This work identifies NONRECOGNITION OF BTH4 (NRB4), which acts in SA signaling in defense and development, finding that NRB4 encodes a predicted subunit of the Mediator complex, which connects specific transcription factors to the general transcription machinery. NRB4 functions downstream of NONEXPRESSER OF PATHOGENESIS-RELATED1in SA signaling.
Abstract
Salicylic acid (SA) signaling acts in defense and plant development. The only gene demonstrated to be required for the response to SA is Arabidopsis thaliana NON-EXPRESSER OF PATHOGENESIS-RELATED GENE 1 (NPR1), and npr1 mutants are insensitive to SA. By focusing on the effect of analogs of SA on plant development, we identified mutants in additional genes acting in the SA response. In this work, we describe a gene necessary for the SA Non-Recognition-of-BTH4 (NRB4). Three nrb4 alleles recovered from the screen cause phenotypes similar to the wild type in the tested conditions, except for SA-related phenotypes. Plants with NRB4 null alleles express profound insensitivity to SA, even more than npr1. NRB4 null mutants are also sterile and their growth is compromised. Plants carrying weaker nrb4 alleles are also insensitive to SA, with some quantitative differences in some phenotypes, like systemic acquired resistance or pathogen growth restriction. When weak alleles are used, NPR1 and NRB4 mutations produce an additive phenotype, but we did not find evidence of a genetic interaction in F1 nor biochemical interaction in yeast or in planta. NRB4 is predicted to be a subunit of Mediator, the ortholog of MED15 in Arabidopsis. Mechanistically, NRB4 functions downstream of NPR1 to regulate the SA response.
INTRODUCTION
Plants mount several types of resistance against different pathogens. Some types of defense consist of preexisting barriers, and others are inducible. The hormone salicylic acid (SA) is key for inducible defenses against biotrophic pathogens (as reviewed by Vlot et al., 2009). Upon pathogen perception, SA biosynthesis is increased, which induces the appropriate defense responses. In addition, other hormones are involved in plant defense, including jasmonic acid (JA) and ethylene, and there are complex interactions among hormone responses (reviewed in Robert-Seilaniantz et al., 2011). Although methyl jasmonate (MeJA) is applied exogenously in the laboratory, it has been shown that the active form is JA-Ile (reviewed in Browse, 2009). Broadly speaking, JA-Ile and ethylene act synergistically, but SA and JA-Ile negatively regulate each other.
A key player in the recognition of SA is NON-EXPRESSER OF PATHOGENESIS-RELATED GENE 1 (NPR1) (reviewed in Dong, 2004). Various genetic screens aiming to identify components of the SA response have exclusively found mutations in NPR1 (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997), suggesting that it is the only gene responsible for the SA response or the only one accessible through mutagenesis. A search for protein–protein interactions in yeast identified components that interact with NPR1, including NIM1-INTERACTING PROTEIN1 (Weigel et al., 2001) and TGAs (for TGACG motif binding factor; Zhang et al., 1999; Després et al., 2000). NPR1 is sensitive to SA in yeast, activating the expression of genes in a stimulus-dependent fashion (Maier et al., 2011), and it has been defined as an SA receptor, either binding to SA (Wu et al., 2012) or interacting with two paralogs, NPR3 and NPR4, which bind SA (Fu et al., 2012).
Benzothiadiazole (BTH) is an analog of SA and is used in the laboratory because it is not as phytotoxic as SA (Lawton et al., 1996). Repeated applications of BTH decrease the size and weight of treated plants (Canet et al., 2010a); this difference was used to screen for nonrecognition of BTH mutants (NRBs). The first complementation group derived from this screen, not surprisingly, was NPR1 (Canet et al., 2010b). The next complementation group, Non-Recognition-of-BTH4 (NRB4), is the focus of this article.
As mentioned, SA is central to pathogen-induced responses, and many of these responses involve alterations in gene expression. Some of these changes in gene expression are regulated by DNA repair proteins (Song et al., 2011), identified as suppressors of npr1, and by chromatin remodeling factors (Wang et al., 2010), identified as suppressors of the suppressors. There are also sets of transcription factors, notably TGAs (Jakoby et al., 2002) and WRKYs (named after the conserved domain WRKYGQK; Eulgem et al., 2000), that act downstream of the SA response to regulate gene expression leading to defense. However, the mechanism by which these specific transcription factors sets interact with RNA Pol II remains unclear.
In yeast, the Mediator complex functions as a bridge between specific transcription factors and the core transcriptional machinery (Kelleher et al., 1990; Flanagan et al., 1991). Mediator is a complex of ∼22 proteins, divided in four modules: head, middle, tail, and a detachable kinase domain. The tail module interacts with the specific transcription factors, and the head module interacts with RNA Pol II (Cai et al., 2009). The Mediator complex has been found in all eukaryotes tested (Chadick and Asturias, 2005), including in Arabidopsis thaliana (Bäckström et al., 2007). Indeed, several reports of Mediator subunits of Arabidopsis affecting a specific signaling process have appeared (reviewed in Kidd et al., 2011).
The plants with nrb4 missense mutations we identified in the screen are only affected in SA response, not in other tested phenotypes; however, plants with null mutations in NRB4 express severe defense and developmental phenotypes. NRB4 is predicted to be a subunit of the Mediator complex located in the tail module, and in this work, we show that the missense mutations are clustered in the KIX domain (named due to its interaction with the kinase inducible activation domain of CREB; Chrivia et al., 1993). Importantly, the orthologs in other species interact with different receptors, and some of these receptors bind salicylates. From the phenotypes presented, we infer an essential role for NRB4 in plants. This essential function could reflect a role for SA in normal development, as previously suggested (Vanacker et al., 2001).
RESULTS
NRB4 Is Required for the SA Response
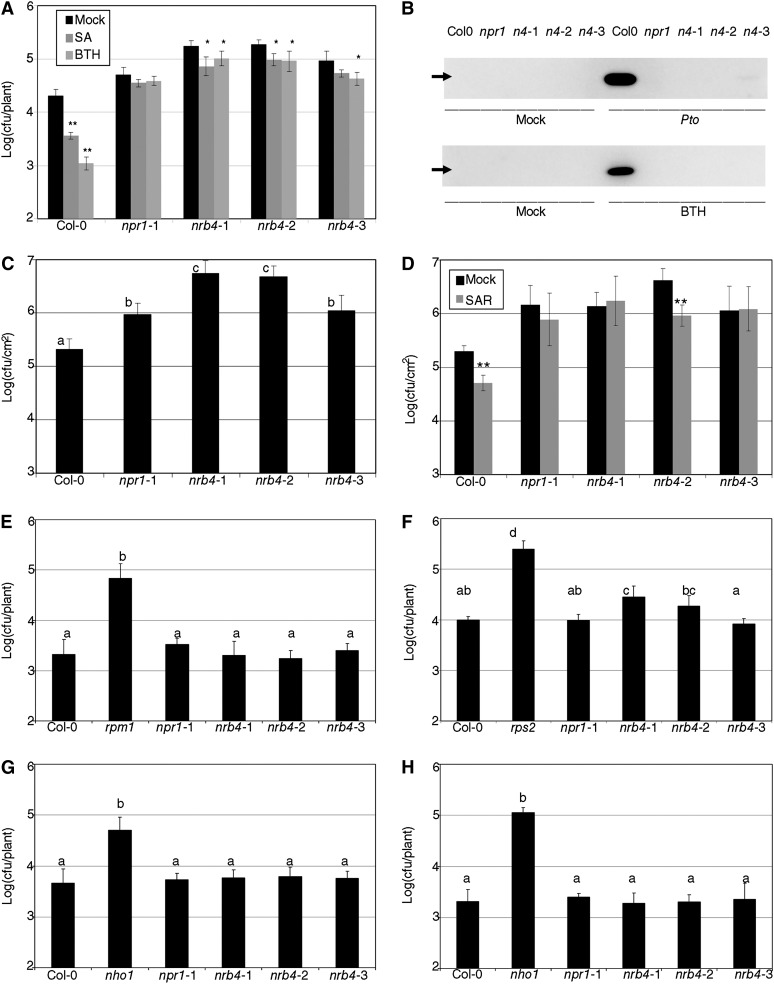
We previously performed a genetic screen for genes involved in the SA response; the first locus we found was NPR1, with 43 alleles (Canet et al., 2010b). The fourth locus (by number of alleles) from this screen was named NRB4, and the rest of the loci will be described elsewhere. NRB4 is defined by three alleles, which came from independent ethyl methanesulfonate (EMS) mutagenesis events. The plants with these alleles were almost as insensitive to BTH as npr1-1 plants, at least in terms of diminished fresh weight when grown in the presence of BTH (Figure 1A, the letters above the bars in the figures indicate different homogeneous groups with statistically significant differences [Fisher’s LSD test, P < 0.05]). The nrb4-1 plants had a less severe phenotype than the nrb4-2 and nrb4-3 plants (Figure 1A). The mutants were recessive (Figure 1A; see Supplemental Table 1 online), although in both nrb4-2 and nrb4-3, F1s with Columbia-0 (Col-0) showed an effect on the SA response, as happens with the F1s with npr1-1 (Figure 1A). There was no genetic interaction with npr1 in the F1, and the F1s between the nrb4 alleles were as insensitive to BTH as their parents (Figure 1A).
Figure 1.
SA-Related Phenotypes of nrb4 Plants.
(A) Plants were treated with either mock or 350 µM BTH four times over 3 weeks, their weights recorded, and the ratio between the BTH and mock-treated plants calculated (15 plants in three groups of five). The ratio is expressed as percentage of fresh weight (%FW).
(B) Plants were grown on Murashige and Skoog plates supplied with 0, 400, and 500 µM SA; the picture shows the 500 µM SA plate at day 14.
(C) The chlorophyll content of plants growing in the plates described in (B) was measured as an indication of the response to SA (30 plants in three groups of 10). The experiments were repeated three times with similar results, and the data represent the average, with the error bars plotting the sd. The letters above the bars indicate different homogeneous groups with statistically significant differences (Fisher’s LSD test, P < 0.05). In (C), the differences were evaluated between genotypes grown at either 400 µM SA or 500 µM SA (marked with a prime symbol).
