This work characterizes factors involved in the predominantly distal location of meiotic crossovers in barley. Recombination initiates first in the distal regions and later in the interstitial regions; manipulating meiotic progression with higher temperatures produced more interstitial crossovers that could improve mapping of agronomical traits and reduce linkage drag.
Abstract
Meiosis involves reciprocal exchange of genetic information between homologous chromosomes to generate new allelic combinations. In cereals, the distribution of genetic crossovers, cytologically visible as chiasmata, is skewed toward the distal regions of the chromosomes. However, many genes are known to lie within interstitial/proximal regions of low recombination, creating a limitation for breeders. We investigated the factors underlying the pattern of chiasma formation in barley (Hordeum vulgare) and show that chiasma distribution reflects polarization in the spatiotemporal initiation of recombination, chromosome pairing, and synapsis. Consequently, meiotic progression in distal chromosomal regions occurs in coordination with the chromatin cycles that are a conserved feature of the meiotic program. Recombination initiation in interstitial and proximal regions occurs later than distal events, is not coordinated with the cycles, and rarely progresses to form chiasmata. Early recombination initiation is spatially associated with early replicating, euchromatic DNA, which is predominately found in distal regions. We demonstrate that a modest temperature shift is sufficient to alter meiotic progression in relation to the chromosome cycles. The polarization of the meiotic processes is reduced and is accompanied by a shift in chiasma distribution with an increase in interstitial and proximal chiasmata, suggesting a potential route to modify recombination in cereals.
INTRODUCTION
Meiosis is a specialized form of cell division required to halve ploidy levels so that chromosome numbers are not doubled following fertilization. It is characterized by an extended prophase I where homologous recombination underpins the formation of genetic crossovers (COs). COs can be visualized cytologically as chiasmata and are essential for correct orientation of the chromosomes on the meiotic spindle and subsequent disjunction at meiosis I (Jones and Franklin, 2006). A consequence of meiotic recombination is the reciprocal exchange of genetic information between the homologous chromosomes distal to the CO site, which results in recombinant chromosomes with different allelic combinations compared with the parental chromosomes. The genetic variation arising from meiotic recombination provides plant breeders with an important resource for the generation of new varieties.
The molecular basis of meiotic recombination has been extensively investigated in budding yeast (Saccharomyces cerevisiae), leading to the current “early decision crossover pathway model” (Bishop and Zickler, 2004). Studies indicate that the process is broadly similar in plants (Hamant et al., 2006; Harrison et al., 2010; Osman et al., 2011). In Arabidopsis thaliana, recombination is initiated by the formation of DNA double-strand breaks (DSBs) catalyzed by the topoisomerase II-like proteins sporulation 11 (SPO11-1) and SPO11-2 (Grelon et al., 2001; Stacey et al., 2006; Hartung et al., 2007). The DSBs are then resected to generate 3′ single-stranded tails on either side of the break site. Based on budding yeast, it is thought that the DNA tails interact with the Arabidopsis RecA homologs radiation sensitive 51 (RAD51) and disrupted meiotic cDNA1 (DMC1) to form nucleoprotein filaments; one of which invades the homologous chromosome to form a stable single-end invasion intermediate. The displaced DNA strand forms a D-loop that extends as strand invasion progresses, allowing the 3′-strand on the other side of the break to anneal. The broken DNA strands are ligated to form a double-Holliday junction (dHj) recombination intermediate. In budding yeast, the resolution of the dHj results in CO formation (Neale and Keeney, 2006). In many species, including plants, only a small proportion (∼5%) of DSBs are repaired as COs (Osman et al., 2011). Based on studies in budding yeast, it is proposed that the majority of non-COs arise through the synthesis-dependent strand annealing pathway (Allers and Lichten, 2001).
Cereals, which include members of the Poaceae family, such as barley (Hordeum vulgare), wheat (Triticum aestivum), rye (Secale cereale), and oat (Avena sativa), are a crucial source of food for mankind. The determination of recombination frequencies between genetic and molecular markers in mapping populations coupled with cytological studies of chiasma distribution along chromosomes indicates that a nonuniform distribution of recombination events is a characteristic feature of these species (Künzel et al., 2000; Sandhu and Gill, 2002). These studies show suppression of recombination in pericentromeric regions with chiasmata/COs predominately occurring in distal regions of the chromosomes. Gene density along the cereal chromosomes is heterogeneous with a majority of genes clustering in relatively gene-rich recombinogenic distal regions; however, studies reveal that a substantial proportion of the gene complement is outside of these regions (Sandhu and Gill, 2002; Mayer et al. 2011). For example, analyses in wheat and barley indicate that >30% of the genes are located in recombinationally cold chromosomal regions (Künzel and Waugh, 2002; Erayman et al., 2004; Mayer et al., 2011). Moreover, the level of recombination within the gene-rich regions can vary severalfold. This gene distribution coupled with the pattern of recombination has several consequences. First, it reduces the genetic variation that is available to breeders. Second, it limits the potential for map-based cloning of genes. In addition, a consequence of this CO distribution may be linkage drag when attempting to introgress new genetic traits.
In this study, we investigated the factors that contribute to the pattern of recombination in barley, a self-compatible, diploid (2n = 14) member of the subgroup Pooideae, which shares close syntenic relationships with wheat, rye, oat, and forage grasses, including Lolium perenne and Festuca pratensis (Moore et al., 1995). An initial survey of chiasma distribution in cv Morex confirmed that chiasma distribution is predominantly confined to the distal regions on all chromosomes. We demonstrate that the initiation and progression of recombination, chromosome axis formation, and synapsis are subject to a marked spatiotemporal asymmetry. It appears that this pattern of events is linked to the relative timing of DNA replication for heterochromatin and euchromatin-rich DNA. When S-phase is perturbed using a modest temperature shift, it results in a disruption of meiotic control leading to a change in the frequency and distribution of chiasmata.
RESULTS
Chiasmata Are Predominantly Distal in Barley
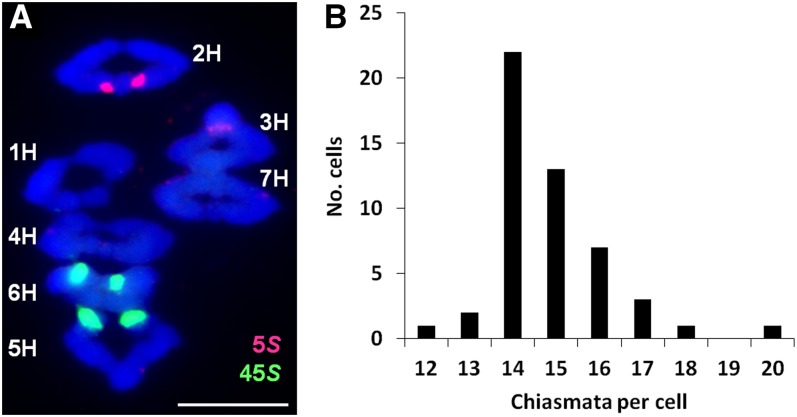
A cytological analysis of chiasma frequency and distribution was first undertaken on the variety ‘Morex.’ Chiasmata, the cytological manifestation of COs, were scored on the basis of chromosome shape (Jones, 1984), and individual chromosomes were identified with the 5S and 45S ribosomal DNA probes using fluorescence in situ hybridization (FISH) (Figure 1A) (Leitch and Heslop-Harrison, 1993; Taketa et al., 1999). The mean chiasma frequency of ‘Morex’ was 14.84 ± 0.19 se per cell (n = 50), and the total number of distal chiasmata (713) was 25-fold greater than the number of interstitial chiasmata (29). Interstitial chiasmata were more frequent in chromosomes 1H (8) and 3H (6) and least in chromosomes 4H (1). Chiasma frequencies varied little between bivalents, ranging from two (chromosome 6H) to 2.26 (chromosome 5H). The majority of bivalents formed rings (338/350) and chromosomes 5H and 6H (nucleolus organizer region [NOR] bearing chromosomes) exhibited the highest number of rods (four and three, respectively) that were lacking chiasmata in the NOR arms. The overall chiasma frequency ranged from 12 to 19 per cell with each chromosome receiving one to three chiasmata (Figure 1B), reflecting that meiotic CO/chiasma formation is a highly controlled process (Jones and Franklin, 2006).
Figure 1.
Chiasma Frequency of Barley ‘Morex.’
(A) Individual bivalents stained with DAPI (blue) can be identified at meiotic metaphase I using the 5S (red) and 45S (green) ribosomal DNA probes with FISH. Chiasma number and position are determined by bivalent shape. Bar = 10 µM.
(B) Histogram of chiasmata per cell. The chiasma frequency has a narrow range overdistributed around the mean (14.84 ± 0.19).
Chromosome Axis Formation and Synapsis Are Initially Polarized to the Distal Regions
During early meiotic prophase I in many species, including barley, the telomeres of the homologous chromosomes attach to the nuclear envelope and cluster together in the so-called bouquet formation (reviewed in Ronceret and Pawlowski, 2010). It is proposed that the telomere clustering may facilitate initial contacts between the homologous chromosomes leading to chromosome pairing and subsequent synapsis. Consistent with this, earlier studies based on the analysis of chromosome spread preparations from meiocytes using electron microscopy indicated that chromosome synapsis is initiated in the distal regions near the telomeres (Albini et al., 1984). The availability of antibodies that recognize components of the chromosome axes and synaptonemal complex (SC) has enabled us to conduct a detailed examination of chromosome axis formation and synapsis in relation to telomere movement (Figure 2; for additional clarity, unmerged images are also presented as Supplemental Figures 2 and 3 online).
