When they move, transposons can cause mutations not only of host genes but also of themselves. This article reports that the maize (Zea mays) element Ac can mutate at a surprisingly high frequency on transposition and documents the several types of self-inflicted changes that it can undergo.
Abstract
The autonomous transposon Activator (Ac) is a powerful mutagen. Ac-induced mutations range from small footprints of host sequences to large rearrangements of transposon or host sequences. These mutations arise by different repair mechanisms of the double-strand break produced by Ac excision: footprints by nonhomologous end joining and rearrangements by various mechanisms, including DNA replication repair. Footprints greatly outnumber other mutations, masking them because they usually share a nonfunctional phenotype. To determine the spectrum and frequencies of host and self-mutations generated by Ac, we used an allele harboring Ac in the 5′ untranslated region bronze (bz). In this system, simple excisions produce purple revertants, whereas deletions of host or transposon sequences produce stable bronze (bz-s) mutants. Internal and terminal deletions of Ac predominated among the 72 bz-s derivatives. Most internal deletions (52 of 54) behaved as nonautonomous Dissociation (Ds) elements. All nine terminal deletions or fractured Ac (fAc) elements had rearrangements of adjacent host sequences. Most Ds and fAc deletion junctions displayed microhomologies and contained filler DNA from nearby sequences, suggesting an origin by DNA repair synthesis followed by microhomology-mediated end joining. All mutations occurred more frequently in pollen, where one in 200 grains carried new Ds or fAc elements.
INTRODUCTION
Activator (Ac) was the first autonomous or self-mobile transposable element (TE) described by McClintock (1949). In addition to itself, Ac can mobilize its nonautonomous Dissociation (Ds) counterparts, which share common ends with it but do not encode a transposase. Ac and Ds belong to the hAT superfamily of DNA TEs (Kunze and Weil, 2002). Ac is 4565 bp long and has 11-bp imperfect terminal inverted repeats (TIRs) and ∼240-bp subterminal regions at both ends, which are essential for transposition (Coupland et al., 1989). The central region of Ac encodes a transposase that is both necessary and sufficient to mobilize Ac and Ds elements in various transgenic organisms that lack Ac-homologous sequences (Coupland et al., 1988).
Ac and Ds are the best-studied plant hAT transposons, and their detailed analysis has allowed a glimpse of some of the genetic interactions that are possible between transposons of that superfamily and their hosts (Dooner and Weil, 2012). Ac is a potent, versatile, and indiscriminate mutagen that has coevolved with its maize (Zea mays) host. It is potent because it causes mutations in a high percentage of gametes, versatile because it can induce various kinds of mutations, and indiscriminate because it does not differentiate between its own DNA and that of the host. Like most other DNA transposons, Ac transposes by a cut-and-paste mechanism. It is the ability to frequently cut itself from the linear continuity of the chromosome by introducing double-strand breaks (DSBs) that makes Ac a powerful mutagen. The subsequent repair of these DSBs by the host’s enzymatic machinery rarely leaves the DNA in its pristine condition, but leads instead to a variety of changes, from the addition or deletion of a few bp to large-scale chromosomal rearrangements, all of which have the potential to generate diversity.
Insertion of Ac causes the duplication of 8 bp of host sequence on either side of Ac (target site duplication [TSD]). Excision of Ac generates a DSB that is repaired predominantly by the nonhomologous end joining (NHEJ) pathway, which leaves behind excision footprints of variable size and composition (Scott et al., 1996; Rubin and Levy, 1997; Huefner et al., 2011). Ac tends to insert in coding regions (Cowperthwaite et al., 2002); therefore, these excision footprints often disrupt the normal reading frame of the gene and lead to nonfunctional alleles.
Other types of mutations can be produced by repair of the DSBs caused by Ac excision, but at much lower frequencies. The most common of these is mutation to Ds. Unlike the highly conserved Ac, Ds elements are structurally diverse, but many are simple internal deletion derivatives of various sizes. McClintock (1956, 1962, 1963) reported several cases at the wx and bz loci in which Ac appeared to have mutated to Ds, and later molecular analysis proved them to be internal deletions of Ac (Pohlman et al., 1984; Dooner et al., 1986; Yan et al., 1999). Similar mutations of Ac to Ds were subsequently described at bz, ps1, and other loci (Yan et al., 1999; Conrad et al., 2007). Less frequently, Ac can cause a large deletion of adjacent host sequences. Derivative bz-s:2114(Ac) from the maize mutable allele bz-m2(Ac) lost 789 bp of bz sequence adjacent to the 5′ end of Ac; however, the transposition frequency of Ac2114, which is not flanked by a TSD, was not affected (Dooner et al., 1988). A second rare Ac rearrangement was reported by Ralston et al. (1989) in the bz-s2094 derivative from bz-m2(Ac), which displayed a remarkable propensity to break chromosomes. The 3′ end of Ac and the adjacent bz sequence in this allele are identical to the parental line, but various rearrangements occurred at the 5′ end. bz-s2094 carries a fractured Ac (fAc), which lost 2 kb from its 5′ end, adjacent to a 37-bp duplication of host sequence and a typical Ac excision footprint. Similar fAc elements retaining only the 3′ end have also been described at P1 by Zhang and Peterson (2004) and at ps1 by Conrad et al. (2007). The spectrum and frequencies of the more rare null mutations are difficult to ascertain because of the overwhelming number of simple excision products that have the same null mutant phenotype. Conrad et al. (2007) analyzed 753 kernels with a mutant phenotype from several Ac donor lines and found only 24 such Ac inactivation events. The system that we report on here is highly efficient for isolating these more rare events, which most likely arise from DSB repair by an error-prone DNA synthesis pathway and from aberrant transpositions.
bz-m39(Ac) is a mutable allele harboring an Ac element in the 5′ untranslated region (UTR) of the bz gene, 32 to 39 bp upstream of the start codon. The transposon footprints generated by Ac39 excision in the bz 5′ UTR do not interfere with gene function; therefore, all simple excisions produce purple (Bz') revertants, rather than stable bronze (bz-s) derivatives. We took advantage of this property to set up an efficient screen for the rare Ac deletion mutations. We isolated 72 exceptional bz-s derivatives and found various types of transposon and host gene mutations. Internal deletions of Ac constituted the majority class. Most of the 54 deletions behaved as new Ds elements, because they could be mobilized by Ac in trans. Surprisingly, given just three sporadic instances in the literature, nine fAcs were found in this study, making them the second most abundant class of bz-s derivatives. The fAc elements, which can retain either the 5′ or 3′ end, were always accompanied by either duplications or deletions of adjacent host sequences. The remaining nine bz-s mutants fell into various classes: seven adjacent deletions with or without Ac, one hypermethylated cycling Ac, and one coincidental Magellan long terminal repeat (LTR) retrotransposon insertion. This study provides a comprehensive view of the types and frequencies of mutations, other than simple excision footprints, that can be produced at a locus by different repair of Ac-induced DSBs.
RESULTS
Simple Excisions versus Complex Mutations of Ac
Complex mutations of Ac-mutable alleles are difficult to identify phenotypically, because they are greatly outnumbered by the Ac simple excision footprints with which they usually share the same loss-of-instability diagnostic phenotype. The bz-m39(Ac) system allows us to efficiently select for such mutations. The Ac element in the bz-m39(Ac) allele excises frequently in somatic tissues, producing heavily variegated kernel and plant phenotypes (Figure 1). The Ac39 excision footprints in the bz 5′ UTR should not interfere with gene function, so all simple germinal excisions should produce purple kernel (Bz′) revertants. By contrast, bz-s mutations should be derived from more complex mutations that disrupt expression of the bz gene.
Figure 1.
bz-m39(Ac) Phenotypes.
Unstable kernel, plant, and anther phenotype of progeny from a cross between bz-m39(Ac) as male and sh-bz-X2 as female. The bronze kernel carries a bz-s mutation.
