The arbuscule-specific expression of two rice PHOSPHATE TRANSPORTER1 (PHT1) genes, PT11 and PT13, suggests that both of these genes play a role in symbiotic phosphate acquisition. Indeed, they are both important for the development of arbuscular mycorrhizal symbioses in rice; however, PT11 is the key player in fungus-delivered phosphate uptake.
Abstract
Pi acquisition of crops via arbuscular mycorrhizal (AM) symbiosis is becoming increasingly important due to limited high-grade rock Pi reserves and a demand for environmentally sustainable agriculture. Here, we show that 70% of the overall Pi acquired by rice (Oryza sativa) is delivered via the symbiotic route. To better understand this pathway, we combined genetic, molecular, and physiological approaches to determine the specific functions of two symbiosis-specific members of the PHOSPHATE TRANSPORTER1 (PHT1) gene family from rice, ORYsa;PHT1;11 (PT11) and ORYsa;PHT1;13 (PT13). The PT11 lineage of proteins from mono- and dicotyledons is most closely related to homologs from the ancient moss, indicating an early evolutionary origin. By contrast, PT13 arose in the Poaceae, suggesting that grasses acquired a particular strategy for the acquisition of symbiotic Pi. Surprisingly, mutations in either PT11 or PT13 affected the development of the symbiosis, demonstrating that both genes are important for AM symbiosis. For symbiotic Pi uptake, however, only PT11 is necessary and sufficient. Consequently, our results demonstrate that mycorrhizal rice depends on the AM symbiosis to satisfy its Pi demands, which is mediated by a single functional Pi transporter, PT11.
INTRODUCTION
Rice (Oryza sativa) is the primary staple food for more than 50% of the human population and, thus, central for future food security. The constantly growing demand for intensifying rice productivity has to match environmental concerns for the sustainability of agriculture in rice growing areas. Conventional farming methods involve the application of fertilizers to compensate for limiting nutrients, such as Pi. Fertilizer application occurs in more than half of the soils used for rice cultivation but can cause undesired environmental pollution when applied in excess (Lumini et al., 2011, and citations therein; MacDonald et al., 2011). On the other hand, rice enters into symbiosis with arbuscular mycorrhizal (AM) fungi of the Glomeromycota, which significantly improve plant Pi nutrition, in particular under aerobic, low-soil Pi conditions, characteristic of upland rice systems where colonization by AM fungi can lead to enhanced biomass production (Sharma et al., 1988; Solaiman and Hirata, 1997; Maiti et al., 2011, and citations therein). When in association with plant roots, AM fungi proliferate extensively within the rhizosphere, thereby significantly enlarging the soil volume for plant nutrient foraging (Bolan, 1991). The fungal intra- and extraradical mycelium functions as a conduit for Pi acquisition from the soil toward the inner root cortex. Upon root invasion, the fungus invaginates inner cortex cells, where it develops highly branched tree-like structures, the arbuscules. Arbuscules are enveloped by the plant plasma membrane, creating an extensive interface between the membranes of both organisms that is ideally adapted to signal and nutrient exchange. Pi is taken up from the soil by fungal hyphae and translocated to the arbuscules, where an unknown mechanism is employed to release Pi into the periarbuscular interface, making it available for uptake by the arbusculated plant cells (recently reviewed in Javot et al., 2007b; Yang and Paszkowski, 2011).
More than 80% of the land plants, including all cereals, enter into AM symbioses and hence may derive a certain amount of Pi via the mycorrhizal uptake pathway. This symbiotic acquisition route most likely has operated ever since the beginning of terrestrial plant life. A comparison of arbuscules from fossil records of rhizomes of early land plants with those from roots of contemporary plants revealed a high degree of morphological resemblance (Remy et al., 1994). Therefore, the symbiotic interaction might be regarded as an evolutionarily ancient and likely ancestral Pi acquisition strategy for plant life on land.
Pi uptake across plant and fungal plasma membranes is mediated by Pi transporters belonging to the PHOSPHATE TRANSPORTER1 (PHT1) class (Poirier and Bucher, 2002) of the plant Pi:H+ symporters of the major facilitator superfamily (Pao et al., 1998). These Pi transporters are membrane intrinsic proteins of comparable size and of high sequence and structure similarity, containing 12 transmembrane (TM) domains that are organized as two groups of six TM domains separated by a hydrophilic loop between TM6 and TM7. Over the past 15 years, numerous plant PHT1 proteins have been isolated, and transport assays in heterologous yeast or Xenopus laevis oocyte systems have confirmed transport activity with high to low affinity for Pi (reviewed in Javot et al., 2007b). Plant PHT1 proteins that exhibit exclusive or induced expression in arbusculated cells have been identified from diverse plant species (recently reviewed in Javot et al., 2007b; Yang and Paszkowski, 2011). Of these, the rice ORYsa;PHT1;11 (PT11) represents the first AM-specific Pi transporter isolated from plants (Paszkowski et al., 2002), and homologous proteins are present across diverse angiosperms hosting AM fungi (Javot et al., 2007b). Importantly, the PT11 homolog of Medicago truncatula (MEDtr;PHT1;4 [PT4]) was shown to localize at the periarbuscular membrane (Harrison et al., 2002) and to be required for AM symbiosis (Javot et al., 2007); its mutation impaired both development of the interaction and symbiotic Pi uptake. By contrast, mutation of the tomato (Solanum lycopersicum) homolog of PT11 (SOLly;PHT1;4) neither affected mycorrhizal Pi uptake nor establishment of the symbiosis, which was attributed to genetic redundancy, since in tomato, AM symbiosis triggers the expression of three PHT1 genes (Nagy et al., 2005). Thus, some plants may employ supplementary Pi transporters in addition to the conserved PT11-like protein; however, their respective contribution to the signaling of symbiotic development and Pi uptake remains to be determined.
In addition to the symbiotic Pi uptake, plants can also employ a direct Pi acquisition pathway. PHT1 proteins that are phylogenetically distant from the AM-associated Pi transporters mediate Pi uptake from the soil in the root periphery and, when disrupted, cause Pi deficiency of the plant (Misson et al., 2004; Shin et al., 2004; Ai et al., 2009; Jia et al., 2011; Sun et al., 2012). Although it could be assumed that both pathways act synergistically to maximize Pi uptake, physiological studies in flax (Linum usitatissimum), tomato, and M. truncatula have demonstrated that for these dicotyledons the mycorrhizal dominates the direct uptake pathway (Smith et al., 2003, 2004). It remains an open question whether a similar dominance of the symbiotic Pi uptake route occurs in monocotyledons. The functional characterization of the interplay between direct and symbiotic Pi uptake, particularly under field conditions, mirroring agricultural practices, would be most valuable and relevant for monocot crops, such as cereals.
The rice genome contains 13 genes coding for PHT1 proteins (Goff et al., 2002; Paszkowski et al., 2002). Of these, ORYsa;PHT1;1 (PT1), PT2, PT6, and PT8 have been implicated in direct Pi uptake (Ai et al., 2009; Jia et al., 2011; Sun et al., 2012). Whereas PT2 and PT6 are transcriptionally induced in roots in response to Pi deficiency, PT1 and PT8 are constitutively expressed. It was recently shown that PT6 and PT8 represent high affinity Pi transporters with predominant expression in the rhizodermis and cortex, while PT2 is a low-affinity transporter with expression in the root vasculature. Knockdown of the Pi starvation-inducible genes PT2 or PT6 via RNA interference (RNAi) results in reduced Pi uptake (Ai et al., 2009), consistent with a function in the direct Pi uptake pathway under low Pi conditions.
PT11 represents the only AM-specific Pi transporter reported from monocotyledons for which expression specific for arbusculated cells and protein localization to the periarbuscular membrane has been reported (Paszkowski et al., 2002; Gutjahr et al., 2008; Kobae and Hata, 2010). In rice, a second AM-inducible PHT1 gene, PT13, was identified that might encode another Pi transporter participating in symbiotic Pi uptake (Güimil et al., 2005), but its role in AM symbiosis is unknown.
Here, we functionally dissect symbiotic Pi acquisition in rice. Surprisingly, we did not find functional redundancy between PT11 and PT13. Instead, both PT11 and PT13 play a central role during the development of AM symbioses, suggesting distinct functions for each PHT1 protein. We show that PT11 is the only transporter involved in symbiotic Pi absorption in rice under the conditions tested. Importantly, we found that mycorrhizal rice receives more than 70% of its Pi via the symbiotic uptake pathway, and, according to our molecular analysis, this is also the case in field soils.
RESULTS
Identification of the PT13 Open Reading Frame
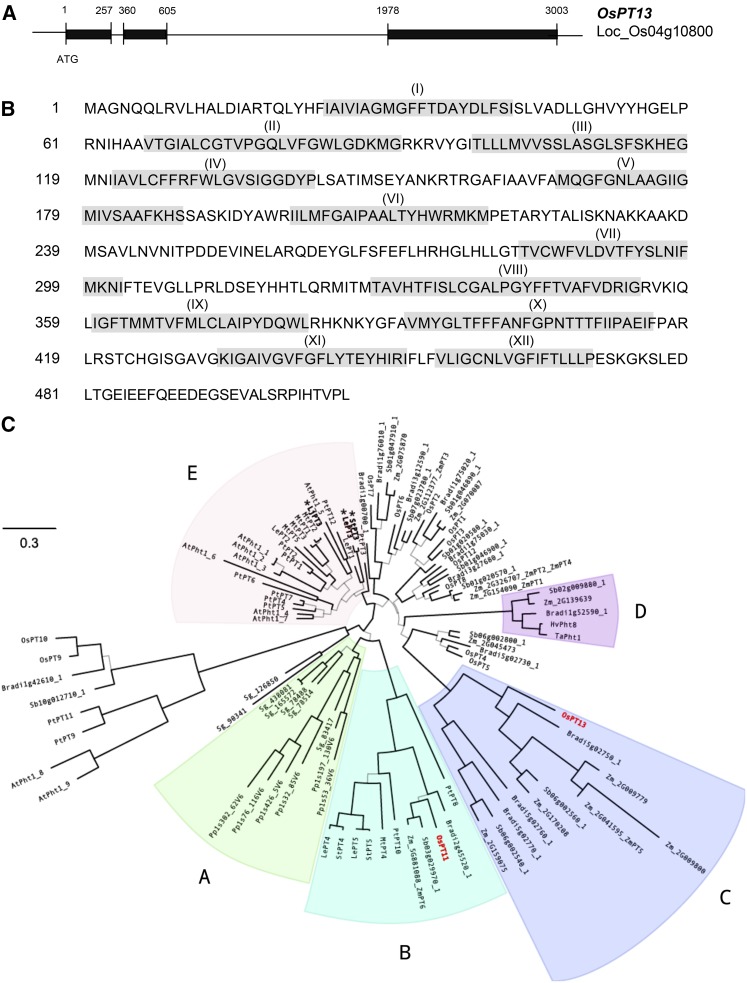
Since the coding region of only rice PT11 was available at the beginning of this investigation (Paszkowski et al., 2002), we cloned the cDNA of PT13 from roots colonized by AM fungi. PT13 contains three exons and two introns (Figure 1A) and encodes a protein of 528 amino acids that corresponds to the predicted gene structure of Os04g10800.1 (http://orygenesdb.cirad.fr/cgi-bin/gbrowse/odb_japonica). In accordance with the topology of other members of the PHT1 family of Pi transporters, PT13 is predicted to contain 12 TM domains, separated by a large hydrophilic loop between TM6 and TM7, and a cytosolic N and C terminus (Figure 1B). The two PHT1 genes, PT11 and PT13, are located on rice chromosomes 1 and 4, respectively, and share 52% amino acid sequence identity.