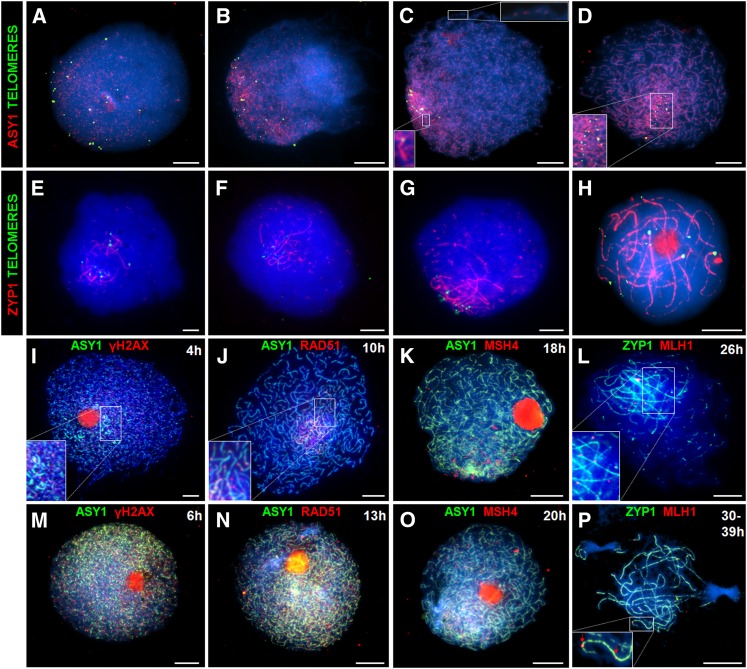
Figure 2.
Immunolocalization of Meiotic Proteins during Prophase I Shows Initial Subtelomeric Loading and Later Distribution throughout the Nucleus.
As a marker for chromosome axis formation, ASY1 foci (red) are initially observed adjacent to the telomeres (green) ([A] and [B]), and as the ASY1 signal progressively linearizes the telomeres cluster into a bouquet ([C] and [D]). Synapsis initiation (marked by SC transverse filament protein ZYP1, red) initially occurs in the vicinity of the telomeres (green) ([E] and [F]) and later in the interstitial regions (G) before complete synapsis (H). Recombination markers γH2AX, RAD51, MSH4, and MLH1 (red) are initially observed in the distal regions ([I] to [L]) and (except for MLH1) later throughout the nucleus ([M] to [P]). Bars = 10 µM.
Arabidopsis asynaptic 1 (ASY1) is an axis-associated protein essential for synapsis and wild-type levels of COs (Caryl et al., 2000; Armstrong et al., 2002; Sanchez Moran et al., 2007). In Arabidopsis and Brassica, ASY1 initially localizes to chromosomes during G2 as numerous foci throughout the nucleus that then associate with the nascent chromosome axes that are elaborated to denote the onset of leptotene. An anti-At-ASY1 antibody (Ab) has previously been used to mark the meiotic chromosome axes in a wide range of plant species, including monocots (Mikhailova et al., 2006; Wang et al., 2009), and as Arabidopsis ASY1 shares 72% amino acid sequence identity with barley ASY1 (see Supplemental Figure 1 online), it was anticipated that the anti-At-ASY1 Ab would be an effective marker for axis formation. Following meiotic S-phase, up to 28 telomeric signals can be detected using FISH with a telomeric repeat sequence probe. Subsequent bouquet formation and homolog pairing then reduce the number of telomeric signals that can be resolved down to 14. In barley, ASY1 becomes detectable as foci in the telomeric hemisphere during early G2 (4 h after S-phase) with an average of 22 telomere foci per cell (n = 10) (Figure 2A; see Supplemental Figures 2A to 2D online). As ASY1 progressively linearizes in the subtelomeric region, bouquet formation nears completion (15.5 telomere foci per cell; n = 10) (Figure 2B; see Supplemental Figures 2E to 2H online). The telomeric bouquet cluster is completed at late G2 (8 h after S-phase) (13.7 foci, n = 10), at which point the subtelomeric ASY1-marked axes are continuous but remain as foci or short stretches in the interstitial regions (Figure 2C; see Supplemental Figures 2I to 2L online). By leptotene (13 h after S-phase), the axes are fully complete and the telomeres remain paired with 13.5 foci cell (n = 10) (Figure 2D; see Supplemental Figures 2M to 2P online).
During zygotene and pachytene, individual pairs of aligned homologous chromosomes are brought together into close apposition mediated by the formation of a tripartite proteinaceous structure, the SC (Zickler and Kleckner, 1999; Page and Hawley, 2004). An Ab raised against the Arabidopsis SC transverse filament protein zipper 1 (ZYP1) allows SC to be monitored in a variety of plant species, including Brassica, wheat, and rye (Higgins et al., 2005; Mikhailova et al., 2006; Osman et al., 2006). Immunolocalization of barley ZYP1 using anti-At-ZYP1b and a telomere FISH-labeled probe enabled investigation of the relationship between telomere clustering and synapsis (Figures 2E to 2H; see Supplemental Figure 3 online). The number of telomeric foci remained constant during zygotene (13.5, n = 10) and pachytene (13.5, n = 10), but by pachytene the paired telomeres no longer clustered (Figure 2H; see Supplemental Figures 3M to 3P online). ZYP1 polymerization was initially observed in the subtelomeric regions from where it extended to form stretches of linear signal. As zygotene progressed, numerous additional synapsis initiation sites appeared in the interstitial regions (Figures 2E and 2G; see Supplemental Figures 3A to 3L online). The mean number of observed synapsis initiation sites was 55 (n = 15), ranging from 76 (early zygotene) to 29 (late zygotene). This gradual reduction in discrete ZYP1 foci probably reflects the fact that as zygotene progresses independent foci form short stretches of SC that will eventually coalesce to form a single continuous signal.
Chronology through Meiosis
The immunolocalization experiments revealed that chromosome axis formation and synapsis were nucleated and developed in the subtelomeric/distal regions of the chromosomes before the interstitial/proximal regions. To better understand this spatial asymmetry in relation to the meiotic program, a time-course experiment was conducted.
Initially, progression through meiosis was monitored using a variation of the method developed for Arabidopsis (Armstrong et al., 2003). 5′-Bromo-2′-deoxyuridine (BrdU) was injected into barley stems before fixing inflorescences at set times and then detecting the appearance of labeled meiotic stages with an anti-BrdU Ab. The first labeled tetrads were observed 43 h after BrdU injection (see Supplemental Figure 4 online). G2 occupied 13 h, followed by an extended prophase I of 27 h. The two meiotic division phases from metaphase I to tetrad were completed in 3 h. We next conducted an immunological time course monitoring ASY1 and ZYP1 polymerization in conjunction with BrdU labeling. ASY1 was first detected as discrete foci 4 h after injection, elongating to form short linear stretches by 6 h (see Supplemental Figure 5 online). At 10 h after BrdU injection, the ASY1 signal was ∼75% continuous. By the onset of leptotene (13 h), ASY1 extended along the entire length of the homologous chromosomes. The linear ASY1 signal remained until diplotene, although detection during zygotene and pachytene was greatly reduced in the synapsed regions. Linear ZYP1 signals were first observed 25 h after BrdU injection, corresponding to the cytologically defined zygotene stage. This persisted through pachytene until the chromosomes desynapsed at diplotene (39 h).
Distribution of Recombination Proteins during Prophase I
We next investigated the progression of meiotic recombination in barley meiocytes (Figure 2; for additional clarity, unmerged images are also presented as Supplemental Figures 6 and 7 online). In budding yeast, it is possible to follow recombination through the direct molecular identification of recombination intermediates (for example, Schwacha and Kleckner, 1995). In multicellular organisms, including plants, this is not yet possible. However, it is possible to monitor recombination progression by immunolocalization of recombination proteins on chromosome spread preparations from meiocytes during prophase I (Osman et al., 2011). In Arabidopsis, recombination is initiated by programmed DSBs followed by phosphorylation of histone H2AX (γH2AX) at the break site (Sanchez Moran et al., 2007). In this study, we used an Ab specifically raised against phosphorylated γH2AX as a marker for DSB formation in chromosome spread preparations. Approximately 200 (n = 25) γH2AX foci were initially detected in the distal chromosomal regions (Figure 2I; see Supplemental Figures 6A to 6D online), coincident with the first appearance of the ASY1 signal (4 h). By late G2 (6 to 8 h after S-phase), the mean number of γH2AX foci had increased to 380 (n = 15), peaking at 465 foci per cell that were distributed throughout the nucleus (Figure 2M; see Supplemental Figures 6E to 6H online). Dual localization of γH2AX with ZYP1 revealed that γH2AX foci reduced in number as synapsis progressed, and by pachytene, there were only few remaining foci (data not shown). Prominent staining of the nucleolus was also apparent at the 4- and 6-h time points. This phenomenon has previously been observed in Arabidopsis meiocytes (Jackson et al., 2006; Vignard et al., 2007). Its basis remains unclear; however, it has been reported that the nucleolus acts as a reservoir sequestering cell cycle proteins, possibly suggesting a similar role for meiotic proteins (Visintin and Amon, 2000). Alternatively, it may be some nonspecific labeling due to the high protein content of the nucleolus.
