This work characterizes the role of cytokinins in lateral root organogenesis, finding that cytokinin activity is likely important to prevent lateral root initiation in close proximity to existing lateral root primordia. The results support the idea that stage-dependent effects of cytokinin are determined by the robustness of the auxin gradient, which might be enhanced in mature primordia.
Abstract
The architecture of a plant’s root system, established postembryonically, results from both coordinated root growth and lateral root branching. The plant hormones auxin and cytokinin are central endogenous signaling molecules that regulate lateral root organogenesis positively and negatively, respectively. Tight control and mutual balance of their antagonistic activities are particularly important during the early phases of lateral root organogenesis to ensure continuous lateral root initiation (LRI) and proper development of lateral root primordia (LRP). Here, we show that the early phases of lateral root organogenesis, including priming and initiation, take place in root zones with a repressed cytokinin response. Accordingly, ectopic overproduction of cytokinin in the root basal meristem most efficiently inhibits LRI. Enhanced cytokinin responses in pericycle cells between existing LRP might restrict LRI near existing LRP and, when compromised, ectopic LRI occurs. Furthermore, our results demonstrate that young LRP are more sensitive to perturbations in the cytokinin activity than are developmentally more advanced primordia. We hypothesize that the effect of cytokinin on the development of primordia possibly depends on the robustness and stability of the auxin gradient.
INTRODUCTION
The fundamental task of the root system is to mediate an interaction between plant and soil and to provide the plant with water and nutrients. The architecture of the root system is determined by endogenous mechanisms that constantly integrate environmental signals, such as availability of nutrients and moisture or salinity, and, accordingly, modulate the primary root growth and branching. The developmental flexibility of root systems, which is extremely important for efficient soil exploitation and survival of plants under less favorable conditions, is largely determined by its postembryonic capacity to branch.
In Arabidopsis thaliana, lateral roots initiate from selected pericycle cells that acquire attributes of founder cells, subsequently undergo a series of anticlinal divisions, and produce a few short initial cells. After initiation, coordinated cell division and differentiation give rise to lateral root primordia (LRP) that continue to grow and emerge through the cortex and epidermal layers of the primary root; finally, the lateral root apical meristems take over the growth regulation (Malamy and Benfey, 1997; Péret et al., 2009; Benková and Bielach, 2010).
Overall, the root system architecture depends largely on the spatiotemporal regulation of individual lateral root initiation (LRI) events. Notably, processes leading to initiation seem to be activated much earlier than the morphological detection of the first anticlinal divisions. Priming, proposed to occur in the basal root meristem by recurrent accumulations of the plant hormone auxin in selected protoxylem cells, precedes founder cell establishment and LRI (De Smet et al., 2007; De Rybel et al., 2010). Several concepts have recently been put forward to assess the mechanisms behind the repetitive initiation events. Either correlation between gravity responses and LRI that regulate root bending (De Smet et al., 2007; Lucas et al., 2008a) or the mechanical stretching of cells as a consequence of the root bending by gravistimulation (Laskowski et al., 2008) have been proposed to underlie the regulation of LRI. Another hypothesis considers regular (gravity-independent) oscillations in a set of genes, including transcriptional regulators, as the primary mechanism recurrently activating processes that result in LRI (Moreno-Risueno et al., 2010). Spatially, LRI is restricted to a developmental window, namely, a distinct region within the primary root with the highest probability for LRI events. This developmental window was defined as a root growth period during which pericycle cells in the young differentiation zone maintain a state permissive for LR founder cell specification. Therefore, the term comprises specific time, space, and cell state (Dubrovsky et al., 2006).
Plant hormones are important endogenous regulatory molecules that control plant growth and development and mediate interactions with the environment to modulate developmental responses. Auxins and cytokinins are two classes of plant hormones that are key in the regulation of lateral root organogenesis. Auxin promotes the earliest events related to lateral root organogenesis, including priming (De Smet et al., 2007), founder cell specification (Dubrovsky et al., 2008), and initiation (Benková et al., 2003; Dubrovsky et al., 2008), as well as the later phases of primordium formation and emergence (Benková et al., 2003; Swarup et al., 2008). Critical for the auxin activity is the dynamic regulation of its distribution mediated through the polar auxin transport machinery (Vanneste and Friml, 2009) and the downstream signaling pathway (Paciorek and Friml, 2006; Quint and Gray, 2006; Calderon-Villalobos et al., 2010). Interference with either of them (polar auxin transport or signaling) leads to severe defects in LRI and development (Casimiro et al., 2001; Fukaki et al., 2002; Benková et al., 2003; Dharmasiri et al., 2005; Okushima et al., 2005; Swarup et al., 2008; De Smet et al., 2010).
By contrast, cytokinin constrains both LRI and development (Li et al., 2006; Laplaze et al., 2007). Plants overproducing cytokinin exhibit reduced lateral root formation, whereas transgenic plants overexpressing the CYTOKININ OXIDASE/DEHYDROGENASE (CKX) gene coding for the enzyme that degrades cytokinin by oxidation have an increased number of lateral roots (Werner et al., 2003; Laplaze et al., 2007). Accordingly, perturbations in either perception or signal transduction of cytokinins affect lateral root organogenesis (Mason et al., 2005; Riefler et al., 2006; To et al., 2007).
The antagonistic roles of auxin and cytokinin in the regulation of lateral root organogenesis imply that their activities must be mutually tightly controlled. In view of the inhibitory effects of cytokinin on LRI, spatiotemporal regulation and restriction of its activity might be crucial for the recurrent LRI. Here, we show that the early phases of lateral root organogenesis, including priming and initiation, take place in the root zone with elevated levels of biologically active cytokinins, but with repressed cytokinin responses. Accordingly, enhanced cytokinin activity prior to the morphologically visible initiation events, in the zone encompassing the basal meristem activity, most efficiently inhibited LRI; by contrast, increased levels of cytokinins in the mature differentiated parts of the roots interfered with the development, but not the initiation, of lateral roots. Enhanced cytokinin responses detected in the pericycle cells neighboring the LRP and the analysis of mutants defective in cytokinin biosynthesis or response indicate that this cytokinin activity might be important to prevent LRI in close proximity to existing LRP. Furthermore, our results demonstrate that primordia in the early phases of development are more sensitive to perturbations in the cytokinin pathway than those in later phases. We hypothesize that the stage-dependent effect of cytokinin on primordium development is determined by robustness and stability of the auxin gradient that might be enhanced in developmentally more mature primordia.
RESULTS
LRI Occurs in the Root Zone with High Active Cytokinin Levels
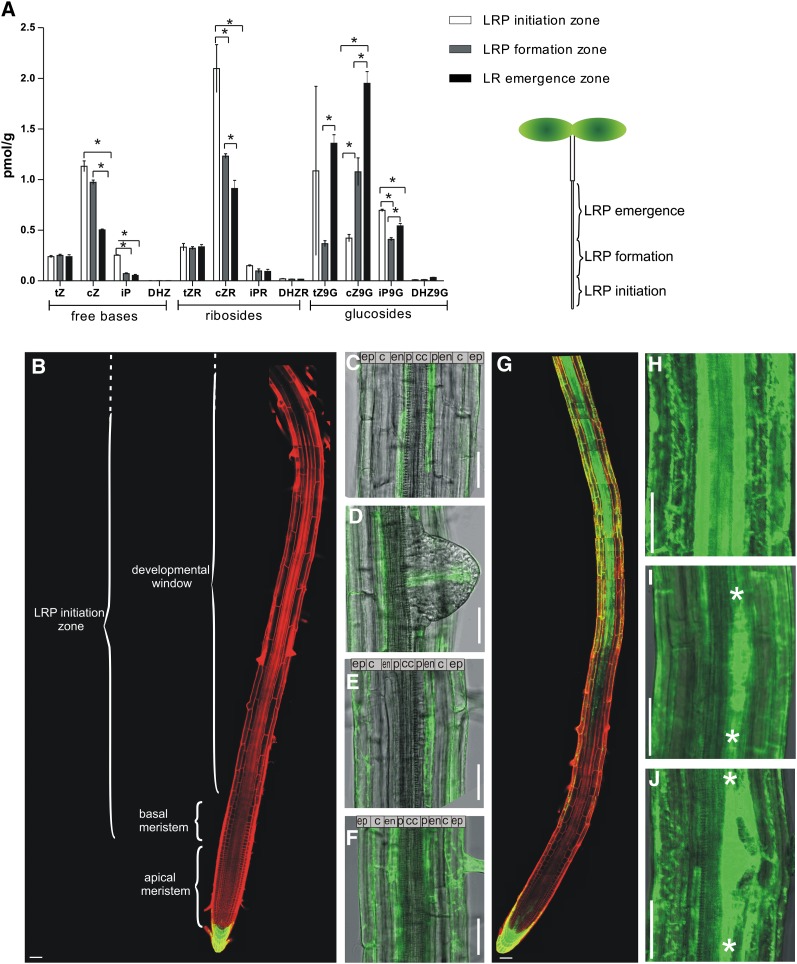
Particular phases of lateral root development take place in distinct zones of the primary root. Priming, an accumulation of auxin in protoxylem cells, and LRI are spatially restricted to the zone near the root meristem, including the basal meristem and developmental window (Dubrovsky et al., 2001, 2011; De Smet et al., 2007), whereas primordium organogenesis and emergence occur in the mature differentiated zones (Swarup et al., 2008). Cytokinin acts as an endogenous inhibitor of lateral root organogenesis and counteracts the stimulatory effect of auxin (Li et al., 2006; Laplaze et al., 2007). However, thus far, the mechanisms behind this crosstalk are unclear. As a first step, we investigated whether cytokinin levels vary along the primary root axis and correlate with lateral root organogenesis. Cytokinin levels were examined in three zones defined by the developmental phases of lateral root organogenesis: the zones of LRP initiation, formation, and LRP emergence through the cortex (Figure 1A). The LRP initiation zone was recognized as the root area where early phases of initiation take place, including priming in the basal meristem, founder cell establishment, and the first stage of LRP development in the developmental window. The LRP formation zone was defined as the zone between the first-stage LRP and the first emerged lateral roots. The root zone from the first emerged lateral roots to the junction between root and hypocotyl was recognized as the LR emergence zone (for details, see Methods). Quantification of cytokinin metabolites in the three different root segments revealed that levels of the cytokinin bases isopentenyladenine and cis-zeatin were slightly higher in the root segments corresponding to the LRP initiation zone than those in the more proximal parts (Figure 1A), whereas trans-zeatin was distributed evenly through the whole root. Cytokinin ribosides are transport forms of cytokinin, which might be activated and therefore represent sources of rapidly available active cytokinins (Kudo et al., 2010). The trans-zeatin riboside, cis-zeatin riboside, isopentenyl adenosine, and dihydrozeatin riboside were distributed similarly to the corresponding free cytokinin bases (Figure 1A). On the contrary, cytokinin-inactive derivatives, such as trans-zeatin-9-glucoside and cis-zeatin-9-glucoside, are conjugated forms that cannot be reactivated into biologically active cytokinins. A high proportion of these biologically inactive derivatives accumulated in the older root segments, corresponding to the LRP emergence zone (Figure 1A).