To select both Bz′ and bz-s mutations, bz-m39(Ac) hemizygotes were backcrossed as either male or female parents to sh-bz-X2, a line carrying a large x-ray–induced deletion of the 2-centimorgansh-bz interval on 9S (Table 1). A total of 72 bz-s mutants were recovered: 11 from 5690 female gametes, and 61 from 11,870 male gametes. By comparison, a similar number of Ac39 simple excision derivatives could be obtained from a much smaller population. A total of 78 Bz′ (purple) revertants with typical transposon footprints were recovered from just more than 1000 bz-m39(Ac) gametes: 25 from 466 female gametes, and 53 from 550 male gametes. Twelve classes of transposon footprints were represented among them (see Supplemental Figure 1 online), the most common ones having either a transversion or a deletion of one of the TSD central bases, as has been observed in other Ac or Ds mutable alleles (Kunze and Weil, 2002). Overall, simple Ac excisions (Bz′) are 21 times more frequent than the more complex Ac mutations (bz-s). Both types of exceptions occur twofold to threefold more frequently in the male than in the female germline. However, the types of mutation generated from bz-m39(Ac) males or females do not seem to differ (see below and in Supplemental Figure 1 online).
Table 1. Frequencies of bz-s and Bz′ Derivatives from bz-m39(Ac) Female and Male Parents.
| Cross | Type of Derivative | Effective Population | No. of Selections | Frequency (×10−3) | Bz′: bz-s | Male: Female |
|---|---|---|---|---|---|---|
| Sh bz-m39(Ac) wx x sh-bz-X2 wx | bz-s | 5690 | 11 | 1.9 | 27.8 | 2.7 |
| sh-bz-X2 wx | Bz′ | 466 | 25 | 53.6 | 1.8 | |
| sh-bz-X2 wx x Sh bz-m39(Ac) wx | bz-s | 11870 | 61 | 5.1 | 18.8 | |
| sh-bz-X2 wx | Bz′ | 550 | 53 | 96.4 |
Characterization of bz-s Derivatives from bz-m39(Ac)
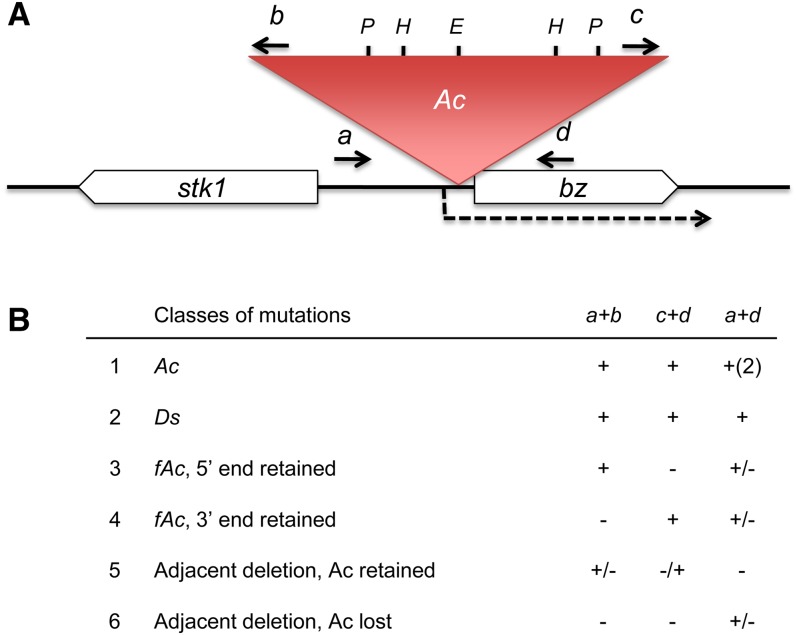
Three diagnostic PCRs were performed on all bz-s derivatives. As shown in Figure 2A, primer pair a+b amplifies the 5′ Ac-bz junction, primer pair c+d amplifies the 3′ Ac-bz junction, and primer pair a+d amplifies the entire transposon plus the adjacent bz sequences in long-range PCR. The bz-s derivatives were then grouped into six categories according to the PCR results (Figure 2B). The parental bz-m39(Ac) allele produces junction PCR products with the a+b and c+d primer pairs and two PCR products with the a+d primer pair: a large one, corresponding to the Ac-occupied site, and a 4.5-kb smaller one, corresponding to the Ac excision or empty site. The derivatives producing nonparental PCR patterns were tentatively classified as follows. Derivatives producing the same two PCR products with the junction primers, but no empty site and a smaller band than the Ac-occupied site with the a+d primer pair, were classified as new Ds elements. Derivatives producing only one of the two junction products were classified as fAc elements or Ac-adjacent deletions. Derivatives that failed to amplify either junction, but yielded a product with the a and d primers were classed as adjacent deletions without Ac.
Figure 2.
PCR Characterization of bz Derivatives.
(A) Structure of bz-m39(Ac), showing Ac inserted in the 5′ UTR of the bz gene. The bz transcript is represented by the dotted arrow; Ac and bz are in the same transcriptional orientation. Primers and their approximate locations are indicated by short arrows. Primers a and d anneal to sequences adjacent to Ac39; primers b and c anneal to the Ac subterminal region. E, EcoRI restriction site; H, HindIII restriction site; P, PvuII restriction site.
(B) Summary of the PCR amplification patterns given by the various derivatives. The derivatives fell into six classes based on the PCR results. +, band present; −, band absent; +/−, band present in some derivatives, absent in others; +(2), two bands amplified.
[See online article for color version of this figure.]
The a+d PCR products from putative Ds elements (class 2) were digested separately with the Ac-cutting enzymes PvuII, HindIII, and EcoRI to identify the deleted region, and deletion junctions were then sequenced. The PCR products from the lines that have undergone an Ac excision and an adjacent deletion were small, so the whole PCR fragments were sequenced directly. To obtain the junction sequences of the fAcs and the Ac-adjacent deletions, sequential PCRs were performed using combinations of an Ac-specific primer and a series of primers based on the adjacent stk1 and stc1 genes and the intergenic regions (not shown in Figure 2A). Alternatively, inverse PCR was used to solve larger genomic alterations involving fAc or Ac-adjacent deletions. Sequencing of the PCR products identified the novel junctions.
In addition, the bz-s derivatives were subjected to a series of genetic tests. They were self-pollinated to confirm heritability of the selected bz-s phenotype and crossed to bz-R wx-m7(Ac) to determine whether a Ds element resided at bz. All non-Ds derivatives and derivatives with small internal deletions were crossed to bz-m2(D1) to determine whether they retained Ac activity.
