This study identifies regions of noncoding DNA in dicot plants that are likely to facilitate complex regulation of genes by binding multiple transcription factors. Regulatory mechanisms that the model organism Arabidopsis is likely to share with crop plants provide a focus for research that has real-world applications.
Abstract
Conserved noncoding sequences (CNSs) in DNA are reliable pointers to regulatory elements controlling gene expression. Using a comparative genomics approach with four dicotyledonous plant species (Arabidopsis thaliana, papaya [Carica papaya], poplar [Populus trichocarpa], and grape [Vitis vinifera]), we detected hundreds of CNSs upstream of Arabidopsis genes. Distinct positioning, length, and enrichment for transcription factor binding sites suggest these CNSs play a functional role in transcriptional regulation. The enrichment of transcription factors within the set of genes associated with CNS is consistent with the hypothesis that together they form part of a conserved transcriptional network whose function is to regulate other transcription factors and control development. We identified a set of promoters where regulatory mechanisms are likely to be shared between the model organism Arabidopsis and other dicots, providing areas of focus for further research.
INTRODUCTION
Plants are sessile organisms, and as such, they rely on their regulatory machinery to recognize, process, and respond to signals from internal and external stimuli, enabling them to cope with environmental change. The success of these response mechanisms affects the future survival and adaptation of the species. Decision making is a complex process, and transcription factors (TFs) are a fundamental part of the regulatory machinery, helping to integrate multiple cues to yield appropriate downstream responses (Lemon and Tjian, 2000). Thus, better understanding of transcriptional networks and how multiple signals are integrated to affect decision making will aid in better understanding of the way organisms adapt and survive. More generally, elucidating transcriptional regulation is vital for understanding cellular and developmental processes at the molecular level. There are in excess of 2000 known TFs in Arabidopsis thaliana (Riechmann et al., 2000; Zhang et al., 2011), and yet the regulatory sequences to which they bind remain largely enigmatic, with only ∼150 plant-specific position-specific scoring matrices (PSSMs) currently identified and deposited in published databases, such as TRANSFAC, JASPAR, and Athena (Wingender et al., 2000; O’Connor et al., 2005; Bryne et al., 2008). There are an additional 469 sequence motifs in PLACE (Higo et al., 1999). The paucity of data is exacerbated, as many of these PSSMs are redundant or based on limited experimental data. Identifying transcription factor binding sites (TFBSs) amid the very noisy background of noncoding sequence is notoriously difficult; simply scanning genomic sequences for PSSM matches results in an exceptionally high false discovery rate.
Due to the action of natural selection on random genomic mutations, noncoding sequences that contain functional regulatory elements evolve more slowly than adjacent nonfunctional DNA, leaving islands of conserved noncoding sequences (CNSs). Therefore, detecting a sequence that has remained conserved across evolutionarily divergent clades implies that the sequence has functional significance; this is the foundation of phylogenetic footprinting (Tagle et al., 1988). Phylogenetic footprinting simplifies the task of finding regulatory elements by identifying CNSs initially using orthologous sequences and then refining the search space to informative regions (Frazer et al., 2003). For CNSs upstream of a gene’s transcription start site (TSS), this conserved function is likely to be regulatory.
Previous attempts at discovery of CNS in Arabidopsis have used paralogous gene pairs (Freeling et al., 2007; Thomas et al., 2007). These studies provide information on intragenomic (paralogous) CNSs associated with homologs (arising from duplication events) but do not attempt to discover intergenomic (orthologous) CNSs using orthologs. Studies considering orthologs in related plant species have so far been of limited scope, focusing on only a handful of specific gene families at a time (Colinas et al., 2002; Guo and Moose, 2003; Inada et al., 2003; Creux and Ranik et al., 2008; Spensley et al., 2009).
For approaches using comparative genomics, the target species must be selected with care; species that are too close together, in evolutionary terms, will yield a large number of false positives, whereas species that are too far apart will show too little conservation (Duret and Bucher, 1997). In this study, we compare orthologous genes from Arabidopsis (Arabidopsis Genome Initiative, 2000) and three dicot genomes, papaya (Carica papaya) (Ming et al., 2008), poplar (Populus trichocarpa) (Tuskan et al., 2006), and grape (Vitis vinifera) (Velasco et al., 2007), that diverged from a common ancestor with Arabidopsis ∼72 million years ago (Mya), 109 Mya, and 117 Mya, respectively (Hedges et al., 2006). It has been suggested that CNSs are nondiscoverable using species as divergent from Arabidopsis as poplar (Thomas et al., 2007). Using phylogenomic comparisons of plant upstream regions, it has been estimated that ∼100 Mya is an appropriate divergence limit to reliable CNS discovery for species within this clade (Reineke et al., 2011). However, the limits of detecting CNS are undoubtedly linked to the methodology used: Previous studies use heuristic alignment methods, such as BLAST, which are insufficiently sensitive to align weakly conserved pairs of sequences (Bray et al., 2003). Instead, to find similarity between orthologous promoters, we use a fast implementation of the alignment plot method (Krusche and Tiskin, 2010) based on the seaweed algorithm of Tiskin (2008). The alignment plot method has been used previously to accurately predict evolutionarily conserved promoter regions in LHY (LATE ELONGATED HYPOCOTYL), TOC1 (TIMING OF CAB EXPRESSION 1), LUX (LUX ARRHYTHMO), CAB2 (CHLOROPHYLL A/B BINDING PROTEIN 2), and ABI3 (ABSCISIC ACID INSENSITIVE 3), all of which matched the key experimentally defined regulatory regions for those genes (Picot et al., 2010; Spensley et al., 2009). These studies demonstrate that alignment plots have performed well at pinpointing key regulatory regions in a handful of tested cases. Our study applies the technique to discover plant CNSs on a genome scale.
Here, we demonstrate that our methods enable the detection of hundreds of high-confidence CNSs in Arabidopsis. Sequence analyses (including TFBS overrepresentation, Gene Ontology (GO) term analysis, and prediction of nucleosome positioning) implicate involvement of the CNS in transcriptional regulation. Our findings are consistent with CNS and their associated genes being components of an ancient regulatory network that is shared between the species studied. We also present the software package Analysis of Plant Promoter-Linked Elements (APPLES) that we developed in order to facilitate comparative genomics of plant noncoding sequences. This is made available as a virtual appliance that has an installation of APPLES and all its dependencies and can be run in a virtual machine. Scripts to reproduce our main results and a manual are provided as a starting point for programmatic use of the tool.
RESULTS
Multispecies Analysis Yields Hundreds of CNSs
We used a comparative genomics approach to identify CNSs in Arabidopsis promoters. We define “promoter” as the 2 kb of DNA upstream from an annotated TSS (this being the length of a typical intergenic space) but apply a rule to stop at the neighboring gene if one is present within this region. In the first step of our analysis, we identified 21,034 Arabidopsis genes that had one or more orthologs in papaya, poplar, and/or grape (see Methods). Orthologs were found in two or more comparator species in 92% of cases (Supplemental Figure 1A online shows the distribution of orthologs across the three species).
Alignment plots of orthologous promoter regions were produced using an implementation of the seaweed algorithm (Krusche and Tiskin, 2010), which computes optimal sequence alignments for all pairs of 60-bp sequence windows, requiring millions of computations for a typical 2-kb promoter. This enables highly sensitive detection of conserved sequences irrespective of their position. To evaluate the significance of the aligned sequences, an equivalent control set was produced, whereby for each of the 21,034 Arabidopsis genes, pseudo-orthologs were assigned at random from each of the three other species on an identical gene-for-gene basis as in the real set, and alignments of the promoter regions were computed as before. Comparison of the alignment scores between Arabidopsis and each of the comparator species in the ortholog and random gene pair sets revealed markedly different distributions, with true orthologs producing a greater number of alignments and higher overall alignment scores (see Supplemental Figure 2 online). High-scoring alignments occur in pairs of orthologous sequences, but do not occur by chance in randomly assigned gene pairs; the randomly assigned gene pairs do not produce alignment scores above 48 (see Supplemental Figure 2A online), 47 (see Supplemental Figure 2B online), and 47 (see Supplemental Figure 2C online) for papaya, poplar, and grape, respectively, whereas ortholog sequences have alignment scores as high as 59 (with the maximum score possible based on a 60 nucleotide window being 60). This demonstrates that a large number of Arabidopsis promoter regions are sequence conserved with orthologous regions in other species and potentially have a conserved function.
To interpret raw alignment scores into a more meaningful scale, we defined a conservation score to determine the significance of an alignment (see Methods). Importantly, this conversion enables the integration of alignment scores across multiple species, which is useful because weak alignments observed in multiple distant species can accumulatively be as biologically meaningful as a stronger alignment in just one pair of species (as demonstrated in Picot et al., 2010). A conservation score has a range from 0 to 1 and signifies how strongly a sequence alignment is expected to reflect conservation (i.e., sequence similarity as a result of evolutionary constraint). The higher the conservation score, the greater is the expectation that the alignment score observed represents true sequence conservation; conversely, alignment scores commonly found by chance determine the lower end of the conservation score range. Also, during the conversion from alignment score to conservation score, the sequences of any overlapping high-scoring window-pair alignments are merged together to form longer contiguous sequence regions. Comparing the distribution of conservation scores from orthologs and random gene pairs, we are able to establish thresholds of increasing stringency and calculate the false-positive discovery rate at these thresholds (Table 1).