[See online article for color version of this figure.]
The screen and the quantification of the fresh weight were performed with BTH. It was possible that these nrb4 plants were impaired in response to BTH but had no effect on the response to SA. To test this possibility, nrb4 plants and controls were grown on Murashige and Skoog plates with 500 µM SA. npr1 plants are unable to grow on these plates, likely because they are unable to detoxify SA (Cao et al., 1997). nrb4-2 and nrb4-3 plants behaved as npr1-1, and nrb4-1 was intermediate between Col-0 and npr1-1 (Figure 1B). This observation was quantified by measuring the amount of chlorophyll per plant for three different treatments (Figure 1C). The quantification corroborated the intermediate phenotype of nrb4-1 plants and showed no difference in chlorophyll in mock treatments. In fact, in the absence of treatment, there were no observable developmental phenotypes in the plants carrying any of the nrb4 alleles; they were indistinguishable from Col-0 in our growth conditions.
The nrb4 plants, like npr1, were also affected in SA-dependent defense (Figure 2). For example, inoculation with Pseudomonas syringae pv tomato strain DC3000 (Pto) after SA and BTH treatments induced a strong resistance in Col-0, but not in npr1-1 (Figure 2A). The nrb4 plants showed some residual resistance, but the difference with respect to Col-0 was always considerable. PR1 is a pathogenesis-related protein used as a marker for stress in plants (Wang et al., 2005), so we produced a PR1 immunoblot blot of plants treated with Pto or BTH (Figure 2B). In both cases, strong accumulation of PR1 was observed only in Col-0. Therefore, even if SA and BTH induce a small amount of resistance in nrb4 plants, this resistance does not result in the accumulation of PR1.
Figure 2.
Pathogenic Phenotypes of nrb4 Plants.
(A) Seventeen-day-old plants were treated with either 500 µM SA, 350 µM BTH, or a mock solution. One day later, the plants were inoculated with Pto at an OD600 of 0.1. Three days after inoculation, the growth of Pto was evaluated as logarithm of colony-forming units per plant.
(B) PR1 immunoblot of the indicated genotypes 3 d after mock or a Pto inoculation (top) and 1 d after mock or 350 µM BTH treatment (bottom). The genotypes are abbreviated as in (A). The arrow indicates the position of PR1 (14 kD).
(C) Plants (32 days old) were treated with Pto as in (A). In these plants, only a sample of the surface area was measured, so the units are log(cfu/cm2).
(D) Three leaves of 30-d-old plants were hand-infiltrated with either Pto(avrRpm1) or a mock solution. Two days later, Pto was inoculated and its growth in systemic leaves measured as in (C).
(E) Pto(avrRpm1) was inoculated in nrb4 as in (A). Resistance to Pseudomonas maculicola 1 (rpm1) is included as a control.
(F) Inoculations with Pto(avrRpt2), with rps2 added as a control.
(G) P. syringae pv phaseolicola isolate NPS3121 was inoculated as in (A), with nho1 used as a control.
(H) Inoculations with P. syringae pv tabaci was inoculated as in (A), with nho1 used as a control.
The experiments were repeated three times with similar results, and the data represent the average, with the error bars plotting the sd of 15 plants in three groups of five in (A) and (E) to (H). In (C) and (D), 12 samples of known size from three plants were taken in three groups of four. The letters above the bars indicate different homogeneous groups with statistically significant differences (Fisher’s LSD test, P < 0.05). Asterisks indicate statistically significant differences from the mock treatment (*P < 0.05; **P < 0.01) using the Student’s t test (one tail).
The similarities between nrb4 and npr1 plants extended beyond the initial characterization. When tested for enhanced disease susceptibility phenotypes (Glazebrook et al., 1996), nrb4 plants were at least as susceptible as npr1 (Figure 2C). Surprisingly, nrb4-2 plants were wild type for pathogen-induced systemic acquired resistance (SAR), but the plants with the other two alleles, like all npr1 plants, were SAR defective (Figure 2D; Cao et al., 1994; Delaney et al., 1995). Since this was an important difference, this experiment was repeated several times, always with the same result. All nrb4 and npr1 plants showed similar effector-triggered immune responses (Figures 2E and 2F) and responses to nonhost pathogens (Figures 2G and 2H). Only in the case of Resistance to Pseudomonas syringae 2–dependent effector-triggered immunity triggered by Pto(avrRpt2) did we observe a decrease in resistance in some plants; these paralleled the responses to Pto (Figures 2A, 2C, and 2D). Other pathogens tested included Pto(hrpC-) (Deng et al., 1998) and Plectosphaerella cucumerina (Ton and Mauch-Mani, 2004). In these cases, the nrb4 plants were not different from the wild type (see Supplemental Figures 1A and 1B, respectively, online).
Plants with npr1 alleles differ in their response to MeJA-induced resistance (Dobón et al., 2011). nrb4 plants were wild type in their response to MeJA (see Supplemental Figure 1C online) and also showed a wild-type phenotype on MeJA plates (see Supplemental Figure 1D online) and in growth of Pto(cor-) (Mittal and Davis, 1995; see Supplemental Figure 1E online).
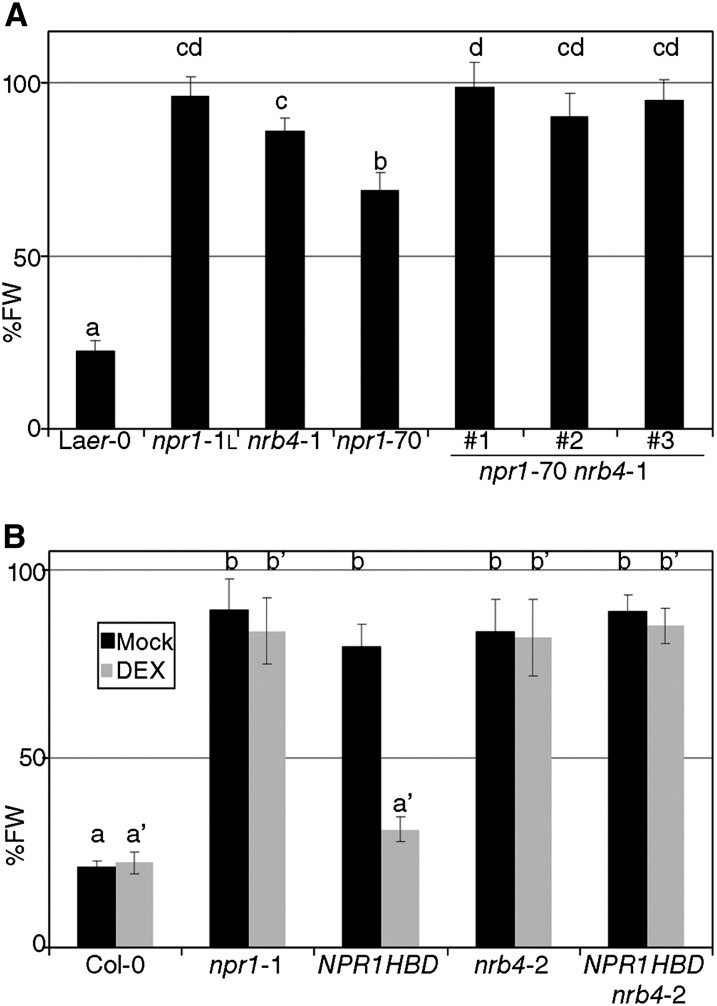
nrb4 and npr1 plants shared most of the phenotypes related to SA-dependent defense and/or response to biotrophic pathogens. Thus, we addressed whether the corresponding genes act in the same pathway by constructing the double mutant nrb4 npr1-1. These double mutant plants showed no additional change in fresh weight in response to BTH (see Supplemental Figure 2A online). Therefore, another line was constructed with npr1-70 (a null allele with an intermediate response to BTH; Canet et al., 2010b) and nrb4-1 since it was the weakest allele (Figure 1A). nrb4-1 npr1-70 plants showed additive phenotypes, since these plants had a stronger phenotype than plants with either weak allele alone (Figure 3A). Similar results were obtained with respect to growth of Pto in response to SA and BTH treatment (see Supplemental Figure 2B online).
Figure 3.
Epistasis of NRB4 with NPR1.
(A) Three double mutants nrb4-1 npr1-70 were tested as in Figure 1A. FW, fresh weight; Laer-0, Landsberg erecta-0.
(B) 35S:NPR1-HBD (NPR1HBD) in an nrb4-2 background was tested with and without dexamethasone (DEX) for its response to BTH. The experiments were repeated three times with similar results, and the data represent the average, with the error bars plotting the sd of 15 plants in three groups of five. The letters above the bars indicate different homogeneous groups with statistically significant differences (Fisher’s LSD test, P < 0.05).
This additive relationship implied that the genes were independent. Mechanistically, this could be translated into several models. NPR1 functions in the nucleus (Kinkema et al., 2000; Maier et al., 2011), and NRB4 could affect its localization and, potentially, that of other proteins acting with NPR1. The trafficking of NPR1 can be manipulated with a transgenic line that overexpresses NPR1 fused to the steroid hormone binding domain of the rat glucocorticoid receptor (NPR1-HBD; Kinkema et al., 2000). Upon application of the glucocorticoid dexamethasone, NPR1-HBD is forced to the nucleus, while in mock conditions, it is excluded from the nucleus. The double nrb4-2 NPR1-HBD plant did not respond to BTH when dexamethasone was applied (Figure 3B). Thus, the presence of NPR1 in the nucleus, in the presence of BTH, was not enough to trigger the response. An alternative explanation for these results could be that NRB4 is a chaperone, required for NPR1 stability. This explanation was ruled out with the help of a line that expressed NPR1 fused to green fluorescent protein (GFP) (Kinkema et al., 2000). In control conditions, NPR1-GFP was detected in the nucleus (see Supplemental Figure 2C online, compare with Figure 2A of Kinkema et al., 2000). The same localization was observed in this transgenic plant in an nrb4-2 background (see Supplemental Figure 2D online). Upon BTH application, NPR1-GFP was also detected in the nucleus, both in NRB4 wild-type and in nrb4-2 backgrounds (see Supplemental Figures 2E and 2F, respectively, online). Therefore, NPR1 was not only stable in nrb4-2 plants, but it is also localized in the nucleus. In spite of this wild-type NPR1 behavior, this line did not respond to BTH (see Supplemental Figure 2G online). Thus, NRB4 functions downstream of NPR1.