Figure 1.
Gene and Protein Structure of Rice PT13 and Phylogenetic Relationships with Plant PHT1 Proteins.
(A) The PT13 gene corresponds to Loc_Os04g10800 and consists of three exons (black boxes).
(B) The encoded membrane intrinsic protein comprises 528 amino acids and 12 TM domains (gray boxes).
(C) Phylogenetic tree of the PHT1 proteins built with the JTT+Γ model of evolution using PhyML. Branches with bootstrap support higher than 85% are shown in black, while branches with bootstrap support lower than 85% are in gray. PT11 and PT13 are in red. The AM-inducible Pi transporters from dicots that group with regular Pi transporters are displayed in bold letters and are marked by an asterisk. The published maize genes are labeled with the locus identifier and the published name. Phylogenetic lineages are colored and marked A to E for visual emphasis.
PT13 Is Conserved across Monocotyledons
To examine the similarity of PT13 relative to other members of the PHT1 class of Pi transporters, a phylogenetic analysis that included individually characterized Pi transporters and sequences from published plant genomes was performed (see Supplemental Data Set 1 online). Both Bayesian inference and maximum likelihood analyses resulted in identical topologies, and only the maximum likelihood analysis will be discussed here. The first reported AM-associated Pi transporter from potato (Solanum tuberosum), St-PT3, is conserved across Solanaceae and Fabaceae plants species and groups together with PHT proteins from dicots involved in direct Pi uptake and translocation throughout the plant (Figure 1C, lineage E; Rausch et al., 2001; Javot et al., 2007b). Rice PT11, however, groups with other AM-associated PHT1 proteins from mono- and dicotyledonous plant species (Figure 1C, lineage B; Javot et al., 2007b). Sequence similarity between PT11 and its lineage members from monocots (>81%) and dicots (>71%) is high, and they are grouped into a single highly supported lineage, which indicates that these proteins may perform related functions. The poplar (Populus spp) POPtr;PHT1;10 (PT10) and POPtr;PHT1;8 (PT8) proteins also group phylogenetically with the PT11 branch, but whereas poplar PT10 displays a PT11-like expression pattern, transcription of poplar PT8 has not been detected under the conditions so far assayed (Loth-Pereda et al., 2011), suggesting PT8 is a pseudogene. Consistently, the predicted PT8 protein clusters at a distance to the expressed proteins within the PT11 lineage (Figure 1C, lineage B). We determined computationally that the genomes of the ancient plants Physcomitrella patens and Selaginella moellendorffii contain six and seven PHT1 genes, respectively. Interestingly, the PT11-lineage is closely related to the homologs from P. patens and S. moellendorffii, supporting its older evolutionary relationship (Figure 1C, lineage A). On the contrary, no ortholog for rice PT13 has been reported from dicot plants. However, the PT13 protein is conserved across monocotyledons (Figure 1C, lineage C). Computational analyses identified 12, 13, and 14 PHT1 genes in the genomes of sorghum (Sorghum bicolor), Brachypodium distachyon, and maize (Zea mays), respectively. While rice has only one copy of PT13, the maize genome harbors five, B. distachyon three, and S. bicolor two paralogs, indicative of recent gene duplication events. Among these, Bradi5g02760 (BRAdi;PHT1;12) and Bradi5g02770 (BRAdi;PHT1;13) group with the rice PT13 homologs, and transcription of both corresponding genes has been detected in mycorrhizal roots of B. distachyon plants (Hong et al., 2012). Another type of PHT1 gene expressed in mycorrhizal roots has been reported from barley (Hordeum vulgare), wheat (Triticum aestivum), and B. distachyon (Glassop et al., 2005; Hong et al., 2012) that is missing from the rice genome but present as a single-copy gene in the genomes of sorghum and maize (Figure 1C, lineage D). It therefore arose in a subset of the Poaceae lineages, namely, the Panicoideae (maize and sorghum) and Pooideae (B. distachyon, wheat, and barley). The absence of a corresponding gene from the rice genome is surprising but might reflect the general lack of this protein across members of the Ehrhartoideae subfamily. Rice is therefore distinct among the monocotyledons, possessing only two phylogenetically distant AM-associated PHT proteins.
Temporal and Spatial Expression Patterns of PT13 Are Similar to Those of PT11
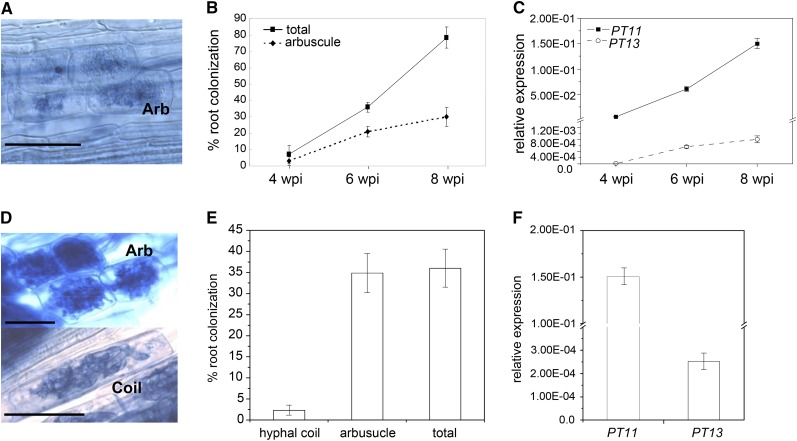
To characterize the expression of PT13 in response to progressing colonization by AM fungi and relative to the arbuscule-specific PT11 (Gutjahr et al., 2008), rice roots were inoculated with Glomus intraradices, which forms highly branched intracellular arbuscules (Figure 2A). Colonization levels and gene expression were measured in a time-course experiment from 4 to 8 weeks postinoculation (wpi). Total root length colonization and arbuscule development by G. intraradices increased over time (Figure 2B). Mirroring the continuous increase in arbuscule number, PT11 transcript levels were low at 4 wpi and enhanced dramatically at 8 wpi (Figure 2C). Interestingly, the kinetics of PT13 transcript accumulation, although showing overall considerably lower values, reflected that of PT11 (Figure 2C). Neither PT11 nor PT13 transcripts were detected in mock-inoculated plants. The lack of PT13 transcript in control roots contradicts our previous, Affymetrix GeneChip-based observation of constitutive root expression (Güimil et al., 2005), which was likely due to cross-hybridization of a subset of probes by mRNA from other root-expressed PHT1 genes.
Figure 2.
Expression Analysis of PT11 and PT13 in G. intraradices and G. rosea Colonized Rice Roots.
(A) Micrograph of G. intraradices arbuscules (Arb).
(B) Total (squares) and arbuscular (diamonds) colonization levels of wild-type roots with G. intraradices at 4, 6, and 8 wpi.
(C) Expression of PT11 (squares) and PT13 (circles) at 4, 6, and 8 wpi with G. intraradices.
(D) Micrograph of G. rosea arbuscules and coils. Bars = 50 μm.
(E) Colonization levels of wild-type rice at 10 wpi with G. rosea.
(F) Expression of PT11 and PT13 at 10 wpi with G. rosea. Gene expression levels are shown relative to the expression of the constitutive rice Cyclophilin 2 gene.
(B) and (E) Mean values of three biological replicates are shown, each consisting of two pooled plants.
(C) and (F) Error bars refer to the se of the mean of three technical repeats. The experiment was repeated twice with similar results.
The robustness of gene induction of PT13 during the interaction of rice with other AM fungi was addressed. Gigaspora rosea belongs to the order of the Diversisporales and is only distantly related to G. intraradices of the Glomales (Schüssler et al., 2001). In rice, G. rosea forms not only arbuscules but also intracellular hyphal coils (Figure 2D; Gutjahr et al., 2008). Rice and G. rosea were cocultivated for 10 weeks to ensure extensive root colonization, including arbuscules and a smaller proportion of hyphal coils (Figure 2E). Both PHT1 genes were induced in colonized roots, with PT11 displaying high and PT13 low but significant mRNA accumulation (Figure 2F). In summary, the expression profile of PT13 was found to match PT11, consistent with a possible role in symbiotic Pi acquisition.
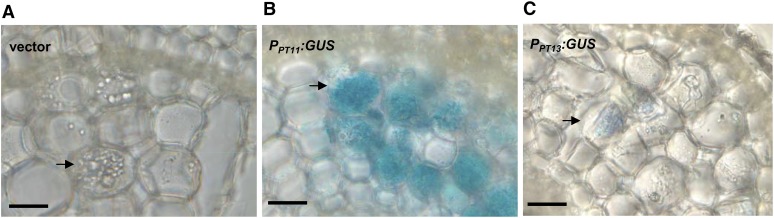
To spatially map the expression pattern of PT13 relative to fungal infection structures and to PT11, transgenic rice roots expressing 2-kb PT11 or PT13 promoter-GUS (for β-glucuronidase) fusions and inoculated with G. intraradices were analyzed. Root cross sections of the reporter lines revealed that whereas empty vector control plants did not show any blue coloration despite visible arbuscules (Figure 3A), a strong blue staining indicative of GUS activity occurred in arbusculated cortical cells of roots expressing the PT11 promoter-GUS construct (Figure 3B). A much fainter blue signal was also observed in arbusculated cells of PT13 promoter-GUS lines (Figure 3C). The arbuscule-specific expression of both promoters provides further support for the involvement of PT11 and PT13 in symbiotic Pi uptake.
Figure 3.
Spatial Expression of PT11 and PT13 Promoters.