Table 1. Numbers of Aligned Regions and Associated Genes from Orthologous Promoters (before and after Filtering for Putative Coding Sequences) and from Promoters of Random Gene Pairs at Different Thresholds of Conservation Score.
| Orthologs |
Random Gene Pairs |
||||||
|---|---|---|---|---|---|---|---|
| Conservation Score Threshold | No. Genes before CDS Filtering | No. Aligned Regions before CDS Filtering | No. Genes after CDS Filtering | No. Aligned Regions after CDS Filtering | No. Genes | No. Aligned Regions | False Positive Rate |
| 1 | 148 | 157 | 136 | 143 | 0 | 0 | 0.0000 |
| 0.9 | 384 | 414 | 365 | 392 | 7 | 7 | 0.0003 |
| 0.8 | 481 | 517 | 460 | 492 | 23 | 23 | 0.0011 |
| 0.7 | 578 | 630 | 554 | 602 | 36 | 36 | 0.0017 |
| 0.6 | 782 | 850 | 758 | 822 | 117 | 119 | 0.0056 |
| 0.5 | 1230 | 1372 | 1202 | 1340 | 412 | 431 | 0.0196 |
| 0.4 | 1319 | 1481 | 1291 | 1448 | 467 | 492 | 0.0222 |
| 0.3 | 1672 | 1902 | 1643 | 1865 | 657 | 700 | 0.0312 |
In the CDS filtering step, a gene and all of its associated conserved regions are removed if one or more of the regions are predicted to be protein coding.
The accuracy of genome annotations is a factor to consider when ascribing meaning to conserved sequence signatures, some of which may represent missed exons, missing genes, or other features. To determine whether any of the aligned sequences could represent a coding feature, we performed BLASTX against the National Center for Biotechnology Information Viridiplantae database. We discarded any gene with an upstream sequence that produced a significant hit (see Methods) to exclude any potentially coding sequences from the data sets used in further analyses, removing 1.9% of the sequences (Table 1). Furthermore, examining the average GC content of the remaining sequences (29.9% [sd 12.4] and 37.7% [sd 9.2] for the 0.3 and 0.7 threshold sets, respectively) shows that it is more similar to the mean noncoding GC content of the Arabidopsis genome (32.7%) than coding (44.1%) (Arabidopsis Genome Initiative, 2000), supporting the conclusion that the sequences are noncoding.
At the 0.3 threshold, we found 1865 CNSs present upstream of the TSS of 1643 Arabidopsis genes (Table 1). Supplemental Figure 1B online shows the distribution of which comparator species contributed to the overall conservation score of each conserved sequence. Alignments from two or more comparator species contributed to the identification of a conserved sequence in 57% of cases at the 0.3 threshold.
The distribution of scores generated by the promoters of random gene pairs (Table 1) informs the threshold to use for further analyses, and a strict threshold helps to select only those regions that have significant conservation and are highly unlikely to occur by chance alone. The false positive discovery rate for the detection of promoter conservation is very low for the stringent thresholds. At a strict threshold (0.9), 392 regions were identified in the upstream regions of 365 genes compared with seven regions (from seven genes) in the control set, giving a false positive error rate of just 0.0003. Even at a lower threshold of 0.6, the numbers of aligned regions in the real orthologs (822) and random gene pairs (119) are significantly different, giving a false positive error rate of only 0.0056. The database of orthologous CNSs developed by this study is provided in Supplemental Data Set 1 online.
Using the conservation score threshold of 0.7, 554 Arabidopsis genes were identified with a total of 602 conserved regions associated with them. We deemed these regions to be suitably significant (false positive error rate of 0.0017) and so selected these as our robust candidate CNS set for subsequent analyses.
CNS Show Positional Bias toward TSS
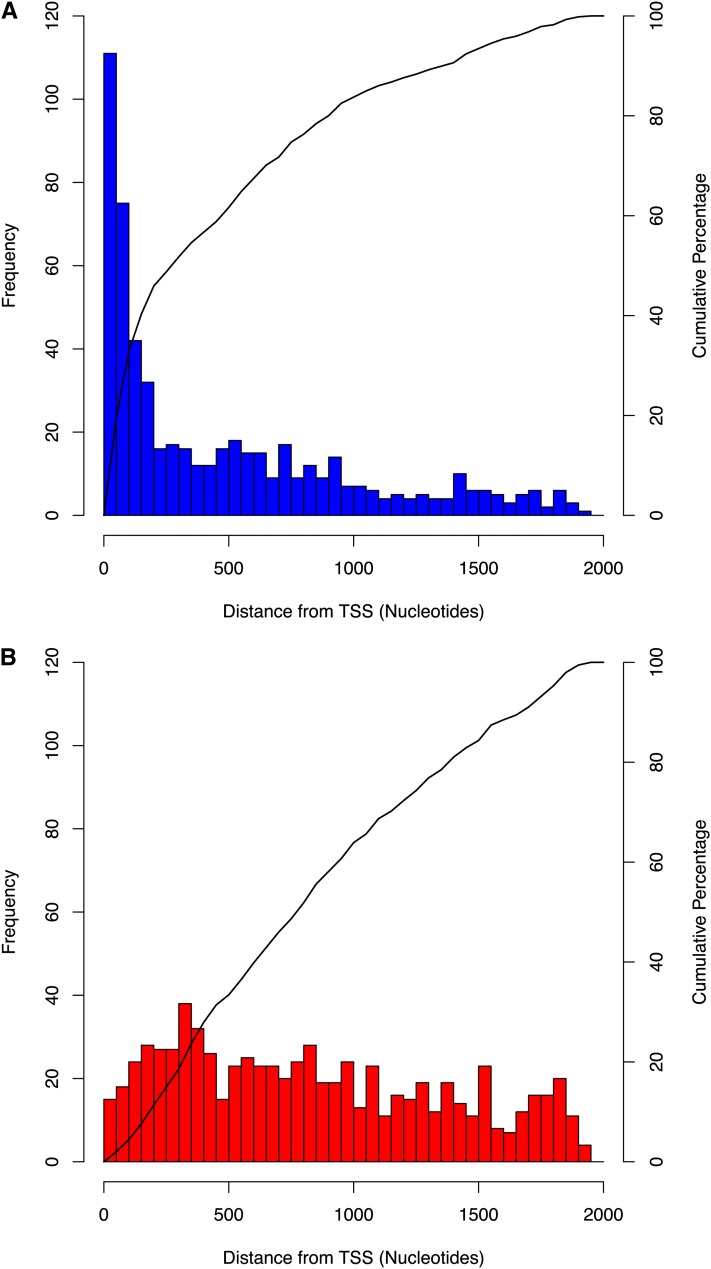
The distance between each Arabidopsis gene’s annotated TSS and the start of the conserved region was recorded for all alignments in orthologs and random gene pairs (Figure 1). For the random gene pair data, it was necessary to apply a relaxed threshold of 0.3 to display a comparable number of alignments to the CNS set (in which the 0.7 threshold was applied). In both plots, a restriction was applied to only plot distances for genes having at least a 500-nucleotide intergenic space before a neighboring gene upstream to limit any bias caused by genes with short intergenic spaces. A clear bias can be observed toward the first 100 to 200 nucleotides upstream from the TSS in the CNS set derived from orthologs (Figure 1A, 0.7 threshold). By contrast, the distribution of distances in the set of random gene pairs (Figure 1B, 0.3 threshold) is uniform, as is expected of randomly occurring alignments whose position is not meaningful. To test for any bias introduced by intergenic length on distance between the CNSs and TSSs, the normalized distances (with respect to intergenic distance) were plotted and show that the regions of CNSs close to TSSs do not all come from short promoters (see Supplemental Figure 3 online). Proximity to the TSS is indicative of a transcriptional function for these regions. In 14% of cases, the CNSs are very close to the TSS (<50 bp) and a TATA box motif is present, consistent with previous studies where the TATA box was positionally biased to the first 50 nucleotides upstream of the TSS (Molina and Grotewold, 2005; Berendzen et al., 2006) and so is likely to correspond to the core promoter region. However, the majority (76%) of conserved regions is further than 50 nucleotides from the TSS and so falls outside of the core promoter region, and 36% of conserved regions are more than 500 nucleotides upstream of the TSS.
Figure 1.
Alignments Produced in Orthologous Promoters Reveal a Positional Bias toward the TSS.
Distribution of distances between conserved regions and the TSS in Arabidopsis promoters. Only distances where the intergenic length is at least 500 nucleotides are plotted. Distances observed in orthologous promoters, 0.7 threshold (566/602 distances plotted) (A), and randomly assigned gene pair promoters, 0.3 threshold (684/700 distances plotted) (B).
CNSs Are Highly Enriched in TFBS Motifs
The set of 602 CNSs in Arabidopsis was tested for the enrichment of known TFBS motifs. We expected that functional CNSs would contain a higher number of known TFBS motifs than randomly selected sequences, and the presence of such motifs would be indicative of a transcriptional function. To reduce redundancy, motifs were firstly clustered using the Hellinger distance, and a representative for each group was selected. The resulting 728 eukaryotic TFBS motifs, represented by PSSMs, were then tested for their presence in the CNS set. As a negative control for this test, sets of control regions were created whereby for each conserved sequence, an intergenic sequence matching the same length and distance from a randomly picked Arabidopsis gene’s TSS was selected. The control sequences are also chosen to include only nonrepetitive regions. Therefore, the control sequences are identical to the CNSs in every respect except conservation. We tested 100 control sets to obtain a robust statistic (see Methods). The control regions help to identify motifs that are generally overrepresented in real biological sequences; each PSSM occurs by chance throughout the genome with a certain frequency, and we accounted for these background distributions by calculating individual scores for each motif against the control regions. Motif redundancy in the test set of 728 is low due to the motif clustering procedure, so the sites identified are less likely to overlap with one another.