Figure 1.
Distribution of Cytokinin and Cytokinin Response in Roots.
(A) Quantification of cytokinin levels in the zones of LRP initiation zone, formation, and emergence. Error bars denote se (*P < 0.05; Student’s t test; n = 3).
(B) to (F) TCS:GFP expression in 8-d-old untreated roots detected in the root cap, but not in the root meristem, basal meristem, and developmental window (B); in the individual xylem pole pericycle cells between developing young primordia (C); in the provasculature tissue in emerging LRPs (D); in the endodermal cells adjacent to early-stage LRPs (E); and continuously in the endodermis of the emerging primordium zone (F).
(G) to (J) TCS:GFP expression in 8-d-old roots treated for 16 h with 10 μM benzyl adenine induced in the provasculature of the root, but not basal, meristem (G) and in all tissues of the differentiation zone ([G] and [H]), including primordia at all developmental stages ([I] and [J]).
Plasma membranes were visualized by propidium iodide staining. c, cortex; cc, central cylinder; cZ, cis-zeatin; cZ9G, cis-zeatin-9-glucoside; cZR, cis-zeatin riboside; DHZ, dihydrozeatin; DHZ9G, dihydrozeatin-9-glucoside; DHZR, dihydrozeatin riboside; en, endodermis; ep, epidermis; iP, isopentenyladenine; iP9G; isopentenyladenine-9-glucoside; iPR, isopentenyl adenosine; p, pericycle; tZ, trans-zeatin; tZ9G, trans-zeatin-9-glucoside; tZR, trans-zeatin riboside. White asterisks indicate borders of LRPs. Bars = 50 μm.
Altogether, the analysis of the cytokinin levels in roots suggests that spatiotemporally regulated metabolic processes, such as degradation or inactivation by conjugation, contribute to a differential distribution of cytokinin derivatives along the primary root. With respect to lateral root organogenesis, the early phases, including initiation and primordium formation, take place in the root zone where biologically active or activatable cytokinin derivatives accumulate, but the later developmental phases, such as emergence, occur in the zone with accumulation of biologically inactive cytokinin conjugates.
Cytokinin Responses Are Minimal in the Basal Root Meristem
The main limitation of analytical approaches, such as liquid chromatography–mass spectrometry-based cytokinin quantifications, is the lack of tissue and cellular resolution. Therefore, reporters sensitive to defined hormones, such as synthetic auxin response element DR5 and domain II (DIIVENUS) for auxin (Ulmasov et al., 1997; Vernoux et al., 2011) or synthetic EIN3-responsive promoter (EBS) to ethylene (Stepanova et al., 2007), have helped elucidate the cellular action of hormones. To monitor the cytokinin responses in cells and tissues involved in lateral root organogenesis (i.e., the basal meristem, the developmental window zone, the pericycle cells, and the individual stages of LRP and cells adjacent to LRPs), we used the cytokinin-sensitive two-component output sensor (TCS) reporter, which harbors concatemerized B-type Arabidopsis response regulator binding motifs and a minimal 35S promoter. Only cytokinins activate TCS-mediated transcription, whereas other plant hormones, such as auxin, abscisic acid, and gibberellic acid, do not (Müller and Sheen, 2008). Cytokinin-dependent expression of a TCS:LUCIFERASE (LUC) reporter was shown to depend on well-established cytokinin signaling components (Müller and Sheen, 2008).
Detailed examination revealed that the expression of the TCS:GREEN FLUORESCENT PROTEIN (GFP) reporter was not or hardly detectable in the basal meristem zone where priming events take place and throughout the root zone corresponding to the developmental window (Figure 1B). In the pericycle, TCS:GFP was expressed in individual xylem pole pericycle cells between the LRP in the same cell file or across existing LRP (Figures 1C, 2A, and 2D; see Supplemental Figures 1A and 1B online). In the proximal root zone, where LRPs are more developed, the TCS signal in the pericycle disappeared (Figure 1F; see Supplemental Figure 1A online). The TCS:GFP expression occurred in the endodermal cells adjacent to the early-stage LRP (Figure 1E) and persisted in the endodermis of the root zone with emerging primordia and mature lateral roots, but no GFP signal was detected in the pericycle (Figure 1F; see Supplemental Figure 1A online). In the LRP body, the cytokinin reporter was expressed the earliest at stage VI, where it was confined to the provascular cells (Figure 1D).
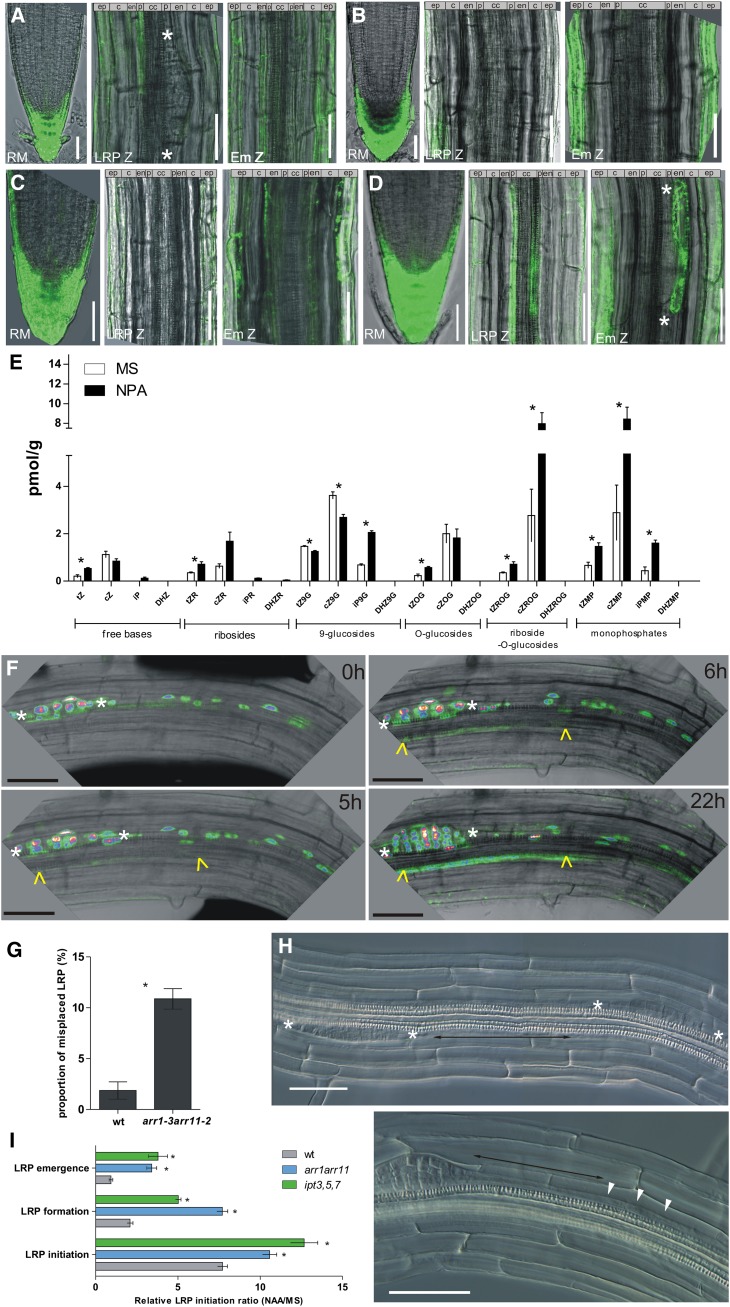
Figure 2.
Cytokinin Response in the Pericycle Cells.
(A) and (B) TCS:GFP signal in pericycle cells of 7-d-old roots without treatment (A) and on media supplemented with 10 μM NPA (B).