Histochemical GUS staining of a proportion of a G. intraradices colonized rice root expressing empty vector (A), PPT11-GUS (B), and PPT13-GUS (C) constructs. Arrowheads represent cortical cells containing arbuscules. Bars = 20 μm.
Generation and Characterization of PT11 and PT13 Mutant Alleles
Single and double mutant lines of PT11 and PT13 were sought to demonstrate the functional relevance of each protein for the symbiosis and to investigate possible genetic redundancy between both proteins as well as to determine the respective contribution of each protein to symbiotic Pi uptake. In the publicly available collections of rice mutants, a Tos17 retrotransposon insertion residing within the promoter region of PT11 was identified (see Supplemental Figure 1A online; Miyao et al., 2003). For PT13, a dSpm element (Kumar et al., 2005) was identified inserted into the first exon of the gene (see Supplemental Figure 1A online). Mutant lines of PT11 and PT13 were designated pt11-1 and pt13-1, respectively. Since only one mutant allele of each AM-specific PHT1 gene was available, additional alleles were created via RNAi and named pt11R1 and pt13R1, respectively. Double RNAi lines, 11/13R1 and 11/13R2, were generated to address functional redundancy or additive effects and the impact of each expression perturbation on transcript levels. Because PT11 and PT13 are not expressed in nonmycorrhizal roots, wild-type and homozygous mutant plants were inoculated with G. intraradices. Relative to the wild type, all single and double mutant lines exhibited significantly reduced transcript levels (see Supplemental Figures 1B to 1D online).
PT11 and PT13 Are Necessary for AM Development in Rice
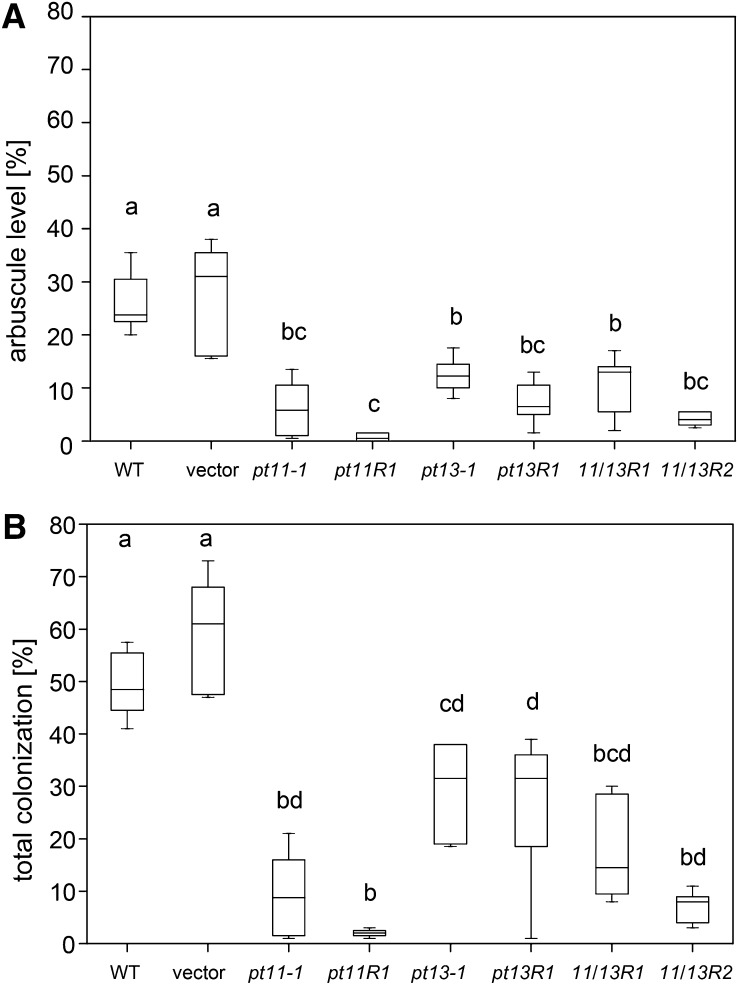
To examine the consequences of mutation of PT11, PT13, or both on the establishment of AM symbiosis, root colonization by G. intraradices was monitored in plants carrying wild-type and mutant alleles. Relative to the controls, pt11-1 and pt11R1 mutant roots showed a dramatically reduced level of arbuscule formation and overall fungal colonization (Figures 4A and 4B). Interestingly, fungal proliferation and arbuscule formation were significantly reduced also in mutant roots of pt13-1 and pt13R1 plants (Figures 4A and 4B). Hence, despite the low level of gene induction, PT13 is indispensable for normal development of the AM symbiosis in rice. The two double RNAi lines, 11/13R1 and 11/13R2, also showed low levels of total colonization and of arbuscule development (Figures 4A and 4B), reaching levels somewhat intermediate to the PT11 and PT13 mutant lines, in line with the comparably low levels of endogenous PT11 and PT13 transcripts relative to the single mutant lines (see Supplemental Figures 1B to 1D online). In summary, PT11 and PT13 are not functionally redundant, and no additive effect was observed when both genes were mutated.
Figure 4.
G. intraradices Colonization Levels in Single and Double Mutants of PT11 and PT13.
Level of G. intraradices arbuscules (A) and total colonization (B) at 8 wpi in control, insertion, and RNAi mutant alleles as indicated. Box plot shows from three to seven biological replicates of each genotype and was generated using the Origin 7 software. The top of the box is the 75th percentile. The bottom of the box is the 25th percentile. The horizontal line intersecting the box is the median value of the group. Horizontal lines above and below the box represent maximum and minimum values. Boxes with dissimilar letters are significantly different at P < 0.01 after one-way analysis of variance (ANOVA) (paired-sample t tests were performed with Bonferroni adjustment). WT, the wild type.
Gene specificity of the RNAi triggers was confirmed by monitoring the respective nontarget PT gene. The absence of PT11 silencing in the PT13 mutant was documented by high PT11 mRNA levels (see Supplemental Figure 2A online). Interestingly, the PT11 mRNA levels in the PT13 mutant remained as high as in the wild type, despite the reduced arbuscule frequency, suggesting a compensatory effect may occur. Transcript levels of PT13 in PT11 mutant lines were reduced relative to the wild type (see Supplemental Figure 2B online); however, this correlated, with the overall decreased fungal colonization (Figures 4A and 4B) and was further demonstrated by the lower expression of the G. intraradices internal transcribed spacer1 (Gi-ITS1; see Supplemental Figure 2C online). In conclusion, no off-target silencing was observed, neither for PT11 nor for PT13 RNAi triggers.
To investigate the requirement of functional PT11 and PT13 for symbiosis with other AM fungi, colonization levels of G. rosea were investigated in wild-type, PT11, and PT13 mutant roots. Relative to wild-type controls, a significant reduction in arbuscules and total colonization levels was found in both mutant genotypes (see Supplemental Figure 3 online), consistent with a central role of PT11 and PT13 in the establishment of symbiosis with diverse AM fungi.
Mutation of PT11 and PT13 Function Affects Arbuscule Morphology
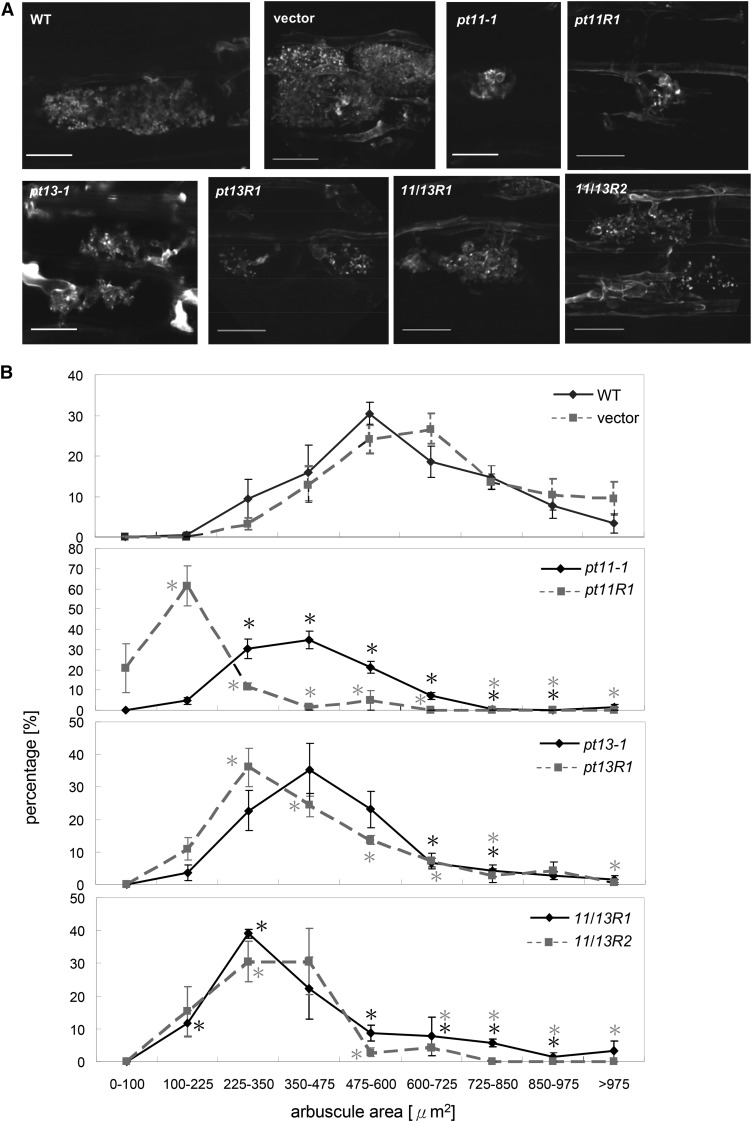
The impact of PT11 and PT13 mutation on the morphology of intraradical fungal structures was examined. In the insertion and RNAi mutants, hyphopodia and intercellularly growing hyphae appeared comparable to the wild type. However, smaller and less intensely branched arbuscules were observed (Figure 5A). To precisely determine the distribution of arbuscule size classes in control and mutant lines, the cross section area of arbuscule populations was measured and size categories were established. The majority of arbuscules in wild-type and empty vector control roots displayed an area of 475 to 725 μm2, while arbuscules in pt11-1 and pt11R1 were significantly smaller at 225 to 475 μm2 and 100 to 250 μm2, respectively (Figure 5B). The transgenic pt11R1 line exhibited a more severe arbuscule phenotype than pt11-1, consistent with the earlier observed lower frequency in arbuscule formation and total root colonization (Figures 4A and 4B), which might be attributed to the lower PT11 mRNA levels in the RNAi relative to the insertion allele (see Supplemental Figures 1B and 1C online). The size distribution was wider in the PT13 and double RNAi lines, with the majority of arbuscules belonging to the class of 224 to 475 μm2. No additive decrease in arbuscule surface area was observed for the double RNAi lines.
