconnected, and it is probably not meaningful to talk about an upstream activator of either growth or division. Rather, at the cell level as at the organ level, the interplay of feedback circuits provides a robust system that links these processes together to ensure orderly growth within the parameters of certain cell sizes. In contrast with auxin, cytokinin appears to link specifically to cell division control and not to cell growth, but again we should note the interconnections between auxin and cytokinin responses (Marhavý et al., 2011).
In complement with chemical signaling, mechanics is also likely to play a role in patterning. Forces generated by differential growth rates and turgor pressure clearly have the potential to generate positional information for the individual cells, but precisely how this occurs is far from clear. This is partially because the tools to probe forces and physical properties of the SAM have yet to be developed.
Integrating Multiple Scales and Multiple Processes
As illustrated throughout this review, the research on meristem development in the past decade has revealed the multiplicity of processes that participate in maintenance and outgrowth of the SAM and the intricate nature of their interactions at different scales. Morphogenesis appears as an emerging characteristic of this interaction, regulated by complex feedback loops between biochemical and mechanical processes, interacting through a multicellular and changing geometry. Because of this complex interaction network, the precise implications at multiple scales--of for example, hypotheses concerning molecular processes--are very difficult to assess, even qualitatively. As we have seen for auxin transport, it is not intuitive at all that an up-the-gradient rule for PIN localization can indeed lead to spiral or whorled phyllotactic patterns at a higher level of organization.
Computational models of tissue development have begun to provide answers to such questions (Jönsson et al., 2012). By making it possible to integrate the effect of local rules throughout tissue structure and time quantitatively, they provide biologists a unique means to estimate the consequences of hypotheses at multiple scales. To this aim, several research groups have started to build virtual tissues or virtual organs (i.e., computational models able to integrate and simulate the key mechanisms regulating shape development within a precise two- or three-dimensional geometric description of the tissues). These models do not only take into account chemical properties, but, importantly, also mechanics. Prototypes of such virtual meristems have helped to make predictions regarding, for example, auxin transport (de Reuille et al., 2006; Jönsson et al., 2006; Smith et al., 2006; Lucas et al., 2008; Bayer et al., 2009). In most of the cases, the predictions could be tested within a short time frame. However, as frequently illustrated in physics, the time lapse between a prediction and its experimental verification may correspond to a much longer period of time as the tools to (in)validate them are not yet available at the moment the prediction is made. As we have seen, predictions regarding auxin distributions could only be tested years later, after novel auxin sensors were developed. In a similar manner, predictions regarding mechanics at the SAM will have to await the development of novel methods to probe these properties.
Acknowledgments
This article was partially funded by the ERASysBio+ initiative under the European Union European Research Area Network Plus scheme in FP7 through the program grant iSAM to A.J., J.A.H.M., J.T., and C.G. and a European Research Council advanced grant (Morphodynamics) to J.T.
AUTHOR CONTRIBUTIONS
All authors contributed to the conception and writing of this review.