Ds Formation
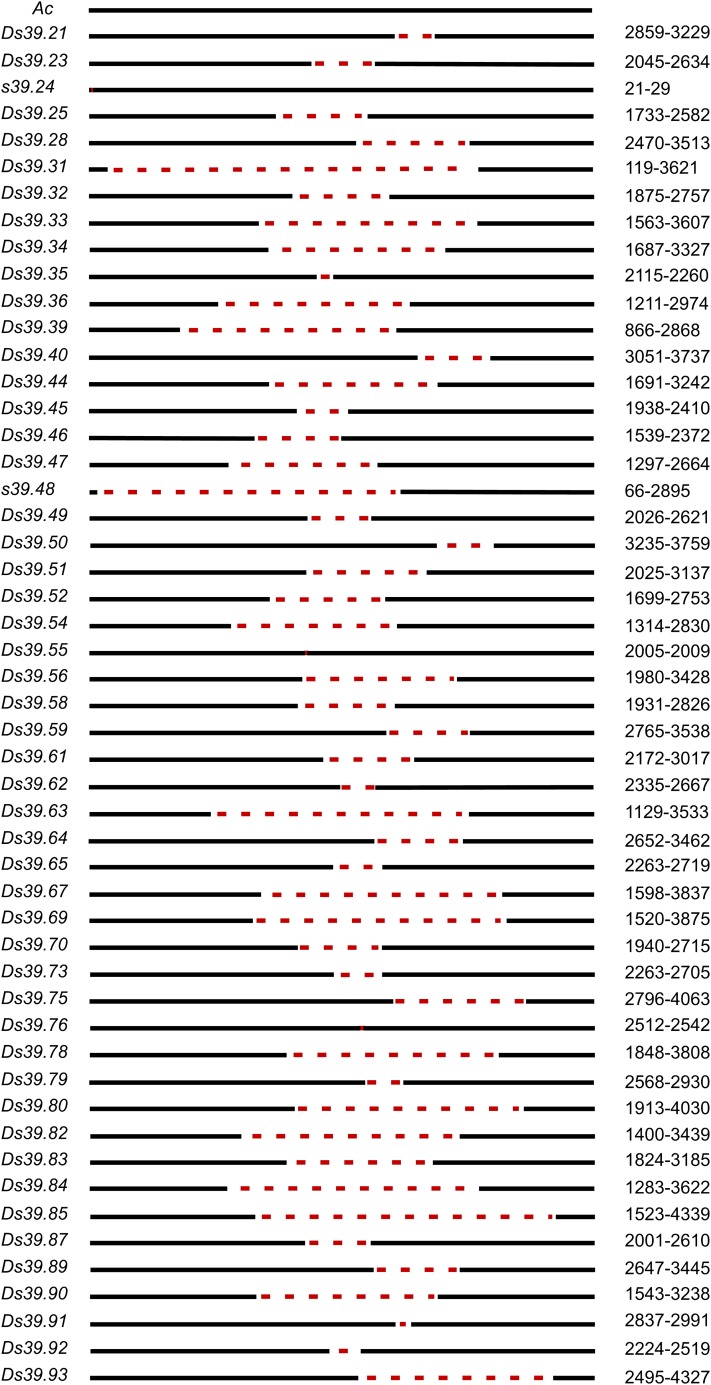
A total of 54 internal deletions were identified among the 72 bz-s selections. All but two of them behaved as Ds elements when crossed to bz-R wx-m7(Ac). They retain mutability in the presence of Ac; therefore, the new mutable alleles are designated bz-m39.x to indicate that they are Ds derivatives of bz-m39(Ac). Six of them apparently arose from three premeiotic events, bringing the number of unique Ds elements to 49. Correcting for these duplicate events, the minimal mutation frequencies of Ac to Ds are 0.9 × 10−3 and 3.7 × 10−3 in female and male gametes, respectively (mutations in which Ac is retained in the genome are not selected). The new Ds elements range in size from 1.1 to 4.6 kb (Figure 3). Although the internal deletions do not seem to occur at any specific location, as seen in previous studies (Yan et al., 1999; Conrad et al., 2007), most Ds deletions extended into the central 2.2- to 3.2-kb segment of Ac, revealing a preferential region for deletion formation. In response to Ac, most internal deletions can excise at a similar frequency as the parental Ac39, with a few exceptions. bz-m39.31 retains just 119 bp of the 5′ end and produces few very fine spots when crossed to Ac, so this deletion of Ac corresponds to a new Ds element with minimal transposition activity (see Supplemental Figure 2 online). The element in bz-s39.48 has only 66 bp left of the 5′ end, and a cross to an Ac source (wx-m7) resulted in only stable bronze kernels, indicating that this element is not a bona fide Ds. The phenotypes of both bz-m39.31 and bz-s39.48 agree with earlier findings in transgenic tobacco (Nicotiana tabacum), in which transposons retaining between 100 and 200 bp of one end had reduced excision frequencies, whereas those retaining less than 100 bp at either end showed no excision (Coupland et al., 1989). bz-m39.55 is the largest Ds element found in this study. It has a 3-bp in-frame deletion in the second exon that results in the loss of Ser-305, a well-conserved residue within the Ac/Tam3 and restless clades of hAT DNA transposases (see Supplemental Figures 3A and 3B online) that is evidently important for activity. Finally, bz-s39.24 has a tiny deletion close to the 5′ end and behaves like an immobile Ac (discussed later).
Figure 3.
Structure of 51 Internal Deletions from Ac39.
Solid lines indicate Ac sequences, and dotted line represent deletions. All but bz-s39.24 and bz-s39.48 are true Ds elements. Short filler DNAs are not shown in the figure, but their sequences can be found in Figure 4.
[See online article for color version of this figure.]
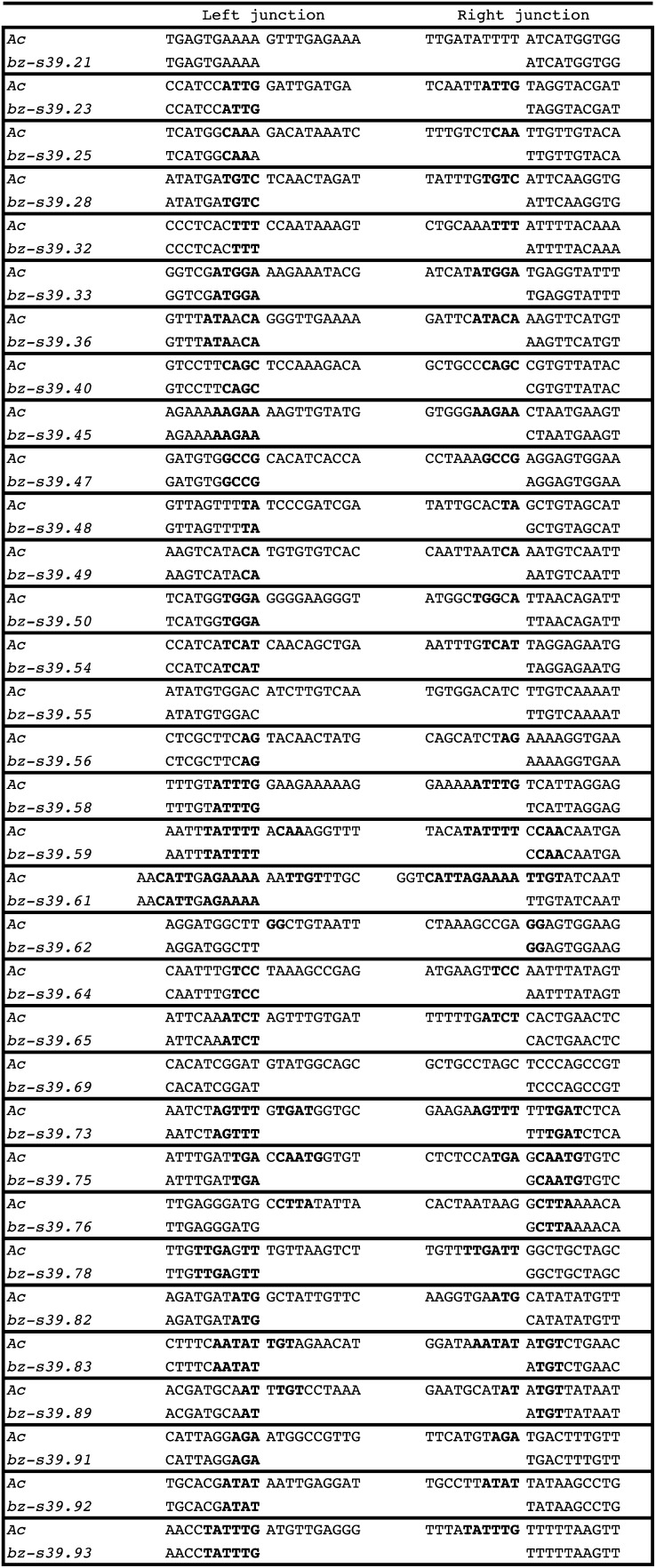
Most Ds deletion junctions occur at short (2- to 9-bp) sequences that are internally duplicated in Ac (Figure 4). Filler DNA is commonly found at the junctions of genetic rearrangements (Roth and Wilson, 1985; Roth et al., 1989; Sainsard-Chanet and Begel, 1990; Wessler et al., 1990) and of approximately one-half of previously characterized Ds elements (Yan et al., 1999; Conrad et al., 2007). Seventeen out of 49 (35%) newly formed Ds39 elements have filler DNA between the deletion endpoints. The size of the filler from this study varies from 2 bp in Ds39.35 to 55 bp in Ds39.63 (Figure 5). In most cases, the filler DNA sequences also appear close to deletion termini in the progenitor Ac sequence, where they are flanked by microhomologies to the Ds sequences flanking the filler DNA (shown in bold in Figure 5). Interestingly, most of the Ds elements with filler DNA (16 of 17) display microhomology at each junction. The filler DNA from eight bz-s39 derivatives originated from more than one location. For example, Ds39.87 has a 32-bp filler derived from three locations within Ac, namely 8 bp from 3799 to 3806, 10 bp from 2036 to 2045, and 14 bp from 2008 to 2021, and all three pieces of filler DNA share microhomology with each other at the junctions.