In Arabidopsis, RAD51 and DMC1 are essential for DSB break repair and CO formation, respectively (Couteau et al., 1999; Li et al., 2004; Kurzbauer et al., 2012; Pradillo et al., 2012). The barley homologs share 92 and 53% amino acid sequence identity with the regions of RAD51 and DMC1 used to make the Arabidopsis Abs, respectively (see Supplemental Figure 1 online). Barley RAD51 and DMC1 foci were first detected on mature stretches of axis (based on the status of ASY1 polymerization) during G2/leptotene corresponding to ∼10 h when ASY1 polymerization was ∼75% continuous throughout the nucleus (Figure 2J; see Supplemental Figures 6I to 6L and 6Q to 6T online). At leptotene (13 h after S-phase), when the ASY1 signal was fully linear, RAD51 and DMC1 foci were also observed in the interstitial regions (Figure 2N; see Supplemental Figures 6M to 6P and 6U to 6X online). This would suggest that the appearance of the foci span a period of around 3 h. The number of RAD51 foci ranged from 175 to 448 per cell (n = 15) and DMC1 foci 115 to 421 per cell (n = 15), reflecting an accumulation of foci as the continuous stretches of chromosome axes extended from the distal to interstitial/proximal regions.
In Arabidopsis, the meiosis-specific MutS homologs, mutS homolog 4(MSH4) and mutS homolog 5 (MSH5), are essential for the majority (∼85%) of COs (Higgins et al., 2004, 2008b). Based on their counterparts in budding yeast and humans, it is thought they function as heterodimer and are required for the formation of dHj recombination intermediates (Snowden et al., 2004). The barley MSH4 and MSH5 homologs share 90 and 82% amino acid sequence identity, respectively, with the regions used to make the Abs (see Supplemental Figure 1 online). Therefore, we used anti-At-MSH4 and anti-At-MSH5 Abs to localize the barley proteins during prophase I. During leptotene (18 h after S-phase), MSH4 foci initially appeared as a cluster associated with the region where the ASY1 signal was most intense, which corresponds to the distal regions of the chromosomes. It then rapidly becomes more widely detectable throughout the nucleus associating with the chromosome axes (20 h) (Figures 2K and 2O; see Supplemental Figures 7A to 7H online). Similar results were obtained for MSH5 (data not shown). During this transition, the number of MSH4 foci per cell ranged from 278 to 411 (n = 15) and similarly for MSH5 from 280 to 428 (n = 15).
The distribution of foci corresponding to the barley MutL homolog MLH1 was then investigated. Based on studies in budding yeast, it is proposed that Mlh1 acts with Mlh3 as a heterodimer to ensure that dHjs are resolved as COs rather than non-COs (Hunter and Borts, 1997; Wang et al., 1999). Immunolocalization studies in a variety of organisms indicate that both MLH1 and MLH3 form foci on pachytene chromosomes that correspond to the sites of COs (Plug et al., 1998; Marcon and Moens, 2003; Jackson et al., 2006; Chelysheva et al., 2010). Dual immunolocalization of MLH1 in conjunction with ZYP1 was conducted on prophase I chromosome spread preparations. MLH1 foci first appeared associated with stretches of SC in the subtelomeric/distal regions from early zygotene, while SC initiation was still occurring in more interstitial regions (Figure 2L; see Supplemental Figures 7I to 7L online). When ZYP1 was fully polymerized at pachytene, 14 to 20 MLH1 foci per cell (n = 30) were observed associated with the SC. The higher number of MLH1 foci than chiasmata observed is consistent with the known underestimation of CO events in barley using chiasma counts given current barley genetic map length estimates (Nilsson et al., 1993; Mayer et al., 2011). In cases where the chromosomes were positioned to the outer region of the spread preparation, it was apparent these foci were located in the distal regions consistent with the chiasma analysis (Figure 2P; see Supplemental Figures 7M to 7P online). MLH3 foci were found to show a similar distribution (data not shown).
Chiasma Localization Is Associated with Early Replicating Euchromatic DNA
The linear differentiation of eukaryotic chromosomes into domains of heterochromatic and euchromatic DNA together with associated histone modifications is well established (Fuchs et al., 2006). Recent studies in budding yeast have revealed that the chromatin mark histone 3 tri-methylated lysine 4 (H3K4me3) is associated with meiotic recombination hot spots (Borde et al., 2009). Histone acetylation is also known to influence meiotic recombination both in budding yeast and Arabidopsis (Perrella et al., 2010). We therefore performed a cytological analysis of barley meiocytes to investigate the chromatin organization in relation to chiasma distribution.
In Arabidopsis, heterochromatin is hypermethylated and associated with silenced regions of the genome and recombination cold spots (Tariq et al., 2003; Zhang et al., 2006). In barley, an anti-5-methylcytosine Ab was used to detect methylated DNA on pachytene chromosome spreads. Anti-5-methylcytosine was abundant throughout the chromosomes except at the centromeres, where it was almost absent (or undetected) (see Supplemental Figure 8A online). Barley Cot-1 DNA was used in conjunction with FISH to estimate the distribution and abundance of repetitive elements (see Supplemental Figure 8B online). Based on the intensity of the FISH signal, the distribution of repetitive DNA appeared greatest in the vicinity of the centromeres but was also present, albeit to a lesser extent, along the chromosome arms. Frequent pairs of bright foci, positioned in register, occurring along the synapsed homologous chromosomes, were also observed. In Arabidopsis, the histone modifications H3K9me2, H3K27me2, and H4K20me1 mark heterochromatin (Naumann et al., 2005). In barley, these marks were generally abundant throughout the chromosomes but had reduced signal intensity in the distal regions. For example, Supplemental Figures 8E to 8L online show the distribution of the H3K9me2 mark.
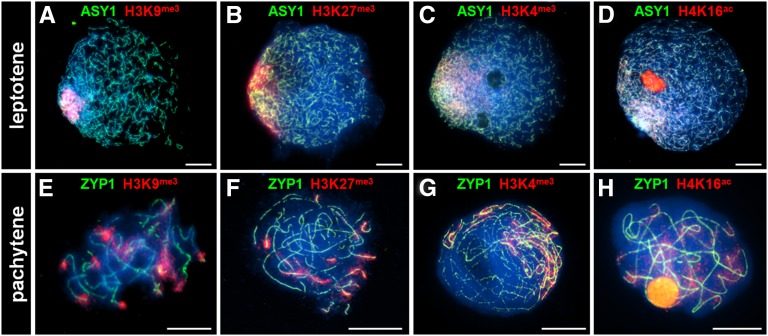
Euchromatin is characterized by gene-rich, actively transcribed regions of the genome. The histone modifications H3K9me3, H3K27me3, H3K4me3, and H4K16ac mark euchromatin in Arabidopsis (Naumann et al., 2005; Zhang et al., 2009). During meiotic prophase I in barley, these marks are extremely abundant in the distal regions and to a lesser extent throughout the rest of the chromosomes (Figures 3A to 3H). Dual pulse labeling of premeiotic S-phase nuclei using BrdU and 5-ethynyl-2′-deoxyuridine (EdU) (see Methods) indicated that whereas the distal euchromatin-rich DNA was replicated first (0 to 4 h), the interstitial regions were replicated later (by 6 h) followed by the proximal/heterochromatin-rich regions (by 13 h) (Figure 4). Thus, it would appear that the spatiotemporal polarization of meiotic progression and eventual chiasma localization reflects the distal distribution of the early-replicating euchromatin.
Figure 3.
Immunolocalization of Euchromatin Histone Marks during Meiotic Prophase I.
Histone markers H3k9me3 ([A] and [E]), H3K27me3 ([B] and [F]), H3K4me3 ([C] and [G]), and H4K16ac ([D] and [H]) (red) are highly abundant in the chromosomal distal regions. Nuclei are stained with DAPI (blue). Bars = 10 µM.
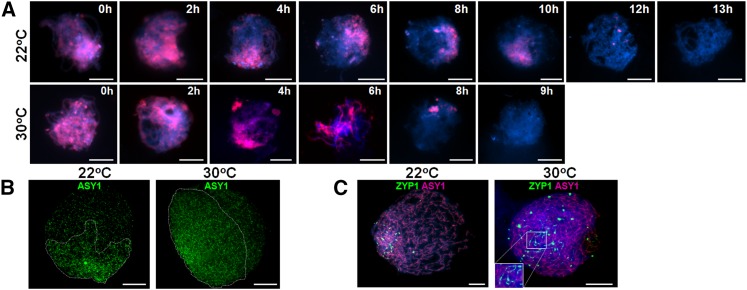
Figure 4.
Distal Chromosomal DNA Is Replicated Prior to the Interstitial/Proximal Regions during Premeiotic S-Phase, Leading to a Meiotic Lag That Is Attenuated at 30°C.