References
- Adler I., Barabe D., Jean R.V. (1997). A history of the study of phyllotaxis. Ann. Bot. (Lond.) 80: 231–244 [Google Scholar]
- Alabadí D., Blázquez M.A., Carbonell J., Ferrándiz C., Pérez-Amador M.A. (2009). Instructive roles for hormones in plant development. Int. J. Dev. Biol. 53: 1597–1608 [DOI] [PubMed] [Google Scholar]
- Bartrina I., Otto E., Strnad M., Werner T., Schmülling T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C.S., Hoth S., Haga K., Philippar K., Aoki N., Hedrich R. (2000). Differential expression and regulation of K(+) channels in the maize coleoptile: Molecular and biophysical analysis of cells isolated from cortex and vasculature. Plant J. 24: 139–145 [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Smith R.S., Mandel T., Nakayama N., Sauer M., Prusinkiewicz P., Kuhlemeier C. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett M.J., Marchant A., Green H.G., May S.T., Ward S.P., Millner P.A., Walker A.R., Schulz B., Feldmann K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Berkowitz O., Jost R., Pollmann S., Masle J. (2008). Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20: 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Landrein B, Schudoma C, Hamant O, Hauser M.T., Persson S. (July 10, 2012). Cracking the elusive alignment hypothesis: The microtubule-cellulose synthase nexus unraveled. Trends Plant Sci. http://dx.doi.org/10.1016/j.tplants2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y., Ikeda Y., Grebe M. (2007). Mechanisms of auxin-dependent cell and tissue polarity. Curr. Opin. Plant Biol. 10: 616–623 [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Chen J.G., Ullah H., Young J.C., Sussman M.R., Jones A.M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O. (2002). Linking cyclins to transcriptional control. Gene 299: 35–55 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (1996). Plant cell enlargement and the action of expansins. Bioessays 18: 533–540 [DOI] [PubMed] [Google Scholar]
- Dale J.E. (1986). Plastic responses of leaves. Symp. Soc. Exp. Biol. 40: 287–305 [PubMed] [Google Scholar]
- de Jager S.M., Scofield S., Huntley R.P., Robinson A.S., den Boer B.G., Murray J.A. (2009). Dissecting regulatory pathways of G1/S control in Arabidopsis: Common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol. Biol. 71: 345–365 [DOI] [PubMed] [Google Scholar]
- de Reuille P.B., Bohn-Courseau I., Ljung K., Morin H., Carraro N., Godin C., Traas J. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J.M., Jacqmard A., Kilby N.J., Murray J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., Murray J.A. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Fernandez R., Das P., Mirabet V., Moscardi E., Traas J., Verdeil J.L., Malandain G., Godin C. (2010). Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat. Methods 7: 547–553 [DOI] [PubMed] [Google Scholar]
- Fleming A., McQueen-Mason S., Madel T., Kuhlemeier C. (1997). Induction of leaf primordia by the cell wall protein expansin. Science 276: 1415–1418 [Google Scholar]
- Gaamouche T., Manes C.L., Kwiatkowska D., Berckmans B., Koumproglou R., Maes S., Beeckman T., Vernoux T., Doonan J.H., Traas J., Inzé D., De Veylder L. (2010). Cyclin-dependent kinase activity maintains the shoot apical meristem cells in an undifferentiated state. Plant J. 64: 26–37 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M., Davies E., Baskin T.I., Staswick P.E. (1996). Association of plant p40 protein with ribosomes is enhanced when polyribosomes form during periods of active tissue growth. Plant Physiol. 111: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrocho-Villegas V., Sánchez de Jiménez E. (2012). TOR pathway activation in Zea mays L. tissues: Conserved function between animal and plant kingdoms. Plant Signal. Behav. 7: 675–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean O., Vernoux T., Laufs P., Belcram K., Mizukami Y., Traas J. (2004). In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16: 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenot B., Bayer E., Kierzkowski D., Smith R.S., Mandel T., Žádníková P., Benková E., Kuhlemeier C. (2012). Pin1-independent leaf initiation in Arabidopsis. Plant Physiol. 159: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Hamant O., Heisler M.G., Jönsson H., Krupinski P., Uyttewaal M., Bokov P., Corson F., Sahlin P., Boudaoud A., Meyerowitz E.M., Couder Y., Traas J. (2008). Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Hamant O., Traas J. (2010). The mechanics behind plant development. New Phytol. 185: 369–385 [DOI] [PubMed] [Google Scholar]
- He S.S., Liu J., Xie Z., O’Neill D., Dotson S. (2004). Arabidopsis E2Fa plays a bimodal role in regulating cell division and cell growth. Plant Mol. Biol. 56: 171–184 [DOI] [PubMed] [Google Scholar]