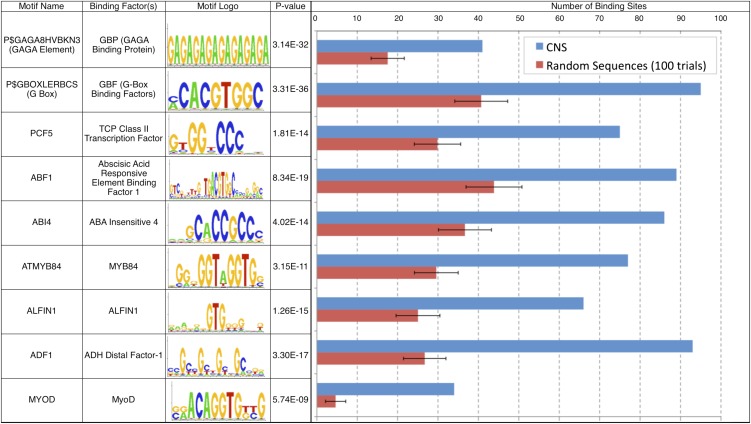
Using the binomial test for overrepresentation (see Methods), 182 of these motifs were found to be overrepresented in the CNS set (whereby in each case the binomial score obtained in the CNS set is strictly lower than the minimum score calculated in the control set after 100 trials). Figure 2 shows a selection of diverse motifs that were significantly overrepresented (see Supplemental Data Set 2 online for a full list of overrepresented motifs). Plant-specific GAGA, GBOX, BZIP, and ABA INSENSITIVE4 (ABI4) binding sites were strongly overrepresented. Binding sites for abscisic acid–responsive factors (ABF1, ABF1/3/4, and AREB1) were also significantly enriched in CNS regions. Abscisic acid is known to be important in both seed development and the mature plants’ response to environmental stress and pathogen attacks (Seo and Koshiba, 2002). Binding sites for ALFIN1, a root growth regulator (Bastola et al., 1998), are overrepresented, as are those for the MYB84 TF. We also included motifs originally found in vertebrates, insects, and fungi in our search, and many of these were found to be overrepresented in the CNS. They include binding sites for PUR, GKLF, and MyoD, all TFs involved in regulating cell proliferation and differentiation in humans. While homologs of these vertebrate TFs are not known to exist in plants, the sites themselves (or ones very similar to) may be bound by other plant-specific TFs. The presence of overrepresented TFBS motifs suggests that the CNSs are able to bind proteins and therefore are likely to play a functional role in transcriptional regulation of their associated genes.
Figure 2.
Arabidopsis CNSs Are Strongly Enriched for Specific TF Binding Sites.
Data for selected TFBS motifs are shown. Numbers of binding site occurrences in CNSs from orthologous promoters (blue) compared with numbers in random control sequences (red; mean of 100 trials and sd shown). Sets of control sequences were picked from Arabidopsis promoter regions and match the CNS in length, number, relative position to TSS, and underrepresentation of known repetitive elements.
There are a total of 10,195 occurrences of the 182 overrepresented motifs in the CNS set, which is an increase of 106% compared with 4958 occurrences found in control sequences (average of 100 trials, sd = 155). These numbers indicate thousands of potential network links where a protein recognizes a conserved binding site and influences expression of the target gene.
Identification of Previously Experimentally Validated Promoter Binding Elements
Experimentally delineating promoter binding elements is an arduous task; consequently, there is little detailed data available with which to directly corroborate the functionality of the CNS identified. To see whether our set of CNSs contains experimentally proven binding regions, we cross-referenced our results against AtProbe, a small database by the Zhang laboratory, focusing on experimentally validated binding elements (see Methods). Of 27,416 protein-coding genes (TAIR version 10), information is provided for 76 of these known genes, using information manually curated from primary literature. One of these genes, AP1 (APETALA 1), is present in our set of 554 genes, and the CNSs identified (430 nucleotides upstream from the TSS) overlap (entirely contains) the experimentally verified binding element LFY that is recognized by LEAFY and functions in controlling flower development (Parcy et al., 1998).
Picot et al. (2010) applied a method analogous to the one used in this study to four genes with well-characterized promoters and showed that it successfully identifies experimentally verified regulatory regions. Our results match the regions identified for TOC1, LUX, and ABI3 (all present in the 0.7 threshold set). For CAB2, orthologs were not identified.
Genes Associated with CNSs Contain a High Proportion of Master Regulators
We compiled a list of 33 so-called plant master regulators described in the scientific literature by searching Web of Science and Google Scholar with the keywords “plant” and “master regulator” (see Supplemental Table 1 online). These are genes, generally TFs, which play a pivotal role in the control of transcriptional regulation hierarchies. Within this set of key genes, seven appear in our 0.7 threshold set and 15 in the 0.3 threshold set of CNS-associated genes. Using a hypergeometric test, these overlaps are highly significant (P values 3.58e-06 and 1.61e-10, respectively).
Prediction of Nucleosome Positioning
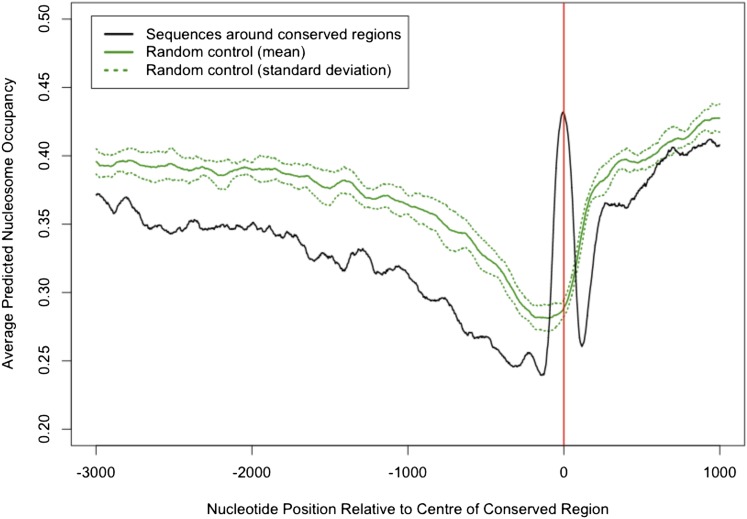
Nucleosome organization has a significant impact on gene regulation, demarcating promoter regions and TSSs and influencing transcription (Jiang and Pugh, 2009). A large part of this organization is determined by the intrinsic DNA sequence preferences of the nucleosome. We investigated the nucleosome occupancy probabilities of sequences surrounding the CNS. The tool used to predict nucleosome positions within genomic DNA is based on a model that determines nucleosome–DNA interactions, reflecting sequence features that are independent of our alignment conservation measure (Kaplan et al., 2009). It provides a probability score for each position along the input sequence. DNA sequences 10 kb long with the CNS positioned centrally were used as input (554 sequences), and the average score at each position for the CNS set was calculated. As a control, 10 comparable sets of sequences were selected from the Arabidopsis genome (see Methods for selection criteria) and scored by the same method. We compared the average nucleosome occupancy probability of the CNS-associated regions against the average of the control sequence sets (mean of 10 trials) (Figure 3).
Figure 3.
Predictions of Nucleosome Occupancy Confirm Significance of CNSs Identified.
Average predicted nucleosome occupancy for 554 10-kb sequences surrounding CNSs (black line) and 10 equivalent sets of control sequences (solid green line represents mean of the 10 control sequence sets; dashed line shows sd) was calculated. The −3 kb to +1 kb regions are plotted for clarity, as values plateau either side of this for the remainder of the −5 kb to +5 kb range examined. Red line at nucleotide position 0 indicates the center position of CNS or control sequences, with TSS therefore being positioned to the right of this.
Studies such as those in yeast (Albert et al., 2007; Kaplan et al., 2009) and human (Ozsolak et al., 2007) show a pattern of nucleosome occupancy that is sharply depleted directly upstream of the TSS, so-called nucleosome-free regions. This decline is reflected in both the control sequences and in the sequences around CNS between coordinates −1500 and 0 (Figure 3, where the red line at position 0 indicates the center of CNS, with the TSS therefore being positioned to the right of this line). In the CNS set, however, there is a clear peak in the averaged predicted nucleosome occupancy that directly coincides with the CNS regions (Figure 3), which indicates that the CNSs tend to have a particularly well-positioned nucleosome in them. The smoothness of the peak is a result of the averaging across all sequences aligned to the center of each conserved sequence. As nucleosome positioning has an effect on gene regulation (Jiang and Pugh, 2009), this result acts as further evidence for CNS playing a role in transcriptional regulation of their associated genes.
GO Term Overrepresentation Unveils Key Biological and Molecular Functions of Genes Associated with CNSs
GO term overrepresentation tests were performed on the genes associated with CNSs to gain further insights into the potential biological meaning of the result. The analysis was performed in three ontological categories, Biological Process, Molecular Function, and Cellular Component, to identify roles these genes may have in common.
For Biological Process terms, two dominant themes arise (see Supplemental Data Set 3 online, BP). First, 10 terms (associated with 134 unique genes) described as being involved in “regulation of…” are strongly overrepresented (P value ≤ 3.70e-24 after multiple testing correction in each case). Interestingly, “regulation of transcription” is highly overrepresented, the annotation being ascribed to 83 genes in the set (P value 1.09e-25). Other “regulation of…” terms include biological process, cellular process, biosynthetic process, and metabolic process. Second, the CNS set is also enriched for many terms (associated with 72 unique genes) relating to developmental processes, for example, organ development (P value 7.22e-26), system development (P value 7.22e-26), shoot development (P value 2.86e-16), flower development (P value 1.33e-11), and meristem development (P value 4.05e-07).
Testing for Molecular Function GO terms revealed transcription regulator activity and associated functions of TF activity and DNA binding to be the most overrepresented processes, with P values of 1.23e-57, 7.43e-51, and 9.79e-48, respectively (see Supplemental Data Set 3 online, MF).
Finally, we examined if there was a specific cellular component that might be overrepresented in the set. “Nucleus” was the most significant overrepresented term (P value 3.53e-11) (see Supplemental Data Set 3 online, CC).