The duration of premeiotic S-phase is determined by applying a pulse of BrdU (data not shown) followed by a second pulse of EdU (red) at various time intervals (A). At 22°C, nuclei no longer incorporate the EdU (red) by 13 h as they have completed S-phase, whereas at 30°C, this occurs by 9 h (A). Axis formation marked by ASY1 (green) loading has a strong bias toward the distal regions at 22°C, whereas at 30°C, this appears more even (B). Chromosome synapsis determined by ZYP1 loading (green) is first observed in the distal regions at 22°C, whereas at 30°C, it is throughout the nucleus (C). Nuclei in (A) and (C) are stained with DAPI. Bars = 10 µM.
Chromosomes Exhibit Distinct Contraction/Expansion Cycles during Prophase I
An appraisal of meiotic and mitotic chromosome behavior across a range of species has revealed that they undergo programmed cycles of chromatin expansion and contraction (Kleckner et al., 2004). Based on this, a mechanical basis for chromosome function model has been developed. This posits that three distinct chronologically conserved chromatin cycles occur during prophase I of meiosis that coordinate four meiotic transitions leading to CO formation via mechanically imposed cyclic stress and relaxation. These events are DSB formation, single end invasion, second-end capture, and dHj resolution, and their occurrence is coincident with a period of chromatin expansion (Kleckner et al., 2004).
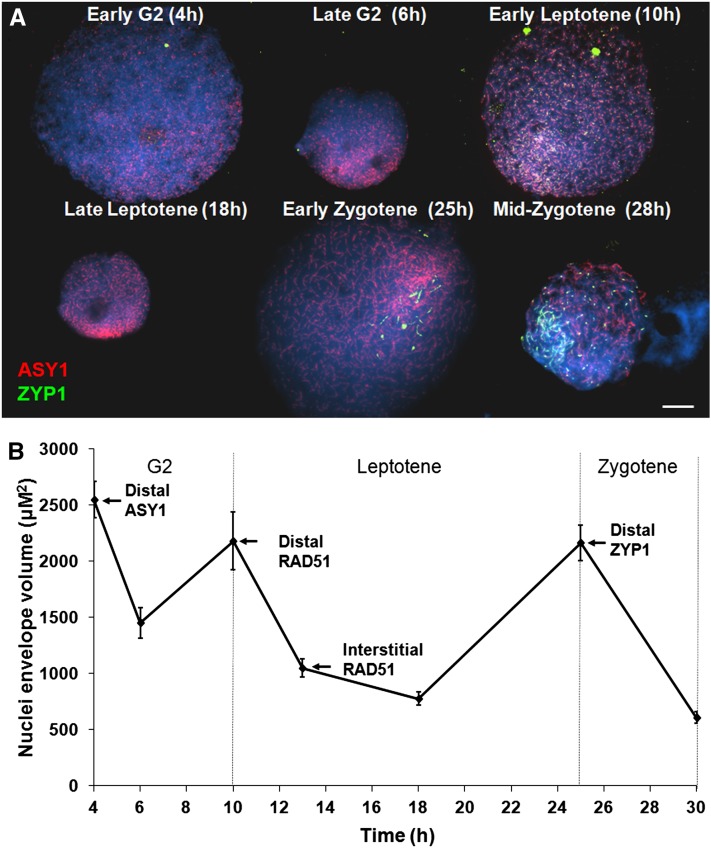
Given the apparent spatiotemporal asymmetry of meiotic pathway progression in barley, we investigated how this might relate to global chromosome dynamics. Meiocyte cell walls were digested so that the nuclei were free to occupy an envelope volume in solution, which is governed by the state of chromatin described by Kleckner et al. (2004). Changes in envelope volume were determined by measuring nuclei sizes at specific meiotic time points defined by the progress of ASY1 and ZYP1 localization. ASY1 loaded as foci in early G2 onto nuclei with the largest envelope volumes (2554 µm2 ± 162 se, n = 25) observed during the course of the analysis (Figures 5A and 5B). As short linear stretches of ASY1 became visible, the mean envelope volume was observed to have significantly decreased to 1454 µm2 ± 135 (n = 23) (P < 0.001, 47 df). Subsequently, an increase in mean nuclear size to 2182 µm2 ± 270 se (n = 13) was observed in nuclei where the linear ASY1 signal had reached 75% completion to mark the end of the first cycle. Thereafter, a second cycle began where the envelope volume underwent another significant reduction to 1027 µm2 ± 76 se (n = 48) (P < 0.001, 57 df) by the time the ASY1 signal was completely linear. However, by zygotene when ZYP1 began to polymerize, the envelope volume was observed to have undergone another significant increase to 2165 µm2 ± 159 se (n = 34) (P < 0.001, 77 df). This was followed by a further contraction such that by mid-zygotene, when ZYP1 polymerization was ∼50% complete, there was a significant reduction in envelope volume to 1025 µm2 ± 107 se (n = 30) (P < 0.001, 66 df). This decrease continued through to pachytene (608 µm2 ± 50 se, n = 33) before a further increase at diplotene (1160 µm2 ± 97 se, n = 20). A summary of these events is presented in Table 1. These data support the existence of chromatin cycles during barley meiotic prophase I similar to that in other species (Kleckner et al., 2004). Moreover, previous studies suggest that the appearance of foci corresponding to meiotic pathway proteins is indicative of the onset of molecular processes (see above; Sanchez Moran et al., 2007; Ferdous et al., 2012). Thus, the timing of appearance of foci corresponding to the meiotic pathway proteins suggests that whereas the meiotic processes, such as strand invasion, in distal regions are coincident with periods of chromatin expansion the corresponding interstitial processes occur during periods of contraction. A similar program of cycles was observed in control experiments using undigested meiocytes (see Supplemental Figure 9 online). However, as the antibodies could not penetrate these cells, it was not possible to localize the meiotic proteins in relation to the cycles.
Figure 5.
Distal Meiotic Events Are in Phase with the Chromatin Cycles of Expansion and Contraction.
During prophase I, nuclei expand at transitional stages and then contract.
(A) Immunolocalization of meiotic proteins ASY1 (red) and ZYP1 (green) in conjunction with a BrdU time course (see Supplemental Figure 5 online) reveals that initial distal meiotic events are synchronized with nuclei expansion periods, whereas the interstitial events occur during periods of nuclei contraction. Bar = 10 µM.
(B) The immunolocalization data are represented, highlighting the difference in loading of meiotic proteins to distal and interstitial chromosomal regions. Data are the mean ± se.
Table 1. A Comparison of Temperature Effects on Meiosis in Barley.
| Phenotype | 22°C | 30°C |
|---|---|---|
| Duration of premeiotic S-phase DNA replication | <13 h | <9 h |
| Duration of meiosis (G2 to tetrad) | 43 h | 43 h |
| Cycles of expansion and contraction | Normal | Normal |
| Axis formation (ASY1 loading) | Polarized | Less polarized |
| Synapsis (ZYP1 loading) | Normal | Loading defect |
| Bivalent type (rings:rods) | 32:1 (n = 50) | 8:1 (n = 128) |
| Ratio of distal:interstitial chiasmata | 25:1 (n = 50) | 11:1 (n = 128) |
| Seed set (mean per stem) | 22.7 (n = 15) | 1.5 (n = 21) |
Adjusting Meiotic Progression Changes the Frequency and Distribution of Chiasmata
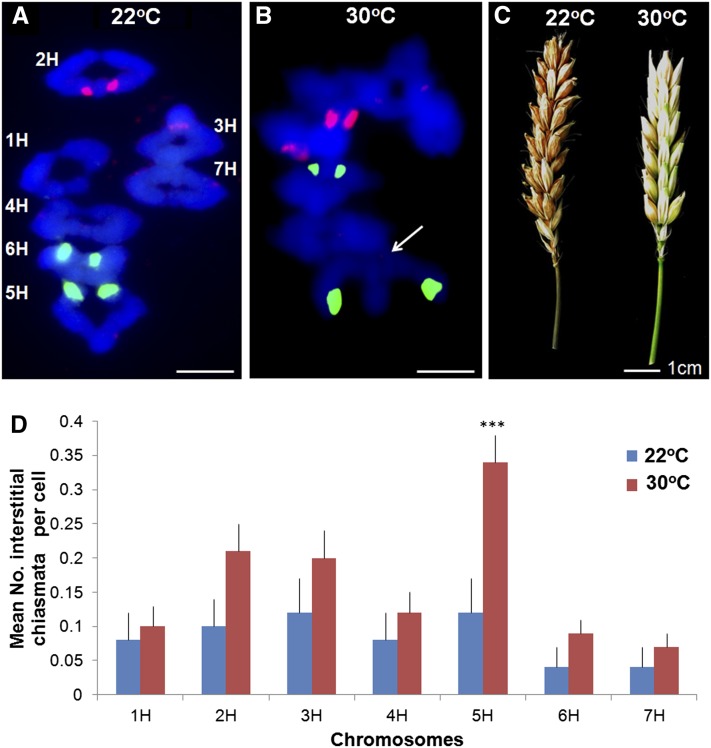
Taken together, our data revealed that the meiotic program during prophase I occurs in the context of programmed global chromosome cycles leading to the formation of COs/chiasmata that are predominantly distal. To explore the factors that contribute to this outcome, we investigated the consequences of perturbing the meiotic program. Previous studies in budding yeast have shown that an increase in temperature affects CO control (Börner et al., 2004). In addition, there are numerous reports of elevated temperature leading to effects on plant meiosis (for example, Jain, 1957; Lin, 1982; Loidl, 1989). Thus, we investigated the effect of increased temperature on barley meiosis. In an initial experiment, exposure of meiocytes to 35°C resulted in meiotic failure with an absence of SC formation and agglomeration of the chromosomes into unusual structures, making it difficult to observe the presence, if any, of chiasmata (see Supplemental Figure 10 online). This finding was consistent with previous studies where extreme temperatures led to a complete disruption in meiosis and subsequent infertility (Loidl, 1989). However, other studies had indicated that smaller elevations in temperature could lead to more subtle effects (Dowrick, 1957). Therefore, we subjected the meiocytes to a more modest increase in temperature to determine if it would be possible to perturb, rather than entirely disrupt, the meiotic program. Analysis of metaphase I chromosome spreads from plants subjected to 30°C during meiosis revealed that chiasma frequency and distribution was distinct from plants grown at 22°C. Overall, we observed a small but significant reduction in mean chiasma frequency from 14.8 ± 0.19 se per cell (n = 50) to 13.5 ± 0.09 se (n = 128) (P < 0.001, 177 df). An interesting feature of these chiasmata was a significant shift in their distribution from distal to interstitial/proximal positions (Figures 6A, 6B, and 6D; see Supplemental Figure 11 online). There was a significant decrease in number of distal chiasmata from 14.3 ± 0.17 se per cell (n = 50) to 12.4 ± 0.12 se (n = 128) (P < 0.001, 177df), coinciding with an increase in number of interstitial chiasmata from 0.58 ± 0.1 se per cell (n = 50) to 1.13 ± 0.09 se (n = 128) (P < 0.001, 177 df). Moreover, it would appear that this effect was not entirely uniform as some chromosomes were more affected than others. In particular, 5H was more likely to gain interstitial chiasmata on the chromosome arm opposite the NOR (P = 0.003, 177 df) but overall had a significant reduction in total number of chiasmata (2.28 to 1.68, P < 0.001, 177 df) (Figure 6D).