Figure 5.
G. intraradices Arbuscule Phenotype in Single and Double Mutants of PT11 and PT13.
(A) WGA-Alexafluor 488 cell wall staining of G. intraradices structures of control and mutant roots as indicated at 8 wpi. Hyphal septa are recognized by the intense, punctuate fluorescence in all genotypes. WT, the wild type. Bars = 20 μm.
(B) Frequency graphs showing the distribution of arbuscule size classes (cross section area) in arbuscule populations measured in control and mutant plants. Mean values of 24 arbuscules per plant and a total of three plants are shown. Error bars refer to se. Asterisks indicate a statistically significant difference from respective wild-type or vector control plants at P < 0.05.
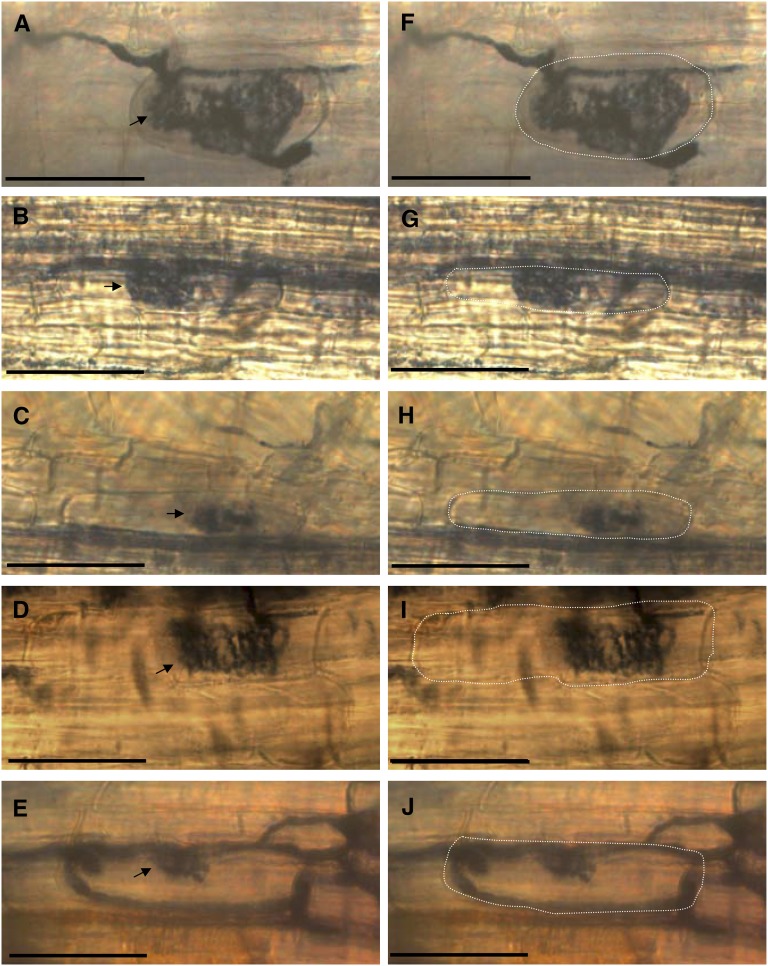
Septate hyphae were observed in a subset of fully developed wild-type and in small mutant arbuscules (Figure 5A), indicating arbuscule collapse and death (Javot et al., 2007). However, in contrast with previous observations made in M. truncatula PT4 mutants (Javot et al., 2007), vital staining of colonized control and mutant roots revealed the presence of live arbuscules in PT11 and PT13 mutants (Figure 6). Although secondary cell wall staining with WGA Alexa 488 proved technically challenging, the vital stain permitted the recapitulation of the typical morphology of reduced intracellular arbuscule expansion within roots of both PT11 and PT13 mutants. In conclusion, mutation of PT11 and/or PT13 not only quantitatively affected intraradical fungal development, but also qualitatively led to altered arbuscule morphology and growth of arbuscules.
Figure 6.
Vital Staining of PT11 and PT13 Mutant Roots Colonized by Glomus intraradices.
Live fungal structures are stained in roots of the wild type ([A] and [F]), pt11-1 ([B] and [G]), pt11R1 ([C] and [H]), pt13-1 ([D] and [I]), and pt13R1 ([E] and [J]) at 7 wpi. In (F) to (J), the cell boundaries of arbusculated cells were computationally marked by dashed white lines to highlight arbuscule expansion relative to plant cortex cells. Bars = 50 μm.
The Symbiotic Pi Uptake Pathway Represents the Primary Route of Mycorrhizal Rice and Requires Functional PT11 but Not PT13
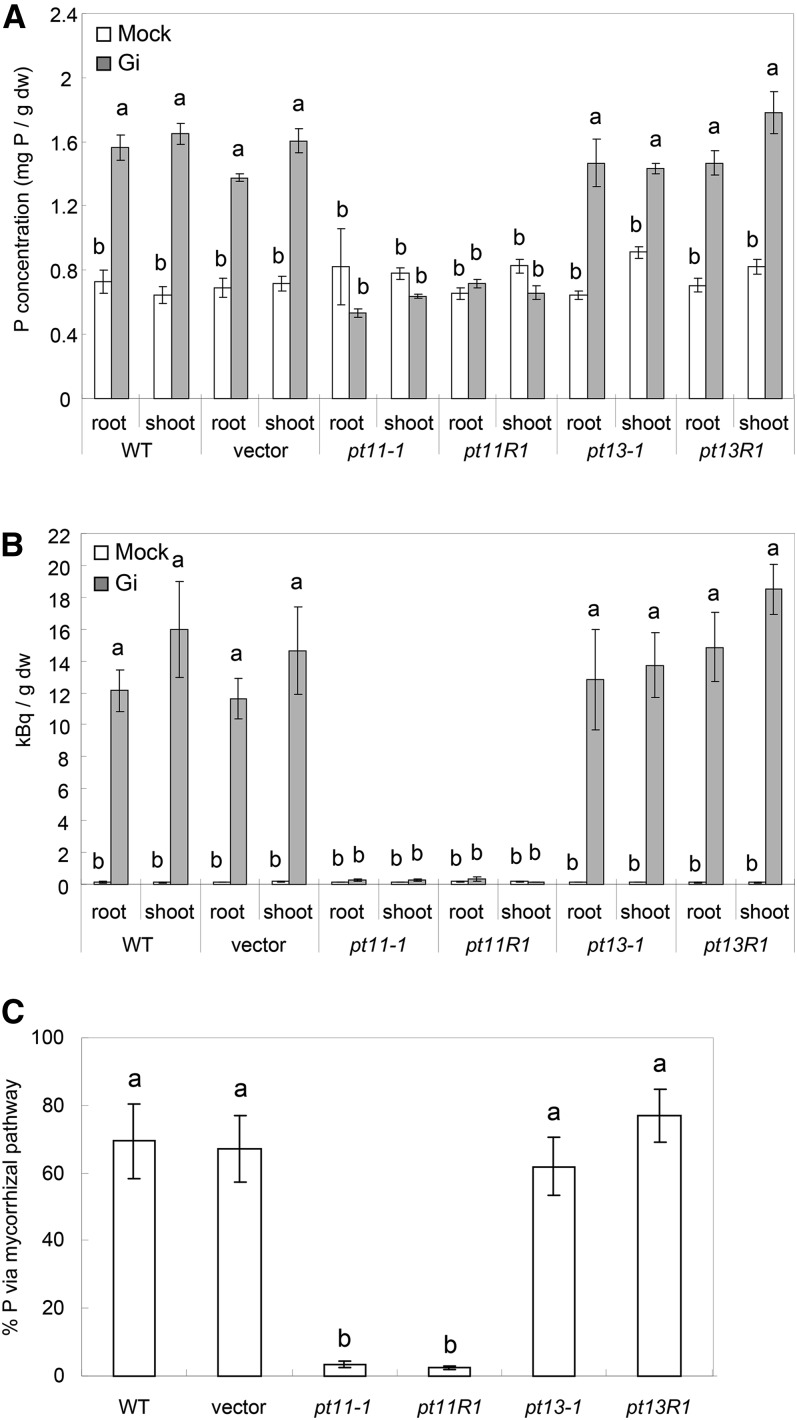
To quantify the contribution of PT11 and PT13 to symbiotic Pi uptake, P uptake measurements were performed (Smith et al., 2004). P concentration was determined in root and shoot tissue of PT11 and PT13 mutant lines when grown under low Pi supply in the presence or absence of G. intraradices. The P concentration in root and shoot tissue of mock-inoculated plants was similar in all genotypes, indicating that loss of PT11 or PT13 function had no effect on Pi nutrition in the absence of symbiosis (Figure 7A). However, in the presence of G. intraradices, control plants showed an enhanced P concentration in roots and shoots, associated with an established AM symbiosis (Figure 7A). By contrast, such an increased amount of tissue P was not observed in roots or shoots of both PT11 mutant lines, suggesting that PT11 is essential for the elevated P concentration of mycorrhizal plants. By contrast, the two PT13 mutant plants consistently displayed an elevated P tissue concentration in colonized plants that reached comparable values to wild-type controls (Figure 7A).
Figure 7.
Quantitative Symbiotic Pi Transfer of PT11 and PT13 Mutants.
(A) and (B) Tissue concentration of P (A) and 33P (B) in the roots and shoots of G. intraradices (Gi) or mock-inoculated control plants at 8 wpi. dw, dry weight; WT, the wild type.
(C) Percentage of contribution of the mycorrhizal pathway to P uptake at 8 wpi in the control and mutant RNAi lines colonized by G. intraradices. The se refers to three to five biological replicates, and each biological replicate consists of a pool of two plants. Bars with dissimilar letters are significantly different at P < 0.01 after one-way ANOVA (paired-sample t tests were performed with Bonferroni adjustment).
To quantify the amount of Pi taken up via the symbiotic pathway, the fungal transfer of 33P from the soil to the plant was measured. For this purpose, radioactive 33P was provided to the fungus in a hyphal compartment (Smith et al., 2003, 2004). The amount of radioactivity in roots and shoots of the two control plants and both PT13 mutants was significantly higher in G. intraradices–inoculated than in the corresponding mock-inoculated control plants. By contrast, 33P uptake in the two PT11 mutant lines (Figure 7B) was almost completely impaired in inoculated plants. The block in symbiotic Pi transfer was confirmed by staining for polyP granules that had accumulated in intraradical fungal vesicles within the PT11 mutant but not in wild-type or PT13 mutant lines (see Supplemental Figure 4 online).