Gene Age Analysis
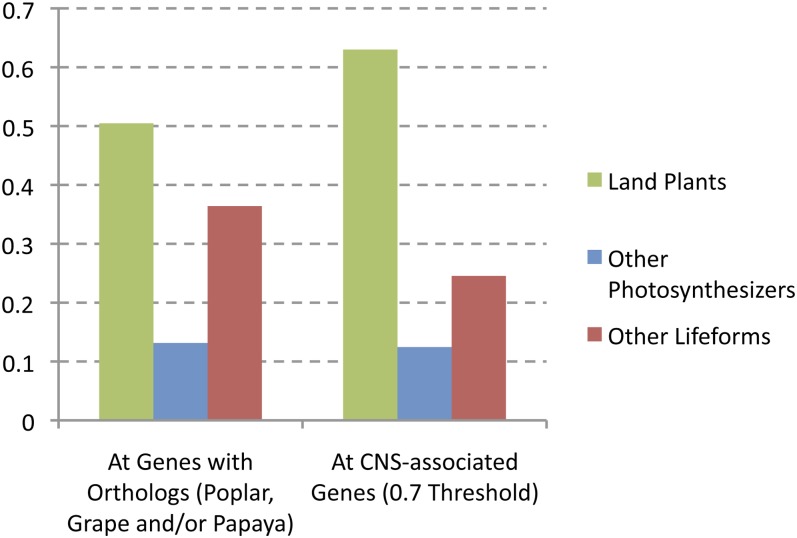
An ortholog age was calculated for each Arabidopsis gene by attempting to identify orthologs in representative species from progressively widening taxonomic groups. Each gene in the subset of 21,034 genes from the genome that had an ortholog in at least one comparator species was then placed into one of three categories: genes specific to Arabidopsis or with orthologs only within land plants, genes with orthologs also found in other photosynthesizing organisms, and genes with orthologs also identifiable in wider taxa (fungi, animals, and Escherichia coli; see Methods). Figure 4 shows the frequency of this age category among genes in the 0.7 threshold CNS set, compared with genes with orthologs found in poplar, grape, and/or papaya. Strikingly, the set of genes associated with CNS are highly enriched for genes with land plant–specific orthologs (P = 1.95e-09, hypergeometric test) and depleted for genes with orthologs found across wider taxonomic distances (P = 1.63e-09, hypergeometric test).
Figure 4.
Genes Associated with CNSs Are More Likely to Be Land Plant Specific and Less Likely to Have Orthologs across Wider Taxonomic Distances.
Histogram compares the frequency of estimated gene age groupings among genes associated with CNSs (554 genes/0.7 threshold set) with all Arabidopsis genes having an ortholog in poplar, grape, and/or papaya.
Comparison of Orthologous and Paralogous CNS Gives Insights into Subfunctionalization of Potential Regulatory Regions
An important aspect in the evolution of Arabidopsis gene regulation is the potential of paralogs to undergo subfunctionalization (partitioning of functions), neofunctionalization (gain of function), and nonfunctionalization (loss of function) at the cis-regulatory level. It is difficult to distinguish between nonfunctionalization and neofunctionalization and not possible to determine either in the absence of detailed expression data for all orthologous genes, which is beyond the scope of this study. However, as subfunctionalization is expected to occur in paralogs but not between orthologs, we can gain some insights into this process by comparing orthologous CNSs with paralogous CNSs.
Thomas et al. (2007) used bl2seq to identify paralogous CNSs in Arabidopsis for a set of 3179 gene pairs retained from the α tetraploidy event. Using our algorithm with modified parameter settings, we were able to identify all the regions previously identified in the 2-kb upstream gene space, plus many additional regions. This methodological comparison is presented in detail in Supplemental Methods 1 and Supplemental Data Set 4 online. However, for consistency within this study, we applied the same parameters and statistical significance framework as used in the discovery of orthologous CNSs to determine paralogous CNSs. Using the list of paralogous pairs produced by Thomas et al., alignment scores were computed for the 2-kb promoter regions upstream from the annotated TSS. A control set was produced by randomly permuting the pairs, and alignments were computed as before. Both sets of alignment scores were converted into conservation scores and thresholded as before. Using this framework, the false positive rates were extremely low at all thresholds (<0.0012; Table 2). At the 0.3 threshold, 1573 regions were found upstream of 1149 genes (Table 2) and have an average length of 98 bp. The paralogous CNSs were then compared against orthologous CNSs. Of 3019 genes with both a paralog and an ortholog (as defined in this study), 565 have paralogous CNSs and 291 have orthologous CNSs above the 0.3 threshold. The overlap of these sets is highly significant, with 133 genes having both types of CNSs (P < 3.79e-29, hypergeometric test; only one paralog of each pair included in the set of 3019). Among this set of 133 genes, paralogous CNSs and orthologous CNSs are overlapping in the promoters of 85 genes. This shows that evolutionary pressure to retain CNSs across genomes is linked with evolutionary pressure to retain CNSs across paralogs within a genome, though a large proportion of paralogs may lose some CNSs. Using GO analysis, these 85 genes are enriched for terms including regulation of biological process, regulation of transcription, and system development (see Supplemental Data Set 5 online). This is consistent with the idea that some types of genes, such as TFs and genes controlling developmental processes, are generally under greater regulatory constraint on the transcriptional level than other genes and that some of this constraint is often maintained after gene duplication.
Table 2. Numbers of Aligned Regions and Associated Genes from Paralogous Promoters and from Promoters of Randomly Paired Paralog Genes at Different Thresholds of Conservation Score.
| Paralogs |
Random Gene Pairs |
||||
|---|---|---|---|---|---|
| Conservation Score Threshold | No. of Genes | No. of Aligned Regions | No. of Genes | No. of Aligned Regions | False Positive Rate |
| 1 | 224 | 243 | 1 | 1 | 0.0003 |
| 0.9 | 479 | 564 | 1 | 2 | 0.0003 |
| 0.8 | 719 | 882 | 1 | 3 | 0.0003 |
| 0.7 | 771 | 964 | 1 | 3 | 0.0003 |
| 0.6 | 952 | 1247 | 1 | 3 | 0.0003 |
| 0.5 | 971 | 1289 | 1 | 3 | 0.0003 |
| 0.4 | 1005 | 1335 | 2 | 4 | 0.0006 |
| 0.3 | 1149 | 1573 | 4 | 6 | 0.0012 |
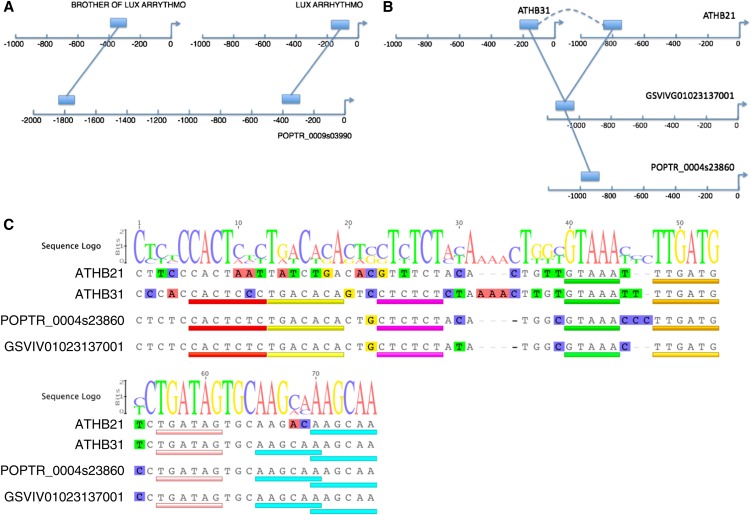
By manually inspecting the positioning and distribution of alignments in this set, we have found interesting examples of potential subfunctionalization at the regulatory region level (illustrated by Figure 5A) and binding site motif level (illustrated by Figure 5C). Figure 5A shows a case of nonoverlapping orthologous conservation in a paralogous pair of genes: LUX ARRHYTHMO and BOA (for Brother of LUX ARRHYTHMO). While the orthologous gene in poplar has two CNSs, we find only one conserved sequence for each of the paralogs in Arabidopsis. We conjecture that each conserved sequence contributes a part of the expression pattern. If these sequences function in a largely independent (additive) manner, then the joint expression pattern of the two paralogs in Arabidopsis may resemble the expression pattern of the single gene in poplar. We also show an example where a paralogous pair of homeobox genes share a common orthologous conserved region in a poplar and grape gene (Figure 5B), but the specific pattern of nucleotide conservation within the conserved region shows potential subfunctionalization of binding motifs (Figure 5C). Within the most conserved 75 nucleotides, the poplar sequence has diverged from the grape sequence at only three positions, while the sequences upstream of ATHB21 (AT2G02540) and ATHB31 (AT1G14440) have diverged at 21 and 15 positions, respectively (Figure 5C). This is consistent with reduced selective pressure at the loci of the Arabidopsis paralogs after gene duplication. The sequence upstream of ATHB21 is particularly diverged, and conserved regions at four sites have been lost in this paralog, suggesting that the set of TFs binding this region has changed.
Figure 5.
Analysis of CNSs Reveals Potential Subfunctionalization in Regulatory Regions of Arabidopsis Paralogs.
(A) and (B) Positions of CNSs upstream of paralogous Arabidopsis genes and their orthologs. Arrows indicate TSS positions. Solid lines joining blocks indicate CNSs between orthologs, and dashed curved line in (B) indicates conservation between paralogs.
(C) Alignment of CNS depicted in (B). Size of letters in the sequence logo indicates conservation of individual nucleotides. Colored bars indicate positions of potential binding sites based on alignment conservation (yellow, purple, green, and orange bars) and matches with known motifs (P300 in red, GATA in pink, and CBNAC in turquoise).
APPLES Software Package Facilitates Sequence Analysis
As exemplified in this study, the identification and characterization of functional regulatory DNA is a key step toward understanding the mechanisms that underpin gene regulation in plants. This requirement has driven the development of an array of bioinformatics tools that attempt to identify regulatory regions, such as promoters and enhancers, together with their smaller constituents, such as TFBS (Lenhard and Wasserman, 2002; Stajich et al., 2002; Tompa et al., 2005; Matys et al., 2006; Bryne et al., 2008). Given the wealth of methods available, it is often desirable for researchers to use several different techniques and link them together into workflows. Such an approach is made difficult, however, when each method operates with subtly different input and output formats, even if they manipulate similar biological entities (e.g., sequences and sequence patterns).