(C) and (D) TCS:GFP expression in pericycle cells of 3-d-old roots without LRPs (C) and 4-d-old roots with one or two initiated LRPs. Weak TCS signal present in xylem pole pericycle cells and in the endodermis adjacent to LRP (D).
(E) Cytokinin measurements in roots, excluding the root meristem of seedlings grown on control medium (white) and in the presence of 10 μM NPA (black). Error bars denote se (*P < 0.05; Student’s t test; n = 3).
(F) Real-time monitoring of TCS:GFP x DR5:3XVENUS-N7 reporter in pericycle cells after LRP initiation. White asterisks and yellow arrowheads indicate LRPs at the early initiation stage in pericycle cells visualized by nuclear-localized DR5:3XVENUS-N7 and the TCS:GFP signal, respectively.
(G) and (H) Defective LRI positioning in 7-d-old seedlings of the arr1 arr11 mutant. Proportion of misplaced per total number of LRPs (wild type, n = 218; arr1-3 arr11-2, n = 267) from 18 roots each. Error bars denote se (*P < 0.05; Student’s t test) (G). One pericycle cell distance between two LRPs (H).
(I) Auxin-induced LRP initiation in arr1 arr11 and ipt3 ipt5 ipt7 mutants. In the mutant background, 25 h of treatment with 1 μM 1-naphthaleneacetic acid (NAA) induces LRP initiation along the whole root, whereas in control roots, induction is restricted mainly to the LRP initiation zone. Error bars denote se (*P < 0.05; Mann-Whitney test; n = 15 roots).
White asterisks indicate borders of LRPs. cZ, cis,zeatin; cZ9G; cis-zeatin-9-glucoside; cZMP, cis-zeatin riboside monophosphate; cZOG, cis-zeatin-O-glucoside; cZR, cis-zeatin riboside; cZROG, cis-zeatin riboside-O-glucoside; DHZ, dihydrozeatin; DHZ9G, dihydrozeatin- 9-glucoside; DHZMP, dihydrozeatin riboside monophosphate; DHZOG, dihydrozeatin-O-glucoside; DHZR, dihydrozeatin riboside; DHRZOG, dihydrozeatin riboside-O-glucoside; DHZR, dihydrozeatin riboside; Em Z, zone of LRP emergence; iP, isopentenyladenine; iP9G; isopentenyladenine-9-glucoside; iPMP, isopentenyl adenosine monophosphate; iPR, isopentenyl adenosine; LRP Z, zone of LRP initiation; RM, root meristem; tZ, trans-zeatin; tZ9G, trans-zeatin-9-glucoside; tZMP, trans-zeatin riboside monophosphate; tZOG, trans-zeatin-O-glucoside; tZROG, trans-zeatin riboside-O-glucoside; tZR, trans-zeatin riboside; wt, the wild type. Bars = 50 μm.
Expression of the TCS reporter was induced by exogenous cytokinin treatment in all cell layers of the mature differentiated root, including LRP at all developmental stages (Figures 1G to 1H), but not or to a very limited extent in the zone corresponding to the basal meristem and the early differentiation zone. These results suggest that the reduced expression of the TCS reporter in the basal meristem and developmental window might result from a local repression of cytokinin signaling that cannot be overcome easily by enhanced cytokinin stimulus (Figures 1G to 1J).
Thus, the TCS:GFP reporter expression revealed that cytokinin signaling was strongly repressed in cells and tissues in which priming and early initiation phases of the lateral root organogenesis took place. By contrast, the cytokinin response was enhanced in the pericycle xylem pole cells between two existing LRP where organogenesis typically was repressed.
Cytokinin Responses Are Enhanced in the Pericycle Cells Near Young LRP
In pericycle cells, a local increase in auxin response had been shown to correlate with founder cell specification (Dubrovsky et al., 2008). In contrast with the findings on auxin, the cytokinin reporter exhibited an enhanced response in the xylem pole pericycle cells between existing LRP (Figure 1C; see Supplemental Figure 1 online), where LRI was compromised (Dubrovsky et al., 2001; Lucas et al., 2008b). The question arises whether enhanced cytokinin signaling might be part of a mechanism preventing new initiation events near the existing primordia. To this end, we compared the expression of the TCS reporter in pericycle cells of 3-d-old roots without detectable LRI, of 4-d-old roots with one, or occasionally, two primordia initiated per root, and of 7-d-old roots with three to four primordia per centimeter root length. In 3-d-old seedlings, no TCS expression was found in the pericycle cells (cf. Figures 2A and 2C); in 4-d-old roots, TCS:GFP was detected in a few pericycle cells in the proximity of the initiated primordia (cf. Figures 2A and 2D); and in 7-d-old roots, the increased TCS:GFP expression correlated with the lateral root formation (Figure 2A; see Supplemental Figure 1 online). To examine the dynamics of the cytokinin responses following LRP initiation, we analyzed the TCS:GFP expression in real time. In parallel, the auxin signaling profile with DR5:3XVENUS-N7 was monitored, the expression of which had been identified as the earliest sign of LRP initiation (Dubrovsky et al., 2008). Within 5 h after initiation, as defined by the expression of DR5:3XVENUS-N7 in nuclei of a few short initials, the TCS-mediated expression of GFP could be detected in pericycle cells neighboring the LRP. This signal gradually increased with time (Figure 2F).
To test whether enhanced cytokinin activity in the pericycle cells was related to primordium formation or to exclude that the enhanced cytokinin response was an inherent characteristic of the pericycle, we examined the TCS:GFP expression in roots lacking LRP due to inhibited auxin transport (Casimiro et al., 2001). The TCS:GFP signal in the pericycle of roots grown for 7 d on 5 μM naphthylphthalamic acid (NPA) was dramatically reduced (cf. Figures 2A and 2B). These results suggested that the enhanced cytokinin response in pericycle cells might be linked to the LRP formation process itself and that inhibition of LRP formation interfered with cytokinin responses in the pericycle. In this respect, the TCS:GFP expression strongly differed from the xylem pole pericycle marker J0121, the expression of which in pericycle cells was not affected by NPA treatment (Casimiro et al., 2001) (see Supplemental Figure 2 online). Additionally, cytokinin measurements revealed an increase in cytokinin metabolites, such as the cytokinin bases trans-zeatin and isopentenyladenine, trans-zeatin riboside-O-glucoside, cis-zeatin riboside-O-glucoside, trans-zeatin riboside monophosphate, cis-zeatin riboside monophosphate, and isopentenyl adenosine monophosphate, in roots of seedlings germinated on NPA. Thus, the reduced TCS:GFP expression might not be the consequence of reduced cytokinin levels in NPA-treated roots (Figure 2E).
In summary, our results suggest that an enhanced cytokinin response in the pericycle is strongly linked to LRP formation and that chemical perturbations of LRP initiation dramatically reduce the TCS-mediated expression in the pericycle, which contrasts with the stable expression of pericycle cell identity markers.
Spacing of LRP Initiation Is Defective in Roots with Compromised Cytokinin Response
In Arabidopsis, LRPs initiate in an acropetal manner with the youngest primordia distally positioned from the older LRPs. This architectural pattern of the root system results from strictly spatiotemporally regulated LRP initiation occurring in defined parts of the root behind the root meristem, designated the developmental window (Dubrovsky et al., 2001, 2011). In this zone, LRPs initiate with the highest probability when compared with the proximal part of the root. Accordingly, pericycle cells in the developmental window are more sensitive to exogenous auxin, which promotes LRP initiation, than are pericycle cells of the older, more proximal part of the root (Dubrovsky et al., 2011). Thus, we hypothesize that enhanced cytokinin activity in the pericycle cells between LRPs might be part of the mechanism restricting new initiation events in the proximity of existing LRPs. Therefore, we examined how genetic perturbations in cytokinin levels and signaling affect the positioning of primordia and the sensitivity of pericycle cells from the developmental window to LRP initiation-triggering signals, such as auxin. The mutant ipt3 ipt5 ipt7 with loss-of-function of the ISOPENTENYL TRANSFERASE (IPT) genes that encode the rate-limiting enzyme in cytokinin biosynthesis exhibits reduced cytokinin levels in the roots (Miyawaki et al., 2006). Detailed analysis of the ipt3 ipt5 ipt7 mutant revealed that LRP initiation was not significantly changed (6.54 ± 0.67 LRP/cm versus 6.03 ± 0.13 LRP/cm in the control). However, when mutant roots were exposed to exogenous auxin, a massive LRP initiation was triggered along the whole root, in contrast with wild-type roots, where the highest LRI occurred in the zone corresponding to the developmental window (Figure 2I). Next, we analyzed mutants lacking the functional B-type Arabidopsis response regulator factors that mediate the signal transfer downstream of the cytokinin receptors (To and Kieber, 2008). Both ARR1 and ARR11 were expressed in roots and pericycle cells, as confirmed by quantitative RT-PCR analysis (see Supplemental Figure 3A online; for details, see Methods). To test the impact of the lack of ARR1 and ARR11 functions on the cytokinin response, we transiently expressed the TCS:LUC reporter in protoplasts isolated from mesophyll cells of the single arr1 and arr11 and double arr1 arr11 mutant plants. The TCS:LUC induction decreased significantly in the mutants when compared with controls, indicating that the cytokinin response depended on the ARR1 and ARR11 activity (see Supplemental Figure 3B online).
