Mutations generating constitutively active (CA) mitogen-activated protein kinases (MAPKs) were identified and used to investigate the role of Arabidopsis MPK4 in plant immunity, revealing functions of MPK4 activity in PTI and ETI, establishing that the generation of CA-MAPKs offers a powerful tool to analyze MAPK functions in plants.
Abstract
Plant mitogen-activated protein kinases (MAPKs) are involved in important processes, including stress signaling and development. In a functional yeast screen, we identified mutations that render Arabidopsis thaliana MAPKs constitutively active (CA). Importantly, CA-MAPKs maintain their specificity toward known activators and substrates. As a proof-of-concept, Arabidopsis MAPK4 (MPK4) function in plant immunity was investigated. In agreement with the phenotype of mpk4 mutants, CA-MPK4 plants were compromised in pathogen-induced salicylic acid accumulation and disease resistance. MPK4 activity was found to negatively regulate pathogen-associated molecular pattern-induced reactive oxygen species production but had no impact on callose deposition, indicating that CA-MPK4 allows discriminating between processes regulated by MPK4 activity from processes indirectly affected by mpk4 mutation. Finally, MPK4 activity was also found to compromise effector-triggered immunity conditioned by the Toll Interleukin-1 Receptor–nucleotide binding (NB)–Leu-rich repeat (LRR) receptors RPS4 and RPP4 but not by the coiled coil–NB-LRR receptors RPM1 and RPS2. Overall, these data reveal important insights on how MPK4 regulates plant defenses and establishes that CA-MAPKs offer a powerful tool to analyze the function of plant MAPK pathways.
INTRODUCTION
Plants are subjected to a large number and variety of environmental challenges and have to adapt their metabolism, growth, and development accordingly. Plants contain genes encoding for proteins involved in signal perception and transduction, among which protein kinases and phosphatases are particularly abundant (Arabidopsis Genome Initiative, 2000). Four percent of Arabidopsis thaliana genes code for protein kinases compared with 1.7% in human and 2% in yeast (Wang et al., 2003), correlating with the fact that at least 30% of proteins are phosphorylated. Mitogen-activated protein kinase (MAPK) pathways are highly conserved between kingdoms, defining key functional signaling modules that are usually composed of three protein kinases that sequentially activate each other by phosphorylation: a MAP3K (MAP kinase kinase kinase) activates a MAP2K (MAP kinase kinase), which in turn activates a MAPK. In plants, MAPK signaling cascades are involved in various processes, including development, hormone signaling, and stress responses (Colcombet and Hirt, 2008). Among the 20 putative MAPKs found in Arabidopsis, extensive studies showed that MPK3, MPK4, and MPK6 are activated by pathogen elicitors and are major regulators of innate immune responses (Asai et al., 2002; Gao et al., 2008). MAPK cascades are branched pathways (Colcombet and Hirt, 2008; Andreasson and Ellis, 2010; Rodriguez et al., 2010) and a given MAPK can be involved in multiple processes. MPK4 for instance, was originally described as a stress-activated protein kinase (Petersen et al., 2000; Droillard et al., 2004; Teige et al., 2004; Brodersen et al., 2006) and was recently shown to be involved in cytokinesis and cytoskeleton organization (Beck et al., 2010; Kosetsu et al., 2010; Zeng et al., 2011).