To circumvent these interoperability issues, sequence analysis software requires a common representation of the biological entities relevant to the research, along with the tools used to study them. Such a strategy is suited to an object-oriented programming approach to software design, which has previously been used to approach similar problems in bioinformatics (Stajich et al., 2002). To perform the large-scale analysis described in this study, we developed the software package APPLES, a collection of object-oriented modules that allow users to investigate noncoding sequences. The purpose of the APPLES software framework is to facilitate research by unifying methods applicable to various aspects of sequence analysis and is structured such that it is possible to link these together. The APPLES software (virtual appliance, instructions for its use, and source code) is available at http://www2.warwick.ac.uk/fac/sci/systemsbiology/staff/ott/tools_and_software/apples/. The APPLES source code is also available at http://sourceforge.net/projects/apples-software/.
Figure 6 describes key functionality offered by APPLES, including some methods that have been used outside of this study (Breeze et al., 2011). The user manual provides more detailed information, and example scripts are supplied within the virtual appliance. APPLES also features a caching system to store results of particular computations together with any specified parameters. This system allows results to be shared among users and redundant computations are automatically avoided. By making use of the collection of APPLES functions, users can program specific applications tailored to their own research questions, while the modular structure facilitates extension of the software through implementation of new methods.
Figure 6.
APPLES Software Facilitates the Analysis of Noncoding Sequences in Plant Genomes.
Users can program scripts to access data from sequence and motif databases and perform sequence analyses using a range of methods. Arrows indicate a typical information flow in APPLES scripts. Technical names of some key APPLES classes are shown in blue boxes. Red boxes represent the functionality of some groups of APPLES methods.
DISCUSSION
This study is a comprehensive genome-scale cross-species examination of upstream CNSs in dicot plants, using the genomes of Arabidopsis, poplar, grape, and papaya. Using an inclusive ortholog map combining synteny and reciprocal best BLAST hits, 21,034 of Arabidopsis’ 27,416 protein coding genes have at least one clearly identifiable ortholog in poplar, grape, and/or papaya, and our analyses are limited to this subset (78%) of the genome. This number may be increased as more sequenced plant genomes become available or as the sensitivity of ortholog assignment methods improve. However, genes without an identifiable ortholog in any related species are unlikely to be informative in identifying CNSs by phylogenetic footprinting methods. Therefore, we considered the majority of the informative gene set for this type of approach, given the current genome data.
Due to large-scale duplication events in plant genomes and subsequent neo-/subfunctionalization of paralogs, similarity between sequences does not implicitly determine orthology in all cases. Therefore, care must be taken when assigning orthologs, and improvements to orthology assignment methodology would enhance the detection of CNSs. Duplicated promoters may also acquire or lose individual cis-elements, meaning that even if orthology between genes is assigned correctly, their individual promoters may have undergone many evolutionary changes since they last shared a common ancestor, rendering CNSs nondiscoverable. The inclusion of multiple species for comparison in our method improves the chances of finding CNSs in at least one species but may not be sufficient in all cases. As they become available, the addition of more comparator species genomes (within an appropriate evolutionary distance from Arabidopsis) would further improve CNS detection using our method. The accuracy of genome annotations is another factor that may affect our ability to detect CNSs, particularly with regard to correctly defining TSS positions. All genome annotations in this study have some degree of EST support, and for Arabidopsis, ∼66% of genes have a defined 5′ untranslated region (UTR) boundary (Chung et al., 2006). In cases where predicted gene models are not supported by full-length ESTs, our TSS position will correspond to the ATG. In this study, if the TSS is correctly annotated in Arabidopsis or correctly annotated in the comparator species, then this is sufficient to exclude discovery of CNSs within 5′ UTR regions of any orthologous genes in question. In some cases, however, a CNS may fall within a 5′ UTR. CNSs and motifs embedded in 5′ UTR may still play a role in transcript regulation, for example, in modulating transcript abundance (Liu et al., 2010; Wang and Xu, 2010).
We have performed a genome-wide analysis of orthologous genes between four dicot plant genomes. Using the premise that functional regions in the genomes of related species evolve at slower rate than nonfunctional regions (Tagle et al., 1988), we used in silico techniques to identify 1865 CNSs present upstream of the TSS of 1643 Arabidopsis genes (Table 1). At a high-confidence 0.7 threshold, we identified a subset of 554 genes with 602 CNSs that we believe to be components of the regulatory machinery shared among dicot plants. The alignment plot method used in this study evaluates millions of alignment scores for all pairs of short sequence fragments, thereby providing comprehensive detection of alignment conservation. The evolutionary distance between Arabidopsis and the different comparator species is reflected in the conversion of raw alignment scores into conservation scores, providing additional enhancement to enable the discovery of even weakly conserved regions between distant organisms.
Our method is strengthened by its statistical approach, which takes into account the distribution of alignment scores generated by randomly assigned promoter pairs from each pair of species. Also, the alignment scores are penalized for repetitiveness using repeat-masked sequences (see Methods). This reduces the occurrence of simple sequence repeats and known repeat sequences (such as transposable elements) in our statistically significant set of CNSs. The conserved regions do not have significant similarity to any known plant proteins and have a GC content similar to that of the mean for noncoding regions of the Arabidopsis genome, which all but eliminates a protein-coding role. While we have not done experimental work as part of this study, our alignment plot method has previously been validated using existing experimental results. Verified regulatory regions were accurately predicted at the loci of LHY, TOC1, LUX, CAB2, and ABI3 (Picot et al., 2010; Spensley et al., 2009).
Sequence conservation implies functional conservation, and several of our results act as independent lines of evidence indicating that many of the identified CNSs are functionally involved in transcriptional regulation. First, the CNSs show a clear and significant positional bias toward the first 100 nucleotides upstream from the TSS, whereas the control set produced alignments with a uniform distribution in the promoter region. For a subset of CNSs (14% that are within 50 bp of the TSS and contain a TATA motif), their positioning corresponds well with the core promoter region. However, the majority (76% >50 bp from TSS) fall outside of the core promoter region. In general, the existence of CNS upstream of TSS is consistent with the hypothesis that they contain embedded cis-regulatory elements to which TFs can bind. Comparing the alignment length distributions, we also found that alignments from orthologs (see Supplemental Figure 4A online) are on average significantly longer than alignments from random gene pair sequences (see Supplemental Figure 4B online), with mean lengths of 93 ± 25 bp and 66 ± 13 bp, respectively. The difference in length distributions is consistent with the view that the former set are meaningful sequences whose lengths are determined by the nature of their biological function (i.e., they contain multiple TFBSs), while the latter set comprises randomly occurring alignments that are expected to be short in length. Transcription complexes are assembled from multiple proteins, and long stretches of conserved sequence will allow enough space for a number of these proteins to bind to DNA in the regulatory region. Therefore, significant CNS length is suggestive of complex function necessitating a large stretch of nucleotides to facilitate binding.
This finding is in contrast with a previous study that identified CNS in Arabidopsis paralogs with a median length of 25 bp (Thomas et al., 2007). The difference is due to the increased sensitivity of our algorithm in detecting longer regions of alignment conservation. At the 0.3 threshold, the average length of paralogous CNSs found by our method is 96 bp. The short regions discovered by Thomas et al. (2007) may correspond to individual binding elements, whereas our CNS regions are longer stretches of conserved sequence, containing multiple binding elements. This is certainly true in the case of TOC1, for example, where the CNS identified upstream of TOC1 contains evening elements (experimentally proven for necessity of circadian activity; Alabadi et al., 2001), as well as G/C-box elements and DOF binding sites (Picot et al., 2010). Our findings are consistent with the “DNA-templated protein assembly” hypothesis put forward by Kaplinsky et al. (2002), whereby long CNSs act as a template for the assembly of regulatory protein complexes that may not associate on their own. Transcription complexes are assembled from multiple proteins, and long stretches of conserved sequence will allow enough space for these multiple proteins to bind in the regulatory region. Therefore, significant CNS length is suggestive of complex function necessitating a large stretch of nucleotides to accommodate multiple binding events. Third, the CNSs are enriched for known TFBSs, when tested with a low redundancy set of 728 eukaryotic motifs represented by PSSMs extracted from TRANSFAC, JASPAR, and PLACE databases. A total of 182 motifs are significantly overrepresented, and CNSs contain 106% more motif matches for these motifs than randomly chosen promoter sequences. Testing CNSs for motif overrepresentation has uncovered a number of diverse TF binding motifs, such as ABF1, ABI4, and GAGA elements (P values < 1e-4; Figure 2). Once again, this result points to the functional nature of the regions being part of the transcription regulatory machinery and regulating the genes with which they are associated. We tested only for the overrepresentation of single motifs, although it is common for a region of 100 nucleotides to contain more than one TF binding site. Further insight could therefore be gained by examining TFBS multiplicity and combinatorics. For example, two TFBS motifs may not be overrepresented individually, but a combination of the two factors binding within a certain degree of proximity might be. We have shown that TFBSs are more common in regions of CNSs (compared with background), and these thousands of discovered binding sites can be interpreted as representing links within the Arabidopsis transcriptional network, though from motif analyses alone we cannot uniquely determine the protein binding each site.
The fourth line of evidence that the CNSs are involved in transcriptional regulation comes from the statistical link found between a clear peak in prediction of average nucleosome occupancy correlating directly with the location of CNSs in Arabidopsis. It is not known why there is an association between CNSs and predicted nucleosome occupancy. It may be that as nucleosomes occlude underlying DNA sequences, and will therefore restrict access to TFBSs present in the CNSs, they are effectively turning off the associated genes. Our findings regarding CNSs being ∼93 bp long and the occurrence of multiple binding sites within them support a hypothesis based on the model of nucleosome-mediated cooperativity between TFs described by Mirny (2010). This article postulates that multiple TF binding sites within a region no longer than the 147 bases occupied by one nucleosome may be required (and in terms of regulatory logic essential) to displace nucleosomes, rendering their associated genes transcriptionally active. Once again, it makes sense for the genes responsible for high-level developmental process and regulation to be themselves tightly regulated. The prediction tool (Kaplan et al., 2009) was claimed to work well for the subset of the genome featuring well-positioned nucleosomes, though not in the majority of genomic sequence where nucleosome positioning is thought not to be determined by DNA motifs (Stein et al., 2010). Hence the predictions are likely to be accurate if the CNS set overall contains features that reflect the intrinsic DNA sequence preferences of the nucleosome.