Detailed phenotype analyses revealed that the arr1 arr11 double mutant was defective in LRP spacing. In 18 examined arr1 arr11 roots, encompassing 267 LRP in total (8.55 ± 0.24 LPR/cm and 2.42 ± 0.2 lateral roots/cm), 31 examples of LRP initiations were found in very close proximity (two LRP one pericycle cell apart), a situation hardly occurring in control seedlings in which four misplaced LRP were detected among 218 LRPs (6.44 ± 0.31 LRP/cm and 1.74 ± 0.05 lateral roots/cm, n = 18 roots) (Figures 2G and 2H; see Supplemental Figure 3D online). Furthermore, similarly to ipt3 ipt5 ipt7, the LRP initiation in arr1 arr11 mutants was significantly increased along the longitudinal root axis in response to auxin treatment and less restricted to the developmental window zone compared with the wild type (Figure 2I). The enhanced sensitivity of pericycle cells in the proximal part of the arr1 arr11 roots to auxin was discernible when roots lacking LRP as a consequence of NPA-inhibited auxin transport were treated with auxin. Whereas the auxin-induced LRP initiation clustered to the zone behind the root tip in the wild-type roots, the corresponding zone in the arr1 arr11 roots was enlarged (see Supplemental Figure 3C online).
Analyses of mutants affected in cytokinin metabolism and signaling indicate that enhanced cytokinin activity in pericycle cells might be important to prevent LRI in close proximity to existing LRP. Decreased cytokinin levels and responsiveness in pericycle cells increased the frequency of defects in LRP spacing and significantly enhanced the sensitivity to LRI-triggering signals, such as auxin, of pericyclecells in the proximal root zone.
Cytokinin and Auxin Responses Are Spatially Complementary
Our results show that the TCS-monitored cytokinin signaling, similarly to the DR5-monitored auxin signaling, was tightly linked to LRP organogenesis. However, a comparison of the cytokinin and auxin responses revealed complementary rather than overlapping patterns. Whereas a strong auxin response was detected in the founder cell identity-acquiring pericycle cells and in stage I LRP (Figure 2F; see Supplemental Figure 4A online) (Dubrovsky et al., 2008), the TCS reporter was expressed in pericycle cells neighboring existing LRP (Figure 2F; see Supplemental Figures 1 and 4A online). Similarly, at later developmental stages, DR5 expression maxima were detected at the LRP tips, and the cytokinin reporter expression was restricted to the provasculature (see Supplemental Figure 4B online). Detailed examination of the TCS reporter in the root tip indicated that the cytokinin response was enhanced in columella cells (see Supplemental Figure 4C online), but, in contrast with DR5 that is strongly expressed in the central columella cells, the TCS:GFP signal was the strongest in the outer columella cells surrounding the DR5 domain (see Supplemental Figure 4C online). As shown previously, inhibition of the auxin transport by NPA enlarged the DR5 expression domain in the columella (Sabatini et al., 1999). In the NPA-treated roots, the cytokinin response domain shifted accordingly toward the cell layers of the outer lateral root cap, thus again surrounding the extended domain of auxin response maxima in the central columella (see Supplemental Figure 4C online). Mutual regulation of the expression of auxin and cytokinin reporters by both auxin and cytokinin was further tested with a transient expression assay in Arabidopsis protoplasts. As expected, expression of the cytokinin reporter TCS:LUC was upregulated by cytokinin and not affected by auxin (see Supplemental Figure 4D online) (Müller and Sheen, 2008). However, the cytokinin stimulatory effect on TCS was significantly compromised by the simultaneous application of auxin. Vice versa, auxin, but not cytokinin, stimulated the expression of the DR5:LUC auxin reporter and simultaneous treatment with cytokinin reduced the auxin induction (see Supplemental Figure 4D online). These data suggest that auxin and cytokinin tend to occupy complementary domains and to modulate their activities mutually.
Cytokinin Accumulation in the Priming Zone Represses LRI
Similarly to inhibition of LRI and development by exogenous cytokinin (Li et al., 2006; Laplaze et al., 2007), endogenous increase of cytokinin levels by ectopic expression of the cytokinin biosynthesis gene IPT in differentiated xylem pole pericycle cells reduces the LRP density by ∼50% (Laplaze et al., 2007). Recent studies have demonstrated that distal root zones, covering the basal meristem and the developmental window, are the sites at which the earliest decisions take place on priming and LRI. Therefore, to test whether an increase in cytokinin levels in these zones might affect LRI, we expressed IPT in a tissue-specific manner with the GAL4>>UAS-based two-component activator-reporter system (Laplaze et al., 2005). Activator lines were selected based on their GFP reporter expression pattern and their insensitivity to exogenous cytokinin to avoid a feedback response due to IPT expression and cytokinin accumulation. An overview of the lines and their expression pattern is summarized in Table 1. Detailed root phenotypes, including root growth, LRI, and development were analyzed in the F1 generation and, as control, activator lines were crossed with the wild type Columbia-0.
Table 1. Overview of the GAL4-GFP Enhancer Trap Line Expression.
| Organ | J2601 | N9193 | J2092 | N9094 | M0028 | J0121 | J0951 | J2351 | J1701 | J1103 | J0671 | J3611 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Root meristem | |||||||||||||
| Col | N.E. | N.E. | xxx N.E. | xxx | N.E. | N.E. | xxx | xxx | N.E. | N.E. | x | ||
| Pvas | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | xxx | xxx | N.E. | N.E. | N.E. | |
| En | N.E. | xx | N.E. | xxx | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | |
| C | N.E. | xxx | N.E. | xxx | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | |
| Ep | N.E. | xxx | xxx N.E. | xx | N.E. | xx | N.E. | N.E. | N.E. | N.E. | N.E. | ||
| Lrc | xx | N.E. | xx | N.E. | N.E. | N.E. | xx | N.E. | N.E. | xx | N.E. | N.E. | |
| Basal meristem | |||||||||||||
| Pvas | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | xx | x-fl | N.E. | N.E. | N.E. | |
| Per | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | |
| En | N.E. | xx | N.E. | xxx | N.E. | N.E. | N.E. | N.E. | N.E. | x | N.E. | N.E. | |
| C | xx | xxx | N.E. | xxx | x | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | |
| Ep | xx | xxx | xxx N.E. | xxx | N.E. | xx | N.E. | N.E. | N.E. | N.E. | N.E. | ||
| Developmental window | |||||||||||||
| Vasc | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | x | x | N.E. | N.E. | N.E. | |
| Per | N.E. | N.E. | N.E. | N.E. | N.E. | xxx | N.E. | N.E. | N.E. | N.E. | N.E. | N.E. | |
| En | xx | xx | x | xxx | N.E. | N.E. | xxx | N.E. | N.E. | xx | N.E. | N.E. | |
| C | xx | xxx | x | xxx | xxx | N.E. | xxx | N.E. | N.E. | N.E. | N.E. | N.E. | |
| Ep | xx | xxx | xxx N.E. | xxx | N.E. | xx | N.E. | N.E. | N.E. | N.E. | N.E. | ||
| Mature root | |||||||||||||
| Vasc | N.E. | N.E. | N.E | N.E | N.E. | N.E. | N.E. | xxx | N.E. | N.E. | N.E. | N.E. | |
| Per | xxx | N.E. | N.E | N.E | N.E. | xxx | xx | N.E. | N.E. | N.E. | xx | N.E. | |
| En | N.E. | xx | xx | xxx | N.E. | N.E. | xx | N.E. | N.E. | xx | xx | Xxx | |
| C | xxx | xxx | xx | xxx | xxx | N.E | xx | N.E. | N.E. | N.E. | xxx | Xx | |
| Ep | xxx | xxx | xxx N.E. | xxx | N.E | xx | N.E. | xx | N.E. | x | N.E. | ||
| LRP | |||||||||||||
| I | N.E. | II on | Em | I on | Up to III | Up to III | IV on | II on | I on | N.E. | N.E. | ||
C, cortex; Col, columella; En, endodermis; Em, emergence; Ep, epidermis; Lrc, lateral root cap; Per, pericycle, Pvas, provasculature; Vasc, vasculature; fl, floem; N.E., not expressed; x, expressed (number of crosses reflects expression intensity).