Plants and metazoans possess pattern recognition receptors that detect conserved pathogen-associated molecular patterns (PAMPs) and initiate PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). Successful pathogens deliver effectors to the plant apoplast and cytoplasm; these effectors suppress PTI and thereby allow host invasion. As a counterpart, plants evolved intracellular receptors with nucleotide binding (NB)–Leu-rich repeat (LRR) domains that sense effectors and mediate effector-triggered immunity (ETI) (Jones and Dangl, 2006). ETI is often associated with the accumulation of the hormone salicylic acid (SA) and a localized programmed cell death known as the hypersensitive response. The bacterial PAMP flg22, a conserved 22–amino acid peptide derived from Pseudomonas syringae flagellin, has provided a very powerful tool to decipher PAMP-induced signaling pathways and revealed the complexity of MAPK cascades. In Arabidopsis, flg22 recognition is mediated by the LRR receptor kinase FLS2 and leads to an array of defense responses, including generation of reactive oxygen species (ROS), callose deposition, ethylene production, and reprogramming of host cell genes (Gómez-Gómez et al., 1999; Zipfel et al., 2004; Ranf et al., 2011). Flg22 recognition also leads to the activation of two MAPK signaling pathways. One of these MAPK cascades is defined by the MAP2Ks MKK4 and MKK5, which act redundantly to activate the MAPKs MPK3 and MPK6 (Asai et al., 2002). The second cascade activated by flg22 is defined by MEKK1, which activates MKK1 and MKK2, which act redundantly on MPK4 (Gao et al., 2008). These protein kinases interact both in yeast (Ichimura et al., 1998; Teige et al., 2004) and in planta (Gao et al., 2008). Additionally, in mekk1 or mkk1mkk2 backgrounds, the flg22-induced activation of MPK4 is abolished (Mészáros et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008). The mkk1mkk2 double mutants and the mekk1 and mpk4 single mutants exhibit similar phenotypes: They are dwarfed plants overaccumulating SA and ROS, show spontaneous cell death on leaves, and constitutive expression of defense genes (Gao et al., 2008). As a consequence, mkk1 mkk2 and mpk4 plants display enhanced resistance to biotrophic pathogens and increased susceptibility to necrotrophic fungi (Petersen et al., 2000; Qiu et al., 2008). These phenotypes are partially suppressed by the expression of the bacterial salicylate hydroxylase NahG or by mutations that impair SA accumulation (Petersen et al., 2000; Ichimura et al., 2006; Suarez-Rodriguez et al., 2007; Qiu et al., 2008). Transcriptome analysis confirmed the similarities of the mutants and their functions in SA and ROS signaling (Petersen et al., 2000; Brodersen et al., 2006). Recently, the activity of a fourth MAPK, MPK11, was shown to be induced by flg22, but its function in plant immunity remains to be clarified (Bethke et al., 2012).
Mutations triggering constitutive activation of protein kinases have been a powerful approach to decipher their roles. This approach has been very successful in the case of plant MAP2Ks; here, we extend this approach to MAPKs. To identify mutations triggering MAPK kinase-independent constitutive activity in Arabidopsis MAPKs, we developed a functional genetic yeast screen and identified several mutations that render MAPKs constitutively active (CA). Importantly, CA forms of MPK3, 4, and 6 maintained their specificity toward their interacting MAP2Ks and substrates. Given that the severe phenotype of mpk4 mutant plants complicates studies of MPK4 function, we decided to use the CA-MPK4 variant to generate Arabidopsis lines with increased MPK4 activity to allow us to investigate MPK4 functions in planta. As expected, Arabidopsis lines with enhanced MPK4 kinase activity displayed reduced SA and resistance levels compared with wild-type plants. We also showed that MPK4 activity negatively regulates PAMP-induced ROS production. Interestingly, MPK4 activity was found to compromise ETI mediated by the Toll Interleukin-1 Receptor (TIR)-type NB-LRR receptors RPS4 and RPP4 but not by the coiled-coil (CC) type NB-LRR receptors RPM1 or RPS2. These results provide further insight into the role of MPK4 in plant defense and show that the CA-MAPKs are an important new tool for investigating signal transduction.