Figure 6.
Significantly More Interstitial Chiasmata Are Observed at 30°C.
A typical ‘Morex’ metaphase I spread at 22°C labeled with rDNA FISH probes (5S, red; 45S, green) to identify individual chromosomes and DAPI (blue) (A) (bar = 10 µM). A metaphase I chromosome spread at 30°C (arrow pointing to interstitial chiasma) (B) (bar = 10 µM). Seed set is reduced at 30°C (C) (bar = 1 cm). Interstitial chiasmata are significantly increased when plants are grown at 30°C compared with 22°C (D) (bars = se, ***P < 0.001).
To ascertain the basis for this effect, we investigated the chronology of meiosis at 30°C. Surprisingly, progression through prophase I took 40 h with the two meiotic divisions occupying a further 3 h. Thus, prophase I progression at 30°C appeared to occur at the same rate as that at 22°C. Nevertheless, dual labeling of meiocytes with BrdU and EdU revealed that the length of premeiotic S-phase at the higher temperature was reduced by around ∼4 h from 12 to 13 h to 8 to 9 h (Figure 4A). There was no obvious effect on the distal regions where replication was completed in 0 to 4 h at both temperatures. However, a clear effect was observed in replication of the interstitial and proximal heterochromatic DNA, which was completed by 13 h at 22°C and 9 h at 30°C (Figure 4A). Immunolocalization of histone euchromatin marks H3K9me3, H3K27me3, and H3K4me3 appeared unchanged at 30°C (see Supplemental Figure 12 online). This suggests that heterochromatin may be more accessible for replication factors at higher temperatures, possibly through reduced occupancy of H2A.Z in the nucleosomes (Kumar and Wigge, 2010). We investigated if increased temperature influenced the distribution of the meiotic proteins during prophase I. In a comparison of initial ASY1 loading at G2 in nuclei with the same envelope volumes, at 22°C the majority of ASY1 foci were observed covering 54% ± 2.8 se (n = 15) of the nucleus stained with 4′,6-diamidino-2-phenylindole (DAPI), whereas at 30°C this was significantly increased to 72% ± 3.3 se (n = 15) (P < 0.001, 29 df) (Figure 4B). This suggested that at the higher temperature, meiotic axis formation and recombination protein loading appeared to be more synchronous across the nucleus. Thus, as a result of elevating the temperature to 30°C, interstitial events were more in phase with the distal events relative to the chromatin cycles. Elevated temperature also resulted in altered loading of ZYP1. The asymmetrical loading of ZYP1 observed at 22°C was no longer evident. Instead, numerous foci of variable size were observed throughout the nucleus, a proportion of which were associated with synapsis initiation sites (Figure 4C). Also these events initiated in nuclei with significantly smaller envelope volumes than at 22°C (1157 µm2 versus 2165 µm2; P = 0.005, 41 df). However, after completing synapsis (573 ± 72 µm2 se, n = 15), they exhibited an increase in nuclear envelope volume at diplotene similar to 22°C (1160 µm2 versus 1188 µm2). Unsurprisingly, ‘Morex’ plants subjected to 35°C during meiosis were entirely infertile. However, even the 30°C treatment reduced seed set to ≤30% (Figure 6C), of which 75% germinated producing healthy plants (see Supplemental Figure 13 online).
DISCUSSION
In this study, we set out to determine the factors that underlie the strong tendency for COs/chiasmata to be localized in the distal regions in barley and by inference in other members of the Poaceae. We established that there is a spatiotemporal bias in the initiation and progression through meiotic prophase I that favors chiasma formation in the distal regions of the chromosomes. This bias is the culmination of a number of factors, including the relative timing of replication of distal versus interstitial/proximal chromosome regions during meiotic S-phase.
Chiasmata Predominantly Occur in Distal Euchromatin-Rich DNA
Previous studies in members of the Poaceae using genetic and cytological approaches have revealed that meiotic COs/chiasmata are highly skewed toward the distal regions of the chromosomes. Consistent with this, we observed that distal chiasmata outnumbered those in interstitial/proximal regions by 25-fold in the barley variety ‘Morex.’ This highly localized formation of chiasmata is not unique and has been reported in other species. One such case is the orthopteran Paratettix meridionalis, which invariably exhibits distal chiasmata (Viera et al., 2009). However, in P. meridionalis and other instances, such as the grasshopper Stethophyma grossum, it appears that the whole meiotic program of chromosome pairing, synapsis, and recombination are restricted to regions where CO/chiasma formation occurs (Calvente et al., 2005). This is not the case in barley as our immunolocalization studies reveal that recombination is initiated throughout the genome and full synapsis is achieved at pachytene. Although synapsis initiates in the subtelomeric/distal regions, later on, interstitial and proximal synapsis initiation sites are also observed, suggesting that potential sites for CO/chiasma formation are present in these regions.
Immunolocalization of H3K9me3, H3K27me3, H3K4me3, and H4K16ac histone marks revealed that the chromosomes were subdivided into euchromatin regions of hyperabundance and low abundance, corresponding to distal and interstitial/proximal regions, respectively. Markers for heterochromatin, such as 5-methylcytosine, H3K9me2, H3K27me2, and H4K20me1, were detected throughout the chromosomes as were barley repetitive DNA elements, although signals appeared reduced in the euchromatin-rich distal regions. A similar distribution of euchromatin and heterochromatin has been observed in barley somatic cells (reviewed in Fuchs et al., 2006). Since the chiasma distribution was coincident with the regions that were highly enriched for euchromatic DNA, it might be assumed that this is the major factor in influencing the distribution of CO sites. Indeed, two independent studies in cereals have shown that location of chiasmata is region specific. In wheat and rye, chiasmata are predominantly formed in distal regions. In a wheat-rye translocation line (1Rinv), the long arm was inverted for most of its length and chiasmata were observed to be proximal to the centromere (Lukaszewski, 2008). Naranjo et al. (2010) studied the meiotic behavior of a rye chromosome pair 5R with a terminal deletion (del5R) in a wheat background having only the proximal 30% of the long arm. Synapsis of the deficient chromosome arm pair del5RL was completed in most meiocytes at pachytene but the number of chiasmata formed was much lower than in the intact 5RL arm (Naranjo et al., 2010). This led the authors from both studies to conclude that there was an apparent division of the chromosome arms, which were either receptive or unreceptive to chiasma formation. Together, these studies could suggest that DSBs forming within the context of euchromatin-rich DNA may be processed to form both COs and non-COs, whereas those occurring in regions of low abundance are predominantly processed via a non-CO route. However, it is likely that euchromatin is an important factor in specifying the global distribution of recombination events, but a recent study suggests that the relationship is more complex. Detailed analysis in budding yeast has indicated that the correlation of the global distribution of the euchromatin histone mark H3K4me3 and DSB sites that initiate recombination is limited (Tischfield and Keeney, 2012). These authors suggest that finer scale features of chromatin structure may be of more significance. Moreover, our studies identified another factor that might influence the choice of repair pathway.