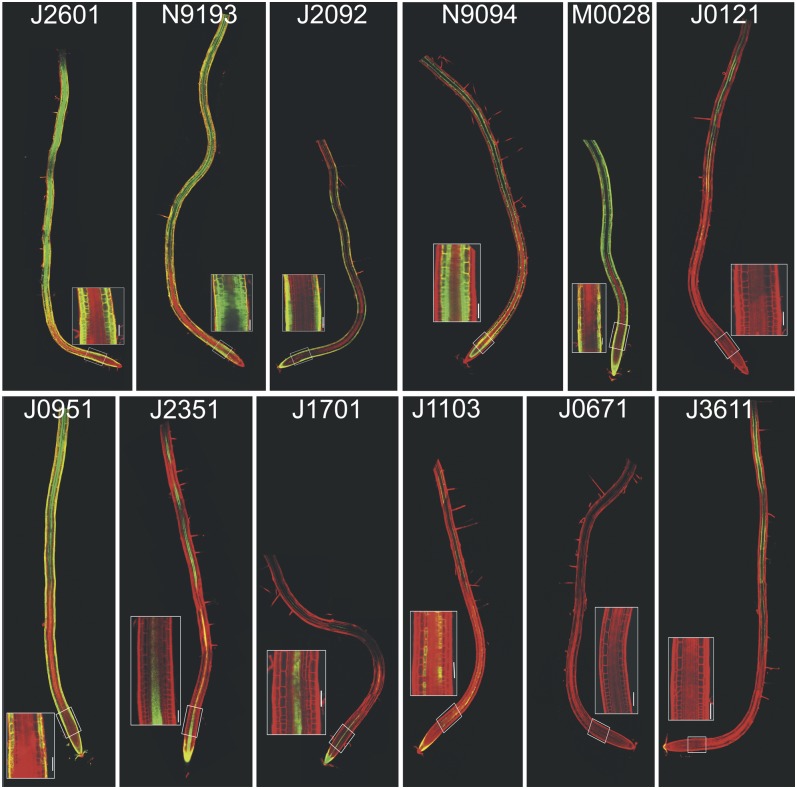
All the lines with IPT expression in the root differentiation and early elongation zones, including epidermis/cortex (J2601>>IPT, N9193>>IPT, J2092>>IPT, M0028>>IPT, and J0951>>IPT), cortex/endodermis (N9094>>IPT), provasculature (J1701>>IPT), or pericycle (J0121>>IPT) exhibited inhibited primary root growth (Figures 3 and 4A, Table 1). By contrast, increased IPT expression in the mature differentiated root zones (J0671>>IPT and J3611>>IPT) did not interfere with root elongation (Figures 3 and 4A, Table 1).
Figure 3.
Expression Pattern of GAL4-GFP Enhancer Trap Lines.
Activator lines exhibit expression in the root differentiation and early elongation zones, including epidermis/cortex (J2601, N9193, J2092,, M0028, and J0951), cortex/endodermis (N9094), provasculature (J1701), or pericycle (J0121), while in lines J0671 and J3611, expression occurs in the mature differentiated root zones. Insets display enlarged basal meristems. Bars = 50 μm.
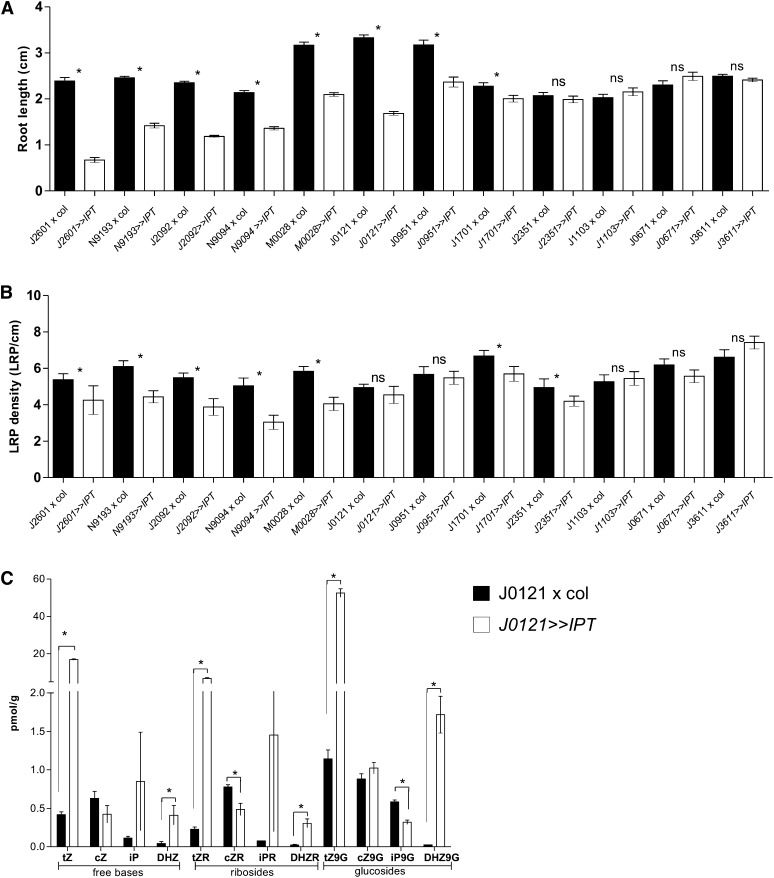
Figure 4.
Spatiotemporal Effect of IPT Expression on Root Growth and LRI.
(A) Inhibited root growth in lines with IPT expressed in the root differentiation and elongation zones (J2601>>IPT, N9193>>IPT, J2092>>IPT, N9094>>IPT, M0028>>IPT, J0121>>IPT, J0951>>IPT, and J1701>>IPT). Error bars denote se (*P < 0.05; Mann-Whitney test, n = 10 roots).
(B) LRP density in lines with IPT expression in the basal meristem. The lines J2601>>IPT, N9193>>IPT, J2092>>IPT, N9094>>IPT, M0028>>IPT, J1701>>IPT, and J2351>>IPT exhibit reduced LRP density. Error bars denote se (*P < 0.05; Mann-Whitney test; n = 10 roots). ns, not significant.
(C) Cytokinin levels in control J0121>>Col and J0121>>IPT roots. Error bars denote se (*P < 0.05; Student’s t test; n = 3).
cZ, cis-zeatin; cZ9G, cis-zeatin-9-glucoside; cZR, cis-zeatin riboside; DHZ, dihydrozeatin; DHZ9G, dihydrozeatin-9-glucoside; DHZR, dihydrozeatin riboside; iP, isopentenyladenine; iP9G, isopentenyladenine-9-glucoside; iPR, isopentenyl adenosine; tZ, trans-zeatin; tZ9G, trans-zeatin-9-glucoside; tZR, trans-zeatin riboside.
Next, we examined the effect of tissue-specific activation of IPT expression on LRI. In all the cases in which IPT was expressed in the distal root zones of the basal meristem encompassing the epidermis/cortex cells (J2601>>IPT, N9193>>IPT, J2092>>IPT, and M0028>>IPT), cortex/endodermis (N9094>>IPT and N9193>>IPT), or provasculature (J2351>>IPT and J1701>>IPT), LRI was inhibited significantly (Figures 3 and 4B, Table 1; see Supplemental Figure 5 online). By contrast, IPT expression in the mature differentiated root zone (J0671>>IPT and J3611>>IPT) did not affect LRI (Figures 3 and 4B, Table 1; see Supplemental Figure 5 online). Surprisingly, the line driving IPT expression directly in the differentiated xylem pole pericycle cells (J0121>>IPT) exhibited a reduced total number of LRP per root (see Supplemental Figure 5 online) but did not exhibit dramatically altered LRI density (LRP per cm of root length; Figure 4B), despite an almost 40-fold increase of free trans-zeatin and a large increase of other cytokinin metabolites, as detected in J0121>>IPT roots (Figure 4C). Detailed analysis of the J0121 expression pattern revealed that under our conditions, the earliest expression could be detected in pericycle cells that had already left the basal meristem (Figure 3). Thus, condition-dependent variability of expression patterns might be the reason for the less efficient inhibition of LRI compared with previous reports (Laplaze et al., 2007).
Notably, in the lines in which the inhibition of LRI was the strongest (N9094>>IPT, N9193>>IPT, J2351>>IPT, and J2092>>IPT) (Figure 4B), the IPT gene was not necessarily expressed at the early stages of LRP, suggesting that cytokinin might act prior to the asymmetric anticlinal division events to inhibit LRP formation. Accordingly, line J1103, exhibiting IPT activation in primordia from stage I on, was not affected in LRI (Figure 4B, Table 1; see Supplemental Figure 5 online). Similarly, lines with IPT expression in the mature part of the root (J0671>>IPT and J3611>>IPT), in which mostly postinitiation phases of lateral root organogenesis take place, did not show any initiation defects (Figures 3 and 4B).
In summary, cytokinin inhibited LRI most efficiently before the morphologically visible initiation events, in the zone encompassing the basal meristem, but did not interfere with LRI when increased in the mature part of t he root or at early stages of LRP. Notably, in no line did ectopic IPT expression fully inhibit LRI, suggesting that mechanisms regulating LRI might be quite robust and markedly resist the cytokinin inhibitory stimulus.
Cytokinin Affects LRP Development in a Stage-Specific Manner
Next, we analyzed the impact of the tissue-specific IPT expression on LRP development. When IPT was expressed prior to LRI (N9193>>IPT and N9094) and/or at the very early stages of LRP development (J2092>>IPT, J2601>>IPT,, J0951>>IPT, M0028>>IPT,, J0121>>IPT, and J1103>>IPT) (see Supplemental Figure 6 online), the inhibition was dramatic, whereas the developmental defects were less strong (J1701>>IPT and J2351>>IPT) when IPT was expressed at later developmental stages (from stage V on). In the endodermis and cortex of mature roots (J0671>>IPT and J3611>>IPT), the activated IPT expression affected primordium development only moderately and delayed LRP outgrowth (see Supplemental Figure 6 online). Accordingly, an overall decrease in cytokinin levels in the ipt3 ipt5 ipt7 loss-of-function mutants enhanced LRP development and increased the proportion of emerged LRP (see Supplemental Figure 3E online).