Figure 4.
Sequence of the Ds Deletion Junctions.
Sequences of the left (5′) and right (3′) deletion junctions and the corresponding sequence in Ac are shown for each Ds element. Microhomology sequences are shown in boldface.
Figure 5.
Sequences of the Ds Filler Junctions.
Sequences of the left (5′) and right (3′) deletion junctions and the corresponding sequence in Ac are shown for each Ds element. Microhomology sequences are bolded. Filler DNA is shown in lowercase, and the Ac sequence from which it originated is shown in underlined uppercase. Numbers refer to coordinates in the Ac sequence (GenBank X05425). Colored filler sequences denote multiple origins in the Ac sequence, where they are represented in the same color.
fAc
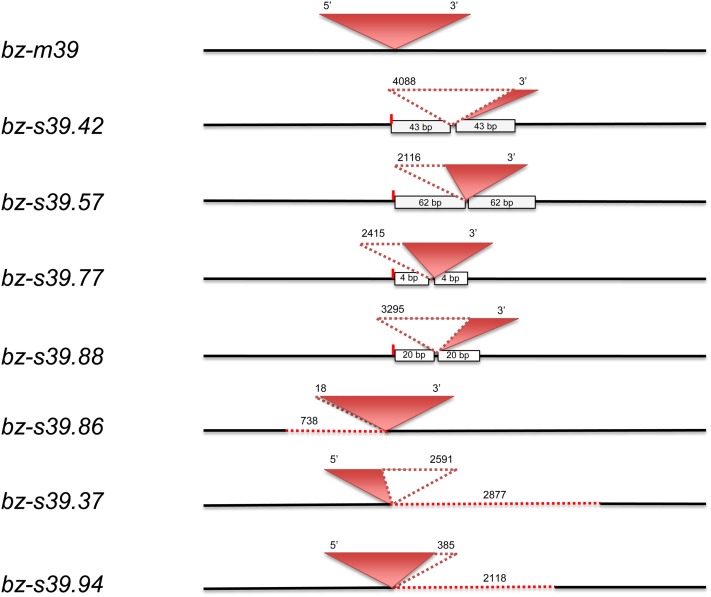
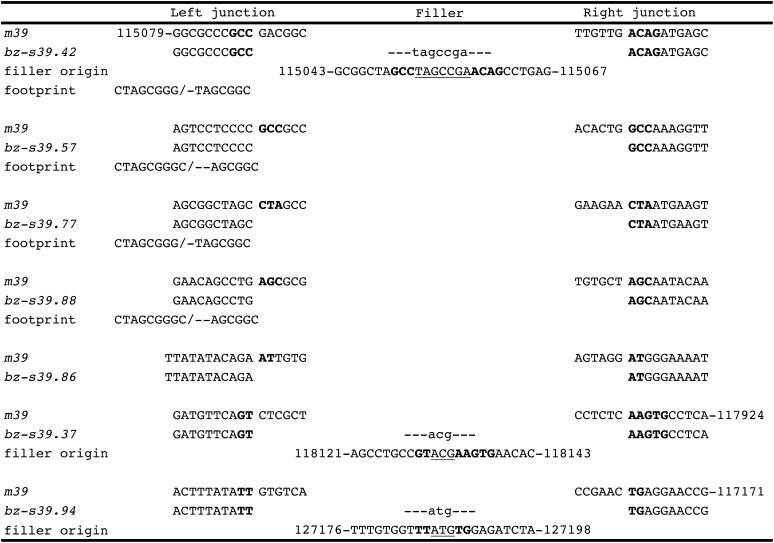
Nine single-ended or fAcs occurred in the collection of stable bronze derivatives, of which six retain the 3′ end of Ac, whereas three retain the 5′ end. The complete structure of seven of them is shown in Figure 6, but two of them had complex rearrangements at the fractured end whose detailed structure could not be elucidated. The size of the fAc elements ranges from 0.5 to 4.5 kb. In every fAc, the terminal Ac deletion is accompanied by changes in the sequence adjacent to Ac in the bz-m39(Ac) progenitor allele. Two types of fAc derivatives were recovered. One type comprises bz-s39.42, bz-s39.57, bz-s39.77, and bz-s39.88. In these four alleles, the 3′ (right) Ac-bz junction is identical to that in the parental allele, but the 5′ (left) side is complex. The 5′ end of Ac has been deleted, leaving a 3′ fAc ranging in size from 0.5 to 2.4 kb. Immediately 5′ of the fAc, a 4- to 62-bp duplication of bz sequences adjacent to the Ac 3′ junction ends in a canonical Ac39 excision footprint. The other type of fAc derivative comprises bz-s39.86, bz-s39.37, and bz-s39.94. These alleles also have one parental Ac-bz junction, but an adjacent deletion, rather than duplication, of bz sequences accompanies the Ac terminal deletion. bz-s39.86 has an intact right Ac-bz junction, but is missing 18 bp from the 5′ Ac end and 738 bp of adjacent upstream bz sequence. bz-s39.37 has an intact left Ac-bz junction, but is missing 2591 bp from the 3′ Ac end and 2877 bp of adjacent downstream bz sequence. Similarly, bz-s39.94 has an intact left Ac-bz junction, but is missing 385 bp from the 3′ Ac end and 2118 bp of adjacent downstream bz sequence. The deletion junctions in all fAc elements occur at sites of direct repeats of a few bases in the bz-m39(Ac) parental allele (Figure 7). Three of these junctions have short filler DNAs with similar properties to those found in Ds deletion junctions.
Figure 6.
Structure of fAcs.
Boxes represent duplicated bz sequences, dotted lines represent deletions, and their sizes (in bp) are shown above each rearrangement. Short vertical lines denote excision footprints at the original insertion site of Ac. All alleles are aligned at this site.
[See online article for color version of this figure.]
Figure 7.
Sequence of Excision Footprints and Deletion Junctions of fAc Elements.
Sequences of the left (5′) and right (3′) deletion junctions and the corresponding sequence in Ac are shown for each fAc element. Microhomology sequences are shown in boldface. Filler is shown in lowercase. The numbers identify the nucleotide positions of the filler DNA origins relative to the sequence of the 226-kb McC bz haplotype contig.
Adjacent Deletions with and without Retention of Ac
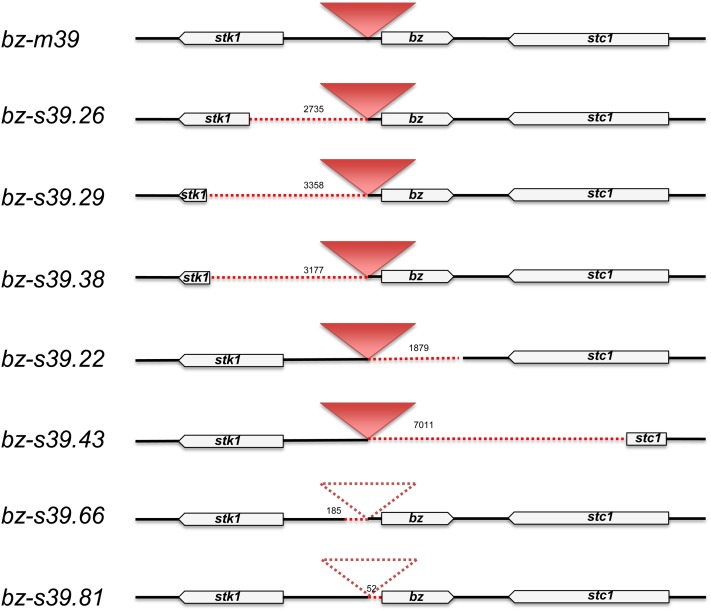
Seven stable bz alleles had deletions of gene sequences adjacent to Ac (Figure 8). The deletions range in size from 52 to 7011 bp and can occur at either end of Ac, often extending into either the proximal stk1 or distal stc1 gene. Five have intact Ac and bz sequences at one end and precise deletions of gene sequences at the other. In this sample, three were next to the 5′ end (bz-s39.26, bz-s39.29, and bz-s39.38), whereas two were next to the 3′ end (bz-s39.22 and bz-s39.43). On subsequent excision of Ac, the former three would result in fusions of a promoterless bz gene to different fragments of the proximal stk1 gene in opposite orientation. The latter two would result in fusions of the bz promoter to different fragments of the distal stc1 gene in opposite orientation, an arrangement that could lead to the formation of antisense RNA. In the remaining two adjacent deletions, bz-s39.66 and bz-s39.81, Ac has also excised, leaving behind one-half of typical excision footprints at bz: a transversion and a deletion of the base nearest Ac, respectively. These deletions are small (185 and 52 bp), but removal of the bz promoter and of part of the coding sequence, respectively, leads to a null bz phenotype.