RESULTS
Functional Complementation of the Yeast pbs2Δhog1Δ Allows the Identification of CA-MPK6 Variants
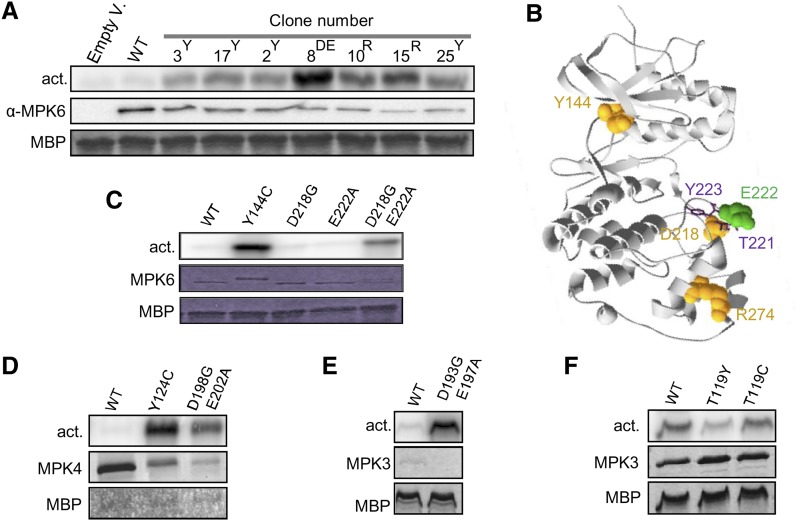
To identify mutations triggering constitutive kinase activation, we adapted a yeast functional screen developed by Engelberg and coworkers (Bell et al., 2001). Our laboratory previously showed that Arabidopsis MPK6, together with the MAP2K MKK2, rescues the salt sensitivity of a yeast strain mutated in the MAP2K PBS2 and the MAPK HOG1 (Teige et al., 2004). In the absence of MKK2, MPK6 does not restore growth of pbs2Δhog1Δ on high salt, indicating that MPK6, without its upstream MAP2K, is not functional. In these conditions, mutations triggering MPK6 constitutive activity should restore the pbs2Δhog1Δ salt growth. We performed a screen on pbs2Δhog1Δ yeast expressing a randomly mutated MPK6 open reading frame (ORF) population. When replicated on 400 to 600 mM NaCl, very few colonies were recovered. To exclude MPK6-independent recovery of salt tolerance, plasmids were extracted, retransformed into pbs2Δhog1Δ, and confirmed by growth on salt. Finally, 27 MPK6 candidate clones were identified and sequenced, revealing mutations in one to six residues in MPK6 (see Supplemental Table 1 online). Y144C, Y144F, and R274H mutations were found in 12, nine, and four clones, respectively, indicating that they are important for pbs2Δhog1Δ complementation.
To confirm that pbs2Δhog1Δ complementation is linked to an increased MPK6 activity, MPK6 protein variants were immunoprecipitated from yeast using a specific anti-MPK6 antibody. The MPK6 kinase activity was subsequently assayed by the ability to phosphorylate myelin basic protein (MBP), a common artificial substrate for MAPKs. Under these conditions, the activity of wild-type MPK6 was barely detectable, whereas all MPK6 mutants phosphorylated MBP efficiently (Figure 1A). This shows that the MPK6 mutants that complement pbs2Δhog1Δ have a higher catalytic activity.
Figure 1.
Characterization of CA-MAPK Activity.
(A) Kinase activity toward MBP of wild-type (WT) MPK6 and candidate mutants (containing Tyr-144 [Y], Arg-274 [R], or Asp-218 Glu-222 [DE] mutations) after MPK6 immunoprecipitation from pbs2Δhog1Δ yeast cells. Clone numbers refer to Supplemental Table 1 online.
(B) Ribbon diagram of ERK2-based MPK6 structure with space field residues identified as CA mutations in the yeast screen. Sticks represent Thr-221 and Tyr-223 of the TEY motif.
(C) Kinase activity toward MBP of recombinant wild-type MPK6 and CA mutants.
(D) to (F) Kinase activity toward MBP of recombinant MPK4 (D) and MPK3 ([E] and [F]) with CA mutations produced as His-tagged ([C], [D], and [F]) or periHisMBP–tagged (E) proteins.
CA-MPK6 Candidates Have MAP2K-Independent Activity
Sequence analysis of the candidate clones pointed to Tyr-144 and Arg-274 as important residues for kinase autoactivity because their mutations were identified in 25 out of 27 clones (Figure 1B). One of the two remaining clones exhibited three mutations, including D218G and E222A, in the kinase activation loop (Figure 1B). Since the double mutants in these residues showed high intrinsic activity after immunoprecipitation from yeast, we chose the mutations Y144C and D218G/E222A for further characterization.
To confirm the importance of these residues, we generated three MPK6 single mutants, MPK6Y144C, MPK6D218G, and MPK6E222A, and the double mutant, MPK6D218G/E222A, without the additional mutations obtained in the yeast screen. The respective MPK6 kinases were purified as recombinant proteins from Escherichia coli cells, an expression system lacking MAPK activators. Wild-type MPK6 did not show significant kinase activity; however, MPK6Y144C and MPK6D218G/E222A strongly phosphorylated MBP (Figure 1C). By contrast, the single mutations D218G and E222A did not increase MPK6 activity, suggesting that MPK6D218G/E222A activity is not a result of an additive effect of each mutation but rather of a synergistic effect.