A fifth line of evidence for the role of CNSs in transcriptional regulation comes from examination of the GO terms ascribed to the CNS-associated genes. Analyzing molecular function GO terms identified overrepresentation in transcriptional regulation and associated processes, such as DNA binding and promoter binding (see Supplemental Data Set 3 online, MF), providing evidence that genes with CNSs are involved in transcriptional regulation of gene expression. There is also a strong bias toward biological process GO terms described as “regulation of….” Interestingly, the term with the strongest significance value is “regulation of transcription.” Cross-referencing the 554 high-confidence CNS-associated genes against a manually curated list of 2468 genes thought to encode Arabidopsis TFs matches 208 genes. Looking at the cellular components showed overrepresentation of “nucleus” only (see Supplemental Data Set 3 online, CC). This is consistent with the involvement of these genes in transcriptional regulation as suggested by the molecular function GO term analysis. In summary, GO term analysis has revealed that the genes that contain CNSs are predominantly involved in transcriptional regulation and developmental processes, and their protein products are localized to or associated with the cell nucleus. Furthermore, the genes associated with CNSs contain a significantly high proportion of genes described as master regulators. Therefore, it seems we have identified not only a set of key transcriptional sequences, but also a set of genes that is likely to play a role at the highest level of the transcriptional hierarchy. This finding that TFs tend to be CNS rich is consistent with previous studies of Arabidopsis paralogous CNSs (Thomas et al., 2007), and it has also been noted in grasses that TF genes are rich in orthologous CNSs relative to genes encoding enzymes or structural proteins (Inada et al., 2003).
Other highly significant GO terms found to be overrepresented among CNS-associated genes are involved in development and morphological processes, relating to organs, systems, flowers, and reproductive structures (see Supplemental Data Set 3 online). It is vital for all organisms to tightly control growth and reproduction, and as a monophyletic taxon, it is to be expected that some of these processes are deep-seated within plants. The abundance of developmental regulatory genes in the highest confidence CNS set (72 genes) mirrors the finding in the human genome that the most highly conserved noncoding sequences associate with developmental regulators, suggesting a key role in the orchestration of early embryo development (Elgar and Vavouri, 2008). Development is a tightly regulated process, and as such there may be strong selective pressure at binding site sequences.
The gene age analysis indicated that the more widely orthologs of a gene are found across taxa, the less likely it is to be associated with CNSs. This is consistent with the idea that genes or gene families that are more recently evolved, and therefore essential to or associated with the nature of land plants, are more likely to have conserved transcriptional regulatory structures within this clade. However, there are large morphological differences between the organisms in this study (a weed, a tropical fruit, a vine, and a nonfruiting tree). Species diversity comes in part from differential regulation of similar sets of genes, combined with regulatory layers beyond transcription (e.g., RNA processing, posttranslational modification, and epigenetics). Our method was unable to find CNSs for many genes, even where a candidate ortholog was identifiable. Therefore, for these genes, it is likely that the sequences involved in transcriptional regulation have diverged, in some cases giving rise to morphological differences.
One previous study of paralogous CNS in Arabidopsis used BLAST to identify CNSs surrounding 3179 paralogous Arabidopsis gene pairs (Thomas et al., 2007). Our study of Arabidopsis CNS considers 21,212 Arabidopsis genes with orthologs and/or paralogs, the overlap between the studies being a subset of 6027 genes. Thomas et al. (2007) found 2170 paralog pairs with CNSs, while we identified 1643 genes with orthologous CNSs and 1149 genes with paralogous CNSs, with an overlapping gene set between both studies of 52% (ortholog and paralog data combined, 0.3 threshold). The Arabidopsis CNSs discovered by each method are largely nonoverlapping with at least 68% of the orthologous CNSs identified by our study being previously unidentified. The two approaches can therefore be considered complementary, capturing CNSs for different subsets of the Arabidopsis genome. Paralogous genes in Arabidopsis arose from large-scale duplication events occurring in the lineage 20 to 70 Mya. By comparison, orthologs in other dicots have experienced a longer period of divergence (in excess of 100 to 145 Mya), so mutations have had longer to accumulate. Therefore, any remaining sequence similarity is strongly indicative of a deep-seated functional role important across plant species. The finding that CNS-associated genes are themselves high up in the regulatory cascade, however, is consistent between both approaches. Focusing on a subset of 3019 genes that have both a paralog and an ortholog, we applied our method to detect both types of conservation. We find a strongly significant statistical link between paralogous conservation and orthologous conservation of CNSs, indicating that regulatory constraints imposed onto genes by CNSs are retained after gene duplication in a significant number of cases.
Not all genes had identifiable upstream CNSs, yet all must be subject to transcriptional regulation. Regulatory regions are not confined to regions upstream of a gene’s TSS but may also occur in the introns (Schauer et al., 2009) or 3′ UTR (Cawley et al., 2004). Although we focused on upstream promoter regions in this study, our method can be applied to any orthologous sequence regions, and doing so may uncover yet more functional CNSs. However, it is also true that a phenomenon called “binding site turnover” means that many regulatory elements mutate rapidly over evolutionary time and are not highly conserved in terms of sequence, yet retain binding functionality (Moses et al., 2006). Hence, our method is not a universal solution to finding the full complement of regulatory elements within plant genomes. However, its loss-free local alignment means that it is able to find all alignment-conserved sequences, given the appropriate orthologous sequence for comparison. Alternative methods using metrics other than sequence alignments (such as the alignment-free model of Koohy et al., 2010) are needed to find the subset of regulatory elements that are functionally but not alignment conserved. Methods that combine comparative genomics with other resources, such as gene expression (as in Vandepoele et al., 2006; Heyndrickx and Vandepoele, 2012; Spangler et al., 2012), are also useful in aiding the discovery and analysis of regulatory modules.
Different types of regulatory regions have been proposed (Arnosti and Kulkarni, 2005). Strictly organized enhanceosomes are thought to provide scaffolds for the formation of large well-defined protein-DNA complexes that exert tight constraints on target gene expression. On the other hand, loosely organized billboard enhancers bind regulatory proteins individually or in small complexes and exhibit regulatory function without strict requirements on the number and order of binding sites. Our method is likely to detect enhanceosome-type regulatory regions that require a number of binding sites in a specific arrangement and therefore have good potential to align with functionally conserved orthologous enhanceosomes. However, billboard-type enhancers are not expected to be detected well using alignments as the number of binding sites may be too low to yield a statistically significant alignment and their arrangement may not be conserved even in the presence of functional conservation. Therefore, while the false negative rate of our method is high if considering the whole genome and all types of transcriptional regulatory regions, if considering only regulatory regions that are (1) of higher complexity and (2) functionally conserved among distant species, then the false negative rate of our method may be low. It is not known what proportion of Arabidopsis promoters fall into this category.
In this study, we predict the involvement of CNSs in transcriptional regulation of their associated genes and find that CNS-associated genes themselves most commonly have a role in transcriptional regulation. The finding that regulatory genes are themselves highly regulated makes biological sense; as plants rely on their regulatory machinery to integrate signals from internal and external stimuli to formulate complex decision-making, it is intuitive to put those genes under strict control. Taking into account CNS length and binding site content, we can predict that each gene is likely to have a few to a dozen regulators that interact directly with the DNA (and others that operate indirectly via protein–protein interactions with DNA-bound regulators). Furthermore, using this method, we have been able to predict several thousand binding sites mediating TF-gene links in the transcriptional regulatory network of Arabidopsis. The implication of our findings is that the strongly maintained CNSs and the genes they are associated with play an intrinsic role in the regulatory network that is shared among dicot plants.
We provided the APPLES software and a virtual appliance that allows users to run APPLES scripts and make programmatic use of APPLES in a virtual machine, without having to install the software or its dependencies. Included is a set of example scripts that make some of the software’s core functionality directly accessible. APPLES can be applied to detect CNSs in other sets of species (not necessarily including Arabidopsis).
Our data are an important resource for the Arabidopsis research community, significantly expanding the set of CNSs and CNS-associated genes. The identification of regulatory sequences experimentally is laborious, typically requiring series of essentially random promoter deletions to be made, fusing these to reporter genes, and subjecting them to expression assays in transgenic organisms from which to infer results. Our method provides a shortcut to identifying key regulatory regions in hundreds of Arabidopsis genes. Through in silico analysis, we identified a high confidence set of previously unidentified candidate regulatory regions that are highly likely to be functional. Our results can be used to inform binding experiments, for example, by refining the design of constructs for yeast one-hybrid experiments, as the regions identified are excellent candidates for containing protein binding sites. We provided examples of how our data can be used to inform studies of subfunctionalization of CNS after gene duplication. Identifying regulatory regions using CNSs as proxies is a valuable exercise, but experimental validation of these regions is required for absolute proof of function. The data we provide are also valuable in that it can enable biologists to focus experimental efforts on regulatory components that are shared between Arabidopsis and other members of the dicot clade (which includes many crop species) so that research results can ultimately be transferred into real-world applications.
METHODS
Databases
Genome databases for Arabidopsis thaliana, grape (Vitis vinifera), and poplar (Populus trichocarpa) were downloaded in MySQL format from Ensembl (http://plants.ensembl.org/index.html), release 62, and installed locally. The papaya (Carica papaya) draft genome sequences (supercontigs.filtered_012808.fasta, contigs.filtered_012808.fasta) and annotation (supercontigs.evm_27950.gff3, contigs.evm_27950.gff3) were downloaded via http://www.life.illinois.edu/plantbio/People/Faculty/Ming.htm and were used to create a local Ensembl-format database using the Ensembl pipeline and customized Perl scripts. All four databases are available to download from the APPLES website given below. The Arabidopsis promoter binding element database (AtProbe) was accessed via http://exon.cshl.org/cgi-bin/atprobe/atprobe.pl.