Figure 8.
Structure of Adjacent Deletions.
Pentagons represent genic sequences, the solid triangle represents Ac, dotted lines represent gene sequence deletions, and the dotted triangle represents an excised Ac. The size of each deletion in bp is shown above the line. Adjacent deletions can occur at either end of Ac and may be accompanied by excision of Ac.
[See online article for color version of this figure.]
Immobilized Ac
bz-s39.24 is a stable bronze mutation that behaved like a Ds derivative in the three diagnostic PCR tests designed to identify structural changes in Ac39 (Figure 2) but did not respond to Ac in genetic crosses to wx-m7(Ac). Surprisingly, the lack of mobility of bz-s39.24 results from a tiny, 7-bp deletion of nucleotides 22 to 28 in the 5′ subterminal region (Figure 3). The immobilized Ac (Ac-im) element described by Conrad and Brutnell (2005) lacks 10 bp from the 5′ TIR, which was known to be essential for excision (Hehl and Baker, 1989; Kunze and Weil, 2002). The 7-bp deletion in the 5′ subterminal region of bz-s39.24 leads to a similar immobility, revealing a new cis requirement for Ac transposition. The immobile 4558-bp element retains full Ac transposase activity and can transactivate excision of the Ds element in the bz-m2(DI) reporter as well as the intact Ac element in wx-m7(Ac); therefore, we have called it Ac-im2.
bz-s39.86 is a single-ended fAc derivative that has lost the 5′ terminal 18 bp of Ac and 738 bp of adjacent bz sequence (Figure 6). In agreement with previous findings (Hehl and Baker, 1989; Xiao and Peterson, 2002; Conrad and Brutnell, 2005), the small terminal deletion completely eliminated the ability of Ac to transpose. The fAc element in bz-s39.86 is similar to Ac-im (Conrad and Brutnell, 2005), having lost an additional 8 bp from the 5′ end, and like Ac-im and Ac-im2, it can also transactivate excision of Ds from a reporter allele. We have termed this 4547-bp element Ac-im3.
Methylated Ac
bz-c39.27 was selected as a stable bronze mutation and behaved like a Ds derivative in the PCR assay (category 2 in Figure 2B). However, sequencing of the entire element revealed no changes at the nucleotide level. After two generations of selfing, this allele regained a weak Ac transposase activity. Crosses to bz-m2(DI) produced many very fine spotted kernels together with a few heavy spotted kernels, and a somatic excision product could be detected by PCR with primers a and d that was absent in the previous generation (see Supplemental Figure 4A, i, online). PCR assays indicated that the internal PvuII sites of Ac were methylated in the first two generations but were hypomethylated in the third (see Supplemental Figures 4A, ii and iii, online), and a DNA gel blot confirmed that the PvuII sites were unmethylated on reactivation (see Supplemental Figure 4B online). Thus, like other Acs (Schwartz and Dennis, 1986; Chomet et al., 1987; Brutnell and Dellaporta, 1994), Ac39 can undergo cycles of activity that correlate with its methylation state. Although this derivative was originally labeled bz-s, because of its stable bronze phenotype, we have renamed it post facto as bz-c39.27 to indicate its cycling nature.
Retrotransposon Insertion Mutation
bz-s39.71 is a most unusual derivative. It has an intact Ac at the original insertion site of bz-m39, but it heritably produces stable bz kernels. Further analysis revealed that the loss of instability was not related to an Ac-induced change but to the chance insertion of a Magellan LTR retrotransposon 65 bp upstream of the Ac insertion site. The retrotransposon insertion not only stabilizes bz but also interferes with Ac function, as evidenced in a cross with a bz-m2(DI) tester that produced very few purple spots.
DISCUSSION
Frequencies of Ac Mutation
On transposition, TEs cause mutations not only of host genes but of themselves. However, because most TEs are followed phenotypically by their effect on host genes, their mutations are detected only indirectly as changes in the mutable phenotypes produced by the host genes into which they are inserted. In mutable alleles where Ac is inserted within the coding sequence, loss-of-instability mutations of Ac usually have the same phenotype as the much more numerous Ac simple excision footprints. Among those mutations, Ds elements originating de novo can be identified, because their stable phenotype is reversible; therefore, putative mutations of Ac to Ds can be established by a series of rather tedious genetic tests. Estimates of Ds mutation frequencies in the literature are scant. McClintock (1963) isolated two Ds derivatives from a combined total of 4700 wx-m9(Ac) male and female gametes produced in Ac/+ heterozygotes. Therefore, the Ac to Ds mutation frequency from this small experiment would be 4 × 10−4. A caveat here is that, although the male and female gamete populations were reported separately, the actual source of the two mutations was not identified, so the above frequencies represent combined estimates from the two sexes. Yan et al. (1999) obtained two bz-m2(Ds) derivatives from 3867 bz-m2(Ac) gametes produced in Ac/+ heterozygous females, for a frequency of 5 × 10−4. Conrad et al. (2007) isolated five new Ds elements from four different loci out of a total population of 19,923 gametes produced in Ac/Ac homozygous females, for an average frequency estimate of 2.5 × 10−4. The one conclusion that can be drawn from the above is that the frequency with which Ac mutates to Ds from one generation to another is on the order of 10−4, but whether the mutation frequency varies from locus to locus, within a locus, or between sexes is unclear. Other loss-of-instability mutations of Ac can only be sorted out from the much more numerous excision footprints by molecular tests, so neither their types nor frequencies have been well documented to date.
Here we report a high frequency of Ac mutations at the bz-m39(Ac) allele, including a much higher frequency of mutation to Ds than reported previously: 0.9 × 10−3 on the female side and 3.7 × 10−3 on the male side. These two frequencies differ very significantly from each other (χ2 = 11.1, 1 df, P < 0.001), indicating that Ac mutates more frequently in the male than in the female germline. The same is true for Ac excisions, which are 1.8 times more abundant in the male (9.6%) than in the female (5.4%). However, the types of mutation generated in the two sexes do not seem to differ.
The bz-m2(Ac) and bz-m39(Ac) alleles arose in the same Bz-McC haplotype (Fu et al., 2001), providing us with the opportunity to compare the frequency of Ac excision from two different locations of the same Bz allele and in the same genetic background. Ac is inserted in the 5′ UTR in bz-m39(Ac) versus the second exon in bz-m2(Ac) (Fedoroff et al., 1984). Ac excision from both alleles generates Bz′ and bz-s derivatives, although the Bz′:bz-s ratio in bz-m2(Ac) is much lower (0.48), because most Ac excisions fail to restore gene function. The combined frequency of Bz′ and bz-s derivatives from bz-m2(Ac)/bz heterozygous female parents in an experiment involving 5650 bz-m2 gametes was 2.1% (Dooner and Belachew, 1989), less than one-half of that from bz-m39(Ac)/bz heterozygotes in a similar sized experiment (Table 1). The difference between these two frequencies is highly significant (χ2 = 73; 1 df; P < 0.001), indicating that the location of Ac within the gene affects its transposition frequency. Most likely, the proximity to the bz promoter accounts for the enhanced transposition of Ac39.
Estimates of spontaneous mutation frequencies in different organisms range from 10−5 to 10−6, 1000-fold lower than the Ac mutation frequency, suggesting that the DSBs caused by Ac transpositions play an important role in the formation of the mutations. Furthermore, in somatic tissues of transgenic tobacco, only Ac, but not a nearly identical Ds element, is capable of generating internal deletions, indicating that their formation is transposition-dependent (Rubin and Levy, 1997). In the mechanisms for the formation of different Ac mutations proposed below, all Ac mutations are accompanied by transposition events. DNA ends produced by Ac excision are often ligated without the need for homology (NHEJ pathway). More rarely, they are repaired via an error-prone DNA synthesis pathway (Yan et al., 1999) that shares features with a synthesis-dependent microhomology-mediated end joining (MMEJ) pathway recently proposed to explain DSB repair in Drosophila (Yu and McVey, 2010).