- Healy J.M., Menges M., Doonan J.H., Murray J.A. (2001). The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J. Biol. Chem. 276: 7041–7047 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Hamant O., Krupinski P., Uyttewaal M., Ohno C., Jönsson H., Traas J., Meyerowitz E.M. (2010). Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Magyar Z., Monardes A., Khan S., Zalejski C., Orellana J., Szabados L., de la Torre C., Koncz C., Bögre L. (2010). Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K., Boucheron E., Vanneste S., de Almeida Engler J., Inzé D., Beeckman T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B.M., Magyar Z., Zhang Y., Hamburger A.W., Bakó L., Visser R.G., Bachem C.W., Bögre L. (2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Sheen J., Müller B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- John P.C.L., Zhang K., Dong C., Diedrich L., Wightman F. (1993). p34cdc2 related proteins in control of cell cycle progression, the switch between division and differentiation in tissue development, and stimulation of division by auxin and cytokinin. Aust. J. Plant Physiol. 20: 503–526 [Google Scholar]
- Jönsson H., Gruel J., Krupinski P., Troein C. (2012). On evaluating models in computational morphodynamics. Curr. Opin. Plant Biol. 15: 103–110 [DOI] [PubMed] [Google Scholar]
- Jönsson H., Heisler M.G., Shapiro B.E., Meyerowitz E.M., Mjolsness E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Tyers M. (2004). How cells coordinate growth and division. Curr. Biol. 14: R1014–R1027 [DOI] [PubMed] [Google Scholar]
- Jurado S., Abraham Z., Manzano C., López-Torrejón G., Pacios L.F., Del Pozo J.C. (2010). The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.R. (2001). Fundamental concepts of leaf morphology and morphogenesis: A contribution to the interpretation of molecular genetic mutants. Int. J. Plant Sci. 162: 465–474 [Google Scholar]
- Kaplan D.R., Hagemann W. (1991). The relationship of cell and organism in vascular plants: Are cells the building blocks of plant form? Bioscience 41: 693–703 [Google Scholar]
- Keegstra K. (2010). Plant cell walls. Plant Physiol. 154: 483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D., Nakayama N., Routier-Kierzkowska A.L., Weber A., Bayer E., Schorderet M., Reinhardt D., Kuhlemeier C., Smith R.S. (2012). Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Korner C., Menendezriedl S.P., John P.C.L. (1989). Why are bonsai plants small - A consideration of cell-size. Aust. J. Plant Physiol. 16: 443–448 [Google Scholar]
- Kwiatkowska D., Dumais J. (2003). Growth and morphogenesis at the vegetative shoot apex of Anagallis arvensis L. J. Exp. Bot. 54: 1585–1595 [DOI] [PubMed] [Google Scholar]
- Laskowski M., Grieneisen V.A., Hofhuis H., Hove C.A., Hogeweg P., Marée A.F., Scheres B. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc N., Perrot-Rechenmann C., Barbier-Brygoo H. (1999). The auxin-binding protein Nt-ERabp1 alone activates an auxin-like transduction pathway. FEBS Lett. 449: 57–60 [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Leyser H.M.O., Pickett F.B., Dharmasiri S., Estelle M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2006). Dynamic integration of auxin transport and signalling. Curr. Biol. 16: R424–R433 [DOI] [PubMed] [Google Scholar]
- Li Y., Jones L., McQueen-Mason S. (2003). Expansins and cell growth. Curr. Opin. Plant Biol. 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lucas M., Guédon Y., Jay-Allemand C., Godin C., Laplaze L. (2008). An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE 3: e3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z., De Veylder L., Atanassova A., Bakó L., Inzé D., Bögre L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Marhavý P., Bielach A., Abas L., Abuzeineh A., Duclercq J., Tanaka H., Pařezová M., Petrášek J., Friml J., Kleine-Vehn J., Benková E. (2011). Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21: 796–804 [DOI] [PubMed] [Google Scholar]
- Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S., Durachko D.M., Cosgrove D.J. (1992). Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Samland A.K., Planchais S., Murray J.A. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C.A., Jacobs H.W., Datar S.A., Du W., Edgar B.A., Lehner C.F. (2000). Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 19: 4533–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani P., Gholamirad M., Traas J., Arnéodo A., Boudaoud A., Argoul F., Hamant O. (2011). In vivo analysis of local wall stiffness at the shoot apical meristem in Arabidopsis using atomic force microscopy. Plant J. 67: 1116–1123 [DOI] [PubMed] [Google Scholar]
- Mitchison G. (1981). The polar transport of auxin and vein pattern in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 295: 461–471 [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull E.A., Riou-Khamlichi C., Murray J.A.H. (2002). Plant D-type cyclins (CycDs) and the control of G1 progression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P.J., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S.A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21: 1720–1726 [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J.N., Höfte H., Laufs P., Pelloux J., Mouille G. (2008). Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 18: 1943–1948 [DOI] [PubMed] [Google Scholar]