To follow and evaluate the short-term cytokinin effects on primordium development, we established time-lapse experiments. Primordia at defined developmental stages were exposed to 2 μM cytokinin and followed for 12 h. Typically, under control conditions, 90% of the LRP progressed from stage I/II (Malamy and Benfey, 1997) to stage III/IV (n = 20; Table 2). After cytokinin treatment, 90% of the LRP were arrested (n = 20). Although in 20% of the LRP one to two divisions could still be detected, the primordia did not progress into the next developmental stage. Similarly, when applied to LRP at stage III/IV, cytokinin dramatically inhibited their development, whereas, upon mock treatment, LRP from stage III/IV progressed to developmental phase VII/emergence (n = 14) and 70% of the cytokinin-treated primordia were fully arrested in their development (n = 20; Table 2). However, the behavior differed when LRP at stage V/VI were exposed to cytokinin: Under control conditions, such primordia rapidly developed and emerged through the cortex (n = 12; Table 2), but application of cytokinin did not cause an arrest and, consequently, all primordia continued to develop (n = 10; Table 2). Altogether, these data show that increased cytokinin levels prior to initiation and during early developmental phases have the most pronounced impact on the progress of LRP organogenesis, in agreement with previously published data (Laplaze et al., 2007).
Table 2. Real-Time Analysis of LRP Development without MS or after Cytokinin (Benzyl Adenine) Application.
|
MS |
Cytokinin (2 µM Benzyl Adenine) |
||
|---|---|---|---|
| Progress of LRP Development After Application | No. of Events | Progress of LRP Development after Application | No. of Events |
| From I to I | 1 | From I to I | 12 |
| From I to II | 3 | From II to II | 7 |
| From I to III | 5 | From II to III | 1 |
| From I to IV | 1 | From I to IV | 0 |
| From I to V | 1 | From I to V | 0 |
| From II to II | 1 | From II to II | 0 |
| From II to III | 1 | From II to III | 0 |
| From II to IV | 5 | From II to IV | 0 |
| From II to V | 1 | From II to V | 0 |
| From II to Em | 1 | From II to Em | 0 |
| Fully arrested/total | 2/20 | Fully arrested/total | 19/20 |
| From III to III | 1 | From III to III | 7 |
| From III to V | 1 | From III to IV | 2 |
| From III to Em | 2 | From IV to IV | 7 |
| From IV to IV | 1 | From IV to V | 1 |
| From IV to VII | 3 | From IV to E | 3 |
| From IV to Em | 6 | From IV to Em | 0 |
| Fully arrested/total | 2/14 | Fully arrested/total | 14/20 |
| From V to VI | 1 | From V to VII | 1 |
| From V to VII | 0 | From V to Em | 7 |
| From V to Em | 5 | From VI to VII | 1 |
| From VI to Em | 2 | From VI to Em | 1 |
| Fully arrested/total | 0/8 | Fully arrested/total | 0/10 |
Em, emergence.
A graded distribution of auxin with a maximum at the primordium tips is crucial for proper LRP organogenesis, and, during primordium formation, the auxin maximum is gradually established as the result of a coordinated modulation of the auxin flux toward the tips of newly formed primordia (Benková et al., 2003; Kleine-Vehn et al., 2009). Therefore, we tested whether cytokinin affected LRP development by influencing the auxin response. In vivo time-lapse monitoring of the DR5:RFP (for red fluorescent protein) auxin reporter during LRP organogenesis revealed that cytokinin rapidly reduced the auxin response and prevented auxin accumulation at the LRP tips when applied on the primordia at the early developmental phases, whereas older LRP with established DR5 maxima at the tips were more resistant to cytokinin treatment (Figures 5A versus 5B). These results indicate that the cytokinin might have a stronger impact on the LRP in the early phases without established auxin maxima than on the developmentally more progressed LRP with an established and stabilized auxin gradient.
Figure 5.
Cytokinin Effects on LRP Development.
Real-time monitoring of LRP development on control medium (Mock) and in the presence of 0.1 μM benzyl adenine, when cytokinin was applied at stage I (A) or stage IV (B). DR5:RFP reporter was used to visualize auxin maxima and developmental stages of LRPs. Bars = 50 μm.
DISCUSSION
Auxin and Cytokinin Activities during Lateral Root Organogenesis
The activity of the auxin signaling pathway and the auxin distribution along the root determines stem cell niche and root meristem activity (Sabatini et al., 1999; Blilou et al., 2005), and recurrent local accumulation of auxin in protoxylem cells in the basal meristem has been linked to priming (De Smet et al., 2007). Auxin plays a role in founder cell specification, LRP initiation (Dubrovsky et al., 2008), primordium patterning and development (Benková et al., 2003), and noninvasive emergence of primordia through surrounding tissues (Swarup et al., 2008). In addition, auxin minima have been linked with the developmental window (Dubrovsky et al., 2011). In general, auxin and its proper distribution have been correlated with organ initiation and proper organ patterning.
Cytokinin has a function in the regulation of root meristem maintenance (Dello Ioio et al., 2007, 2008) and lateral root organogenesis (Li et al., 2006; Laplaze et al., 2007). Interestingly, monitoring of tissues and cells exhibiting enhanced TCS reporter expression has revealed that cytokinin and auxin response maxima scarcely overlap within the root. For example, whereas DR5 is detected in pericycle founder cells (Dubrovsky et al., 2008), the TCS reporter is expressed in pericycle cells between two, or across, existing LRP. Cytokinin response maxima in the pericycle are strongly attenuated in young roots or roots without primordia because of inhibited auxin transport, linking cytokinin activity in the pericycle with LRP organogenesis. Perturbations in cytokinin signaling in arr1 arr11 loss-of-function mutants increased the frequency of abnormally positioned LRP, hinting at a role for cytokinin in preventing LRI near existing LRP. Accordingly, defects in LRP positioning occur in lines overexpressing CKX (Werner et al., 2003). Reduction in cytokinin levels and responsiveness in pericycle cells significantly enhanced the sensitivity to LRI-triggering signals, such as auxin, of pericycle cells in the proximal root zone when compared with controls. At this point, we can only speculate about the mechanism that locally stimulates cytokinin responses in LRP in the vicinity of pericycle cells. Possibly, cytokinin production itself in the primordia might be triggered and diffused laterally afterward. Alternatively, an originally noncytokinin signal could be released from primordia, stimulating cytokinin production and activity in pericycle cells in the vicinity of LRPs. Complex expression patterns of a whole set of genes involved in cytokinin metabolism, such as IPT (Miyawaki et al., 2004), the cytokinin-activating gene LONELY GUY (Kuroha et al., 2009), or CKX (Werner et al., 2003), imply that levels of active cytokinin might be under rapid tissue-specific metabolic regulation.
Besides in the pericycle, in other root tissues, auxin and cytokinin response maxima are also rather complementary. In the primordia, DR5 is typically expressed at the primordium tips from the early stages on (Benková et al., 2003), whereas the cytokinin response is strongly suppressed during the early stages of development and is detectable only in provascular cells of emerging LRP. Similarly, in the primary root tip, DR5 expression exhibits response maxima in the central columella cells, whereas TCS occupies the expression domain in the outer columella cells (Müller and Sheen, 2008). Enhancement of the auxin maxima by chemical treatment shifts the cytokinin response zone toward the outer columella cells, suggesting that both pathways might mutually delimit their activities.
Role of Cytokinin in the Spatiotemporal Control of LRI
The acropetal architecture of the root system in Arabidopsis results from initiation events limited to a restricted zone behind the root tip in the simultaneously growing primary root. As a consequence, new initiation events occur distally from the previous ones toward the root apex. Although the mechanisms ensuring such an initiation pattern are still not understood, it might be anticipated that to restrict initiation to a certain root zone, pericycle cells are regulated spatiotemporally at a stage competent for cell divisions and/or recurrent LRI-inducing signals. Both mechanisms might indeed contribute to early initiation events. Previous research on the characterization of xylem pole pericycle cells (Beeckman et al., 2001) and on the identification of the priming zone (De Smet et al., 2007; De Rybel et al., 2010) and developmental window (Dubrovsky et al., 2006) suggest that during primary root growth, pericycle cells in a certain root zone are maintained in an LRI-competent stage. In addition, recurrent enhancement of auxin signals contributes to the initiation of the lateral root developmental program (De Smet et al., 2007; Moreno-Risueno et al., 2010).
In view of its inhibitory effect on LRI, the overall cytokinin activity might be expected to be attenuated in the zone where the earliest phases of LRI take place. Indeed, as demonstrated by the TCS reporter analysis, the cytokinin response is minimal in the priming and developmental window zones. Interestingly, analysis of cytokinin derivatives has not revealed any dramatic decrease in cytokinin levels in the parts encompassing the priming zone and the developmental window. Although detection of such a tissue-specific reduction in cytokinin levels might be under the resolution limit of the analytical approach used, the observation that exogenous cytokinin could not enhance the TCS reporter expression suggests that an important part of the regulation mechanism strongly suppresses the cytokinin signaling pathway.
LRI is inhibited severely by an increase in cytokinin activity in the zone encompassing the basal meristem and the developmental window, but elevated cytokinin levels in more distant root zones are insufficient, although they still interfere with lateral root organogenesis. These data suggest that cytokinin might act prior to any visible signs of LRP initiation to counteract positive LRI signals. Moreover, cytokinin inhibition of LRI was in most of the cases accompanied by root growth reduction, indicating tight interconnection between these two developmental processes. Interestingly, despite the strong IPT overexpression, none of the ectopic increases in endogenous cytokinin levels in defined root zones fully prevented LRI, implying that LRI-regulating mechanisms markedly resist the inhibitory effects of cytokinin.