Software
Sequence analysis (ortholog identification, sequence alignments, and TFBS enrichment tests) was performed with APPLES and the ENSEMBL API (Hubbard et al., 2009). The APPLES virtual appliance, instructions for its use, and the APPLES source code can be found at http://www2.warwick.ac.uk/fac/sci/systemsbiology/staff/ott/tools_and_software/apples/. The APPLES source code is also available at http://sourceforge.net/projects/apples-software/.
APPLES is predominantly written in Perl, with some R and C extensions for computationally intensive tasks, and consists of >20,000 lines of code organized in more than 80 classes. Users need to familiarize themselves with the key concepts of the APPLES software and connect APPLES to the relevant genome databases. For genome-wide applications, installing APPLES on a cluster machine should be considered as otherwise the time requirement (using a virtual machine on a single node) is likely to be in the order of months, depending on parameters. Computing CNS for one pair of species, using a 50 CPU machine, took ∼4 d. APPLES follows an object-oriented software design that facilitates future use and development of the software.
Ortholog and Paralog Identification
A pan-rosid syntenic gene set created by Haibao Tang was obtained from CoGePedia (http://genomevolution.org/wiki/index.php/Syntenic_gene_sets). This uses the QUOTA-ALIGN algorithm (Tang et al., 2011) to identify inferred syntenic regions when no homologous gene is present and enforce a set syntenic relationship based on the whole-genome duplication history of each genome (1:1:2:4 in grape, papaya, poplar, and Arabidopsis). This data set was combined with a set of orthologous genes identified using an implementation of the reciprocal best hit method (Moreno-Hagelsieb and Latimer, 2008). In summary, a FASTA file of all proteins in each genome is made and formatted into a BLAST database. BLASTP (Altschul et al., 1990) is performed between each set of proteins, selecting the best hit for each protein. The BLAST results are compared, and where the best match of protein A in genome 1 is protein B in genome 2 and vice versa (i.e., reciprocal best BLAST hit), an ortholog assignment is made. Using this method produced three lists of Arabidopsis genes with a corresponding orthologous gene from each of the target genomes, which were merged to produce a single list of 15,386 Arabidopsis genes that had at least one orthologous gene in one target species.
An accurate list of manually curated paralogous pairs was obtained from Thomas et al. (2007). For each member of a paralog pair, and where the synteny-based map indicated multiple Arabidopsis genes are orthologs, each Arabidopsis gene is also assigned the orthologs of its gene-duplicate partner(s). In the combined ortholog map for Arabidopsis against poplar, grape, and papaya, 21,034 Arabidopsis genes have at least one ortholog assigned in at least one species.
To determine the phylogenetic age of each gene, the Inparanoid method (Ostlund et al., 2010) was used to identify putative orthologs in the following more distant taxa, chosen to span the range of evolutionary divergence from Arabidopsis: poplar, rice (Oryza sativa), sorghum (Sorghum bicolor), Physcomitrella patens, Cyanidioschyzon merolae, Ostreococcus tauri, Chlamydomonas reinhardtii, Thalassiosira pseudonana, Neurospora crassa, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, and Escherichia coli K12. To identify the most distant ortholog of each gene, putative orthologs were first filtered to remove suggested orthologs in very distant taxa where none were found in more closely related taxa. The main effect of this filtering was to remove a number of putative orthologs between Arabidopsis and animal genes, where no orthologs were found in any other sampled plant species.
Sequence Alignments
The TSSs and upstream sequences for each Arabidopsis gene and its ortholog(s) were retrieved from sequence databases (see above). For Arabidopsis, poplar, and grape, TSSs correspond to Ensembl annotations, and for papaya, these correspond to the 5′-most feature (mRNA or CDS) in the gff3 file. A maximum of 2000 and minimum of 200 nucleotides were taken, but truncating the sequence to the neighboring gene if within 2 kb. Sequence alignment scores were calculated using an implementation of the seaweed algorithm (Krusche and Tiskin, 2010) in C, with a sliding window length of 60 nucleotides. The scoring mechanism applied in the seaweed algorithm is +1 for a match, 0 for a mismatch, and −0.5 for a gap. Thus, for a 60-bp window, the highest score possible is 60.
Converting Alignment Scores to Conservation Scores
The alignment score is converted into a conservation score using a sigmoidal function with upper and lower thresholds. The upper threshold indicates that any alignment score found above this threshold is assigned a conservation score of 1. Conversely, the lower threshold indicates that any alignment score found below this threshold is assigned a conservation score of 0. The upper and lower thresholds were calculated for each species pairing using the distribution of alignment scores from randomly assigned gene pairs. The upper threshold was established by manual inspection of the alignment score histograms (see Supplemental Figure 2 online), taking the score above which no random gene pair produced a significant alignment. The lower bound is chosen as the point where the control set and the ortholog set begin to show significantly different numbers of alignments. These thresholds were used for the real orthologs to find the conservation score of each CNS. Repetitive sequences are penalized in the conversion procedure. A region is called repetitive if it is annotated as a repeat in the Ensembl sequence database (identified by RepeatMasker, based on species-specific libraries of repeats). Repetitive sequence in a window shifts the sigmoidal curve proportionally to the right, so a region containing repeats requires a higher alignment score than a window of nonrepetitive sequence to obtain the same conservation score. During the conversion procedure, where significantly high-scoring window pairs positionally overlap, they are merged into a single contiguous region. In the multispecies analysis, the conservation scores between each of the three target species and Arabidopsis (where available) are combined into a single conservation score using the formula:
where P is the maximum conservation score for a region in one species pair, and i is each species pair. For example, in the case of three species with conservation score 0.2 (P1 = P2= P3 = 0.2), the overall conservation score is 0.488, whereas a conservation score of 0.5 in just one species (P1 = 0.5, P2 = P3 = 0) yields an overall conservation score of 0.5.
Filtering Out Potential Protein Coding Regions
To exclude potential protein coding regions from the set of conserved regions, we removed any gene and all of its associated conserved regions from the 0.3 threshold set and above if any of its regions had a significant BLASTX hit to any Viridiplantae sequence in the National Center for Biotechnology Information database. To establish an appropriate e-value cutoff for a significant hit, we randomly permuted each sequence in the 0.3 threshold set and performed the BLASTX search using this set of sequences to obtain the distribution of e-values for random sequences. We then performed the same BLASTX search on the real sequences, using the minimum e-value from the random set (4.00e-08) as the cutoff for a significant hit.
Motif Overrepresentation
A total of 2595 known position specific weight matrices were retrieved, including all matrices from TRANSFAC v2009.4 (Wingender et al., 2000), JASPAR (Bryne et al., 2008), and PLACE (Higo et al., 1999) databases. To reduce redundancy of this set of weight matrices and the computational load of performing the binomial overrepresentation test for all motifs, the matrices were clustered into 728 clusters using the Hellinger distance function with a threshold of 2.3, and a single representative motif was selected from each. This set of 728 motifs was used in the binomial overrepresentation tests, which test each motif against the set of CNSs (or against a set of randomly selected regions). The best 100 matches of the motif in the sequence set are identified, and the binomial distribution is used to compute an overall overrepresentation score, taking into account the strength of motif matches and the overall length of sequences in the set. The score is computed for the top n motif matches where 1 ≤n ≤ 100 is chosen to optimize the overrepresentation score. For each of the 602 CNSs from the Arabidopsis 0.7 threshold set, genes were randomly assigned from the same genome to make background sets for comparison as described in the main text. The overrepresentation test was run 100 times to assess the distribution of motif overrepresentation scores in the random sets, and it was run once on the set of CNSs. Using the distribution of binomial overrepresentation scores associated with each motif, we removed motifs with insignificant scores. Individual P values were calculated using the pnorm function in R. Known repeats in all sequences were masked using the repeat annotations in the sequence databases.
GO Term Analysis
GO term analysis was performed using the BiNGO plugin (version 2.3) (Maere et al., 2005) for Cytoscape (version 2.6) (Shannon et al., 2003). The set of 554 Arabidopsis genes was compared for overrepresentation using a hypergeometric test statistic using the set of Arabidopsis genes with an identifiable ortholog as the reference set. Benjamini and Hochberg false discovery rate correction for multiple testing was applied, with significance level of 0.05 (5%). The tests were performed using three ontology files that come as part of BiNGO (updated August 2010): “GO_Biological_Process,” “GO_Molecular_Function,” and “GO_Cellular_Component.”
Prediction of Nucleosome Positioning
To compute the nucleosome occupancy probabilities, we used the nucleosome prediction software of Kaplan et al. (2009) with default parameters (http://genie.weizmann.ac.il/software/nucleo_prediction.html). Sequences of 10 kb, with the CNS positioned centrally in each sequence, were used as input. Where a gene was associated with more than one CNS, one was selected. Nucleosome occupancy probabilities were calculated at each nucleotide position and the results averaged across the CNS set (554 sequences). Ten sets of control sequences were created, whereby for each conserved sequence, regions of 10 kb were selected upstream of 10 randomly picked genes in the Arabidopsis genome, such that the center of the selected region is at the same position relative to the TSS as the center of its comparable conserved sequence and not allowing the sequence in the center to contain repetitive sequences. Average nucleosome occupancy probabilities were calculated for each of the 10 control sets of 554 sequences, and the mean and sd of these averaged values were plotted. As the prediction software does not tolerate input sequences containing non-ACGT characters, up to two sequences were omitted from each set prior to analysis (<0.4% of sequences).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TOC1 (AT5G61380), LUX (AT3G46640), BOA (AT5G59570), ATHB31 (AT1G14440), ATHB21 (AT2G02540), ABI3 (AT3G24650), CAB2 (AT1G29920), AP1 (AT1G69120), ABF1/3/4 (AT1G49720, AT4G34000, and AT3G19290), ABI4 (AT2G40220), ANAC076 (AT4G36160), and ANAC037 (AT2G18060).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of Arabidopsis Orthologs and CNSs across Three Comparator Species, Papaya, Grape, and Poplar.