Distal and Interstitial Chromosomal Regions Exhibit Temporal Differentiation in Recombination Initiation Corresponding to the Relative Timing of Replication
The BrdU time course in conjunction with immunolocalization of the meiotic proteins indicated a difference in the timing of initiation of meiotic events in distal and interstitial/proximal regions by 2 to 3 h. Thus, based on the detection of γH2AX foci, DSB formation in the distal regions was occurring before the elaboration of a continuous chromosome axis in the interstitial/proximal regions. This temporal separation of events along the chromosomes is in contrast with observations in plants such as Arabidopsis and Brassica. In these species, the formation of the chromosome axes at late G2/leptotene is very rapid and recombination initiation, as far as can be discerned, is essentially synchronous throughout the chromosomes (Sanchez Moran et al., 2007). Moreover, although distal chiasmata outnumber those in interstitial regions, this is far less pronounced than in barley with a typical distal v interstitial ratio in Arabidopsis of 3:1 for the non-nucleolar organizer associated chromosomes (J.D. Higgins and F.C.H. Franklin, unpublished data).
It appeared that the temporal differentiation may be established during premeiotic S-phase. A dual BrdU and EdU pulse experiment revealed that the distal, euchromatin-rich chromosomal regions were replicated first (0 to 4 h). The interstitial DNA was completed next (by 6 h), followed later by the proximal heterochromatin (by 13 h). This pattern has previously been observed in barley somatic cells (Jasencakova et al., 2001) and is consistent with the association of pairing initiation with chromatin remodeling at the bouquet stage in wheat (Colas et al., 2008). A relationship between premeiotic S-phase DNA synthesis and chromatin transition at DSB sites has previously been observed in budding yeast where origins of replication were deleted on the left arm of chromosome III leading to delays in the formation of DSBs (Murakami et al., 2003). This is consistent with the early replicating DNA in barley being the first to initiate programmed DSBs.
Factors That Influence a Bias toward Distal COs
There is a strong body of evidence to indicate that the CO/noncrossover decision is made early, likely prior to stable strand exchange (Bishop and Zickler, 2004; Börner et al., 2004). Assuming this mechanism is conserved in barley, it is likely that the CO/noncrossover decision is coincident with the appearance of RAD51/DMC1 foci. As the distal RAD51/DMC1 foci localize ∼2 to 3 h before the interstitial foci, it seems possible that the decision is made before the interstitial events occur. CO designation at a distal site would then, through interference, channel other events along the bivalent into a noncrossover pathway. That said, it is questionable whether this is the only factor at play. Previous studies in a variety of species indicate that complete interference extends over a distance of 25 to 30% of a chromosome arm, before gradually reducing over the next 30% (Jones, 1984). Consistent with this, we observed some bivalents with two chiasmata in the same chromosome arm. Although the additional chiasma may arise via the noninterference route (Copenhaver et al., 2002; de los Santos et al., 2003; Higgins et al., 2008a), in some cases at least, we observed two MLH1 foci on the same chromosomal arms, suggesting these are interfering COs. Thus, interference alone may not account for the marked absence of interstitial/proximal COs.
A hallmark feature of meiotic recombination is that it is controlled to ensure that each bivalent receives at least one obligate CO with additional CO formation along the bivalent subject to interference (Jones and Franklin, 2006). Various models have been proposed to account for this control, but the underlying mechanism remains elusive (Berchowitz and Copenhaver, 2010). Among these models, the mechanical stress model posits that CO control is governed by programmed cycles of chromosome expansion and contraction (Kleckner et al., 2004). Although this model has yet to be verified experimentally, it is attractive in that it predicts many aspects of CO control, including CO assurance and CO homeostasis (Berchowitz and Copenhaver, 2010). Our data provide evidence that the chromosome cycles are conserved during prophase I in barley. Moreover, immunolocalization studies suggest that the chronology of foci corresponding to the meiotic proteins in the distal regions of the chromosomes relative to these cycles is consistent with them coordinating CO formation. Intriguingly, the appearance of foci in interstitial/proximal regions 2 to 3 h later, which are predominantly destined to be repaired without CO formation, are out of phase, relative to distal events, with the cycles. It is tempting to speculate that this may result in such intermediates being somehow channeled via a route that leads to repair without CO formation.
Thus, it may be that both factors act in conjunction to favor distal COs. Early CO designation would establish interference that prevents DSBs in regions nearby progressing to form COs. While DSBs further away in proximal regions cannot progress to form COs as they occur too late in relation to the chromosome cycles and/or length of prophase I, which appears tightly regulated (see below).
CO Control Is Affected by Moderate Heat Stress
Previous studies have shown that elevated temperature can disrupt plant meiosis. For example, exposure of Allium ursinum to 35°C for a period of 30 h was found to have a detrimental effect on chromosome synapsis with the majority of meiocytes arresting in early prophase I (Loidl, 1989). Consistent with this, in barley, synapsis did not occur at 35°C, and the majority of chromosomes formed disorganized, agglomerated structures. However, a study in Tradescantia bracteata and Uvularia perfoliata indicated that a moderate increase in temperature could result in a change in chiasma distribution (Dowrick, 1957). Hence, we surmised a similar approach in barley might perturb meiotic recombination rather than entirely disrupting it and thereby provide further insight into the factors underlying the differences in CO formation in the distal and interstitial chromosomal regions. Exposure to 30°C led to a change in chiasma distribution and frequency. A significant increase in interstitial/proximal chiasmata (0.58 to 1.13 chiasmata per cell) was observed, but overall chiasmata per cell were significantly reduced from 14.8 to 13.5. Chromosome-to-chromosome variation also existed, with 5H being the most affected and receiving significantly more interstitial/proximal chiasmata in the arm opposite to the NOR. Surprisingly, increased temperature did not alter the duration of meiosis, ∼43 h at both 22 and 30°C. This value is similar to that previously determined in barley (∼39.4 h) (Bennett and Finch, 1971). The chromatin cycles also appeared unchanged. However, the duration of premeiotic S-phase was reduced by ∼4 h, from 12 to 13 h (at 22°C) to 8 to 9 h (at 30°C). Replication in the distal regions did not seem affected and was completed by 4 h at both temperatures, but at 30°C the interstitial and proximal heterochromatic regions appeared to be replicated faster. This suggests that at higher temperatures the heterochromatin-rich regions may be more accessible for replication. A recent study by Kumar and Wigge (2010) showed that the histone H2A.Z perceives ambient temperature. At higher temperatures, H2A.Z has reduced occupancy of the nucleosomes, allowing expression of genes such as HSP70 but may have an overall effect on chromatin conformation. Nevertheless, the elevated temperature did not obviously affect the global pattern of euchromatic histone marks detectable at the cytological level, although subtle local changes cannot be excluded.
Elevated temperature had two clear effects on the behavior of the meiotic proteins that potentially underlie the changes in chiasma distribution and frequency. At 30°C, ASY1 loading in the interstitial and distal regions appeared to be more synchronous such that at G2 there was a 33% increase in the nuclear distribution of ASY1 relative to that at 22°C. This suggests that the spatiotemporal differentiation of meiotic progression between distal and interstitial regions observed at 22°C is less pronounced at the higher temperature. A possible consequence of this is that the pattern of CO designation may be altered, such that both distal and interstitial recombination events could progress to form COs. That the increase in interstitial chiasmata is significant but not dramatic may reflect the fact that the temperature increase does not entirely remove the spatiotemporal differentiation across the nucleus.
In addition to a change in chiasma distribution at 30°C, there was also a small yet significant overall reduction in chiasma frequency. We propose that this arises from a defect in SC assembly at this temperature. Although full synapsis is achieved at 30°C, the dynamics of this process are abnormal. ZYP1 polymerization along the chromosome axes was delayed, occurring in nuclei with significantly smaller envelope volumes than at 22°C (1157 µm2 versus 2165 µm2; P = 0.005, 41 df). Moreover, this is observed in conjunction with the presence of numerous irregular sized ZYP1 foci similar to polycomplexes (Chen et al., 2011). Studies in budding yeast have revealed that installation of the ZYP1 homolog Zip1 is critical for normal CO formation (Storlazzi et al., 1996). It is proposed that following CO designation, imposition of the CO fate requires Zip1 loading (Börner et al., 2004). Similarly, in Arabidopsis, depletion of ZYP1 by RNA interference or T-DNA insertion into one of the two functionally redundant genes ZYP1a or ZYP1b results in a reduction in chiasma frequency and a loss of CO control (Higgins et al., 2005). Nevertheless, it is possible that the reduction in chiasmata arises for other reasons. For instance, the increase in temperature may deleteriously affect the activity of one or more proteins required to convert DSBs into COs or affect the turnover of the recombination complexes.