Role of Cytokinin in the Regulation of Early LRP Development
Overall, the cytokinin response, as monitored with the TCS reporter, was strongly repressed during the early phases of lateral root organogenesis and detected in the primordium provasculature only later in older primordia. Increased cytokinin responses during the early phases of lateral root organogenesis, including priming, either by endogenous expression of IPT or exogenous treatment, revealed that primordia are more sensitive to increased cytokinin levels in the early phases of development than in the later phases. Similar developmental phase-dependent cytokinin sensitivity has been reported previously, demonstrating that endogenous cytokinin levels fluctuate during the early stages of primary root development and that a developmental window (between 2 and 3 d after germination) specifies the increased sensitivity of the root toward endogenous cytokinin (Kuderová et al., 2008).
During primordium formation, an auxin maximum is gradually established at the primordium tips, as a result of the coordinated repolarization of the auxin flux toward the tips of newly formed primordia (Benková et al., 2003; Kleine-Vehn et al., 2009). The auxin gradient formation is critical for the proper development of the primordia, and genetic or chemical interference with their auxin distribution results in defective primordium organogenesis (Benková et al., 2003). Spatiotemporal manipulation of cytokinin levels at defined stages of primordium development revealed that enhancement of the cytokinin activity during the early phases strongly interferes with the auxin gradient formation, whereas at the later stages, cytokinin only mildly affects the established auxin gradients. Thus, stage-dependent effects of cytokinin on primordium development might rely on the robustness and stability of the possibly higher auxin gradient in more developed primordia.
METHODS
Plant Lines and Growth Conditions
The transgenic Arabidopsis thaliana lines were DR5:GFP, DR5:3XVENUS-N7 (Heisler et al., 2005), and DR5:RFP (Marin et al., 2010). The GAL4-GFP enhancer trap lines J2601, N9193, J2092, N9094, M0028,, J0951, J1701, J0671,, J1103, J2351, ,and J3611 and the cytokinin signaling mutant arr1-3 arr11-2 (N6980) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/). The mutant line ipt3 ipt5 ipt7 had been described previously (Miyawaki et al., 2006) as well as the ProUAS:IPT, J0121, TCS:GFP, and TCS:LUC lines (Laplaze et al., 2007; Müller and Sheen, 2008). The GAL4 enhancer trap lines were crossed with ProUAS:IPT and the wild type Columbia-0. The F1 generation was analyzed. Seeds were sterilized with chloral gas, plated in Petri dishes on 0.8% agar 0.5× Murashige and Skoog (MS) medium containing 1% Suc, stored for 2 d at 4°C, and grown vertically at 18°C under a long-day photoperiod. Seedlings were harvested and processed 3, 4, or 7 d after germination.
Phenotypic Analysis, Microscopy, and Statistics
For the confocal laser scanning microscopy, a TCS SP2 AOBS microscope (Leica) was used. Seedlings were stained for 5 min in 0.01 mg mL−1 propidium iodide. Images were processed in Adobe Photoshop. Plant material was cleared as described (Malamy and Benfey, 1997). Root lengths were measured on scanned slides with the ImageJ software (NIH). LRPs were counted with a differential interference contrast microscope (BX51; Olympus). At least 15 seedlings were analyzed, and experiments were repeated twice independently. LRP density was calculated as the number of LRP per total root length. The Excel statistical package (Microsoft) and GraphPad Prism software were used for data analysis, and Mann-Whitney nonparametric and Student’s t tests were applied.
For real-time analysis of LRP development, 6-d-old seedlings were placed in chambered cover glasses (Nunc Lab-Tek) and covered with 0.2-mm-thin square blocks of solid MS (mock) media with or without cytokinin. LRPs were scanned at 3- or 5-min time intervals for 8 to 12 h by a confocal microscope (FV10 ASW; Olympus). The frequency of the positioning-defective LRPs was calculated as the percentage of primordia initiated at distances shorter than those of the control situation from the total initiation events per root.
For the phenotypic analysis after auxin treatment, 7-d-old seedlings grown on MS medium or 10 μM NPA were treated with liquid MS medium with or without 1 µm NAA for 25 h. Seedlings were cleared and the lateral root number was scored in the zones of LRP initiation, formation, and LRP emergence (see below), and the ratio was calculated between the LRP induced by auxin and during incubation in MS medium.
Cytokinin Measurements
Approximately 150 mg of plant tissue was ground in liquid nitrogen and 1 pmol of deuterium-labeled cytokinin internal standards was added to each sample prior to the analysis (Olchemin). After overnight extraction, the samples were purified with a cation exchanger (SCX cartridge; Agilent) and by immunoaffinity extraction with a broad-spectrum monoclonal anticytokinin antibody (Novák et al., 2003). The eluates from the immunoaffinity columns were evaporated to dryness and redissolved in 20 μL of the initial mobile phase. Cytokinins were separated by ultraperformance liquid chromatography (Acquity UPLC; Waters) on a C18 reversed-phase chromatographic column (BEH 18; 1.7 µm; 2.1 × 50 mm; Waters). The mobile phase consisted of 15 mM ammonium formate, pH 4 (A) and methanol (B). For the cytokinin separation, a gradient elution was used (0 min, 10% B; 0 to 8 min, 50% B; flow rate 0.25 mL min−1) with a column temperature of 40°C. The column eluate was introduced into a triple-stage quadrupole mass spectrometer (Xevo TQ MS; Waters), quantified by multiple reaction monitoring of [M+H]+ and the appropriate product ion by Masslynx software (Waters) with a standard isotope dilution method (Novák et al., 2003). The optimal dwell time, cone voltage, collision energy, and exact transition for each cytokinin had been optimized previously (Novák et al., 2008).
For cytokinin measurements in the zones defined by the developmental phases of lateral root organogenesis, namely LRP initiation, formation, and LRP emergence through the cortex, the borders between the three zones were recognized under a confocal microscope for five seedlings 7 d after germination. The zone lengths were measured and an average value was used. The zone of the LRI corresponded to ∼6 mm from the root tip, the zone between 6 and 12 mm from the root tip to the LRP formation zone and above 12 mm from the root tip to the lateral root emergence zone.
Fluorescence-Activated Cell Sorting of Plant Protoplasts
Cells were sorted as described (Birnbaum et al., 2005). Approximately 5000 seeds (per replicate) of each GFP line used in the experiments were sterilized with bleach and plated on MS media. To allow rapid harvesting, seeds were arranged in rows on square plates at a density of ∼500 seeds per row on top of nylon mesh (Nitex 03 100/47; Sefar). Five-day-old roots were cut off ∼1 cm from their tip. Dissected roots were placed in protoplasting solution B (50 mL of solution A [see below], 750 mg cellulase, and 50 mg pectolyase) inside 70-μm cell strainers placed in small Petri dishes and incubated for 1 h at room temperature with agitation. Protoplasted cells were collected from Petri dishes and concentrated by spinning down (at ∼120 relative centrifugal force). The supernatant was aspirated and the cell pellet was resuspended in 1.5 mL of solution A (2 mL of 1 M KCl 1 M, 400 μL of 1 M MgCl2, 400 μL of 1 M CaCl, 0.2 g BSA, 78 mg MES, and 21.86 g mannitol; final volume, 200 mL, pH 5.5, with 1 M Tris).
GFP-expressing cells were isolated on a fluorescence-activated Epics Altra cell sorter (Beckman Coulter) fitted with a 100-μm nozzle at a rate of 2000 to 5000 events per second and a fluid pressure of 30 p.s.i. Protoplasts from Columbia wild-type plants that did not express GFP were used as a negative control for establishing sorting criteria based on the following cell properties: live protoplast clusters with intact membranes were selected based on a high forward to side scatter ratio and GFP-positive cells by their emission intensity in the green channel (∼530 nm) above the negative controls. Cells were sorted directly into the RLT lysis buffer (Qiagen), mixed, and immediately frozen at −80°C for later RNA extraction. An autofluorescence filter was established by eliminating cells that fluoresced at an equal intensity in the green and orange (∼575 nm) channels.
RNA Extraction of Sorted Cells and Quantitative RT-PCR
RNA was extracted with the RNeasy Micro kit (Qiagen) from sorted cells. A DNase treatment with the RNase-free DNase Set (Qiagen) was performed for 15 min at 25°C. Poly(dT) cDNA was prepared from 1 μg total RNA with the iScript cDNA synthesis kit (Bio-Rad) and quantified with a LightCycler 480 (Roche) and SYBR Green I Master (Roche) according to the manufacturer’s instructions. PCR was performed in 384-well optical reaction plates heated for 10 min to 95°C to activate the hot-start Taq DNA polymerase, followed by 40 cycles of denaturation for 60 s at 95°C and annealing/extension for 60 s at 58°C. Targets were quantified with specific primer pairs designed with the Beacon Designer 4.0 (Premier Biosoft International). Expression levels were normalized to UBIQUITIN10 (UBQ10) expression levels. All qRT-PCR experiments were done at least in triplicate. The following primers were used: 5′-CACACTCCACTTGGTCTTGCGT-3′ and 5′-TGGTCTTTCCGGTGAGAGTCTTCA-3′ for UBQ10; 5′-TTGAAGAAACCGCGTGTCGTCT-3′ and 5′-CCTTCTCAACGCCGAGCTGATTAA-3′ for ARR1; and 5′-TAATGATGTCGGTGGACGGCGAAA-3′ and 5′-AAGCTCCGTGTTGCACTCCCTTCA-3′ for ARR11.