Corresponding CA Mutations in Other Arabidopsis MAPKs Trigger Hyperactivity
The alignment of the amino acid sequences of the 20 Arabidopsis MAPKs (see Supplemental Figure 1 online) showed that most MAPKs of the groups A, B, and C have a Tyr residue at the equivalent position of MPK6 Tyr-144. Despite the fact that the activation loop sequences are not very conserved among MAPKs, we identified the equivalent positions of the MPK6 D218 and E222 for the groups A, B, and C. In order to test if the corresponding mutations trigger autoactivity in the other MAPKs, we mutated the corresponding residues of MPK3 and MPK4, which are among the most studied MAPKs in plants (see Supplemental Figure 1 online for positions). Hexa-His–tagged, wild-type MPK3 and MPK4 and mutant proteins were expressed in E. coli for purification. We failed to purify the 6xHIS-MPK3D193G/E197A, and we expressed it as PERIPLASMIC-6xHIS-MALTOSE BINDING PROTEIN (periHisMBP)–tagged protein. CA-MPK4 forms (Figure 1D), and the periHisMBP-MPK3D193G/E197A variant (Figure 1E) showed higher activity than the respective wild-type proteins, whereas MPK3 activity was not increased by the T119C mutation (Figure 1F). Since MPK3 possesses a Thr instead of a Tyr residue in the MPK6 Tyr-144 homologous position, we hypothesized that substitution of this Thr, like the Cys and Phe substitutions of Tyr-144 in MPK6, allows MAP2K-independent MPK3 activity. To test this hypothesis, we created the MPK3T119Y mutant, mimicking the wild-type MPK6 form, which was expected to have decreased kinase activity. Kinase assay showed a reduced MPK3T119Y intrinsic activity compared with wild-type MPK3 and MPK3T119C (Figure 1F).
These results indicate that the residues identified in the yeast screen to activate MPK6 provide a strategy to render MAPKs constitutively active.
CA-MAPKs Phosphorylate Conventional MAPK Sites
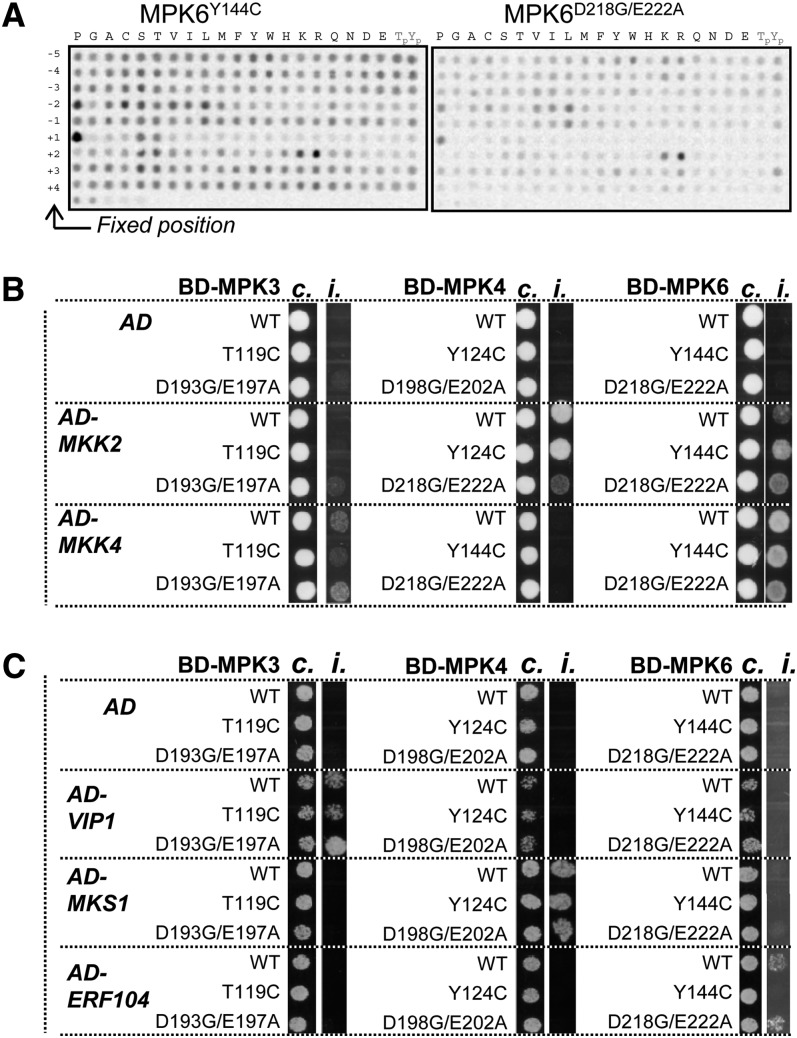
To test whether the kinase preference for the residues surrounding the phosphorylation site was not affected by the CA mutations, we took advantage of a semidegenerate peptide array tool (Vlad et al., 2008). This array consists of 198 peptide pools, and each pool has as a putative phosphorylation site (Ser/Thr) at its central position and is degenerate for all other positions except a fixed residue at one of the nine positions surrounding the phosphosite (between −5 and +4). The level of phosphorylation of each peptide