Our results identify at least some of the factors involved in promoting the distal location of chiasmata in barley. We have shown that there is a spatiotemporal disparity between recombination initiation events in distal and interstitial regions and their subsequent relative timing with the chromatin cycles of contraction and expansion. Distal chiasmata correlate with regions of euchromatin hyperabundance, but we hypothesize that this is an indirect relationship as these regions are replicated first during premeiotic S-phase and subsequently enter meiosis first. Our hypothesis was supported by changing the dynamics of premeiotic S-phase replication and ZYP1 loading using elevated temperatures. There was no apparent change in euchromatin abundance but an effect on chiasma frequency and distribution with chromosomes receiving significantly more interstitial and proximal chiasmata. Finally, as viable pollen and seeds were recovered from these plants, we suggest that application of a modest temperature increase could be useful for plant breeding. However, our data suggest that it would be necessary to select an appropriate temperature that balances changes in chiasma distribution with potentially deleterious reductions in chiasma frequency.
METHODS
Plant Material
Barley (Hordeum vulgare cv Morex) was grown to maturity in a soil-based compost in a greenhouse under supplementary light (16-h-light/8-h-dark cycle). Pollen mother cells (PMCs) were harvested from the inflorescences and leaf samples taken for genomic DNA extraction. For standard growing conditions, plants were grown at 22°C. For the elevated temperature experiments, they were grown at 22°C until the plants reached a height of 30 to 35 cm and then transferred to a growth chamber at 30 or 35°C with the same light/dark cycle.
Cytological Procedures
PMCs were examined by light microscopy after staining with DAPI, and chiasmata were recorded after FISH with the 5S and 45S probes to identify individual bivalents (Leitch and Heslop-Harrison, 1993; Taketa et al., 1999; Sanchez Moran et al., 2001). The telomere repeat (Richards and Ausubel, 1988) was used to generate a probe according to Armstrong et al. (2001). Immunolocalization and other cytological techniques were performed according to Higgins (2012). Briefly, PMCs were digested for 2 min at 37°C with 0.4% cytohelicase, 1.5% Suc, and 1% polyvinypyrrolidone and fixed in 4% paraformaldehyde. Antibodies used in this study, including At-ASY1 (rat), At-RAD51 (rabbit), At-DMC1 (rabbit), At-ZYP1 (rat), At-MSH4 (rabbit), At-MSH5 (rabbit), and At-MLH1 (rabbit), have previously been described (Armstrong et al., 2002; Jackson et al., 2006; Higgins et al., 2004, 2005, 2008b; Sanchez Moran et al., 2007). γH2AX (rabbit) and H4K16ac (rabbit) were obtained from Millipore, while H3K9me2 (rabbit), H3K9me3 (rabbit), H3K27me2 (rabbit), H3K27me3 (rabbit), H4K20me1 (rabbit), H3K4me3 (rabbit), and 5-methylcytosine (mouse) were obtained from Diagenode. Undigested nuclei to monitor changes in nuclear volume were obtained by transversally slicing off the ends of the anthers and squeezing them with a large mounted needle into 2:1 0.01 M citrate buffer:water. They were then fixed with 4% paraformaldehyde and counterstained with DAPI.
Meiotic Time Course
For the cytological time course, 10 mM BrdU (0.5 mL) was injected into barley inflorescences, which were then fixed in 3:1 ethanol:acetic acid at various time points and detected with BrdU labeling and detection kit I (Roche Diagnostics). For the immunolocalization time course, BrdU was injected as above and fresh anthers were used to make slides incubated with either At-ASY1 (rat) or At-ZYP1 (rat) antibodies. For determining the length of premeiotic S-phase, BrdU was injected as above and then a second pulse of EdU was injected at various time intervals (0 to 13 h). Inflorescences were fixed 36 h after the initial BrdU injection and detected as above and with the Click-iT EdU Alexa Fluor 594 HCS assay kit (Invitrogen). Staging of the distribution of the recombination proteins was determined in relation to ASY1 localization (as foci, short linear stretches, or a linear signal) based on a BrdU time-course experiment and also on the nuclear volume measurement.
Microscopy
Fluorescence microscopy was performed using a Nikon Eclipse 90i microscope. NIS elements software (Nikon) was used to record numbers of foci and to determine nuclear volume. The distribution of ASY1 signal in the temperature shift experiments was determined by tracing the region of the ASY1 signal and using NIS elements to convert this to a percentage of the overall nuclear volume. Meiotic staging was based on nuclear volume. Statistical analysis was performed using Minitab and one-way analysis of variance. Numbers after ± are se.
Nucleic Acid Extraction and Cloning
Cot-1 DNA was isolated and prepared as described by Zwick et al. (1997) except genomic DNA was extracted with a Nucleon Phytopure kit (GE Life Sciences) and sheared to ∼200 bp by sonication. Total RNA was extracted from optic inflorescences at meiotic stage (0.5- to 1.5-mm anthers) using the RNAqueous kit (Ambion; catalog number AM1912). First-strand cDNA library was synthesized with the GeneRacer kit (Invitrogen; catalog number L1502-02). The 5′ and 3′ ends of the cDNA of interest were amplified by PCR according the GeneRacer kit protocol. PCR fragments were cloned into pGEM-Teasy vector (Promega) and sequenced. These 5′ and 3′ sequences were used to design primers for full-length cDNA amplification. Primer sequences are shown in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Hv-RAD51 (JQ855496), Hv-DMC1 (JQ855497), Hv-MSH4 (JQ855498), Hv-MSH5 (JQ855499), Hv-MLH1 (JQ855500), Hv-ZYP1 (JQ855502), and Hv-ASY1 (JQ855503).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Arabidopsis and Barley Meiotic Protein ClustalW Alignments.
Supplemental Figure 2. Axis Morphogenesis in Relation to Telomere Clustering (Unmerged Images from Figure 2).
Supplemental Figure 3. Chromosome Synapsis Initiates from Subtelomeric Regions (Unmerged Images from Figure 2).
Supplemental Figure 4. A Cytological Meiotic Time Course.
Supplemental Figure 5. An Immunological Time Course of Barley Meiotic Prophase I.
Supplemental Figure 6. Axis Morphogenesis in Relation to Early Recombination Markers (Unmerged Images from Figure 2).
Supplemental Figure 7. Axis Morphogenesis in Relation to Late Recombination Markers (Unmerged from Figure 2).
Supplemental Figure 8. Heterochromatin, Methylated DNA, and Repetitive Cot DNA Are Abundant throughout the Barley Chromosomes.
Supplemental Figure 9. Undigested Meiocytes Squeezed out of the Anther Locules Show That Nuclear Volume Changes in Vivo but Not as Much as Cell Wall Free Nuclei.
Supplemental Figure 10. Meiotic Defects at 35°C.
Supplemental Figure 11. Subtelomeric Euchromatic Histone Marks Are Indistinguishable at 22 and 30°C.
Supplemental Figure 12. Metaphase I Chromosome Spreads Reveal Increased Interstitial Chiasmata in Plants Grown at 30°C Compared with 22°C.
Supplemental Figure 13. Postmeiotic Fertility.
Supplemental Table 1. Primers Used for Cloning Barley Meiotic cDNAs.
Supplementary Material
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council for financial support with the LoLa Grant BB/F019351/1. Valuable technical support was provided by Stephen Price and Karen Staples. We thank Gareth Jones for his insightful comments and helpful ideas in this work. We also thank the reviewers for their constructive comments.
AUTHOR CONTRIBUTIONS
F.C.H.F., S.J.A., J.D.H., R.W., and C.H. designed research. J.D.H., A.B., L.R., and R.M.P. performed research. F.C.H.F., J.D.H., S.J.A., R.W., C.H., A.B., and L.R. analyzed data. F.C.H.F., S.J.A., and J.D.H. wrote the article.
Glossary
- CO
crossover
- DSB
double-strand break
- dHj
double-Holliday junction
- FISH
fluorescence in situ hybridization
- NOR
nucleolus organizer region
- SC
synaptonemal complex
- BrdU
5′-bromo-2′-deoxyuridine
- EdU
5-ethynyl-2′-deoxyuridine
- DAPI
4′,6-diamidino-2-phenylindole
- PMC
pollen mother cell
References
- Albini S.M., Jones G.H., Wallace B.M.N. (1984). A method for preparing two-dimensional surface-spreads of synaptonemal complexes from plant meiocytes for light and electron microscopy. Exp. Cell Res. 152: 280–285 [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C., Jones G.H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C.H., Jones G.H. (2003). A meiotic time-course for Arabidopsis thaliana. Sex. Plant Reprod. 16: 141–149 [Google Scholar]
- Bennett M.D., Finch R.A. (1971). Duration of meiosis in barley. Genet. Res. 17: 209 [Google Scholar]
- Berchowitz L.E., Copenhaver G.P. (2010). Genetic interference: Don’t stand so close to me. Curr. Genomics 11: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K., Zickler D. (2004). Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15 [DOI] [PubMed] [Google Scholar]
- Börner G.V., Kleckner N., Hunter N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Borde V., Robine N., Lin W., Bonfils S., Géli V., Nicolas A. (2009). Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvente A., Viera A., Page J., Parra M.T., Gómez R., Suja J.A., Rufas J.S., Santos J.L. (2005). DNA double-strand breaks and homology search: Inferences from a species with incomplete pairing and synapsis. J. Cell Sci. 118: 2957–2963 [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., Franklin F.C.H. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Grandont L., Vrielynck N., le Guin S., Mercier R., Grelon M. (2010). An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129: 143–153 [DOI] [PubMed] [Google Scholar]
- Chen Z., Higgins J.D., Li Hui J.T., Li J., Franklin F.C.H., Berger F. (2011). Retinoblastoma protein is essential for early meiotic events in Arabidopsis. EMBO J. 30: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G.P., Housworth E.A., Stahl F.W. (2002). Crossover interference in Arabidopsis. Genetics 160: 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas I., Shaw P., Prieto P., Wanous M., Spielmeyer W., Mago R., Moore G. (2008). Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proc. Natl. Acad. Sci. USA 105: 6075–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M.P. (1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M. (2003). The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowrick G. (1957). The influence of temperature on meiosis. Heredity 11: 37–49 [Google Scholar]
- Erayman M., Sandhu D., Sidhu D., Dilbirligi M., Baenziger P.S., Gill K.S. (2004). Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Res. 32: 3546–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous M., Higgins J.D., Osman K., Lambing C., Roitinger E., Mechtler K., Armstrong S.J., Perry R., Pradillo M., Cunado N., Franklin F.C.H. (2012). Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8: e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J., Demidov D., Houben A., Schubert I. (2006). Chromosomal histone modification patterns—From conservation to diversity. Trends Plant Sci. 11: 199–208 [DOI] [PubMed] [Google Scholar]
- Grelon M., Vezon D., Gendrot G., Pelletier G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Ma H., Cande W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57: 267–302 [DOI] [PubMed] [Google Scholar]
- Harrison C.J., Alvey E., Henderson I.R. (2010). Meiosis in flowering plants and other green organisms. J. Exp. Bot. 61: 2863–2875 [DOI] [PubMed] [Google Scholar]
- Hartung F., Wurz-Wildersinn R., Fuchs J., Schubert I., Suer S., Puchta H. (2007). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D. (2012). Analyzing meiosis in barley. In Methods in Molecular Biology: Plant Meiosis. W.P. Pawlowski, M. Grelon, and S.J. Armstrong, eds (New York: Humana Press, Springer), in press.