Models for Ac Mutations
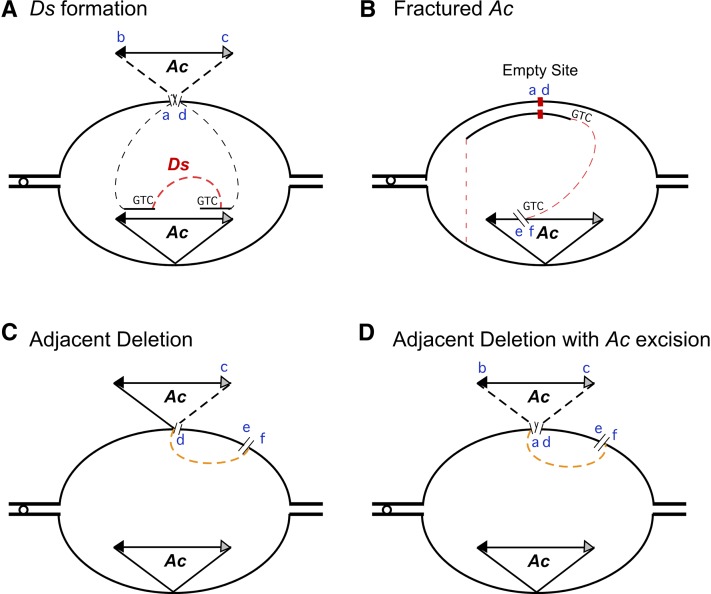
Nearly all (70 of 72) stable bz derivatives from bz-m39(Ac) can be grouped into one of two general categories: either with or without rearrangements of transposon sequences. The former include internal and terminal deletions of Ac and constitute formal mutations of Ac. The most frequent mutations of Ac are internal deletions, most of which behave as Ds elements. More than two-thirds (68.5%) of Ac mutations fall under this category. Out of the 49 newly formed Ds elements, 45 (90%) have 2- to 9-bp microhomologies at the deletion junction, suggesting an origin by a mechanism involving strand invasion and synthesis from both ends, followed by MMEJ and, therefore, deletion of the sequence between the two sites (Figure 4). As illustrated in Figure 9A, repair of the DSB caused by Ac excision would be initiated by strand invasion from both ends to begin copying the Ac sequence in the sister chromatid. MMEJ between two short direct repeats (GTC in the example in Figure 9A) results in the deletion of one repeat and the sequence between the two repeats, yielding a new Ds element.
Figure 9.
Models for the Origin of the Various Ac Mutations.
The diagrams illustrate DNA replication bubbles during chromosome replication and the aberrant repair of Ac-induced DSBs that leads to the formation of Ac mutations. The TE 5′ ends are represented as solid arrowheads, and the 3′ ends are represented as hatched ones. In each transposition reaction, the Ac transposase makes three cuts, one at each TE end to be mobilized and one at the receptor site, and generates six cut ends (a to f), as follows: a, host DNA adjacent to TE 5′ end; b, TE 5′ end; c, TE 3′ end; d, host DNA adjacent to TE 3′ end; e and f, host target site. The GTC trinucleotide sequences represent sites of microhomology in (A) and (B). Red dotted lines represent synthesis-dependent MMEJ–mediated ([A] and [B]) mutational events, and orange dotted lines represent NHEJ–mediated ([C] and [D]) mutational events. The dotted lines in the Ac triangle ([A] and [D]) indicate discontinuity between Ac and the host chromosome.
(A) DSB repair of the Ac excision site is initiated with strand invasion of the sister chromatid by the a and d cut ends to begin copying Ac. MMEJ between the two GTC repeats leads to the formation of an internally deleted Ds element.
(B) In an aborted Ac transposition to the Ac element in the sister chromatid, the f end of the receptor site cut invades the sister chromatid at a site of microhomology (GTC) near the NHEJ-repaired a to d excision site. DNA repair synthesis and eventual return to the original chromatid result in the formation of a fractured Ac and a duplication of Ac-adjacent DNA.
(C) An aberrant transposition resulting from noncleavage or rejoining of the a to b site leads to the formation of a typical c to f transposition junction, but loss of the fragment between Ac and the e to f receptor site.
(D) An aberrant ligation of the host a and f cut ends at the excision and receptor sites, respectively, leads to loss of Ac and the fragment between Ac and the receptor site.
More than one-third of newly formed Ds39 elements (17 of 49) have filler DNA between the deletion endpoints (Figure 5). The presence at the deletion junctions of filler DNAs from nearby sequences suggests that the filler insertions are templated. They likely arise from misannealing to the sister chromatid during repair DNA synthesis. Multiple cycles of strand invasion occur during repair of the DSB: a short repair synthesis tract dissociates from its template, probes for its complementary sequences, and reinvades a template for strand extension when a complementary sequence is not found (McVey et al., 2004). When nascent DNA misanneals at a microhomology site, DNA synthesis would slow down, promoting the activity of alternative end joining DNA ligase I (Liang et al., 2008; Huefner et al., 2011), which ligates the two partially synthesized ends. In Drosophila, θ DNA polymerase has been speculated to create microhomologies that can be used during the annealing stage of the end joining when suitable microhomologies are not present (Chan et al., 2010). The above model for the formation of deletions and filler DNA is similar to one proposed by Yan et al. (1999) for the formation of new Ds elements. In that model, slip-mispairing during repair DNA synthesis between two short direct repeats would result in the deletion of one repeat and the sequence between the two repeats, yielding a new Ds element. A second slip-mispairing would explain the appearance of filler DNA (Yan et al., 1999), and sequential slips would explain filler DNA originating from multiple locations (Conrad et al., 2007).
A synthesis-dependent strand-annealing pathway was originally proposed to explain the formation of internal deletion derivatives of P after excision (Nassif et al., 1994) and the retention of a single LTR from a copia retrotransposon inserted between the two ends of a P element (Kurkulos et al., 1994). This mechanism has also been proposed to explain the formation of internal deletions of the Mutator transposon in maize (Lisch et al., 1995; Hsia and Schnable, 1996). However, the original synthesis-dependent strand-annealing model does not readily explain the frequent occurrence of filler DNA at the Ds and fAc deletion junctions reported here.
Terminal deletions of Ac or fAc elements are the second most abundant class of bz-s derivatives. Nine such alleles were recovered in our study, accounting for 15% of Ac mutations. The three instances of fAc elements reported in the literature retained the terminal 1.5- to 2.5-kb 3′ portion of Ac (Ralston et al., 1989; Zhang and Peterson, 1999; Conrad et al., 2007). Six of the nine fAcs recovered in this experiment also retained the 3′ end, but three retained the 5′ end instead, indicating that either end of Ac can be terminally deleted, although the 5′ end seems to be deleted more frequently. Four alleles have short host sequence duplications of 4 to 62 bp, whereas two others have larger adjacent deletions of either side. Both types can be explained by the model presented in Figure 9B. We propose that a fAc arises from an attempted, but failed, transposition to the Ac element on the sister chromatid, followed by microhomology-mediated repair synthesis of the DSB at the intended target site. There is no Ac template on the sister chromatid; therefore, repair of the DSB may begin with one end invading the sister chromatid at a site of microhomology near the Ac excision site, followed by DNA synthesis and eventual return to the original chromatid. This would give rise to a fractured or terminally deleted Ac and either a duplication or deletion of adjacent DNA, depending on the location of the microhomology relative to the empty site. If the proposed transposition to the Ac element in the sister chromatid had resolved normally, a double Ac element would have resulted. Alleles carrying double Ac elements, such as o2-m55 (Michel et al., 1994), produce mutable phenotypes, so a similar derivative of bz-m39 would not have been selected in our experiment.