Cloning of NRB4 and Phenotypes of Null Alleles
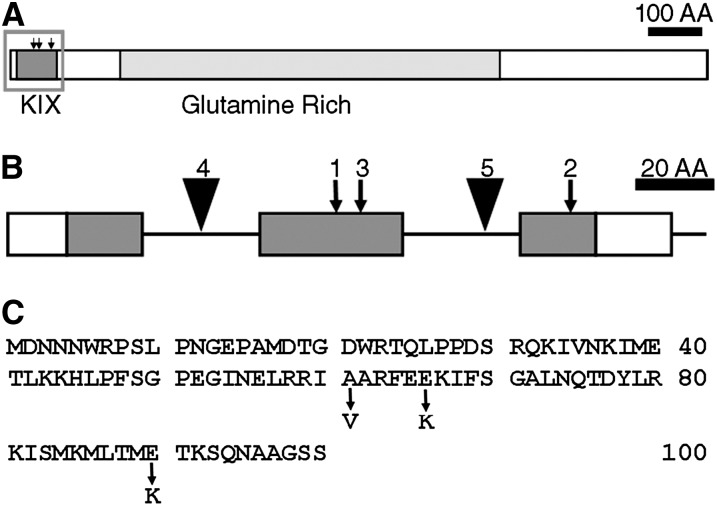
Conventional mapping showed that NRB4 is encoded by At1g15780, a gene labeled as “unknown” in The Arabidopsis Information Resource (version 9; Swarbreck et al., 2008). The predicted protein contains a KIX domain (Radhakrishnan et al., 1997) at the very beginning and a Gln-rich region (Guo et al., 2007) in the middle (Figure 4A). The sequences of the three alleles revealed that each allele had a single point mutation in the KIX domain (Figure 4). The mutations were not extreme in terms of physiochemical distances (Grantham, 1974). In fact, the mutations introduced did not change the prediction of an alpha helix structure (Radhakrishnan et al., 1997). Therefore, mutations that introduced small changes in the protein produced plants with a considerable change in phenotype.
Figure 4.
Predicted Structure of NRB4.
(A) Drawing of the predicted NRB4 protein, showing the conserved KIX domain and the region rich in Gln. AA, amino acids.
(B) Magnification of the KIX domain, showing the introns (horizontal lines), the point mutations (arrows), and the T-DNA insertions (triangles) found in NRB4 (At1g15780). The number above the mutation indicates the number of alleles. Only a section of the NRB4 gene is shown; the region shown corresponds to the gray rectangle in (A).
(C) Sequence of the first 100 amino acids of NRB4, indicating the point mutations.
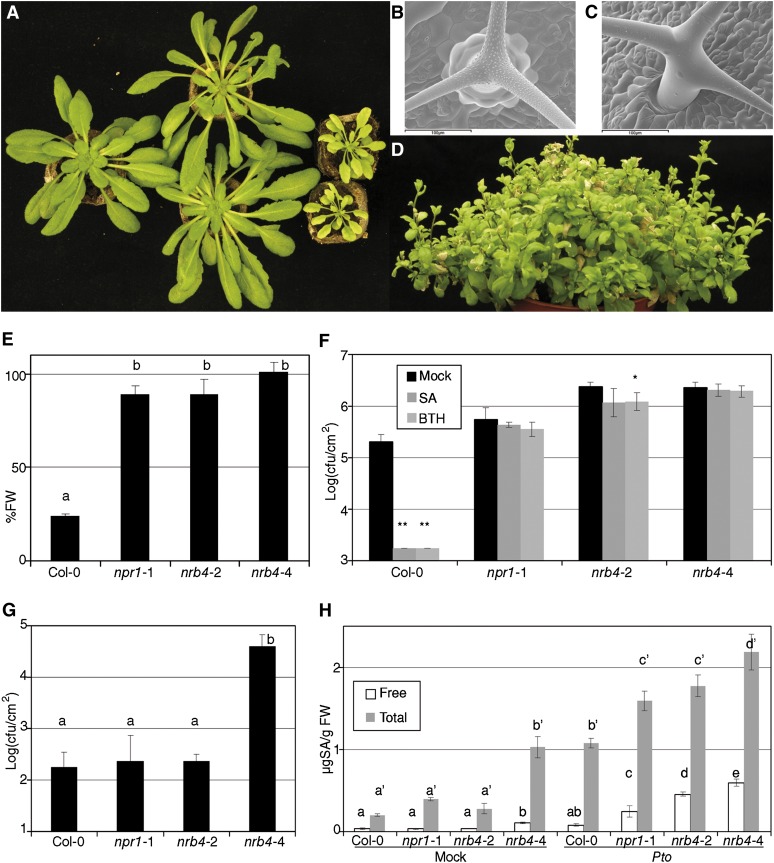
Three independent T-DNAs insertions in NRB4 were found in the databases; two of them are in introns (nrb4-4 and nrb4-5; Figure 4B). In the case of the third T-DNA, we did not find any insertion (see Methods for details). One-quarter of the progeny of plants heterozygous for nrb4-4 (see Supplemental Table 1 online) was smaller in size and more chlorotic than the wild type, while heterozygous plants were wild type (Figure 5A). The smaller plants were confirmed to be homozygous nrb4-4 by PCR, and they grew very slowly in comparison with wild-type plants (Figure 5A; see Supplemental Figures 3A and 3B online). nrb4-4 plants did not seem to be affected in leaf anatomy (see Supplemental Figure 4 online), but we did observe differences in the trichomes. Wild-type plants had trichomes with papillae on their surface and prominent cells at their base (Figure 5B), but nrb4-4 plants lacked these two elements. Additionally, the arms of the trichomes were different, irregular, and chaotically arranged (Figures 5C; see Supplemental Figure 4F online). Several types of staining showed no difference in cell death or callose deposition (see Supplemental Figure 5 online), but 4′,6-diamidino-2-phenylindole staining revealed differences in the nuclei (Kubista et al., 1987). NPR1 and the SA response are necessary for appropriate DNA content in the nucleus, with npr1-1 plants having more endoreplication than the wild type (Vanacker et al., 2001). The point mutations in nrb4 produced plants with normal endoreplication using this assay, but nrb4-4 plants had the same or more DNA per cell than npr1-1 (see Supplemental Figure 6A online). Therefore, NPR1 and NRB4 share a role in controlling endoreplication of nuclear DNA.
Figure 5.
nrb4-4 Is a Null Allele.
(A) From left to right, wild-type plant, NRB4/nrb4-4 plants, and nrb4-4 homozygous plants. Picture was taken after 6 weeks in short-day conditions.
(B) Cryo-scanning electron microscopy pictures of wild-type trichomes. The leaves sampled were ∼7 mm, and plants were 5 weeks old.
(C) Cryo-scanning electron microscopy of nrb4-4 trichomes, although plants were 7 weeks old to sample leaves of roughly the same size. Bar = 100 µm.
(D) Picture of nrb4-4 plants taken after 18 weeks (seven in short day and 11 in long day).
(E) Plants were tested as in Figure 1A, but with one more week of growth and two more treatments. FW, fresh weight.
(F) Response of nrb4-4 plants to SA and BTH in growth curves. The inoculations were done as in Figure 2C, except that the nrb4-4 plants were 7 weeks old.
(G) P. syringae pv maculicola CR299 was inoculated and its growth measured as in (F).
(H) The amount of SA (both free and total) was measured 3 days after a mock or a Pto inoculation.
The experiments were repeated three times with similar results, and the data represent the average, with the error bars plotting the sd of 15 plants in three groups of five, except in the case of (H), where three samples of 100 mg were taken. The letters above the bars indicate different homogeneous groups with statistically significant differences (Fisher’s LSD test, P < 0.05). The differences in free SA and total SA (marked with the prime symbol) were evaluated between genotypes. Asterisks indicate statistically significant differences from the mock treatment (*P < 0.05; **P < 0.01) using the Student’s t test (one tail).
[See online article for color version of this figure.]
When transferred to long-day conditions to induce flowering, nrb4-4 plants bolted, but did not produce any seeds (Figure 5D; see Supplemental Figure 3 online). In most plants, there was no production of flowers at all, but in a few plants some flowers did appear (see Supplemental Figures 3D and 3E online). These flowers did not have stamens, and the carpels did not completely enclose the ovules (see Supplemental Figure 3F online). The growth habit of nrb4-4 plants was normal until several days after bolting. Then several additional stems appeared, and afterwards a next generation of stems appeared in the previous stems in a pattern similar to a fractal (Figure 5D; see Supplemental Figure 3C online). Some plants kept growing up to 23 weeks, and when they died, they did not seem to be following the normal program of senescence.
The phenotypes of nrb4-4 homozygous plants were reproduced by nrb4-5 homozygous plants (see Supplemental Figure 6B online). Similarly, the ratio of wild type versus no response to BTH was 1:1 in all cases of F1s from heterozygous nrb4-4 plants and any of the plants that were homozygous for an EMS allele (see Supplemental Table 1 online), while the plants with EMS alleles and the nulls were fully recessive (Figure 1A; see Supplemental Table 1 online). F1s between heterozygous nrb4-4 plants and heterozygous nrb4-5 plants had a ratio of wild type versus no response to BTH of 3:1 (see Supplemental Table 1 online). Therefore, the phenotypes observed in plants with the alleles nrb4-4 and nrb4-5 were caused only by the insertions of the T-DNAs in the NRB4 gene, and one copy of the missense mutation was enough to complement the phenotypes of nrb4-4 plants, besides the response to BTH.