- Higgins J.D., Armstrong S.J., Franklin F.C.H., Jones G.H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Buckling E.F., Franklin F.C.H., Jones G.H. (2008a). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C.H. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Vignard J., Mercier R., Pugh A.G., Franklin F.C.H., Jones G.H. (2008b). AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55: 28–39 [DOI] [PubMed] [Google Scholar]
- Hunter N., Borts R.H. (1997). Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582 [DOI] [PubMed] [Google Scholar]
- Jackson N., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C.H. (2006). Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain H. (1957). Effect of high temperature on meiosis in Lolium: Nucleolar inactivation. Heredity 11: 23–36 [Google Scholar]
- Jasencakova Z., Meister A., Schubert I. (2001). Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110: 83–92 [DOI] [PubMed] [Google Scholar]
- Jones G.H. (1984). The control of chiasma distribution. SEB Symp. 38: 293–320 [PubMed] [Google Scholar]
- Jones G.H., Franklin F.C. (2006). Meiotic crossing-over: Obligation and interference. Cell 126: 246–248 [DOI] [PubMed] [Google Scholar]
- Kleckner N., Zickler D., Jones G.H., Dekker J., Padmore R., Henle J., Hutchinson J. (2004). A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101: 12592–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Künzel G., Korzun L., Meister A. (2000). Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzel G., Waugh R. (2002). Integration of microsatellite markers into the translocation-based physical RFLP map of barley chromosome 3H. Theor. Appl. Genet. 105: 660–665 [DOI] [PubMed] [Google Scholar]
- Kurzbauer M.-T., Uanschou C., Chen C.B., Schlogelhofer P. (2012). The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis Plant Cell 24: 2058–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch I.J., Heslop-Harrison J.S. (1993). Physical mapping of four sites of 5S rDNA sequences and one-site of the a-amylase-2 gene in barley (Hordeum vulgare). Genome 36: 517–523 [DOI] [PubMed] [Google Scholar]
- Li W.X., Chen C.B., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J. (1982). Temperature and chiasma formation in Rhoeo-spathaceavar variegata. Genetica 60: 25–30 [Google Scholar]
- Loidl J. (1989). Effects of elevated-temperature on meiotic chromosome synapsis in Allium ursinum. Chromosoma 97: 449–458 [Google Scholar]
- Lukaszewski A.J. (2008). Unexpected behavior of an inverted rye chromosome arm in wheat. Chromosoma 117: 569–578 [DOI] [PubMed] [Google Scholar]
- Marcon E., Moens P. (2003). MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165: 2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F., et al. (2011). Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailova E.I., Phillips D., Sosnikhina S.P., Lovtsyus A.V., Jones R.N., Jenkins G. (2006). Molecular assembly of meiotic proteins Asy1 and Zyp1 and pairing promiscuity in rye (Secale cereale L.) and its synaptic mutant sy10. Genetics 174: 1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G., Devos K.M., Wang Z., Gale M.D. (1995). Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Murakami H., Borde V., Shibata T., Lichten M., Ohta K. (2003). Correlation between premeiotic DNA replication and chromatin transition at yeast recombination initiation sites. Nucleic Acids Res. 31: 4085–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo T., Valenzuela N.T., Perera E. (2010). Chiasma frequency is region specific and chromosome conformation dependent in a rye chromosome added to wheat. Cytogenet. Genome Res. 129: 133–142 [DOI] [PubMed] [Google Scholar]
- Naumann K., Fischer A., Hofmann I., Krauss V., Phalke S., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Reuter G. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24: 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Keeney S. (2006). Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson N.O., Säll T., Bengtsson B.O. (1993). Chiasma and recombination data in plants: Are they compatible? Trends Genet. 9: 344–348 [DOI] [PubMed] [Google Scholar]
- Osman K., Higgins J.D., Sanchez-Moran E., Armstrong S.J., Franklin F.C.H. (2011). Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 190: 523–544 [DOI] [PubMed] [Google Scholar]
- Osman K., Sanchez-Moran E., Higgins J.D., Jones G.H., Franklin F.C.H. (2006). Chromosome synapsis in Arabidopsis: Analysis of the transverse filament protein ZYP1 reveals novel functions for the synaptonemal complex. Chromosoma 115: 212–219 [DOI] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558 [DOI] [PubMed] [Google Scholar]
- Perrella G., Consiglio M.F., Aiese-Cigliano R., Cremona G., Sanchez-Moran E., Barra L., Errico A., Bressan R.A., Franklin F.C.H., Conicella C. (2010). Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J. 62: 796–806 [DOI] [PubMed] [Google Scholar]
- Plug A.W., Peters A.H., Keegan K.S., Hoekstra M.F., de Boer P., Ashley T. (1998). Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 111: 413–423 [DOI] [PubMed] [Google Scholar]
- Pradillo M., López E., Linacero R., Romero C., Cuñado N., Sánchez-Morán E., Santos J.L. (2012). Together yes, but not coupled: New insights into the roles of RAD51 and DMC1 in plant meiotic recombination. Plant J. 69: 921–933 [DOI] [PubMed] [Google Scholar]
- Richards E.J., Ausubel F.M. (1988). Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136 [DOI] [PubMed] [Google Scholar]
- Ronceret A., Pawlowski W.P. (2010). Chromosome dynamics in meiotic prophase I in plants. Cytogenet. Genome Res. 129: 173–183 [DOI] [PubMed] [Google Scholar]
- Sanchez Moran E., Armstrong S.J., Santos J.L., Franklin F.C.H., Jones G.H. (2001). Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9: 121–128 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J.L., Jones G.H., Franklin F.C.H. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D., Gill K.S. (2002). Gene-containing regions of wheat and the other grass genomes. Plant Physiol. 128: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. (1995). Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791 [DOI] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R. (2004). hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Stacey N.J., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48: 206–216 [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Xu L.Z., Schwacha A., Kleckner N. (1996). Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc. Natl. Acad. Sci. USA 93: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa S., Ando H., Takeda K., von Bothmer R. (1999). Detection of Hordeum marinum genome in three polyploid Hordeum species and cytotypes by genomic in situ hybridization. Hereditas 130: 185–188 [DOI] [PubMed] [Google Scholar]
- Tariq M., Saze H., Probst A.V., Lichota J., Habu Y., Paszkowski J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield S.E., Keeney S. (2012). Scale matters: The spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 11: 1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera A., Santos J.L., Rufas J.S. (2009). Relationship between incomplete synapsis and chiasma localization. Chromosoma 118: 377–389 [DOI] [PubMed] [Google Scholar]
- Vignard J., Siwiec T., Chelysheva L., Vrielynck N., Gonord F., Armstrong S.J., Schlögelhofer P., Mercier R. (2007). The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 3: 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Amon A. (2000). The nucleolus: The magician’s hat for cell cycle tricks. Curr. Opin. Cell Biol. 12: 372–377 [DOI] [PubMed] [Google Scholar]
- Wang C.-J.R., Carlton P.M., Golubovskaya I.N., Cande W.Z. (2009). Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics 183: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.F., Kleckner N., Hunter N. (1999). Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W.L., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33: 603–754 [DOI] [PubMed] [Google Scholar]
- Zwick M.S., Hanson R.E., Islam-Faridi M.N., Stelly D.M., Wing R.A., Price H.J., McKnight T.D. (1997). A rapid procedure for the isolation of Cot-1 DNA from plants. Genome 40: 138–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.