Of the few stable bz derivatives without rearrangements of transposon sequences, the most numerous are Ac-adjacent deletions. Transposon-adjacent deletions are common in bacteria and were first described for IS1 by Reif and Saedler (1975). Seven such alleles were recovered in our study, accounting for less than 10% of the bz-s mutations. Five of them retained Ac at the original location. They most likely arose from an incomplete Ac transposition of only one end of the transposon to a nearby site that results in the loss of sequences between that site and the original insertion site (Figure 9C). As in bz-s:2114(Ac) (Dooner et al., 1988), Ac is not flanked by an 8-bp TSD in any of these five bz-s alleles. Peculiarly, although adjacent inversions occur as frequently as adjacent deletions in bacteria, none were recovered in this study. Two alleles have adjacent deletions accompanied by an Ac excision. Such deletions constitute the most common type of excision product of the P element in Drosophila (Engels, 1989). They likely arose from an aborted transposition event to a nearby site followed by NHEJ (Figure 9D). The transposase cleaves at the original insertion site and the potential reinsertion site; however, the transposon fails to reinsert, and ligation of the two cut sites via NHEJ leads to loss of the sequences between them.
Identification of Additional Requirements for Ac Transposition
The analysis of loss-of-function derivatives from bz-m39(Ac) has uncovered previously unknown trans and cis requirements for Ac transposition. In derivative bz-m39.55, Ac has suffered a tiny 3-bp deletion that removes a Ser at position 305 and renders the transposase almost inactive, indicating that Ser-305 is important for activity. This residue is well conserved among hAT transposases of the Ac/Tam3 clade (Robertson, 2002) and is found in some nonplant transposases, such as the human Tramp and the fungal restless clade (see Supplemental Figures 3A and 3B and Supplemental Data Set 1 online). The 4562-bp element in bz-m39.55 transposes very rarely by itself, but normally in the presence of Ac, so only its trans function has been affected by the deletion.
In derivative bz-s39.24, Ac has undergone a 7-bp deletion of nucleotides 22 to 28 (AAAATCC) in the 5′ subterminal region that prevents it from transposing. It can still encode a functional transposase, behaving as an Ac-im (Conrad and Brutnell, 2005). The 11-bp TIR and multiple copies of the transposase binding A/TCGG subterminal repeat were known to be essential for excision (Kunze and Starlinger, 1989; Becker and Kunze, 1997; Kunze and Weil, 2002), but the deleted sequence is not a part of either. Thus, the analysis of the nontransposing element in bz-s39.24 identifies a new cis requirement for Ac transposition, which could be either the deleted sequence itself or simply a change in the spacing between the TIR and the subterminal repeats where the transposase binds.
METHODS
Genetic Stocks
All the stocks used in this study shared the common genetic background of the inbred W22. The bronze alleles and the aleurone phenotypes of the various stocks are described below.
bz-m39(Ac) (heavy purple spots on a bronze background): a mutable allele harboring an Ac element in the 5′ UTR of the Bz-McC allele, at a position 39 to 32 bp upstream of the start codon. The 8-bp span refers to the location of the bz TSD on either side of Ac. The simple transposon excisions from this site restore gene function. This allele was recovered from a reinsertion of Ac2094 into the bz locus in the same experiment described by Yan et al. (1999) for the isolation of bz-m41 and bz-m43.
Bz-McC (purple): the normal progenitor allele of the bz-m39(Ac) and bz-m2(Ac) mutations.
bz-m2(Ac) (purple spots on a bronze background): an allele that arose from the insertion of the 4.6-kb Ac element at position 755 to 762 in the second exon of Bz-McC (McClintock, 1955; Ralston et al., 1988). Most transposon excisions from this site fail to restore gene function (Dooner and Belachew, 1989).
bz-m2(D1) (bronze in the absence of Ac, spotted in its presence): the first derivative from bz-m2(Ac), harboring a 3.3-kb internally deleted Ds element at the same position as Ac in bz-m2(Ac) (McClintock, 1962; Dooner et al., 1986).
sh-bz-X2 (shrunken, bronze): an x-ray–induced deletion of a large chromosomal fragment that includes sh, bz, and other loci between sh and bz, such as stc1 (Mottinger, 1973; Fu et al., 2001).
wx-m7(Ac) (waxy endosperm with sectors of nonwaxy revertant tissue): an unstable wx allele described by McClintock (1964). It arose by insertion of the 4.6-kb Ac element in the 5′ UTR of the Wx gene (Müller-Neumann et al., 1984; Klösgen et al., 1986).
Selection and Analysis of Bz′ and bz-s Derivatives
Bz′’ and bz-s derivatives were isolated from testcrosses of Sh bz-m39(Ac) wx/sh-bz-X2 wx heterozygotes to sh-bz-X2 wx. In these crosses, the two parental kernel classes are plump, spotted and shrunken, bronze. sh and bz cannot recombine in the hemizygous deletion parent; therefore, exceptional plump, purple (Bz′) kernels represent Bz′ derivatives from bz-m39(Ac), and plump, stable bronze (bz-s) kernels represent bz-s derivatives. bz-m39(Ac) as female parent produces very heavily spotted kernels, making it hard to differentiate purple from spotted kernels. Therefore, the whole population was planted, and those seedlings with solid purple coleoptiles were selected as Bz′ derivatives. bz-s derivatives were first screened by three diagnostic PCR reactions (as described in Results) to identify different types of deletion mutations and were then crossed to wx-m7(Ac) to determine the mobility of the element. A selected group was crossed to bz-m2(D1) to score for Ac activity (see Results).
DNA Extraction, Blotting, and Hybridization
Leaf DNA for DNA gel blot hybridization was isolated by a urea extraction procedure (Greene et al., 1994). A modified cetyltrimethylammonium bromide extraction method (Huang and Dooner, 2008) was used for the large number of DNA preparations required for PCR and sequencing analysis. Restriction digested DNA (10 μg) was resolved on 0.8% agarose gels and transferred to Hybond XL nylon membranes (Amersham Biosciences). 32P-labeled probes were generated with Ready-To-Go DNA labeling beads (Amersham Biosciences). The 740-bp Ac probe was amplified from a genomic clone using primers in exon 2 (CACACTGGCCAAAGGTTATCACA) and exon 3 (TCATTGCAACGGCCATTCTCCTAA) of the Ac transposase gene.
PCR, Inverse PCR, and Sequencing
PCR was performed according to the protocol of QiaTaq (Qiagen). Long-strand DNA fragment amplification was performed according to the protocol of Roche Expand Long Range (Hoffmann-La Roche). PCR products were cloned into pGEM-T Easy Vector (Promega) and transformed into XL-Blue competent cells. Plasmids were purified with a Qiagen Spin Miniprep Kit. DNA sequencing of plasmids or PCR products was performed in an ABI 3730 sequencer (Perkin-Elmer) following the manufacturer’s instructions. Primers used in this work are shown in Supplemental Table 1 online.
Phylogenetic Analysis
The Ac sequence was queried against the GenBank nr protein databases, excluding searches from Zea. Sample hits from different species were chosen for further analysis. Some hAT transposons of the more distant groups (Robertson, 2002; Xu and Dooner, 2005) were also incorporated. To reduce the redundancy within the data, sequences with a high degree of similarity to another sequence from the same species were eliminated so that only one of them was included in the comparison. The full-length amino acid sequences were aligned by Clustal Omega (http://www.clustal.org/omega/). A phylogenetic tree was constructed using neighbor joining in MEGA version 5.05 (http://www.megasoftware.net/), with 1000 bootstrap replicates and the pairwise-deletion option for handling gaps.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Ac sequence (GenBank X05425), McC bz haplotype contig (GenBank AF391808), bz-m39(Ac) and its bz derivatives (GenBank JX910919 to JX910943).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Excision Footprints among Bz′ Revertants Recovered from Female and Male Gametes.
Supplemental Figure 2. bz-m39.31 Phenotype.
Supplemental Figure 3. Sequence Conservation among hAT Transposases.
Supplemental Figure 4. Methylation of Inactive Ac in bz-c39.27.
Supplemental Table 1. Primers Used in This Work.
Supplemental Data Set 1. Text File of Alignment Corresponding to Phylogenetic Tree in Supplemental Figure 3 Online.
Supplementary Material
Acknowledgments
We thank Yubin Li and Qinghua Wang for comments on the article and Yubin Li for part of Figure 1. This research was supported by a Charles and Johanna Busch Predoctoral Fellowship and a Busch-Waksman Postdoctoral Fellowship to J.T.H. and by National Science Foundation grant DBI-0929350 to H.K.D.