by the kinase indicates the preferred residues for each position surrounding the phosphorylation site. Active MPK6Y144C and MPK6D218G/E222A were first tested and both showed an increased phosphorylation on peptides with Pro at position +1 and to a lesser extent to peptides with Pro at the −2 position (Figure 2A). Moreover, we saw a preference for charged amino acids at position +2. This result is in agreement with wild-type MPK6 specificity determined previously (Stulemeijer et al., 2007). Assays for wild-type MPK3, MPK3T119C, and MPK4Y124C gave similar results, indicating that the CA mutations do not change the phosphorylation motif preference of the MAPKs (see Supplemental Figure 2 online).
Figure 2.
Substrate Preferences and Interaction Specificities of the Wild Type and CA-MAPKs.
(A) Phosphorylation of the semidegenerate peptide array by MPK6Y144C and MPK6D218G/E222A.
(B) and (C) Combinatory interaction in yeast two-hybrid assays of wild-type (WT) and CA forms of MPK3, 4, and 6 with MKK2 and MKK4 (B) and with VIP1, MKS1, and ERF104 (C). Cotransformed single yeast colonies were spotted on control medium (c.) and selective medium (i.) supplemented with 65 mM (B) or 100 mM 3-amino-1,2,4-triazole (C).
CA-MAPKs Retain Binding Specificity toward Substrates and Activators
We next used yeast two-hybrid assays to examine whether the specific interactions between MAPKs and known interacting substrates and activating MAP2Ks are affected by the CA-MAPK versions. Wild-type and CA forms of MPK3, 4, and 6 were fused to the GAL4 binding domain and coexpressed in the reporter strain together with several known substrates fused to the GAL4 activation domain. In accordance with previous reports (Andreasson et al., 2005; Djamei et al., 2007; Bethke et al., 2009), we observed that CA-MPK3 exclusively interacts with VIRE2-INTERACTING PROTEIN 1 (VIP1), CA-MPK4 with MAP KINASE SUBSTRATE 1 (MKS1), and CA-MPK6 with ETHYLENE RESPONSE FACTOR 104 (ERF104). In some cases, CA mutations appeared to affect the strength of the interactions but not its specificity (Figure 2C). Similar results were obtained with their corresponding activating MAP2Ks (Figure 2B): CA-MPK3 specifically interacted with MKK4, CA-MPK4 with MKK2, and CA-MPK6 with both MKK2 and MKK4. Taken together, these data indicate that the CA-MAPK versions do not change the specificity of the kinases toward their cognate MAP2Ks nor to their specific target substrates.