The nrb4-4 homozygous plants were easily distinguished at 2 or 3 weeks, and some experiments could be performed or adapted to this circumstance. nrb4-4 plants did not perceive BTH, either in terms of fresh weight (Figure 5E) or Pto growth (Figure 5F). The levels of symptoms in nrb4-4 plants after inoculation with Pto indicated that these lines were more susceptible than any other genotype, but perhaps the growth of Pto was already reaching a maximum. This extra susceptibility could be quantified with a weak pathogen, P. syringae pv maculicola CR299 (Ritter and Dangl, 1995). Thus, P. syringae pv maculicola CR299 was unable to grow in Col-0, npr1-1, or nrb4-2, but grew two log units (100-fold) in nrb4-4 plants (Figure 5G).
Since plants with nrb4-4 had an extreme susceptibility to pathogens, we wondered if they had also an extreme phenotype with SA. The responses to SA/BTH of the plants with null alleles were similar to the plants with EMS alleles (Figures 5E and 5F), so we searched for another phenotype. SA content was considered a promising one since it increases in plants under biotic stress, like Pto inoculation, but it is also increased in npr1 plants with respect to the wild type. nrb4-2 plants behaved like npr1-1, accumulating roughly the same SA amounts as the wild type in control conditions and more than the wild type after Pto inoculation (Figure 5H). nrb4-4 plants behaved differently, accumulating SA in both free and total form (free plus conjugated) in control conditions. Upon Pto inoculation, levels of both forms of SA were strongly increased (Figure 5H). It was possible to identify nrb4-4 homozygous plants in vitro (see Supplemental Figure 6C online) and to test their growth on plates with 500 µM SA. Growth of these plants was severely affected on SA plates, while heterozygous or wild-type siblings were largely unaffected by SA (see Supplemental Figure 6D online).
NRB4 Is an Ortholog of MED15
NRB4 was coimmunoprecipitated in Arabidopsis with MED6 (Bäckström et al., 2007), a subunit of the Mediator complex (reviewed in Taatjes, 2010). Due to its homology to subunits of Mediator in other species, it was labeled MED15, and an in silico search claims that it is one of the three MED15 loci in Arabidopsis (Mathur et al., 2011). However, the role of MED15 in plants may be divided among these three genes, since the expression of NRB4 in yeast lacking a functional GAL11/MED15 did not complement the mutant phenotypes (see Supplemental Figure 6E online). MED15 belongs to the tail module of the Mediator, a module that interacts with specific transcription factors (Taatjes, 2010). Since NRB4 is a subunit of the Mediator complex, and the Mediator complex is critical for global regulation of transcription (Boube et al., 2002), it seemed logical to test the behavior of other Mediator subunits in the SA response. The Arabidopsis genome encodes 51 additional potential Mediator complex–encoding genes (The Arabidopsis Information Resource, version 9; Swarbreck et al., 2008), and we tested the plants with T-DNAs insertions available. Six genes had no insertion, 15 had one or more heterozygous insertions, and 30 had one or more homozygous insertions. These populations were tested in the same conditions that allowed the identification of nrb4-4 homozygous plants, yet none of them produced a phenotype different from the wild-type control (see Supplemental Table 2 online). Therefore, the role of NRB4 in the SA response was unique among the Mediator subunits.
Molecular Footprint of nrb4
The additional phenotypes of the plants with null alleles compared with the EMS alleles of NRB4 were striking, but they did not point to any obvious process (e.g., auxins or light) that was altered, besides the SA response. A transcriptomic analysis was performed in nrb4-4 plants to identify possible physiological processes affected by the null mutation. Thus, RNAs from three biological replicates of nrb4-2 plants and Col-0 3-week-old plants (without treatment or inoculation), plus nrb4-4 of the same size (five weeks old), were isolated and hybridized to a commercial oligonucleotide microarray (see Methods). Interestingly, the molecular footprints of the two nrb4 plants were quite different.
nrb4-2 plants had a very small impact on transcription, with eight genes significantly downregulated and only one upregulated (see Supplemental Data Set 1A online). By contrast, nrb4-4 plants had 243 genes significantly downregulated and 106 upregulated (see Supplemental Data Set 1B online). Among the genes upregulated, there were genes related to SA biosynthesis (Enhanced disease susceptibility 5 and Salicylic acid induction deficient 2,) and to defense (e.g., PR1, PR2, and PR3), although the levels of induction of the defense genes were quite low in comparison to pathogen inoculation of wild-type plants (see Supplemental Data Set 1 online). This induction, though significant, was not strong enough to detect PR1 protein by immunoblot (see Supplemental Figure 7A online). The software package MapMan (Usadel et al., 2005) is able to identify groups of genes that are altered in one situation with respect to the control. In the case of nrb4-2 plants, there were only two main groups (e.g., “signaling”) strongly altered (P < 0.001), but in nrb4-4, there were up to 10 main groups strongly altered (see Supplemental Data Set 1B online), thus reflecting the severity of the pleiotropic phenotype of the mutant plants.
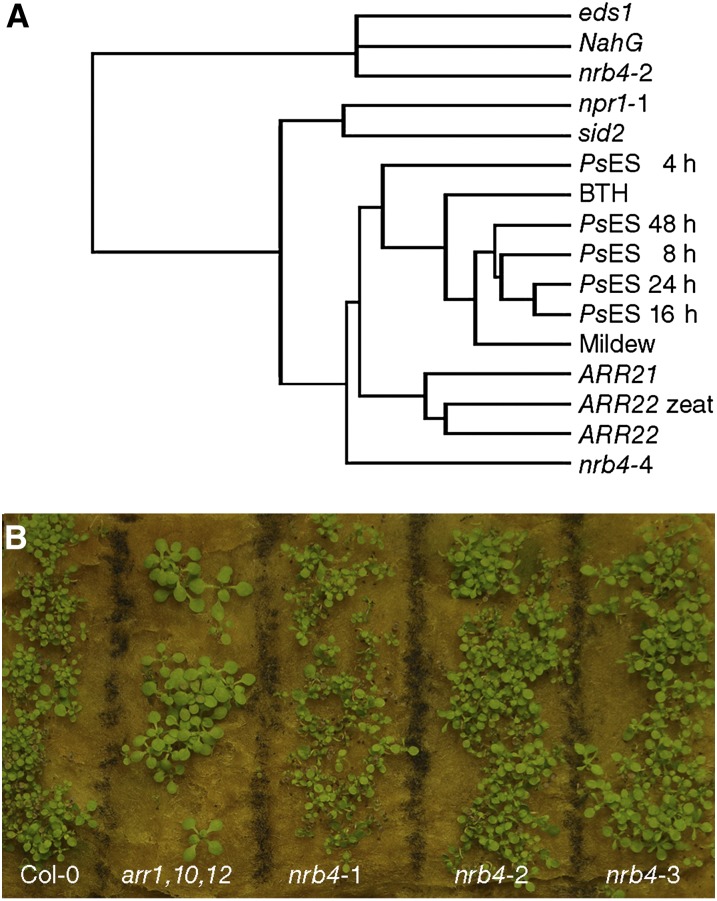
In spite of the main groups suggested by these and other analyses (see Methods), there was no evidence of specific processes being altered, other than SA and defense. Using the global footprint of the transcriptome, we searched for other mutants or treatments that could give us any hint about other processes regulated by NRB4. AtCAST is a software package that “enables the identification of unknown relations among experiments to uncover the underlying biological relationships” (Sasaki et al., 2011), and, with the default settings, AtCAST indicated a weak correlation (Spearman’s rank-order correlation coefficients) of the nrb4-4 transcriptome with the transcriptome of plants overexpressing ARR22 and ARR21 (for ARABIDOPSIS RESPONSE REGULATOR; Kiba et al., 2004, 2005). To put these correlations into perspective, several transcriptomic experiments were clustered with Cluster 3.0 (de Hoon et al., 2004; genes filtered for values >1 with centroid linkage and hierarchical clustering) along the nrb4-2 and nrb4-4 transcriptome and visualized with JavaTreeView (Saldanha, 2004) (Figure 6A). nrb4-2 data were closer to both eds1 (Falk et al., 1999) and NahG (a transgenic that overexpresses salicylate hydroxylase from Pseudomonas putida; Lawton et al., 1995), two mutations that produced a decrease in defense, but nrb4-4 data were closer to the treatments that induced defenses and to ARR21 and ARR22 overexpression. The correlations with ARR21 and ARR22 overexpressor plants, although not very strong, might indicate altered cytokinin signaling in nrb4-4 plants. When the nrb4 plants were grown in presence of trans-zeatin, there were no phenotypes of cytokinin insensitivity (Figure 6B), so even if the overexpression of genes involved in cytokinin signaling produced the data closest to nrb4-4, there was no visible cytokinin-related phenotype caused by the nrb4 alleles. The application of high amounts of cytokinins has been reported to induce SA biosynthesis and, therefore, defenses (Choi et al., 2010). There is also a negative regulation of cytokinins by SA, which may help to fine-tune the amplitude of the defense output (Argueso et al., 2012). We did not detect any difference between the plants with EMS alleles of nrb4 and the wild type in this regard (see Supplemental Figure 7B online) nor did the nrb4-4 plants show cytokinin-related phenotypes (see Supplemental Figure 7C online). Therefore, there was no evidence for a role of NRB4 in cytokinin response or in any other specific process besides the SA response.
Figure 6.
Transcriptome Analysis of nrb4 Mutants.
(A) The transcriptomes of nrb4-2, nrb4-4, and Col-0 plants were determined and then compared with different transcriptomic experiments by means of hierarchical clustering with Cluster 3.0 (de Hoon et al., 2004) and visualized as a dendogram with JavaTreeView (Saldanha, 2004). The references of the experiments used are specified in Methods, and the parameters used were the default settings.
(B) Growth response to cytokinins. Seeds of the indicated genotypes were grown in rock wool imbibed with 5 µM trans-zeatin. This picture was taken after 21 d of growth. arr1,10,12 stands for the triple mutant arr1 arr10 arr12, used as a control for lack of response to cytokinins.
[See online article for color version of this figure.]