Next, the contribution of the symbiotic Pi acquisition pathway to overall Pi uptake and the percentage of P transferred via the mycorrhizal pathway were determined. In wild-type, vector control, pt13-1, and pt13R1 plants, between 67% ± 9% and 77% ± 7% of P was taken up via the mycorrhizal pathway, demonstrating that the majority of the Pi of mycorrhizal rice had been symbiotically acquired (Figure 7C). By contrast, in pt11-1 and pt11R1, only between 2.2% ± 0.5% and 5.1% ± 2.3% of P was derived from AM fungi (Figure 7C). The double PT11/PT13 RNAi line phenocopied PT11 mutants throughout the P transfer experiment (see Supplemental Figure 5 online), further supporting the importance of PT11 and the insignificance of PT13 for symbiotic Pi uptake. In summary, these results demonstrated that PT11 but not PT13 is required for symbiotic Pi uptake, which is the primary route for obtaining Pi from aerobic soil in rice.
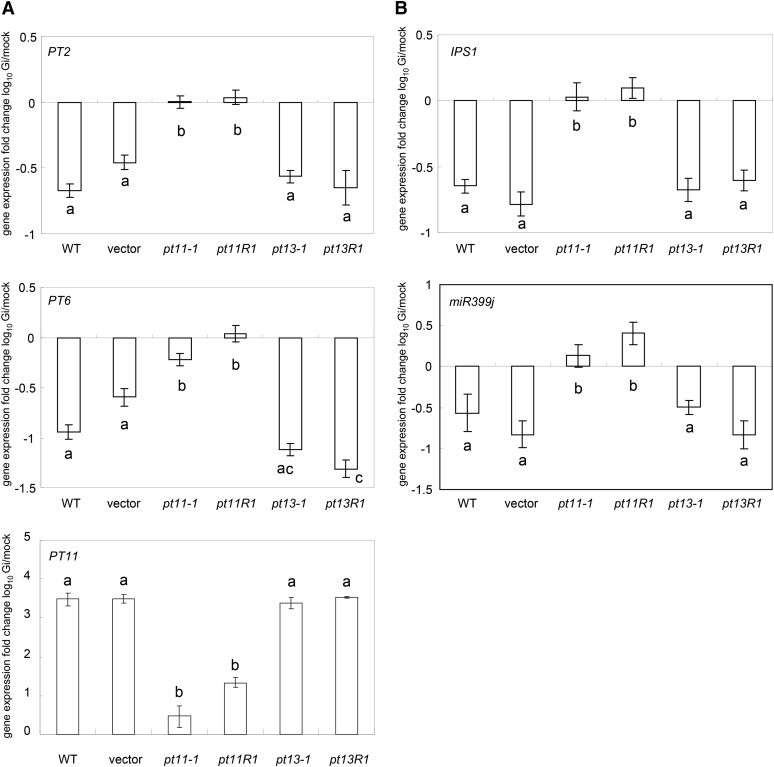
To molecularly examine the activity of the direct and symbiotic Pi uptake pathway across the single and double mutants of PT11 and PT13, we used PT11 for the symbiotic pathway and the marker genes PT2 and PT6, which both play a role in direct Pi acquisition and are Pi starvation inducible (Ai et al., 2009). Transcript levels of PT2 and PT6 were downregulated in the wild type, empty vector control, and PT13 mutant roots (Figure 8A) that had efficiently accumulated higher P tissue levels during symbiosis (Figure 7A) and strongly expressed PT11 (Figure 8A). By contrast, in G. intraradices–inoculated mutant roots of PT11, which maintained low levels of tissue P, no reduction of PT2 and PT6 mRNA was observed, consistent with the continuous functioning of the direct uptake pathway (Figure 8A). Furthermore, to molecularly monitor possible Pi starvation of mutant and wild-type plants in the presence or absence of symbiosis, the different mutant genotypes were examined for the expression of well-established Pi starvation response marker genes, namely IPS1 and one of the rice miR399 precursors, miR399j (Wasaki et al., 2003; Hou et al., 2005; Bari et al., 2006; Wang et al., 2009; Hu et al., 2011). Similar to PT2 and PT6, the expression of both marker genes was reduced in mycorrhizal wild-type and PT13 mutant plants (Figure 8B), thereby demonstrating that the functional establishment of the AM symbiosis ameliorated the Pi status of control and PT13 mutant plants, but not of PT11 mutant genotypes.
Figure 8.
Expression Patterns of Marker Genes for the Direct and the Symbiotic Uptake Pathway and the Pi Starvation Response in PT11 and PT13 Mutant Lines.
Real-time RT-PCR–based expression analysis of PT2 and PT6 for the direct and PT11 for the symbiotic Pi uptake pathway (A) and IPS1 and miR399j for Pi starvation (B) in mock and mycorrhizal roots of control and mutant rice genotypes as indicated. Mean and se values of three biological replicates are shown. The fold changes in gene expression levels of G. intraradices (Gi) relative to mock-inoculated rice are depicted. As PT11 is not expressed in the absence of symbiosis, the mock value was artificially floored to 2.00E-05. Bars with dissimilar letters are significantly different at P < 0.01 after one-way ANOVA (paired-sample t tests were performed with Bonferroni adjustment). WT, the wild type.
According to our earlier work, PT11 encodes a functional Pi transporter (Paszkowski et al., 2002). To verify transporter activity of PT13, the yeast Pi uptake mutant pam2, which lacks the high-affinity, proton-coupled phosphate transporter PHO84 and the high-affinity, sodium-coupled transporter PHO89 (Martinez and Persson, 1998), was transformed with the rice PT13 gene under the control of the constitutive yeast plasma membrane ATPase (PMA1) promoter. Whereas the ectopic expression of the yeast PHO84 gene resulted in significant Pi uptake, no transport activity was measured for yeast strains expressing the rice PT13 gene, despite high mRNA and protein levels (see Supplemental Figures 6A and 6B online).
To investigate the association of PT13 gene activity with the mineral status of the plant, wild-type rice was grown under increasing Pi, nitrate, and sulfate regimes. Marker genes were selected, namely, the Pi transporter PT6 (Paszkowski et al., 2002; Ai et al., 2009), nitrate transporter NRT2.1 (Orsel et al., 2002; Araki and Hasegawa, 2006), and sulfate transporter Sultr1.1 (Rouached et al., 2008; Kumar et al., 2011), and their suitability to mark starvation conditions was confirmed in noncolonized roots of plants cultured at low, medium, and high concentrations of the respective mineral (see Supplemental Figure 7A online). PT11 is a well-established and highly specific marker gene for monitoring arbuscule development (Gutjahr et al., 2008). An induction of PT13 gene activity in response to changing fertilizer conditions would thus result in diverging expression patterns from PT11. However, fertilization of rice plants with increasing amounts of Pi, nitrate, or sulfate did not result in detectable levels of PT11 or PT13 transcripts in the absence of symbiosis (see Supplemental Figure 7A online). Roots inoculated with G. intraradices showed high levels of PT11 mRNA in response to low Pi that decreased with higher fertilizer Pi concentration (see Supplemental Figure 7B online), reflecting the well-known suppression of colonization by high Pi supplement (Smith and Read, 2008; Breuillin et al., 2010). Also, the addition of higher levels of sulfate reduced PT11 transcript accumulation, indicative of lower colonization levels (see Supplemental Figure 7B online) and confirming recent observations in M. truncatula (Casieri et al., 2012). Interestingly, although the increased concentration of nitrate resulted in reduced NRT2.1 expression, it did not lead to diminished fungal root colonization, as mirrored by the continuously high level of PT11 transcripts (see Supplemental Figure 7B online). By contrast, comparable N regimes led to suppression of colonization in M. truncatula (Javot et al., 2011), suggesting differences between rice and M. truncatula regarding the control of AM symbiosis development in response to increasing N availability. The expression profile of PT13 reproduced that of PT11 throughout the experiment (see Supplemental Figure 7B online), suggesting gene expression to be tightly associated with AM development and independent of the P, N, or S nutritional status of the plant.
Marker Gene Expression Analysis Suggests Dominance of Mycorrhizal Pi Uptake also under Field Conditions
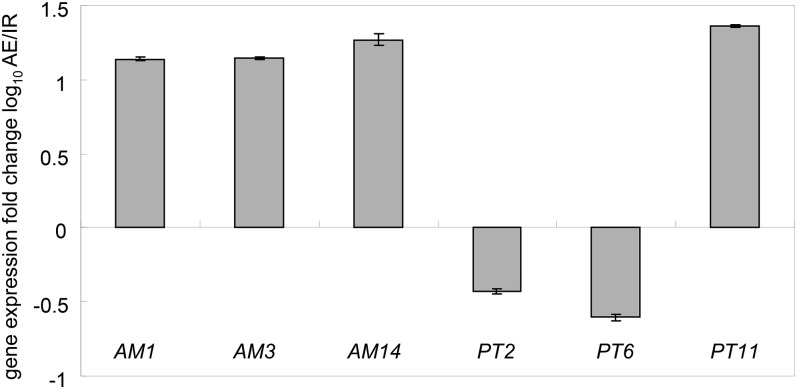
We intended to obtain an estimate of the coordination between direct and symbiotic Pi uptake pathways under field conditions. For this purpose, we compared noncolonized and mycorrhizal rice plants grown in soil from agricultural rice fields. Accordingly and to mimic standard agricultural practices, we used soil from irrigated and aerobic fields to produce low/noncolonized and mycorrhizal rice, respectively (Solaiman and Hirata, 1997; Vallino et al., 2009; Lumini et al., 2011), without further addition of inoculum or fertilizer. The rice variety IR66 was selected for this study as it is known for its tolerance to diverse growth conditions (Carpenter, 2005). Roots of 6-week-old plants were analyzed for the presence of fungal colonization using the well-characterized rice marker genes AM1 and AM3, which show an early and continuous induction during the interaction, and AM14, which is activated at later stages of the symbiosis (Gutjahr et al., 2008, 2012; Mukherjee and Ané, 2011). A more than 10-fold increase in marker gene mRNA accumulation in plants grown in soil from two different aerobic fields relative to soil from an irrigated field indicated high and low fungal colonization, respectively (Figure 9). Therefore, whereas continuous submersion had reduced AM fungal inoculum in the soil, aerobic cultivation maintained elevated levels of AM fungal propagules that permitted efficient natural inoculation of rice. Next, we addressed the activity of the direct and mycorrhizal Pi acquisition pathway by quantifying the expression of the corresponding Pi transporter marker genes PT2 and PT6 as well as PT11. As expected, the expression profile of PT11 matched that of the AM marker genes (Figure 9), suggesting symbiotic Pi uptake to be particularly active in rice cultivated on soil from aerobic fields. By contrast, both Pi transporters of the direct uptake pathway, PT2 and PT6, were expressed at lower levels in rice plants grown in aerobic soil compared with irrigated soil (Figure 9). Therefore, the expression profile of marker genes for the direct and symbiotic Pi uptake route are identical in two very distinct rice varieties, the japonica-type variety Nipponbare and the indica-type rice IR66, grown under defined experimental laboratory conditions as well as in natural soil collected from rice fields (Figures 8 and 9), suggesting an important role of the symbiotic uptake pathway for field-grown rice.