Transient Expression in Protoplasts
The DR5rev promoter was amplified from the original plasmid (Friml et al., 2003) using the primers DR5_FOR (5′-ATCCCCCGACGGTATCGCAGCCCAGG-3′) and DR5_REV (5′-TGTAATTGTAATTGTAAATAGTAATTG-3′). The amplified fragment was extracted and purified from gel and cloned in the pENTR5′-TOPO vector using the TA Cloning kit (Invitrogen) according to manufacturer’s instructions. The DR5:LUC construct was created by combining pEN-L1-L-L2 and an entry clone holding the DR5 sequence combined by a MultiSite Gateway LR reaction with pm42GW7 as destination vector. TCS:LUC was created by PCR amplification of the TCS, minimal 35S promoter, and TMVΩ sequence fragment from TCS:GFP (Müller and Sheen, 2008), using the primers 5′-GTGCCAAGCTTTGCTAGCAAAATC-3′ and 5′-TGTAGGCCTGTAATTGTAATTGTAAATAG-3′. The resulting fragment was digested with StuI and HindIII. A second PCR amplification including the LUC-coding sequence and nopaline synthase 3′-untranslated region sequence of GCC:LUC:NOS was performed using the primers 5′-ACAGGCCTACATGGAAGACGCCAAAAACATAAAG-3′ and 5′-TACGAATTCTCATGTTTGACAGCTTATCATC-3′. The resulting fragment was digested with StuI and EcoRI, and both digested PCR fragments were then ligated into the HindIII-EcoRI–digested pUC18 to result in TCS:LUC.
Mesophyll protoplasts were isolated from rosette leaves of 4-week-old Arabidopsis plants grown on soil under controlled environmental conditions in continuous light at 22°C (arr1 and arr11 versus wild-type control analysis) or in a 16-h/8-h light/dark cycle (hormone treatments for TCS and DR5 reporter genes). Protoplasts were isolated as described (Wu et al., 2009) with modifications (Wehner et al., 2011): The surface of Arabidopsis leaves was cut with a razor blade and placed in a Petri dish with 5 mL enzyme solution (1% cellulase Onozuka R10 [Yakult], 0.3% macerozyme Onozuka R10 [Yakult], 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, and 20 mM MES, pH 5.7). After a 4-h digestion, the released protoplasts were filtered into a 50-mL tube and collected by centrifugation. The supernatants were discarded and the protoplasts were resuspended in 5 mL W5 (154 mM NaCl, 125 mM CaCl2, and 5 mM KCl). The protoplasts were counted with a hemocytometer under a light microscope and kept on ice for at least 30 min. For polyethylene glycol transfection, protoplasts were adjusted to a final concentration of 3 × 105 mL with MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7) (Yoo et al., 2007). Protoplasts (3 × 104 cells) were cotransfected with 20 µg of a reporter plasmid containing the LUC reporter gene and 2 µg of a normalization construct expressing Renilla LUC (rLUC). Transfections were done by adding an equal volume of a freshly prepared solution of 40% (w/v) polyethylene glycol 4000 (Fluka) with 0.2 M mannitol and 0.1 M CaCl2. After 10 min of incubation, 0.44 mL W5 solution was added and mixed by pipetting. Transfected protoplasts were collected by centrifugation and gently resuspended in 50 μL W5 solution (for hormone treatments, 0.5 µM benzyl adenine and/or 1 µM NAA was added to the buffer), then transferred to 12-well plates coated with 1% BSA and incubated during 16 h in darkness. Protoplasts were collected and LUC was measured with the dual luciferase reporter assay system (Promega). Luciferase cell culture lysis reagent (Promega) was added to the protoplasts and mixed by pipetting. After 5 min of incubation on ice, 100 μL lysate was used to measure the LUC activity according to the manufacturer’s protocol. The mean value (±sd) was calculated from two measurements, and each experiment was repeated at least three times. To normalize the values, rLUC was used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT3G63110.1 (IPT3), AT5G19040.1 (IPT5), AT3G23630.1 (IPT7), AT3G16857.1 (ARR1), AT1G67710.1 (ARR11), and EF090415 (GCC1:LUC:NOS).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Spatial Distribution of Cytokinin Response along the Root.
Supplemental Figure 2. Xylem Pole Pericycle Cell Identity in Control and NPA-Treated Roots.
Supplemental Figure 3. LRP Initiation Spacing Defective in Roots with Compromised Cytokinin Responses.
Supplemental Figure 4. Cytokinin and Auxin Response Distribution in Roots.
Supplemental Figure 5. Effect of IPT Expression on LRI.
Supplemental Figure 6. Spatiotemporal Effect of IPT Expression on LRP Development.
Supplementary Material
Acknowledgments
We thank Jen Sheen, Dolf Weijers, Tatsuo Kakimoto, Stephen Depuydt, and Laurent Laplaze for sharing published material, Jiri Friml for discussions, and Martine De Cock and Annick Bleys for help in preparing the manuscript. This work was supported by a Starting Independent Research grant from the European Research Council (ERC-2007-Stg-207362-HCPO) and the project CZ.1.07/2.3.00/20.0043 to the Central European Institute of Technology to E.B. and grants from the Ministry of Education, Youth, and Sports of the Czech Republic (MSM 6198959216) and the Centre of the Region Haná for Biotechnological and Agricultural Research (ED0007/01/01) to P.T.
AUTHOR CONTRIBUTIONS
A.B. and E.B. designed the project. A.B., K.P., P.M., J.D., C.C., B.M., W.G., and P.T. performed the experiments and analyzed data. A.B. and E.B. wrote the article.
Glossary
- LRP
lateral root primordia
- LRI
lateral root initiation
- GFP
green fluorescent protein
- NPA
naphthylphthalamic acid
- MS
Murashige and Skoog
- NAA
to be defined
References
- Beeckman T., Burssens S., Inzé D. (2001). The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52(Spec Issue): 403–411 [DOI] [PubMed] [Google Scholar]
- Benková E., Bielach A. (2010). Lateral root organogenesis - From cell to organ. Curr. Opin. Plant Biol. 13: 677–683 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Jung J.W., Wang J.Y., Lambert G.M., Hirst J.A., Galbraith D.W., Benfey P.N. (2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2: 615–619 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Calderon-Villalobos L.I., Tan X., Zheng N., Estelle M. (2010). Auxin perception—Structural insights. Cold Spring Harb. Perspect. Biol. 2: a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inzé D., Sandberg G., Casero P.J., Bennett M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 2705–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Gambetta G.A., Hernández-Barrera A., Shishkova S., González I. (2006). Lateral root initiation in Arabidopsis: Developmental window, spatial patterning, density and predictability. Ann. Bot. (Lond.) 97: 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky J.G., Napsucialy-Mendivil S., Duclercq J., Cheng Y., Shishkova S., Ivanchenko M.G., Friml J., Murphy A.S., Benková E. (2011). Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 191: 970–983 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Rost T.L., Colón-Carmona A., Doerner P. (2001). Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214: 30–36 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benková E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuderová A., Urbánková I., Válková M., Malbeck J., Brzobohatý B., Némethová D., Hejátko J. (2008). Effects of conditional IPT-dependent cytokinin overproduction on root architecture of Arabidopsis seedlings. Plant Cell Physiol. 49: 570–582 [DOI] [PubMed] [Google Scholar]
- Kudo T., Kiba T., Sakakibara H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52: 53–60 [DOI] [PubMed] [Google Scholar]
- Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., Fukuda H., Sugimoto K., Sakakibara H. (2009). Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L., Parizot B., Baker A., Ricaud L., Martinière A., Auguy F., Franche C., Nussaume L., Bogusz D., Haseloff J. (2005). GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Laskowski M., Grieneisen V.A., Hofhuis H., Hove C.A., Hogeweg P., Marée A.F., Scheres B. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mo X., Shou H., Wu P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Lucas M., Godin C., Jay-Allemand C., Laplaze L. (2008a). Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J. Exp. Bot. 59: 55–66 [DOI] [PubMed] [Google Scholar]
- Lucas M., Guédon Y., Jay-Allemand C., Godin C., Laplaze L. (2008b). An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE 3: e3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marin E., Jouannet V., Herz A., Lokerse A.S., Weijers D., Vaucheret H., Nussaume L., Crespi M.D., Maizel A. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., M atsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno M.A., Van Norman J.M., Moreno A., Zhang J., Ahnert S.E., Benfey P.N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O., Hauserová E., Amakorová P., Doležal K., Strnad M. (2008). Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 2214–2224 [DOI] [PubMed] [Google Scholar]
- Novák O., Tarkowski P., Tarkowska D., Doležal K., Lenobel R., Strnad M. (2003). Quantitative analysis of cytokinins in plants by liquid chromatography–single-quadrupole mass spectrometry. Anal. Chim. Acta 480: 207–218 [Google Scholar]
- Okushima Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T., Friml J. (2006). Auxin signaling. J. Cell Sci. 119: 1199–1202 [DOI] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. (2009). Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Quint M., Gray W.M. (2006). Auxin signaling. Curr. Opin. Plant Biol. 9: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- To J.P.C., Deruère J., Maxwell B.B., Morris V.F., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. (2007). Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To J.P.C., Kieber J.J. (2008). Cytokinin signaling: Two-components and more. Trends Plant Sci. 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner N., Hartmann L., Ehlert A., Böttner S., Oñate-Sánchez L., Dröge-Laser W. (2011). High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J. 68: 560–569 [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.