NRB4 Expression and Localization
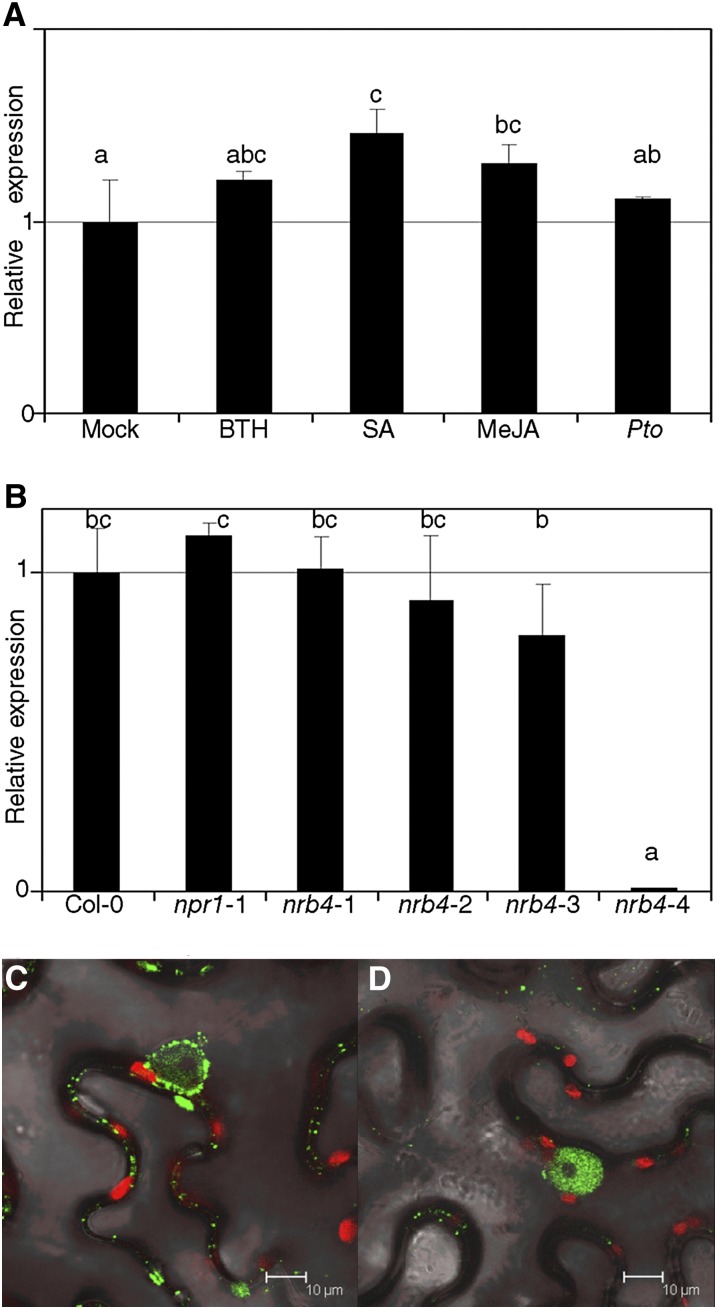
NRB4 expression is apparently unaltered by stimuli covered in the available microarray data (Hruz et al., 2008). We confirmed this using quantitative RT-PCR (RT-qPCR) to measure the levels of NRB4 after several treatments, including Pto inoculation and chemical treatments (Figure 7A). There was a reproducible increase in NRB4 expression after several treatments, but even in the best conditions (SA) it was quite low (1.46-fold induction). We did not detect any interaction between NRB4 and NPR1 (or its interactors) in yeast two-hybrid assays, regardless of the presence of SA in the media (see Supplemental Figure 8 online). The EMS alleles did not produce a measurable instability in the mutated mRNA, but the nrb4-4 mutation rendered the mRNA below the threshold of detection (Figure 7B). The expression of NRB4 was unaltered in npr1-1 plants (Figure 7B), and NPR1 was detectable in nrb4-2 at normal levels (see Supplemental Figure 9A online).
Figure 7.
Expression of NRB4 and Subcellular Localization.
(A) NRB4 expression was measured 1 d after treatment with mock, 350 µM BTH, 500 µM SA, and 100 µM MeJA or 3 d after a Pto inoculation (RNA extracted and qRT-PCR from three independent samples of 100 mg each). The levels of expression are normalized to three reference genes and to the level of Col-0.
(B) RNA was extracted from 3-week-old plants (five weeks for nrb4-4), and transcript levels for NRB4 were measured by RT-qPCR as in (A).
(C) Agrobacterium tumefaciens with the construct 35S:NRB4-GFP was infiltrated into leaves of N. benthamiana, and the expression was detected by confocal microscopy 4 d later.
(D) Similar to (C) with the construct 35S:GFP-NRB4.
The experiments were repeated three times with similar results, and the data represent the average, with the error bars plotting the sd of three samples. The letters above the bars indicate different homogeneous groups with statistically significant differences (Fisher’s LSD test, P < 0.05).
Although the Mediator complex is described to act in the nucleus, NRB4 did not contain any obvious nuclear localization signal. To determine the localization of the protein, we transiently overexpressed the NRB4 cDNA fused to the GFP coding sequence at the 3′ or 5′ terminus in Nicotiana benthamiana (Figures 7C and 7D, respectively). In both cases, there was a strong localization in the nucleus, with NRB4-GFP accumulating also outside the nucleus. The nuclear localization did not change with the application of 350 µM BTH (see Supplemental Figure 9B online). The predicted size of NRB4 was 146 kD, well above the free diffusion limit into the nucleus of 50 kD (Talcott and Moore, 1999).
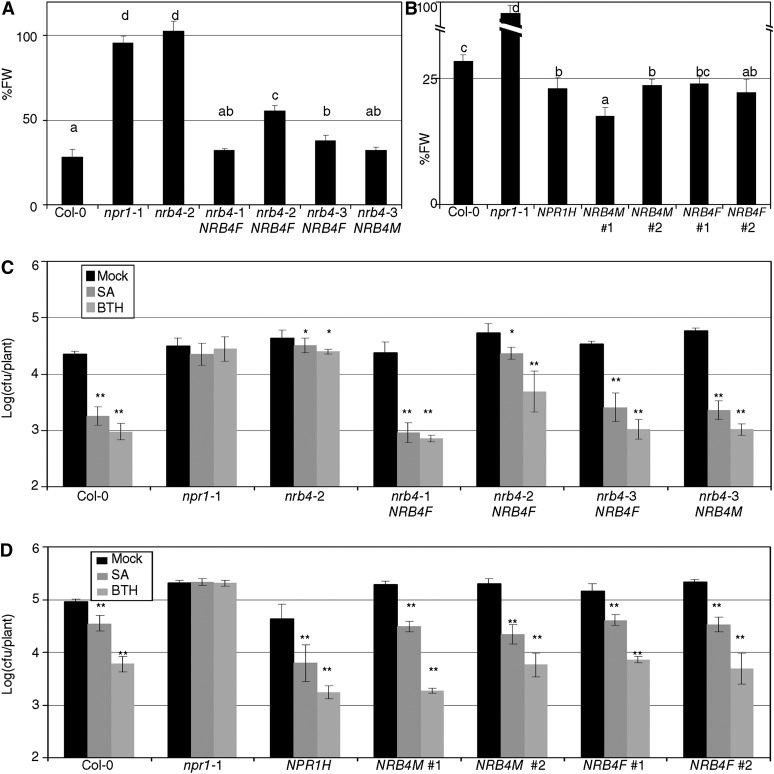
These constructs were transformed into nrb4 mutant Arabidopsis; none of the GFP-NRB4 constructs complemented the nrb4 mutants, but the NRB4-GFP plants complemented the EMS alleles with some variation when the response to BTH in terms of fresh weight was considered (Figure 8A). This variation was representative of the transgenic lines obtained regardless of the background. A version of NRB4 containing the first 670 amino acids was also able to complement the mutations in some lines (Figure 8A), but the version of NRB4 containing only the first 112 amino acids did not. In the complemented lines, GFP was not detectable by means of confocal microscopy or immunoblot (see Supplemental Figure 10A online). However, since there was detectable function, we also transformed the wild type Col-0 to study the effects of NRB4 overexpression. In this experiment, 35S:NPR1 was included as a control, since it is more sensitive to SA (Cao et al., 1998). The transgenic lines that overexpressed both versions (1335 and 670 amino acids) of NRB4 had an enhanced SA response as measured by fresh weight after BTH applications (Figure 8B). We additionally complemented the wild type restriction of Pto growth after SA or BTH application using these lines (Figure 8C, also checked for PR1 expression; see Supplemental Figure 10B online). Note that the overexpression of NRB4 did not produce a strong defense response under control conditions, but when SA or BTH was applied, there was an enhanced response to SA (Figure 8D). Therefore, the effect of NRB4 was specific and limited to SA response.
Figure 8.
Phenotypes of Transgenic Lines.
(A) Transgenic plants homozygous for the construct 35S:NRB4-GFP (NRB4F) or for the equivalent construct with only the first 670 amino acids (NRB4M) were obtained in the mutant alleles. The panel shows the response to BTH in fresh weight (FW) of the mentioned lines tested as in Figure 1A.
(B) The constructs described in (A) were transformed into Col-0, and their response to BTH in fresh weight recorded. The number indicates an independent line.
(C) Response of the transgenic lines described in (A) to SA and BTH in growth curves, as described in Figure 2A.
(D) Response of the transgenic lines described in (B) to SA and BTH in growth curves, as described in Figure 2A.
The experiments were repeated three times with similar results, and the data represent the average, with the error bars plotting the sd of 15 plants in three groups of five. The letters above the bars indicate different homogeneous groups with statistically significant differences (Fisher’s LSD test, P < 0.05). Asterisks indicate statistically significant differences from the mock treatment (*P < 0.05; **P < 0.01) using the Student’s t test (one tail).