- Péret B., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K., Fuchs I., Luthen H., Hoth S., Bauer C.S., Haga K., Thiel G., Ljung K., Sandberg G., Bottger M., Becker D., Hedrich R. (1999). Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc. Natl. Acad. Sci. USA 96: 12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S., Wyrzykowska J., McQueen-Mason S., Smart C., Fleming A. (2001). Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S., Samland A.K., Murray J.A. (2004). Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 38: 616–625 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A., Murray J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Menges M., Healy J.M., Murray J.A. (2000). Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C., Thomas M., Meyer C. (2012). Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15: 301–307 [DOI] [PubMed] [Google Scholar]
- Robert S., et al. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck A., Palme K., Venis M.A., Napier R.M., Felle H.H. (1993). Patch-clamp analysis establishes a role for an auxin-binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J. 4: 41–46 [Google Scholar]
- Sachs T. (1969). Polarity and the induction of organized vascular tissues. Ann. Bot. (Lond.) 33: 263 [Google Scholar]
- Sanz L., et al. (2011). The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23: 641–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Balla J., Luschnig C., Wisniewska J., Reinöhl V., Friml J., Benková E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20: 2902–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Francis P., Berleth T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131: 3445–3455 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schrader J., Nilsson J., Mellerowicz E., Berglund A., Nilsson P., Hertzberg M., Sandberg G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Murray J.A. (2008). A model for Arabidopsis class-1 KNOX gene function. Plant Signal. Behav. 3: 257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Smith R.S., Guyomarc’h S., Mandel T., Reinhardt D., Kuhlemeier C., Prusinkiewicz P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R., Carmichael J.P., Shah Z.H., Murray J.A. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7: 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dewitte W., Murray J.A., Benfey P.N. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R., Maggio C., Varotto S., Canova S., Bergounioux C., Albani D., Cella R. (2006). Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T.A., Sussex I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Stoma S., Lucas M., Chopard J., Schaedel M., Traas J., Godin C. (2008). Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLOS Comput. Biol. 4: e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Bennett M. (2003). Auxin transport: The fountain of life in plants? Dev. Cell 5: 824–826 [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Hayashi K., Kinoshita T. (2012). Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Turck F., Zilbermann F., Kozma S.C., Thomas G., Nagy F. (2004). Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 134: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttewaal M., Burian A., Alim K., Landrein B., Borowska-Wykręt D., Dedieu A., Peaucelle A., Ludynia M., Traas J., Boudaoud A., Kwiatkowska D., Hamant O. (2012). Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451 [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K., Vuylsteke M., Florquin K., Rombauts S., Maes S., Ormenese S., Van Hummelen P., Van de Peer Y., Inze D., De Veylder L. (2003). Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J. Cell Sci. 116: 4249–4259 [DOI] [PubMed] [Google Scholar]
- Wabnik K., Kleine-Vehn J., Balla J., Sauer M., Naramoto S., Reinöhl V., Merks R.M., Govaerts W., Friml J. (2010). Emergence of tissue polarization from synergy of intracellular and extracellular auxin signaling. Mol. Syst. Biol. 6: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P.B., Ruzicka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Worley C.K., Zenser N., Ramos J., Rouse D., Leyser O., Theologis A., Callis J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21: 553–562 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Sheen J. (2012). Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287: 2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J.G., Wu M.J., Perrot-Rechenmann C., Friml J., Jones A.M., Yang Z. (2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Mandel T., Kuhlemeier C. (2011). Stem cell activation by light guides plant organogenesis. Genes Dev. 25: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]