Supplemental Figure 2. A Large Number of Arabidopsis Promoters Contain Evolutionarily Conserved Noncoding Sequences.
Supplemental Figure 3. Distribution of Distances between CNSs and TSSs in Arabidopsis, Normalized by Intergenic Length.
Supplemental Figure 4. The Distribution of Alignment Lengths in Orthologs and Randomly Assigned Gene Pairs.
Supplemental Table 1. Manually Curated List of Plant Master Regulators Derived from Current Literature.
Supplemental Methods 1. Methodological Comparison for Discovery of Paralogous CNSs.
Supplemental Data Set 1. Database of Arabidopsis Cross-Species CNSs.
Supplemental Data Set 2. 182 Overrepresented Motifs in Arabidopsis CNSs.
Supplemental Data Set 3. GO Analysis of the CNS-Associated Gene Set.
Supplemental Data Set 4. Database of Arabidopsis Paralogous CNSs (0.3 Threshold).
Supplemental Data Set 5. GO Analysis for 85 Genes with Overlapping Paralogous and Orthologous CNSs.
Supplementary Material
Acknowledgments
L.B., J.M., V.B.-W., J.B., K.D., and S.O. were funded by the grant “Plant Responses to Environmental Stress in Arabidopsis (PRESTA)” (BB/F005806/1), which was awarded by the Biotechnology and Biological Sciences Research Council (BBSRC). A.J., R.H., and C.B. are members of the Engineering and Physical Sciences Research Council/BBSRC–funded Warwick Systems Biology Doctoral Training Centre. We thank all members of the PRESTA project for valuable discussions. We also thank the three anonymous reviewers for their valuable comments and feedback to improve the article. The computing facilities were provided by the Centre for Scientific Computing of the University of Warwick with support from the Science Research Investment Fund.
AUTHOR CONTRIBUTIONS
All authors had a role in discussion of results. L.B. and R.H. developed the core of the APPLES software platform with support from N.P.D. J.M. developed an in-house job-handling system needed for running large-scale computations and a cache database to store the results. C.B. implemented the conservation score conversion procedure. L.B. supervised the development of this conservation score and integration into APPLES. P.K. developed a highly optimized implementation of the seaweed algorithm, facilitating the genome-wide application. L.B. and A.J. wrote the majority of the article, with input from S.O., K.D., J.M., R.H., and J.B. L.B. and A.J. performed a majority of the computations and data analysis with input from J.M., R.H., and S.O. A.T. provided expertise in algorithm development and supervised the development of the alignment plot software. S.O. conceived and planned the APPLES project, supervised the software development, and defined the conservation score. K.D. and S.O. cosupervised the data analysis. S.O., K.D., V.B.-W., and J.B. designed and supervised the research as part of the PRESTA project.
Glossary
- TF
transcription factor
- PSSM
plant-specific position-specific scoring matrix
- TFBS
transcription factor binding site
- CNS
conserved noncoding sequence
- TSS
transcription start site
- Mya
million years ago
- GO
Gene Ontology
- UTR
untranslated region
References
- Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. (2007). Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arnosti D.N., Kulkarni M.M. (2005). Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94: 890–898 [DOI] [PubMed] [Google Scholar]
- Bastola D.R., Pethe V.V., Winicov I. (1998). Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol. 38: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Berendzen K.W., Stüber K., Harter K., Wanke D. (2006). Cis-motifs upstream of the transcription and translation initiation sites are effectively revealed by their positional disequilibrium in eukaryote genomes using frequency distribution curves. BMC Bioinformatics 7: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N., Dubchak I., Pachter L. (2003). AVID: A global alignment program. Genome Res. 13: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. (2008). JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res. 36(Database issue): D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S., et al. (2004). Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Chung B.Y., Simons C., Firth A.E., Brown C.M., Hellens R.P. (2006). Effect of 5’UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinas J., Birnbaum K., Benfey P.N. (2002). Using cauliflower to find conserved non-coding regions in Arabidopsis. Plant Physiol. 129: 451–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creux N.M., Ranik M., Berger D.K., Myburg A.A. (2008). Comparative analysis of orthologous cellulose synthase promoters from Arabidopsis, Populus and Eucalyptus: Evidence of conserved regulatory elements in angiosperms. New Phytol. 179: 722–737 [DOI] [PubMed] [Google Scholar]
- Duret L., Bucher P. (1997). Searching for regulatory elements in human noncoding sequences. Curr. Opin. Struct. Biol. 7: 399–406 [DOI] [PubMed] [Google Scholar]
- Elgar G., Vavouri T. (2008). Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 24: 344–352 [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Elnitski L., Church D.M., Dubchak I., Hardison R.C. (2003). Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Rapaka L., Lyons E., Pedersen B., Thomas B.C. (2007). G-boxes, bigfoot genes, and environmental response: characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Moose S.P. (2003). Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15: 1143–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B., Dudley J., Kumar S. (2006). TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972 [DOI] [PubMed] [Google Scholar]
- Heyndrickx K.S., Vandepoele K. (2012). Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 159: 884–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T.J., et al. (2009). Ensembl 2009. Nucleic Acids Res. 37(Database issue): D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada D.C., Bashir A., Lee C., Thomas B.C., Ko C., Goff S.A., Freeling M. (2003). Conserved noncoding sequences in the grasses. Genome Res. 13: 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Pugh B.F. (2009). Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., Segal E. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky N.J., Braun D.M., Penterman J., Goff S.A., Freeling M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99: 6147–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koohy H., Dyer N.P., Reid J.E., Koentges G., Ott S. (2010). An alignment-free model for comparison of regulatory sequences. Bioinformatics 26: 2391–2397 [DOI] [PubMed] [Google Scholar]
- Krusche P., Tiskin A. (2010). Computing alignment plots efficiently: CoRR, abs/0909.2000. In Advances in Parallel Computing 19: Parallel Computing: From Multicores and GPUs to Petascale, B. Chapman, F. Desprez, G.R. Joubert, Al, Lichnewsky, F. Peters, and T. Priol, eds (Amsterdam: IOS Press), pp. 158–165
- Lemon B., Tjian R. (2000). Orchestrated response: A symphony of transcription factors for gene control. Genes Dev. 14: 2551–2569 [DOI] [PubMed] [Google Scholar]
- Lenhard B., Wasserman W.W. (2002). TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics 18: 1135–1136 [DOI] [PubMed] [Google Scholar]
- Liu W.X., Liu H.L., Chai Z.J., Xu X.P., Song Y.R., Qu Q. (2010). Evaluation of seed storage-protein gene 5′ untranslated regions in enhancing gene expression in transgenic rice seed. Theor. Appl. Genet. 121: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Matys V., et al. (2006). TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34(Database issue): D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny L.A. (2010). Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. USA 107: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C., Grotewold E. (2005). Genome wide analysis of Arabidopsis core promoters. BMC Genomics 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G., Latimer K. (2008). Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324 [DOI] [PubMed] [Google Scholar]
- Moses A.M., Pollard D.A., Nix D.A., Iyer V.N., Li X.-Y., Biggin M.D., Eisen M.B. (2006). Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ostlund G., Schmitt T., Forslund K., Köstler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L.L. (2010). InParanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38(Database issue): D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., Song J.S., Liu X.S., Fisher D.E. (2007). High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 25: 244–248 [DOI] [PubMed] [Google Scholar]
- Parcy F., Nilsson O., Busch M.A., Lee I., Weigel D. (1998). A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Picot E., Krusche P., Tiskin A., Carré I., Ott S. (2010). Evolutionary analysis of regulatory sequences (EARS) in plants. Plant J. 64: 165–176 [DOI] [PubMed] [Google Scholar]
- Reineke A.R., Bornberg-Bauer E., Gu J. (2011). Evolutionary divergence and limits of conserved non-coding sequence detection in plant genomes. Nucleic Acids Res. 39: 6029–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Schauer S.E., Schlüter P.M., Baskar R., Gheyselinck J., Bolaños A., Curtis M.D., Grossniklaus U. (2009). Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J. 59: 987–1000 [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler J.B., Subramaniam S., Freeling M., Feltus F.A. (2012). Evidence of function for conserved noncoding sequences in Arabidopsis thaliana. New Phytol. 193: 241–252 [DOI] [PubMed] [Google Scholar]
- Spensley M., Kim J.Y., Picot E., Reid J., Ott S., Helliwell C., Carré I.A. (2009). Evolutionarily conserved regulatory motifs in the promoter of the Arabidopsis clock gene LATE ELONGATED HYPOCOTYL. Plant Cell 21: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J.E., et al. (2002). The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Takasuka T.E., Collings C.K. (2010). Are nucleosome positions in vivo primarily determined by histone-DNA sequence preferences? Nucleic Acids Res. 38: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle D.A., Koop B.F., Goodman M., Slightom J.L., Hess D.L., Jones R.T. (1988). Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203: 439–455 [DOI] [PubMed] [Google Scholar]
- Tang H., Lyons E., Pedersen B., Schnable J.C., Paterson A.H., Freeling M. (2011). Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics 12: 102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Rapaka L., Lyons E., Pedersen B., Freeling M. (2007). Arabidopsis intragenomic conserved noncoding sequence. Proc. Natl. Acad. Sci. USA 104: 3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiskin A. (2008). Semi-local string comparison: Algorithmic techniques and applications. Math. Comput. Sci 1: 571–603 [Google Scholar]
- Tompa M., et al. (2005). Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnology 23: 137–144
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Casneuf T., Van de Peer Y. (2006). Identification of novel regulatory modules in dicotyledonous plants using expression data and comparative genomics. Genome Biol. 7: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.T., Xu Y.N. (2010). The 5′ untranslated region of the FAD3 mRNA is required for its translational enhancement at low temperature in Arabidopsis roots. Plant Sci. 179: 234–240 [Google Scholar]
- Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Prüss M., Reuter I., Schacherer F. (2000). TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 28: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jin J., Tang L., Zhao Y., Gu X., Gao G., Luo J. (2011). PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 39(Database issue): D1114–D1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.