DISCUSSION
A Role for Mediator in SA Response
The Mediator complex interacts with RNA Pol II, but mutations in specific Mediator subunits typically impact specific phenotypes, rather than general transcription (reviewed in Taatjes, 2010). This observation has lead to the hypothesis that the Mediator complex performs both general and specific roles to regulate gene expression (Taatjes, 2010). In plants, the Mediator complex is emerging as a crucial component of transcriptional regulation in response to specific signals (reviewed in Kidd et al., 2011). The components of the Mediator complex have been identified using biochemistry and genetics. Thus, the immunopurification of MED6 in Arabidopsis led to the identification of 19 Mediator subunits, NRB4 among them (Bäckström et al., 2007). A null mutation in SWP/MED14 produces sterile plants with reduced growth, small leaves, and an increase in endoreplication (Autran et al., 2002), similar to our observations with nrb4-4 plants. The main difference in swp/med14 mutants was the size of the cells; by contrast, in nrb4-4 plants, the size of cells was similar to the wild type (see Supplemental Figure 4 online), and in 35S:NRB4, the plants were macroscopically similar to the wild type. In the case of SWP, both the knockout and the overexpressor produced plants and cells of smaller size than the wild type.
Mutations in MED21 (Dhawan et al., 2009), PFT1/MED25, and SETH10/MED8 (Kidd et al., 2009) affect disease resistance against necrotrophic pathogens to different degrees. Specifically, null homozygous mutants in MED21 have an embryo-lethal phenotype, and RNA interference plants with low levels of MED21 are more susceptible to necrotrophic pathogens (Dhawan et al., 2009). PHYTOCROME AND FLOWERING TIME1 (PFT1) was first described as a gene required for the shade avoidance response and flowering (Cerdán and Chory, 2003). Once PFT1 was identified as a Mediator subunit (Bäckström et al., 2007), a screen for similar phenotypes in the rest of subunits identified a mutant in SETH10/MED8 as required for both wild-type flowering time and resistance to necrotrophic pathogens (Kidd et al., 2009). Following this logic, we tested the available T-DNAs insertions but found no additional Mediator subunits with a measurable phenotype in SA response (see Supplemental Table 2 online).
It is striking that mutations in three subunits of Mediator cause defense phenotypes in response to necrotrophic pathogens that are related to JA-Ile response, whereas mutations in only one subunit, NRB4, cause a phenotype related to SA-dependent defense responses. A plausible explanation would be that NRB4 is a negative regulator of JA-Ile, and its removal would increase JA-Ile response. Then, this increase in JA-Ile signaling could be observed as loss in SA signaling, since both signals crosstalk negatively (Robert-Seilaniantz et al., 2011). However, we did not observe a specific phenotype related to JA-Ile in the plants with nrb4 EMS alleles [MeJA plates, MeJA-induced resistance, P. cucumerina infection, and growth of Pto(cor-); see Supplemental Figure 1 online]. In any case, in plants there are four subunits of Mediator involved in biotic stress. Plants with mutations in MED25 are more sensitive to salt stress (Elfving et al., 2011), so there is clearly an overrepresentation of stress phenotypes in the described mutations of Mediator subunits.
MED15 also plays a role in stress responses alongside its roles in other processes. MED15 in Drosophila melanogaster was identified in a mosaic screening where the effect of the mutation was limited to the wings (Terriente-Félix et al., 2010). Homozygous null mutations were lethal at embryogenesis, and the weak point mutations found were lethal at later stages (Terriente-Félix et al., 2010). MDT-15 is the ortholog of MED15 in Caenorhabditis elegans, and the knockdown by RNA interference of MDT-15 produced multiple deleterious effects (reduced life span, sterility, etc.; Taubert et al., 2006). A reduction in functional MDT-15 protein leads to animals being hypersensitive to xenobiotics, thus affecting selectively stress response related to ingestion (Taubert et al., 2008). GAL11 is the ortholog of MED15 in yeast, and the deletion of this gene is not lethal but is essential for growth on nonfermentable carbon sources, for sporulation, and for mating (Mylin et al., 1991, and references therein). The deletion of GAL11 renders yeast more sensitive to cycloheximide (Shahi et al., 2010). Using this phenotype, we introduced NRB4 in yeast Δgal11, but it did not complement the growth in cycloheximide (see Supplemental Figure 6E online). NRB4 may not be the correct MED15 ortholog, since an in silico analysis predicts that there are three MED15s in Arabidopsis (Mathur et al., 2011). The existence of more than one ortholog is not new in the Mediator complex in plants (Kidd et al., 2011) or other organisms (Bourbon et al., 2004). If NRB4 is one of three MED15 subunits of Arabidopsis, then its role in SA response is nonredundant, since the two null alleles of NRB4 produced plants that did not respond to SA.
How Specific Is NRB4?
Mediator is a complex required for the normal transcription of genes. In a high-throughput screening for genes involved in any process, there could be a point when elements of the general transcriptional machinery would start to appear. Our data regarding NRB4 do not fit this concept, but point toward a specific role for NRB4 in the SA response. First, we did not see any noticeable phenotype in the plants with the three hypomorphic EMS alleles, besides the response to SA. It is true that the first selection was done with BTH, but in the case of npr1, different alleles diverge in their response to MeJA (Dobón et al., 2011). Second, the phenotypes of nrb4-4 and nrb4-5 plants, although dramatic, did not resemble mutants generally impaired in signaling (e.g., hormones and light). Third, the transcriptomes of both nrb4-2 and nrb4-4 plants were not indicative of perturbation of any specific process compared with untreated wild-type plants (see Supplemental Data Set 1 online). Fourth, NRB4 has not been found in other screenings for hormone responses, and some have been performed en masse in plate format. It is not a small protein (1335 amino acids), and it has a Gln-rich region. EMS is the most frequent mutagen used in Arabidopsis, and its effect in Gln is introducing stop codons (two possible stop codons and one silent mutation; Martinez-Zapater and Salinas, 1998). Therefore, it is more likely to have stop codons introduced by EMS than the average coding sequence. Fifth, the overexpression of NRB4 did not produce any noticeable phenotypes except an increase in response to SA (Figure 8). The specificity of NRB4 should be localized in the KIX domain, since the three missense alleles were localized there. It is the more conserved domain, and half of the protein can be deleted without major loss of function (Figure 8).
A Model of the SA Response
There are several genes that act downstream of NPR1. Among the genes found to be relevant in the SA response, there are several that are involved in DNA repair (Song et al., 2011) and chromatin remodeling (Wang et al., 2010) genes. Since these proteins play a role in forming a complex relevant for transcription (Durrant et al., 2007), perhaps NRB4 is required for the proper function of these proteins.
We have shown that NRB4 is necessary for the SA response, and the pivotal role of NPR1 in this signaling has been abundantly reported (Maier et al., 2011; Fu et al., 2012; Wu et al., 2012). In spite of the importance of these genes in the response to SA, we did not detect any interaction between the genes or their proteins. The F1s between the mutant alleles were wild type (Figure 1A), no protein–protein interaction was detected in yeast or in planta, and the overexpression of NPR1 in an nrb4 mutant background did not restore the response to SA (Figure 3B; see Supplemental Figure 2G online). Such an effect could have occurred if the corresponding proteins worked together. As a consequence, with the necessary precautions for the evaluation of negative results, it seems that NRB4 and NPR1 act at different points in SA signaling, which also explains the phenotypes of the double mutants, both with strong and weak alleles (Figure 3A; see Supplemental Figure 2A online). A version of NPR1 tagged with GFP became localized in the nucleus, both in an nrb4 and in a wild-type background (see Supplemental Figure 2 online), but did not rescue the altered response to SA. Therefore, NRB4 does not play a role in the stability of NPR1 (i.e., it does not act as chaperone), the concentration of NPR1 in the nucleus is NRB4 independent, and NRB4 functions downstream of NPR1.
It is possible that NRB4 interacts with NPR1 only in special conditions. An interaction between NPR1 and SA has been detected only recently, since the SA-NPR1 complex is quite labile (Wu et al., 2012). Alternatively, the interaction could happen with a complex that would include SA, NPR1, NPR3, and/or NPR4 (Fu et al., 2012). There are ample precedents of MED15 interacting with a nuclear receptor. In yeast, GAL11/MED15 is necessary for the expression of genes required for growth in Gal media (Suzuki et al., 1988). But other functions include the use of fatty acids (see below) and the regulation of multidrug resistance. Thus, Δgal11 yeast does not grow in media with small amounts of ethidium bromide (Mylin et al., 1991) or cycloheximide (Shahi et al., 2010), while the wild type grows unaffected. This pathway is the same used by Candida glabrata to pump ketoconazole out of the cell (Thakur et al., 2008). The mechanism is that Pdr1p and Pdr3p are xenobiotic nuclear receptors that bind GAL11 (specifically in the KIX domain) in a xenobiotic-dependent manner (Thakur et al., 2008). This is not a unique case, since in C. elegans, NHR-49 binds MDT-15/MED15 also in the KIX domain (Taubert et al., 2006). In this and other organisms, MED15 regulates the metabolism of fatty acids, with a proposed model that NHR-49 and other nuclear receptors are binding a hormone-like small molecule(s) present in the food (Taubert et al., 2006).
Although none of the previous examples involve a SA receptor, similar molecules have been found to participate in these pathways. Oaf1P is a yeast nuclear receptor that, upon binding fatty acids, interacts with GAL11 and activates the transcription of genes required for the use of fatty acids (Thakur et al., 2009). A similar function is performed by NHR-49 in C. elegans (Taubert et al., 2006) and by PPARα in vertebrates (Issemann and Green, 1990). These three receptors bind fatty acids but also bind nonsteroidal anti-inflammatory drugs, such as salicylates. Therefore, the orthologs of MED15 interact with receptors in the KIX domain that bind salicylates and fatty acids. There is a strong representation of proteins related to lipids among the Arabidopsis defense mutants (eds1, Phytoalexin deficient 4, and Senescence associated gene 101 Wiermer et al., 2005; Defective in induced resistance 1, Maldonado et al., 2002; Suppressor of sai1, Kachroo et al., 2003; Suppressor of Fatty Acid Desaturase Deficiency 1, Nandi et al., 2004; etc.), so it is plausible that this connection is maintained in Arabidopsis. There are no genes in Arabidopsis with significant homology to Oaf1P, NHR-49, or PPARα (see Supplemental Table 3 online), so it is possible that NPR1, NPR3, and NPR4 have evolved independently from the aforementioned receptors.