AUTHOR CONTRIBUTIONS
J.T.H. and H.K.D. designed the work, performed the research, analyzed the data, and wrote the article.
Glossary
- TE
transposable element
- TIR
terminal inverted repeat
- DSB
double-strand break
- TSD
target site duplication
- NHEJ
nonhomologous end joining
- UTR
untranslated region
- LTR
long terminal repeat
- MMEJ
microhomology-mediated end joining
References
- Becker H.A., Kunze R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254: 219–230 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Dellaporta S.L. (1994). Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.H., Yu A.M., McVey M. (2010). Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6: e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet P.S., Wessler S., Dellaporta S.L. (1987). Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad L.J., Bai L., Ahern K., Dusinberre K., Kane D.P., Brutnell T.P. (2007). State II dissociation element formation following activator excision in maize. Genetics 177: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad L.J., Brutnell T.P. (2005). Ac-immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of Dissociation elements in maize. Genetics 171: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland G., Baker B., Schell J., Starlinger P. (1988). Characterization of the maize transposable element Ac by internal deletions. EMBO J. 7: 3653–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland G., Plum C., Chatterjee S., Post A., Starlinger P. (1989). Sequences near the termini are required for transposition of the maize transposon Ac in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 86: 9385–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M., Park W., Xu Z., Yan X., Maurais S.C., Dooner H.K. (2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Belachew A. (1989). Transposition pattern of the maize element Ac from the bz-m2(Ac) allele. Genetics 122: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., English J., Ralston E.J. (1988). The frequency of transposition of the maize element Activator is not affected by an adjacent deletion. Mol. Gen. Genet. 211: 485–491 [DOI] [PubMed] [Google Scholar]
- Dooner H.K., English J., Ralston E., Weck E. (1986). A single genetic unit specifies two transposition functions in the maize element activator. Science 234: 210–211 [DOI] [PubMed] [Google Scholar]
- Dooner H.K., Weil C.F. (2012). Transposons and gene creation. In Molecular Genetics and Epigenetics of Plant Transposons: Sculpting Genes and Genomes, N.V. Fedoroff, ed (Hoboken, NJ: John Wiley & Sons), in press
- Engels W.R. (1989). P elements in Drosophila melanogaster. In Mobile DNA, D. Berg and M. Howe, eds (Washington, D.C.: American Society for Microbiology Press), pp. 437–484
- Fedoroff N.V., Furtek D.B., Nelson O.E., Jr(1984). Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc. Natl. Acad. Sci. USA 81: 3825–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Park W., Yan X., Zheng Z., Shen B., Dooner H.K. (2001). The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98: 8903–8908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B., Walko R., Hake S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl R., Baker B. (1989). Induced transposition of Ds by a stable Ac in crosses of transgenic tobacco plants. Mol. Gen. Genet. 217: 53–59 [DOI] [PubMed] [Google Scholar]
- Hsia A.P., Schnable P.S. (1996). DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics 142: 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T., Dooner H.K. (2008). Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell 20: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huefner N.D., Mizuno Y., Weil C.F., Korf I., Britt A.B. (2011). Breadth by depth: Expanding our understanding of the repair of transposon-induced DNA double strand breaks via deep-sequencing. DNA Repair (Amst.) 10: 1023–1033 [DOI] [PubMed] [Google Scholar]
- Klösgen R.B., Gierl A., Schwarz-Sommer S., Saedler H. (1986). Molecular analysis of the waxy locus of Zea mays. Mol. Gen. Genet. 203: 237–244 [Google Scholar]
- Kunze R., Starlinger P. (1989). The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8: 3177–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R., Weil C.F. (2002). The hAT and CACTA superfamilies of plant transposons. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: American Society for Microbiology Press), pp. 565–610
- Kurkulos M., Weinberg J.M., Roy D., Mount S.M. (1994). P element-mediated in vivo deletion analysis of white-apricot: Deletions between direct repeats are strongly favored. Genetics 136: 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Deng L., Nguyen S.C., Zhao X., Maulion C.D., Shao C., Tischfield J.A. (2008). Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 36: 3297–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., Chomet P., Freeling M. (1995). Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics 139: 1777–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. (1949). Mutable loci in maize. Carnegie Inst. Wash. Yrbk. 48: 142–154 [Google Scholar]
- McClintock B. (1955). Controlled mutation in maize. Carnegie Inst. Wash. Yrbk. 54: 245–255 [Google Scholar]
- McClintock B. (1956). Mutation in maize. Carnegie Inst. Wash. Yrbk. 55: 323–332 [Google Scholar]
- McClintock B. (1962). Topographical relations between elements of control systems in maize. Carnegie Inst. Wash. Yrbk. 61: 448–461 [Google Scholar]
- McClintock B. (1963). Further studies of gene-control systems in maize. Carnegie Inst. Wash. Yrbk. 62: 486–493 [Google Scholar]
- McClintock B. (1964). Aspects of gene regulation in maize. Carnegie Inst. Wash. Yrbk. 63: 592–602 [Google Scholar]
- McVey M., Adams M., Staeva-Vieira E., Sekelsky J.J. (2004). Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D., Salamini F., Motto M., Döring H.-P. (1994). An unstable allele at the maize Opaque2 locus is caused by the insertion of a double Ac element. Mol. Gen. Genet. 243: 334–342 [DOI] [PubMed] [Google Scholar]
- Mottinger J.P. (1973). Unstable mutants of bronze induced by premeiotic X-ray treatment in maize. Theor. Appl. Genet. 43: 190–195 [DOI] [PubMed] [Google Scholar]
- Müller-Neumann M., Yoder J.I., Starlinger P. (1984). The DNA sequence of the transposable element Ac of Zea mays L. Mol. Gen. Genet. 198: 19–24 [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W.R., Gloor G.B. (1994). Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlman R.F., Fedoroff N.V., Messing J. (1984). The nucleotide sequence of the maize controlling element Activator. Cell 37: 635–643 [DOI] [PubMed] [Google Scholar]
- Ralston E.J., English J., Dooner H.K. (1989). Chromosome-breaking structure in maize involving a fractured Ac element. Proc. Natl. Acad. Sci. USA 86: 9451–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E.J., English J.J., Dooner H.K. (1988). Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics 119: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif H.J., Saedler H. (1975). IS1 is involved in deletion formation in the gal region of E. coli K12. Mol. Gen. Genet. 137: 17–28 [DOI] [PubMed] [Google Scholar]
- Robertson H.M. (2002). Evolution of DNA transposons in eukaryotes. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: American Society for Microbiology Press), pp. 1093–1110
- Roth D.B., Chang X.B., Wilson J.H. (1989). Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 9: 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B., Wilson J.H. (1985). Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc. Natl. Acad. Sci. USA 82: 3355–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E., Levy A.A. (1997). Abortive gap repair: Underlying mechanism for Ds element formation. Mol. Cell. Biol. 17: 6294–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsard-Chanet A., Begel O. (1990). Insertion of an LrDNA gene fragment and of filler DNA at a mitochondrial exon-intron junction in Podospora. Nucleic Acids Res. 18: 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D., Dennis E. (1986). Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol. Gen. Genet. 205: 476–482 [Google Scholar]
- Scott L., LaFoe D., Weil C.F. (1996). Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142: 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S., Tarpley A., Purugganan M., Spell M., Okagaki R. (1990). Filler DNA is associated with spontaneous deletions in maize. Proc. Natl. Acad. Sci. USA 87: 8731–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.L., Peterson T. (2002). Ac transposition is impaired by a small terminal deletion. Mol. Genet. Genomics 266: 720–731 [DOI] [PubMed] [Google Scholar]
- Xu Z., Dooner H.K. (2005). Mx-rMx, a family of interacting transposons in the growing hAT superfamily of maize. Plant Cell 17: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Martínez-Férez I.M., Kavchok S., Dooner H.K. (1999). Origination of Ds elements from Ac elements in maize: Evidence for rare repair synthesis at the site of Ac excision. Genetics 152: 1733–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.M., McVey M. (2010). Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 38: 5706–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T. (1999). Genome rearrangements by nonlinear transposons in maize. Genetics 153: 1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T. (2004). Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167: 1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.