Figure 9.
Gene Expression Analysis of Marker Genes from Roots of Rice Grown in Irrigated and Aerobic Soils.
Transcript levels of marker genes were determined from roots of 6-week-old IR66 roots. The genes represent markers for mycorrhizal colonization (AM1, AM3, and AM14) and Pi transporters involved in the direct (PT2 and PT6) and symbiotic Pi uptake pathway (PT11). The fold changes in gene expression levels of rice grown in aerobic (AE) relative to irrigated (IR) soil are depicted. Mean and se values of three biological replicates are shown.
DISCUSSION
The rice PT11 type of PHT1 proteins forms a monophyletic group within mycorrhizal angiosperms that indicates functional conservation across its two classes, the di- and the monocotyledons. This group is closely related to the lineage that includes all P. patens and most S. moellendorffii PHT1 proteins, which are evolutionarily old species and considered remnants of early land (Rensing et al., 2008) and vascular plants, respectively (Banks et al., 2011). The branch point between the PT11 and P. patens/S. moellendorffii lineages demonstrates that these proteins share an ancient last common ancestor, supporting the evolutionary antiquity of proteins involved in symbiotic Pi uptake (Wang et al., 2010). However, the acquisition of mycorrhizal function for the PT11-like protein only occurred after this ancient common ancestor or was alternatively lost in some lineages because P. patens is not capable of forming mycorrhizal symbiosis. Evolutionarily more recent, the grasses acquired a second AM-specific PHT1 protein, PT13, which is phylogenetically distant from PT11. Except for rice, the PT13 homologs of the currently sequenced cereals underwent gene duplication, resulting in a small gene family of two to four members. Two of the three corresponding B. distachyon genes (PT12 and PT13) are induced during AM symbiosis (Hong et al., 2012), similar to rice PT13, indicative of a related function.
Interestingly, a number of mycorrhizal plant species or families have evolved variations to the PT11 theme. Maize PT6 and B. distachyon PT7, for instance, represent unusual members of the PT11 lineage, as transcripts accumulate not only in mycorrhizal but also in noncolonized roots and leaves of Pi-starved plants (Nagy et al., 2006; Hong et al., 2012), suggesting that these proteins play an additional role during asymbiosis. In tomato, gene duplication of the PT11 ortholog PT4 gave rise to PT5, an AM-induced Pi transporter with expression in the absence of symbiosis (Nagy et al., 2005). The lack of an AM-related phenotype in the tomato PT4 mutant was therefore attributed to the genetic redundancy among its AM-induced PHT1 gene (Nagy et al., 2005). In rice, the tight association of PT11 and PT13 expression with arbusculated cells pointed to a similar genetic redundancy of a Pi transporter as observed in tomato. Therefore, the clear functional separation between PT11 and PT13 represents a novel and surprising division of labor of AM-specific PHT1 proteins in plants. It became apparent that PT11 alone is necessary and sufficient to mediate the entire symbiotic Pi uptake in rice, whereas mutation in PT13 did not affect this process. Lack of functional PT13 resulted in the overall decrease in intraradical fungal development of either G. intraradices or G. rosea, but interestingly, in the absence of detectable alterations in Pi transfer. Thus, each of these two rice PHT1 genes is unexpectedly indispensable for the formation of AM symbioses. Mutations in either gene resulted in underdeveloped arbuscules, suggesting deficiencies in signals that coordinate normal expansion of arbuscules. According to a previous hypothesis (Javot et al., 2007; Oldroyd et al., 2009; Yang and Paszkowski, 2011), symbiotically delivered and acquired Pi might itself serve as a signal for the plant for the reprogramming of cortex cells, which is necessary for the proper formation of the sophisticated arbuscular structures. Moreover, spatial discrimination between the acquisition of Pi ions deep inside the root cortex and at the root peripheral cell layers might be necessary for the plant’s decision to enter and support symbiosis. The PT11 mutants phenocopied M. truncatula deficient for PT4 (Javot et al., 2007), which is in line with both hypotheses, implying a general and, thus, possibly ancient regulatory mechanism of symbiotic interaction associated with active Pi acquisition.
However, our functional studies on rice PT13 demonstrate that extrapolations across plant families or even species cannot always be made. Although intact PT13 is necessary for arbuscule development in rice, it is unlikely that PT13 encodes a functional Pi transporter in the plasma membrane, as revealed by our experiments in plants and yeast. Instead, it is more probable that PT13 plays a central role in symbiotic signaling that directs developmental decisions in symbiosis. There is a growing body of evidence for the importance of environmental sensors related to nutrient transporters, the so-called transceptors (Holsbeeks et al., 2004; Thevelein and Voordeckers, 2009), which include not only transporting receptors but also their nontransporting close homologs. Both types could be represented by PT11 and PT13, respectively, and only their concerted action might allow for the full culmination of the symbiosis. The role PT13 plays in this process is still enigmatic, but it might be involved in monitoring ambient Pi levels at the periarbuscular interface, which is needed for adjusting arbuscule development to maximize harnessing the desired mineral. In the absence of PT13, development of arbuscules terminates prematurely due to the lack of a stimulus that is required to reach maximal expansion. Interestingly, the level of PT11 mRNA present in PT13-deficient roots seems to be constant relative to wild-type roots. Therefore, in PT13 mutants, constant PT11 gene activity could account for stable rates of symbiotic Pi uptake.
Finally, our study demonstrated that mycorrhizal rice receives more than 70% of its overall Pi via the symbiotic pathway and therefore largely depends on the AM symbiosis. Dominance of the symbiotic Pi uptake route was molecularly confirmed and demonstrated to similarly occur in field soil-grown rice plants upon establishment of AM symbiosis. Also, the dicotyledons flax, M. truncatula, and tomato, when living in association with G. intraradices, received most of their Pi via the mycorrhizal route (Smith et al., 2003). However, maintenance of this dominance in members of the genus Oryza is particularly surprising, as they evolved from a semiaquatic ancestor, well adapted to anaerobic growth conditions that are nonpermissive for efficient colonization by the aerobic AM fungi (Barea, 1991). Furthermore, generations of intentional breeding of rice varieties, including Nipponbare and IR66 used in this study, targeted improved plant performance under anaerobic paddy field conditions, generally well supplied with Pi fertilizer. Yet, even these modern varieties depend on AM fungi or Pi uptake under aerobic conditions, which is achieved by the single functional and evolutionary conserved Pi transporter protein, PT11.
Currently, rice varieties adapted to aerobic conditions are becoming favorable alternatives to irrigated lowland rice cultivars due to their tolerance to less intense agricultural practices and water scarcity. The data presented in this study enhance our understanding of symbiotic Pi uptake in rice, which might be directly relevant for the development of rice cultivars better adapted to a low-input rice agro-ecosystem.
METHODS
Plant Material
The rice (Oryza sativa ssp japonica) mutant genotypes used in the laboratory study arose in the cv Nipponbare background. Homozygous seeds of the rice insertional mutant line NG6473 of PT11 were obtained from the Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan (Miyao et al., 2003). Segregating seeds of the PT13 insertional mutant (RdSpm1154_3.1) were provided by the Department of Plant Biology, University of California, Davis (Kumar et al., 2005). Experiments with field soil were conducted with the O. sativa spp indica variety IR66.
Plant Growth Conditions and Inoculation with AM Fungi
For standard laboratory conditions, rice seeds were surface sterilized and inoculated with 200 aseptically grown Glomus intraradices spores or Gigaspora rosea purchased from Biorhize (strain BEG9) as previously reported (Gutjahr et al., 2008). Inoculated plants were grown in a phytochamber with a 12-h day/night cycle at 28/22°C and 60% air humidity. The plants were regularly watered for the first 2 weeks and thereafter fertilized every second day with a mix of one-half Hoagland solution containing 25 μM Pi and 0.01% (w/v) Sequestren Rapid (Syngenta).
Nutrient Regime Experiment
After 2 weeks, the plants were supplied every second day with a mix of one-half Hoagland solution containing a low, medium, or high concentration of Pi, nitrate, or sulfate (see Supplemental Table 1 online). Increasing nitrate and sulfate regimes were applied in the presence of low Pi.
Quantitative P Uptake Assay
The compartmented pot design followed the protocol by Smith et al. (2004), where the root plus hyphal compartment (RHC) consisted of a plastic bag–lined pot, containing 910 g of growth medium. The hyphal compartment (HC) was a small plastic vial, containing 59 g of the growth medium and capped with a 25-μm nylon mesh at both ends. The growth medium in both RHC and HC was a 1:1 mixture of sand and low P irradiated soil (10 kGy, 10 MeV electron beam), which was mixed with basal nutrients and 10 mg KH2PO4-P kg−1 (Pearson and Jakobsen, 1993). N was added as 30 mg NH4NO3-N kg−1 at the start of the experiment. Supplemental N was added periodically during plant growth, to provide an additional 90 mg N per pot by the end of the experiment. Soil for the HCs was well mixed with carrier-free H333PO4 to provide 187.8 kBq mg −1 bicarbonate-extractable P (Olsen et al., 1954). Sufficient water was added to bring the soil to a water content of 0.16 g per g oven-dried soil. Two pregerminated seedlings were planted per pot. Dry sand inoculum of G. intraradices was mixed into the soil in the RHC of mycorrhizal pots only.
Field Soil Experiments
Soil was collected from fields that had been continuously irrigated or were maintained in an aerobic state for many years at the international rice research institute field station (Metro Manila, Philippines). Pregerminated seeds of IR66 were surface sterilized as described above and transplanted into the respective soil conditions. The plants were grown in a glasshouse for 6 weeks and regularly watered without fertilizer. Representative root samples for gene expression analysis were harvested in triplicate from individual plants at 6 weeks.