There is a striking difference in phenotype between plants with the EMS mutations and the null mutations (Figure 5). The null mutants produce a stronger phenotype in defense than the EMS alleles and a severe phenotype in development. The phenotype in defense is even stronger than that caused by the npr1 alleles so far described (Canet et al., 2010b). Psm CR299 grows 100-fold in nrb4-4 plants, while it does not grow in the rest of genotypes (Figure 5G). More strikingly, SA itself is upregulated in npr1 and nrb4 plants, but in nrb4-4, the levels reach a maximum. Interestingly, this implies that the metabolism of SA is partially independent of NRB4 and NPR1. It also implies that SA itself does not affect Pto directly, since the levels of SA in nrb4-4 plants clearly exceed the levels of SA in wild-type plants (Figure 5H). This increased phenotype caused by the null mutants could be explained if the EMS alleles were not completely devoid of function. The point mutations in the KIX domain do not impair nrb4-1 metabolism of SA in plates (Figure 1B) or nrb4-2 to develop SAR (Figure 2D). The npr1 null alleles do not cause the same phenotype as the nrb4 null alleles, perhaps because the rest of NPR1 paralogs are able to compensate for its loss in the SA response (Canet et al., 2010b). The strong phenotype caused by the null nrb4 in development could be due to additional signals that are lost in these mutants. However, so far we have found no indication of such signals.
SA plays an important role in different plant processes besides disease resistance (Rivas-San Vicente and Plasencia, 2011), and one possible explanation could be that the phenotypes of nrb4 null plants are only due to a lack of response to SA, therefore supporting the postulated essential role of SA in normal plant development (Vanacker et al., 2001). Thus, npr1-1 and nrb4-4 plants showed increased endoreplication of the nuclear DNA (see Supplemental Figure 6A online), reflecting a role of SA in this process. The available plants with less SA do not show any of the previous phenotypes (Vanacker et al., 2001), but several analyses show that these plants still have some SA (Rivas-San Vicente and Plasencia, 2011), and plants that lack several SA biosynthetic genes are not viable (Garcion et al., 2008).
METHODS
Plant Growth and Inoculation
Arabidopsis thaliana was sown and grown as described (Canet et al., 2010a) in controlled environment rooms with days of 8 h at 21°C, 150 µmol m−2 s−1 of light intensity, and nights of 16 h at 19°C. The treatments, inoculations, and sampling started 30 min after the initiation of the artificial day to ensure reproducibility. The following genotypes were used: npr1-1 (Cao et al., 1997), 35S:NPR1-HBD and 35S:NPR1-GFP (Kinkema et al., 2000), rpm1 (Grant et al., 1995), rps2 (Mindrinos et al., 1994), nho1 (Lu et al., 2001), and arr1-3 arr10-5 arr12-1 (Argyros et al., 2008). nrb4-4 was SAIL_792_F02, and nrb4-5 was GABI_955_E02. The line in which we did not find any insertion was SALK_106110C. Pto was grown, inoculated, and measured as described (Tornero and Dangl, 2001). Briefly, plants of 18 d were inoculated by spray with Pto at OD600= 0.1 with 0.02% Silwet L-77 (Crompton Europe). Three days later, the amount of colony-forming units (cfu) per plant was quantified and represented on a logarithmic scale. When inoculations of older plants were measured, a sample of known surface was taken, and the resulting unit was log(cfu/cm2). Other strains used were Pto(avrRpm1) (Ritter and Dangl, 1996) and Pto(avrRpt2) (Debener et al., 1991). Pseudomonas syringae pv tabaci and pv phaseolicola NPS3121 were obtained from Jeff Dangl (University of North Carolina, Chapel Hill, NC). P. syringae pv maculicola CR299 has been described (Ritter and Dangl, 1995). SAR was measured as reported (Macho et al., 2010), inoculating leaves with Pto(avrRpm1) or a mock treatment using a blunt syringe. For all the experiments, at least three independent treatments were performed (three independent sets of plants sown and treated on different dates). Pto was maintained as described (Ritter and Dangl, 1996).
Chemical Treatments
Primers and chemical products were purchased from Sigma-Aldrich unless otherwise stated. BTH (CGA 245704), in the form of commercial product (Bion 50 WG; a gift from Syngenta Agro), was prepared in water for each treatment and applied with a household sprayer. The response to BTH in terms of fresh weight was done as reported (Canet et al., 2010a). Briefly, plants were treated with mock or 350 µM BTH four times over three weeks. Then, the fresh weight of the plants was recorded and expressed as the ratio between BTH and mock-treated plants. A total of 100 µM MeJA (Duchefa) was applied by spray with 0.1% DMSO and 0.02% Silwet L-77. Dexamethasone was applied at 2 µM, diluted in water from a stock of 20 mM in ethanol. SA (in the form of sodium salicylate) was applied at 500 µM. For the treatments with cytokinins, trans-zeatin at 5 µM was used to imbibe pieces of wool rock (from a local gardening shop). Seeds were sown directly in the wool rock, and additional water was added to compensate for evaporation.
SA in Plates and in Planta
Arabidopsis seeds were surface sterilized for 10 min in 70% ethanol and for 10 min in commercial bleach. Then, five washes were done with distilled water and the seeds were distributed on agar plates. The medium contains 0.5× Murashige and Skoog salts (Duchefa), 0.6% (w/v) Phyto Agar (Duchefa), 2% (w/v) Suc, with 0, 400, or 500 µM SA (final concentration). The results were evaluated 14 d after transferring to growing conditions. The chlorophyll was extracted with ethanol for 2 h at 65°C and quantified as described by Frye et al. (2001). Three replicates of 10 plants each per treatment and genotype were measured. For the measurement of SA in planta, three samples of ∼100 mg were frozen in liquid nitrogen. SA extraction was performed as described (Huang et al., 2005; Defraia et al., 2008).
Expression in Planta and Microscopy
NRB4 was cloned in pDONR221 (Invitrogen) from an RT-PCR product and then transferred to pMDC43 (Curtis and Grossniklaus, 2003; GFP-NRB4) and pB7FWG2 (Karimi et al., 2002; NRB4-GFP) for expression in planta. Nicotiana benthamiana leaf tissue was mounted in water under a cover slip 4 d after infiltration with Agrobacterium tumefaciens containing the constructs. The Arabidopsis plants containing NPR1-GFP were 3 weeks old at the time of the pictures. A Leica TCS SL confocal laser scanning microscope using an HCX PL APO CS ×40/1.25 oil objective was used to study the subcellular localization of the fluorescence-tagged proteins. GFP was visualized by 488-nm excitation with an argon laser, and its emissions were examined with a band-pass filter for 500 to 530 nm. The primers used are included in Supplemental Table 4 online. The scanning electron microscopy pictures were taken with a JSM-5410 scanning electron microscope (JEOL) at the Electron Microscopy Service (Universidad Politécnica de Valencia, Spain).
Immunoblot and RT-qPCR
Immunodetection of PR1 protein was performed as described (Wang et al., 2005) using Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare). The second antibody was a 1:25,000 dilution of anti-rabbit IgG horseradish peroxidase conjugate (Promega). Chemiluminescent signals were detected using a LA-3000 luminescent image analyzer (Fujifilm Life Science).
Total RNA was extracted with Trizol (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized with a RevertAid first-strand cDNA synthesis kit (Fermentas), and the quantitative PCR performed with LuminoCt Sybr Green qPCR Ready Mix (Sigma-Aldrich) in a 7000 RT-PCR system (Applied Biosystems) following the manufacturer’s instructions. For each measurement, three biological replicates were done. The obtained values were referred to the geometric average of three reference genes (At3G18780, At1G49240, and At5G60390), as described (Vandesompele et al., 2002), and normalized with mock-treated Col-0 equal to one. The list of primers used is provided in Supplemental Table 4 online.
Microarrays and Software Used
RNA was isolated as described above and purified with the RNeasy mini kit (Qiagen). Array hybridization to an Arabidopsis GeneChip ATH1 (Affymetrix) was performed following the manufacturer’s recommendations. The hybridization was performed in the “Sección de Chips de DNA-S.C.S.I.E.,” University of Valencia (Valencia, Spain). Three biological replicates of each genotype were hybridized, with no technical replicates (three replicates of three genotypes and nine microarrays). The original hybridization data files were submitted to the European Bioinformatics Institute ArrayExpress repository, and the accession number E-MEXP-3602 was assigned to this experiment. The analysis of the microarrays was accomplished with Robin 1.1.7 (Lohse et al., 2010). The robust multiarray averaging normalization was used. Both mutants were compared with the wild-type control, and the P value cutoff was set at 0.05 with the Benjamini and Hochberg P value correction (Benjamini and Hochberg, 1995). After the P value adjustment, a nested F test was used to classify the comparisons as significant (Lohse et al., 2010). Then, the following software was used: MapMan (Usadel et al., 2005), Sample Angler (http://142.150.214.117), AtCAST (Sasaki et al., 2011), Cluster 3.0 (de Hoon et al., 2004), and JavaTreeView (Saldanha, 2004). For the statistic analysis, we used Excel 2003 (Microsoft) and Statgraphics 5.1 (Statpoint Technologies). The data analyzed corresponded with the following experiments: eds1, E-MEXP-546; NahG, E-GEOD-5727; npr1-1, E-GEOD-5745; sid2, and BTH, E-GEOD-9955; PsES, E-GEOD-5685; Mildew, E-GEOD-431; ARR21, GSE5699; and ARR22, GSE5698.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NRB4, At1g15780; NPR1, At1g64280; ACT2, At3g18780; ACT8, At1g49240; and ELF, At5G60390.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Additional Phenotypes of nrb4 in Defense.
Supplemental Figure 2. Epistasis of NRB4 with NPR1.
Supplemental Figure 3 Additional Pictures of nrb4-4.
Supplemental Figure 4. Additional Pictures of Cryo-SEM.
Supplemental Figure 5. Stainings of nrb4.
Supplemental Figure 6. Characterization of nrb4 Null Alleles.