Root Staining and Mycorrhizal Quantification
Roots were stained with Trypan blue, and mycorrhizal colonization was examined and quantified with a modified gridline intersect procedure as described (Paszkowski et al., 2006). Images were taken with a Leica DM 5000 B microscope equipped with a Leica DFC420 camera. Images of roots stained with WGA were taken with a Zeiss LSM 710 NLO with a Zeiss AxioExaminer Z1 microscope and two-photon laser at an excitation of 960 nm.
Monitoring Tissue Phosphorus Content and Radioactive Phosphate Transfer
At 8 wpi, total dry weight was determined on subsamples of root and shoot tissue. For monitoring P concentration and specific activity of 33P, dried ground material was digested in a solution of nitric and perchloric acids (4:1, v/v). Total P concentration in the digest was measured by the molybdate blue method (Murphy and Riley, 1962) on a Technicon Autoanalyser II (Analytical Instruments Recycle). Digests were also analyzed for 33P content in a Packard TR 1900 liquid scintillation counter using the scintillation cocktail Ultima Gold (Perkin-Elmer) and corrected for isotopic decay. The percentage of contribution of the mycorrhizal pathway to total P uptake was calculated as follows: 100 * (SA33P plant/SA33P HC) * (P in pot/P in HC) * (HLD in RHC/HLD in HC); SA refers to the specific activity, P is bicarbonate-extractable P, and HLD is hyphal length density. Based on previous studies with G. intraradices (Smith et al., 2004), it was assumed that HLD was similar in the RHC and the HC.
Identification of Insertion Mutant Lines
The Tos17 retrotransposon mutant collection (Miyao et al., 2003) was screened by PCR and an insertion 226 bp upstream of the start codon of PT11 was identified (see Supplemental Figure 1A online). For 5′-rapid amplification of cDNA ends, the Gene-Racer kit (Invitrogen) was used according to the manufacturer’s instructions. The transcriptional start site was placed 84 bp upstream of the ATG start codon (see Supplemental Figure 1A online). For PT13, a dSpm element (Kumar et al., 2005) was found to have inserted into the first exon of PT13 at 88 bp downstream of the start codon (see Supplemental Figure 1A online). Insertions into PT11 and PT13 were confirmed by PCR amplification (see Supplemental Table 2 online) and subsequent sequencing.
Generation of Constructs for Promoter-GUS and RNAi Rice Lines
The 3′-untranslated region (UTR) region of PT11 and PT13 was amplified (see Supplemental Table 3 online) from genomic DNA of rice roots and cloned into the pENTR/D-TOPO vector (Invitrogen) for subsequent introduction into the Gateway-compatible pANDA vector (Miki and Shimamoto, 2004). For generating the concatameric double RNAi construct, the 3′-UTR region of PT11 was subcloned into the pGEM-T-easy vector (Promega), digested with NotI (Promega), and ligated into the pENTR/D-TOPO vector containing the 3′-UTR region of PT13 at NotI. Finally, the fragment containing the 3′-UTR region of both PHT1 genes was introduced into the pANDA vector. Segregating rice transformants were genotyped by PCR (see Supplemental Table 4 online). From five to 10 independent transformants, the line exhibiting the strongest downregulation was selected for further analyses.
A 1967-bp fragment of the promoter region of PT11 and 2000 bp of PT13 (see Supplemental Table 3 online) were subcloned into the pENTR/D-TOPO vector and cloned into a Gateway pHGWFS7.0 vector containing an enhanced green fluorescent protein-GUS fusion reporter gene (Karimi et al., 2002).
Rice Transformation
The rice transformation protocol was adopted from Sallaud et al. (2003) and Toki et al. (2006). Briefly, scutellar calli were generated from mature seeds of wild-type rice cv Nipponbare. Callus production was induced, maintained on N6D media, and kept in the dark at 30°C. Three-week-old calli were incubated with Agrobacterium tumefaciens EHA105 containing plasmid DNA. Transformants were selected by keeping the calli on N6D media supplemented with hygromycin (50 mg/L) and carbenicillin (400 mg/L) in the dark for 3 weeks and then transferring them to PRAG media for 5 d. Resistant colonies were transferred to regeneration media.
RNA Extraction, cDNA Synthesis, RT-PCR, and Real-Time RT-PCR
RNA extraction, cDNA synthesis, RT-PCR, and real-time RT-PCR were performed as previously reported (Gutjahr et al., 2008). Briefly, RNA was extracted from 100 mg ground root tissue with the TRIzol reagent, and residual DNA was removed by treatment with DNaseI (Invitrogen) following the manufacturer’s instructions. Reverse transcription was performed using Superscript II reverse transcriptase (Invitrogen) and oligo (dT) primer. The absence of contaminating genomic DNA was confirmed by performing a control PCR on RNA not reverse transcribed (-RT). Real-time RT-PCR was performed as described (Güimil et al., 2005) with gene-specific primers (see Supplemental Table 5 online). Transcript levels were normalized to the constitutively expressed Cyclophilin2 or Ubiquitin gene and displayed as a function of Cyclophilin2 or Ubiquitin expression.
Histochemical GUS, Vital, and Polyphosphate Staining
Published reports were followed for GUS (Scarpella et al., 2003), vital (Schaffer and Peterson, 1993), and polyphosphate staining (Ezawa et al., 2003).
Phylogenetic Analyses
A BLASTO search (Zhou and Landweber, 2007) using the keyword KOG0252 (inorganic Pi transporter) as a query was performed to retrieve the PHT1 sequences of Physcomitrella patens, Selaginella moellendorffii, maize (Zea mays), sorghum (Sorghum bicolor), and Brachypodium distachyon from the Phytozome v7.0 database (http://www.phytozome.net/). The identified sequences were downloaded from GenBank. Phylogenetic analyses were performed under both maximum likelihood and Bayesian inference. The software PhyML was used for maximum likelihood, and the phylogenetic trees were reconstructed using the JTT+Γ model of amino acid evolution. Confidence in the topology reconstructed was assessed by running 1000 bootstrap replicates. Bayesian inference was performed with the software MrBayes using the JTT+Γ model of amino acid evolution. Four heated Markov chain Monte Carlo chains were run for 50 million generations, and the full analyses was repeated twice. The convergence of the Markov chain was assessed by checking the model parameters and priors in the software Tracer.
Heterologous Protein Expression in Saccharomyces cerevisiae
PT13 was expressed under the plasma membrane ATPase (PMA1) promoter (vector pDR) in S. cerevisiae strain pam2. Yeasts were cultivated in SD medium with 7.4 mM phosphate. Transport studies were performed essentially as described (see Supplemental Table 6 online; Dohmen et al., 1991; Rentsch et al., 1995; Martinez and Persson, 1998). For immunoblot analysis, the open reading frames of PT13 and PHO84 were transferred into vector pDR-Sp-GFP6 (Daram et al., 1998). Total protein was extracted according to Komarova et al. (2012), and protein was detected using anti–green fluorescent protein antibodies (Invitrogen).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Os03g05640 (PT2), Os08g45000 (PT6), Os01g46860 (PT11), Os04g10800 (PT13), Os03g05334 (IPS1), Os04g04750 (AM1), Os01g57400 (AM3), Os11g26140 (AM14), Os02g02170 (NRT2.1), Os03g09970 (Sultr1.1), Os02g02890 (Cyclophilin2), and Os06g46770 (Ubiquitin).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of PT11 and PT13 Insertion Mutants and Corresponding RNAi Lines.
Supplemental Figure 2. Absence of Off-Target Silencing as Monitored by Real-Time RT-PCR.
Supplemental Figure 3. Colonization of the PT11 and PT13 Mutant Lines by Gigaspora rosea.
Supplemental Figure 4. Polyphosphate Stain of Mycorrhizal Control, PT11, and PT13 Mutant Roots.
Supplemental Figure 5. Quantitative Symbiotic Pi Transfer in Double Mutant of PT11 and PT13.
Supplemental Figure 6. Transcript and Protein Levels of Ectopically Expressed Yeast PHO84 and Rice PT13 in Yeast.
Supplemental Figure 7. Expression Analysis of PT11 and PT13 in Roots in Response to Increasing Regimes of Phosphate, Nitrate, or Sulfate Fertilization.
Supplemental Table 1. Concentration of Compounds in One-Half Strength Hoagland Solution.
Supplemental Table 2. Primer Sequences Used for Genotyping of Wild-Type and Insertion Mutant Alleles of PT11 and PT13.
Supplemental Table 3. Primer Sequences Used for RNAi and Reporter Lines of PT11 and PT13.
Supplemental Table 4. Primer Sequences Used for Genotyping of RNAi Segregating Populations.
Supplemental Table 5. Real-Time RT-PCR Primers for Gene Expression Analysis of PHT1 Genes, Pi Starvation-Induced Genes, Mycorrhizal Marker Genes, Nitrate and Sulfate Transporters, and Constitutive Genes.
Supplemental Table 6. RT-PCR Primer Sequences Used for Gene Expression Analysis of PHO84 and PT13 in the Yeast pam2 Mutant.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Figure 1C.
Supplementary Material
Acknowledgments
We thank Ruairidh Sawers and Philippe Reymond for their valuable suggestions on the article and Marco Chiapello for assistance with statistics. We also thank Jacqueline Gheyselinck for her excellent technical help with rice transformation. S.-Y.Y. was supported by the National Center of Competence in Research program “plant survival” and by a stipend from the Societé Académique Vaudoise. V.S. received funding from the National Science Foundation and U.P. from the Swiss National Science Foundation “professeur boursier” Grants PP00A_110874 and PP00P3_130704.
AUTHOR CONTRIBUTIONS
S.-Y.Y. did most of the experimental work and analyses and was involved in writing the article. M.G. and I.J. supervised S.-Y.Y. for the experimental work on radioactive Pi transfer and contributed to writing the article. M.S.G. and D.R. performed the yeast uptake and PT13 expression studies. A.M. and H.H. PCR screened the Tos17 library for insertions into PT11. C.S.K. and V.S. provided the insertion mutant of PT13. S.C., N.M., and S.H. conducted the experiments with field soil and performed the molecular analyses. S.H. also contributed to writing the article. U.P. was the principal investigator of the project, designed the work, supervised S.-Y.Y., and wrote the article.
Glossary
- AM
arbuscular mycorrhizal
- TM
transmembrane
- wpi
weeks postinoculation
- GUS
β-glucuronidase
- RNAi
RNA interference
- RHC
root plus hyphal compartment
- HC
hyphal compartment
- UTR
untranslated region
- ANOVA
analysis of variance
References
- Ai P., Sun S., Zhao J., Fan X., Xin W., Guo Q., Yu L., Shen Q., Wu P., Miller A.J., Xu G. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Araki R., Hasegawa H. (2006). Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breed. Sci. 56: 295–302 [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barea J. (1991). Vesicular-arbuscular mycorrhizae as modifiers of soil feritlity. In Advances in Soil Science 15, B. Stewart, ed (New York: Springer), pp. 1–40
- Bari R., Datt Pant B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolan N. (1991). A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant Soil 134: 189–207 [Google Scholar]
- Breuillin F., et al. (2010). Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 64: 1002–1017 [DOI] [PubMed] [Google Scholar]
- Carpenter D. (2005). The in situ conservation of rice plant genetic diversity: A case study from a Philippine barangay. Agric. Human Values 22: 421–434 [Google Scholar]
- Casieri L., Gallardo K., Wipf D. (2012). Transcriptional response of Medicago truncatula sulphate transporters to arbuscular mycorrhizal symbiosis with and without sulphur stress. Planta 235: 1431–1447 [DOI] [PubMed] [Google Scholar]
- Daram P., Brunner S., Persson B.L., Amrhein N., Bucher M. (1998). Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta 206: 225–233 [DOI] [PubMed] [Google Scholar]
- Dohmen R.J., Strasser A.W.M., Höner C.B., Hollenberg C.P. (1991). An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7: 691–692 [DOI] [PubMed] [Google Scholar]
- Ezawa T., Cavagnaro T., Smith S., Smith F., Ohtomo R. (2003). Rapid accumulation of polyphosphate in extraradical hyphae of an arbsucular mycorrhizal fungus as revealed by histochemistry and polyphosphatekinase/luciferase system. New Phytol. 161: 387–392 [DOI] [PubMed] [Google Scholar]
- Glassop D., Smith S.E., Smith F.W. (2005). Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222: 688–698 [DOI] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Güimil S., Chang H.S., Zhu T., Sesma A., Osbourn A., Roux C., Ioannidis V., Oakeley E.J., Docquier M., Descombes P., Briggs S.P., Paszkowski U. (2005). Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. USA 102: 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Banba M., Croset V., An K., Miyao A., An G., Hirochika H., Imaizumi-Anraku H., Paszkowski U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., et al. (2012). The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J. 69: 906–920 [DOI] [PubMed] [Google Scholar]
- Harrison M.J., Dewbre G.R., Liu J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde S., Thevelein J.M. (2004). The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29: 556–564 [DOI] [PubMed] [Google Scholar]
- Hong J.J., Park Y.S., Bravo A., Bhattarai K.K., Daniels D.A., Harrison M.J. (2012). Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta 236: 851–865 [DOI] [PubMed] [Google Scholar]
- Hou X., Wu P., Jiao F., Jia Q., Chen H., Yu J., Song X., Yi K. (2005). Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ. 28: 353–364 [Google Scholar]
- Hu B., Zhu C., Li F., Tang J., Wang Y., Lin A., Liu L., Che R., Chu C. (2011). LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Breuillin F., Bhattarai K.K., Noar R.D., Gomez S.K., Zhang Q., Cook D.R., Harrison M.J. (2011). Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J. 68: 954–965 [DOI] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H., Pumplin N., Harrison M.J. (2007b). Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ. 30: 310–322 [DOI] [PubMed] [Google Scholar]
- Jia H., Ren H., Gu M., Zhao J., Sun S., Chen J., Wu P., Xu G. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kobae Y., Hata S. (2010). Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 51: 341–353 [DOI] [PubMed] [Google Scholar]
- Komarova N.Y., Meier S., Meier A., Grotemeyer M.S., Rentsch D. (2012). Determinants for Arabidopsis peptide transporter targeting to the tonoplast or plasma membrane. Traffic 13: 1090–1105 [DOI] [PubMed] [Google Scholar]
- Kumar C.S., Wing R.A., Sundaresan V. (2005). Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J. 44: 879–892 [DOI] [PubMed] [Google Scholar]
- Kumar S., Asif M.H., Chakrabarty D., Tripathi R.D., Trivedi P.K. (2011). Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct. Integr. Genomics 11: 259–273 [DOI] [PubMed] [Google Scholar]
- Loth-Pereda V., Orsini E., Courty P.E., Lota F., Kohler A., Diss L., Blaudez D., Chalot M., Nehls U., Bucher M., Martin F. (2011). Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol. 156: 2141–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumini E., Vallino M., Alguacil M.M., Romani M., Bianciotto V. (2011). Different farming and water regimes in Italian rice fields affect arbuscular mycorrhizal fungal soil communities. Ecol. Appl. 21: 1696–1707 [DOI] [PubMed] [Google Scholar]
- MacDonald G.K., Bennett E.M., Potter P.A., Ramankutty N. (2011). Agronomic phosphorus imbalances across the world’s croplands. Proc. Natl. Acad. Sci. USA 108: 3086–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti D., Toppo N.N., Variar M. (2011). Integration of crop rotation and arbuscular mycorrhiza (AM) inoculum application for enhancing AM activity to improve phosphorus nutrition and yield of upland rice (Oryza sativa L.). Mycorrhiza 21: 659–667 [DOI] [PubMed]
- Martinez P., Persson B.L. (1998). Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258: 628–638 [DOI] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Misson J., Thibaud M.C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Miyao A., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Ané J.M. (2011). Germinating spore exudates from arbuscular mycorrhizal fungi: Molecular and developmental responses in plants and their regulation by ethylene. Mol. Plant Microbe Interact. 24: 260–270 [DOI] [PubMed] [Google Scholar]
- Murphy J., Riley J.P. (1962). A modified single solution method for determination of phosphate in natural water. Anal. Chim. Acta 27: 495–501 [Google Scholar]
- Nagy R., Karandashov V., Chague V., Kalinkevich K., Tamasloukht M., Xu G., Jakobsen I., Levy A.A., Amrhein N., Bucher M. (2005). The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 42: 236–250 [DOI] [PubMed] [Google Scholar]
- Nagy R., Vasconcelos M.J.V., Zhao S., McElver J., Bruce W., Amrhein N., Raghothama K.G., Bucher M. (2006). Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol. (Stuttg.) 8: 186–197 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Harrison M.J., Paszkowski U. (2009). Reprogramming plant cells for endosymbiosis. Science 324: 753–754 [DOI] [PubMed] [Google Scholar]
- Olsen S., Cole C., Watanabe F., Dean L. (1954). Estimation of available phosphorus in soils by extraction with sodium bicarbonate. In USDA Circ. 939, (Washington, D.C., Government Printing Office), pp. 1–19
- Orsel M., Krapp A., Daniel-Vedele F. (2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao S.S., Paulsen I.T., Saier M.H., Jr (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U., Jakovleva L., Boller T. (2006). Maize mutants affected at distinct stages of the arbuscular mycorrhizal symbiosis. Plant J. 47: 165–173 [DOI] [PubMed] [Google Scholar]
- Paszkowski U., Kroken S., Roux C., Briggs S.P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.N., Jakobsen I. (1993). The relative contribution of hyphae and roots to phosphorus uptake by arbuscular mycorrhizal plants, measured by dual labelling with 32P and 33P. New Phytol. 124: 489–494 [Google Scholar]
- Poirier Y., Bucher M. (2002). Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book 1: e0024, doi/10.1199/tab.0024 [DOI] [PMC free article] [PubMed]
- Rausch C., Daram P., Brunner S., Jansa J., Laloi M., Leggewie G., Amrhein N., Bucher M. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–470 [DOI] [PubMed] [Google Scholar]
- Remy W., Taylor T.N., Hass H., Kerp H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rentsch D., Laloi M., Rouhara I., Schmelzer E., Delrot S., Frommer W.B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Rouached H., Wirtz M., Alary R., Hell R., Arpat A.B., Davidian J.C., Fourcroy P., Berthomieu P. (2008). Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol. 147: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C., et al. (2003). Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor. Appl. Genet. 106: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Rueb S., Meijer A.H. (2003). The RADICLELESS1 gene is required for vascular pattern formation in rice. Development 130: 645–658 [DOI] [PubMed] [Google Scholar]
- Schaffer G., Peterson L. (1993). Modifications to clearing methods used in combination with vital staining of roots colonized with vesiculararbuscular mycorrhizal fungi. Mycorrhiza 4: 29–35 [Google Scholar]
- Schüssler A., Schwarzott D., Walker C. (2001). A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycol. Res. 105: 1413–1421 [Google Scholar]
- Sharma A., Singh R., Singh U. (1988). Effect of vesicular-arbuscular mycorrhiza on uptake of phosphorus and zinc in rice (Oryza sativa L.). Curr. Sci. 57: 901–902 [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smith S., Read D. (2008). Mycorrhizal Symbiosis. (London: Academic Press)
- Smith S., Smith F., Jakobsen I. (2004). Functional diversity in arbuscular mycorrhizal (AM) symbioses: The contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol. 162: 511–524 [Google Scholar]
- Smith S.E., Smith F.A., Jakobsen I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133: 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaiman M., Hirata H. (1997). Effect of arbuscular mycorrhizal fungi inoculation of rice seedlings at the nursery stage upon performance in the paddy field and greenhouse. Plant Soil 191: 1–12 [Google Scholar]
- Sun S., Gu M., Cao Y., Huang X., Zhang X., Ai P., Zhao J., Fan X., Xu G. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in Pi-replete rice. Plant Physiol. 159: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J.M., Voordeckers K. (2009). Functioning and evolutionary significance of nutrient transceptors. Mol. Biol. Evol. 26: 2407–2414 [DOI] [PubMed] [Google Scholar]
- Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., Tanaka H. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Vallino M., Greppi D., Novero M., Bonfante P., Lupotto E. (2009). Rice root colonisation by mycorrhizal and endophytic fungi in aerobic soil. Ann. Appl. Biol. 154: 195–204 [Google Scholar]
- Wang B., Yeun L.H., Xue J.Y., Liu Y., Ané J.M., Qiu Y.L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186: 514–525 [DOI] [PubMed] [Google Scholar]
- Wang C., Ying S., Huang H., Li K., Wu P., Shou H. (2009). Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wasaki J., Yonetani R., Shinano T., Kai M., Osaki M. (2003). Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytol. 158: 239–248 [Google Scholar]
- Yang S.Y., Paszkowski U. (2011). Phosphate import at the arbuscule: Just a nutrient? Mol. Plant Microbe Interact. 24: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Landweber L.F. (2007). BLASTO: A tool for searching orthologous groups. Nucleic Acids Res. 35 (Web Server issue): W678–W682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.