Supplemental Figure 7. Phenotypes from the Transcriptomic Analysis.
Supplemental Figure 8. Yeast n-Hybrid Interactions.
Supplemental Figure 9. Expression of NPR1 and NBR4.
Supplemental Figure 10. Characterization of NRB4 Complementation.
Supplemental Table 1. Evaluation of Segregations.
Supplemental Table 2. Evaluation of Phenotypes in T-DNA Insertion Lines.
Supplemental Table 3. Lack of Homologs in Arabidopsis for Several Nuclear Receptors.
Supplemental Table 4. Primers Used.
Supplemental Data Set 1. Transcriptomic Analysis.
Supplementary Material
Acknowledgments
This work was supported by the “Ministerio de Economía y Competitividad” (MINECO) of Spain (Grant BIO201018896 to P.T., a Junta de Ampliación de Estudios-Consejo Superior de Investigaciones Científicas Fellowship to J.V.C., and a Formación del Personal Investigador-MINECO to A.D.) and “Generalitat Valenciana” of Spain (Grant ACOMP/2012/105 to P.T.). We appreciate the opinions and generous help of Jeff Dangl and Pablo Vera with the article as well as the revision of Philippa Borrill.
AUTHOR CONTRIBUTIONS
J.V.C. and A.D. performed the experiments, analyzed the data, and revised the article. P.T. designed the research, analyzed the data, and wrote the article.
Glossary
- SA
salicylic acid
- JA
jasmonic acid
- MeJA
methyl jasmonate
- BTH
benzothiadiazole
- EMS
ethyl methanesulfonate
- Col-0
Columbia-0
- Pto
Pseudomonas syringae pv tomato strain DC3000
- SAR
systemic acquired resistance
- GFP
green fluorescent protein
- qRT-PCR
quantitative RT-PCR
- cfu
colony-forming units
References
- Argueso C.T., Ferreira F.J., Epple P., To J.P., Hutchison C.E., Schaller G.E., Dangl J.L., Kieber J.J. (2012). Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 8: e1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D., Jonak C., Belcram K., Beemster G.T., Kronenberger J., Grandjean O., Inzé D., Traas J. (2002). Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg B. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57: 289–300 [Google Scholar]
- Boube M., Joulia L., Cribbs D.L., Bourbon H.M. (2002). Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151 [DOI] [PubMed] [Google Scholar]
- Bourbon H.M., et al. (2004). A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14: 553–557 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Cai G., Imasaki T., Takagi Y., Asturias F.J. (2009). Mediator structural conservation and implications for the regulation mechanism. Structure 17: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet J.V., Dobón A., Ibáñez F., Perales L., Tornero P. (2010a). Resistance and biomass in Arabidopsis: A new model for salicylic acid perception. Plant Biotechnol. J. 8: 126–141 [DOI] [PubMed] [Google Scholar]
- Canet J.V., Dobón A., Roig A., Tornero P. (2010b). Structure-function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ. 33: 1911–1922 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95: 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán P.D., Chory J. (2003). Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick J.Z., Asturias F.J. (2005). Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30: 264–271 [DOI] [PubMed] [Google Scholar]
- Choi J., Huh S.U., Kojima M., Sakakibara H., Paek K.H., Hwang I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19: 284–295 [DOI] [PubMed] [Google Scholar]
- Chrivia J.C., Kwok R.P., Lamb N., Hagiwara M., Montminy M.R., Goodman R.H. (1993). Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859 [DOI] [PubMed] [Google Scholar]
- Debener T., Lehnackers H., Arnold M., Dangl J.L. (1991). Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1: 289–302 [DOI] [PubMed] [Google Scholar]
- Defraia C.T., Schmelz E.A., Mou Z. (2008). A rapid biosensor-based method for quantification of free and glucose-conjugated salicylic acid. Plant Methods 4: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., Miyano S. (2004). Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Friedrich L., Ryals J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92: 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W.-L., Preston G., Collmer A., Chang C.-J., Huang H.-C. (1998). Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 180: 4523–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten Scheid O., Mengiste T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobón A., Canet J.V., Perales L., Tornero P. (2011). Quantitative genetic analysis of salicylic acid perception in Arabidopsis. Planta 234: 671–684 [DOI] [PubMed] [Google Scholar]
- Dong X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Durrant W.E., Wang S., Dong X. (2007). Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA 104: 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., Müller D., Nilsson A., Ulfstedt M., Ronne H., Wingsle G., Nilsson O., Björklund S. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Falk A., Feys B.J., Frost L.N., Jones J.D., Daniels M.J., Parker J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan P.M., Kelleher R.J., III, Sayre M.H., Tschochner H., Kornberg R.D. (1991). A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438 [DOI] [PubMed] [Google Scholar]
- Frye C.A., Tang D., Innes R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S.H., Tada Y., Zheng N., Dong X. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J.-P. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147: 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Grantham R. (1974). Amino acid difference formula to help explain protein evolution. Science 185: 862–864 [DOI] [PubMed] [Google Scholar]
- Guo L., Han A., Bates D.L., Cao J., Chen L. (2007). Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc. Natl. Acad. Sci. USA 104: 4297–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Wang H., Zheng H., Huang L., Singer A.C., Thompson I., Whiteley A.S. (2005). Chromosomally located gene fusions constructed in Acinetobacter sp. ADP1 for the detection of salicylate. Environ. Microbiol. 7: 1339–1348 [DOI] [PubMed] [Google Scholar]
- Issemann I., Green S. (1990). Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347: 645–650 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F. bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15: 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kelleher R.J., III, Flanagan P.M., Kornberg R.D. (1990). A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Kiba T., Aoki K., Sakakibara H., Mizuno T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Kiba T., Naitou T., Koizumi N., Yamashino T., Sakakibara H., Mizuno T. (2005). Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His->Asp phosphorelay circuitry. Plant Cell Physiol. 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Kidd B.N., Cahill D.M., Manners J.M., Schenk P.M., Kazan K. (2011). Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22: 741–748 [DOI] [PubMed] [Google Scholar]
- Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubista M., Akerman B., Nordén B. (1987). Characterization of interaction between DNA and 4′,6-diamidino-2-phenylindole by optical spectroscopy. Biochemistry 26: 4545–4553 [DOI] [PubMed] [Google Scholar]
- Lawton K., Weymann K., Friedrich L., Vernooij B., Uknes S., Ryals J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8: 863–870 [DOI] [PubMed] [Google Scholar]
- Lawton K.A., Friedrich L., Hunt M., Weymann K., Delaney T., Kessmann H., Staub T., Ryals J. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Lohse M., et al. (2010). Robin: An intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Tang X., Zhou J.M. (2001). Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P., Guevara C.M., Tornero P., Ruiz-Albert J., Beuzón C.R. (2010). The Pseudomonas syringae effector protein HopZ1a suppresses effector-triggered immunity. New Phytol. 187: 1018–1033 [DOI] [PubMed] [Google Scholar]
- Maier F., Zwicker S., Hückelhoven A., Meissner M., Funk J., Pfitzner A.J., Pfitzner U.M. (2011). NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 12: 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado A.M., Doerner P., Dixon R.A., Lamb C.J., Cameron R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater J.M., Salinas J. (1998). Arabidopsis Protocols. (Totowa, New Jersey: Humana Press).
- Mathur S., Vyas S., Kapoor S., Tyagi A.K. (2011). The Mediator complex in plants: Structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M., Katagiri F., Yu G.-L., Ausubel F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mittal S., Davis K.R. (1995). Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Mylin L.M., Gerardot C.J., Hopper J.E., Dickson R.C. (1991). Sequence conservation in the Saccharomyces and Kluveromyces GAL11 transcription activators suggests functional domains. Nucleic Acids Res. 19: 5345–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A., Welti R., Shah J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I., Pérez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E. (1997). Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91: 741–752 [DOI] [PubMed] [Google Scholar]
- Ritter C., Dangl J.L. (1995). The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol. Plant Microbe Interact. 8: 444–453 [DOI] [PubMed] [Google Scholar]
- Ritter C., Dangl J.L. (1996). Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. (2004). Java Treeview—Extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Sasaki E., Takahashi C., Asami T., Shimada Y. (2011). AtCAST, a tool for exploring gene expression similarities among DNA microarray experiments using networks. Plant Cell Physiol. 52: 169–180 [DOI] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Shahi P., Gulshan K., Näär A.M., Moye-Rowley W.S. (2010). Differential roles of transcriptional mediator subunits in regulation of multidrug resistance gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2469–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Durrant W.E., Wang S., Yan S., Tan E.H., Dong X. (2011). DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 9: 115–124 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Nogi Y., Abe A., Fukasawa T. (1988). GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36(Database issue): D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes D.J. (2010). The human Mediator complex: A versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 35: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talcott B., Moore M.S. (1999). Getting across the nuclear pore complex. Trends Cell Biol. 9: 312–318 [DOI] [PubMed] [Google Scholar]
- Taubert S., Hansen M., Van Gilst M.R., Cooper S.B., Yamamoto K.R. (2008). The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 4: e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Van Gilst M.R., Hansen M., Yamamoto K.R. (2006). A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terriente-Félix A., López-Varea A., de Celis J.F. (2010). Identification of genes affecting wing patterning through a loss-of-function mutagenesis screen and characterization of med15 function during wing development. Genetics 185: 671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J.K., Arthanari H., Yang F., Chau K.H., Wagner G., Näär A.M. (2009). Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 284: 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J.K., et al. (2008). A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452: 604–609 [DOI] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38: 119–130 [DOI] [PubMed] [Google Scholar]
- Tornero P., Dangl J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28: 475–481 [DOI] [PubMed] [Google Scholar]
- Usadel B., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H., Lu H., Rate D.N., Greenberg J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28: 209–216 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang S., Durrant W.E., Song J., Spivey N.W., Dong X. (2010). Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. USA 107: 22716–22721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel R.R., Bäuscher C., Pfitzner A.J., Pfitzner U.M. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46: 143–160 [DOI] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J., Parker J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J.Y., Boyle P., Wang Y., Brindle I.D., De Luca V., Després C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1: 639–647 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.