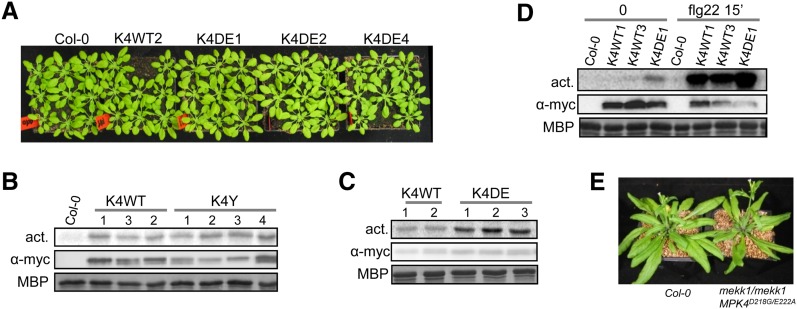
MPK4D198G/E202A Complements the mpk4 and mekk1 Developmental Phenotypes and Is Hyperactive in Planta
mpk4 homozygous plants are dwarfed (Petersen et al., 2000). We took advantage of this phenotype to test whether CA-MPK4 versions can functionally replace the endogenous MPK4. We generated mpk4-2 lines transformed with the MPK4WT, MPK4Y124C, or MPK4D198G/E202A constructs, tagged with c-Myc at the C terminus (further referred to as K4WT, K4Y, and K4DE lines, respectively). Wild-type and CA-MPK4 constructs largely restored a wild-type morphological phenotype in all independent transgenic lines (n = 6, 10 and six lines of K4WT, K4Y, and K4DE, respectively; Figure 3A), suggesting that the cascade specificity is conserved in CA-MPK4 plants. Our first attempts to biochemically characterize these transgenic lines revealed that the antibody raised against the C terminus of the MPK4 native protein does not detect the C-terminally tagged MPK4 fusion proteins. Therefore, to test if the CA-MPK4 kinase is constitutively active when stably expressed in planta, we immunoprecipitated CA-MPK4 from the corresponding lines using anti-c-Myc antibody. All K4DE lines showed increased MPK4 activity compared with K4WT lines (Figure 3C) but, surprisingly, not the K4Y lines (Figure 3B). We determined that the increased MPK4 kinase activity of a K4DE line is ∼10% of the total flg22-induced wild-type MPK4 activity, indicating that the enhanced activity of CA-MPK4 in untreated lines is still lower than the full activation level obtained upon pathway stimulation (Figure 3D). Moreover, we observed that MPK4 was still activatable in the K4DE lines. We then used K4DE plants for further characterization.
Figure 3.
Characterization of Arabidopsis mpk4-2 Mutants Complemented with CA-MPK4 Loci.
(A) Morphological aspect of mpk4-2/mpk4-2 lines complemented with MPK4WT and MPK4D198G/E202A. Plants were grown in short days for 5 weeks. K4WT and K4DE are abbreviations for mpk4 lines complemented with pGREEN0229-MPK4L-PC2 and pGREEN0229-MPK4LD198G/E202A-PC2, respectively. Numbers 1 to 4 refer to line numbers.
(B) and (C) Kinase activity toward MBP of MPK4-myc immunoprecipitated from mpk4-2/mpk4-2 lines complemented with MPK4WT ([B] and [C]), MPK4Y124C (B), and MPK4D198G/E202A (C). K4Y (B) is an abbreviation for mpk4 lines complemented with pGREEN0229-MPK4LY124C-PC2.
(D) Kinase activity of K4WT and K4DE lines upon 15 min flg22 (1 µM) treatment.
(E) Morphological phenotype of 5-week-old mekk1-1/mekk1-1 MPK4D198G/E202A plants grown in long days. mekk1-1/mekk1-1 plants barely survive 2 weeks in pots.
[See online article for color version of this figure.]
As MEKK1 functions upstream of MPK4 (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007), we hypothesized that CA-MPK4 may suppress the mekk1 dwarf phenotype by reactivating downstream events controlled by the MEKK1-MKK1/2-MPK4 cascade. To test this hypothesis, we crossed heterozygous mekk1-1 plants with the K4DE1 line. The generated mekk1-1/mekk1-1 plants expressing MPK4D198G/E202A produced viable seeds and developed normally but showed curlier leaves than wild-type plants (Figure 3E). This result confirms that MPK4 acts downstream of MEKK1 and that CA-MPK4 activity is sufficient to suppress MEKK1 deficiency.
CA-MPK4 Lines Are Impaired in Resistance to P. syringae
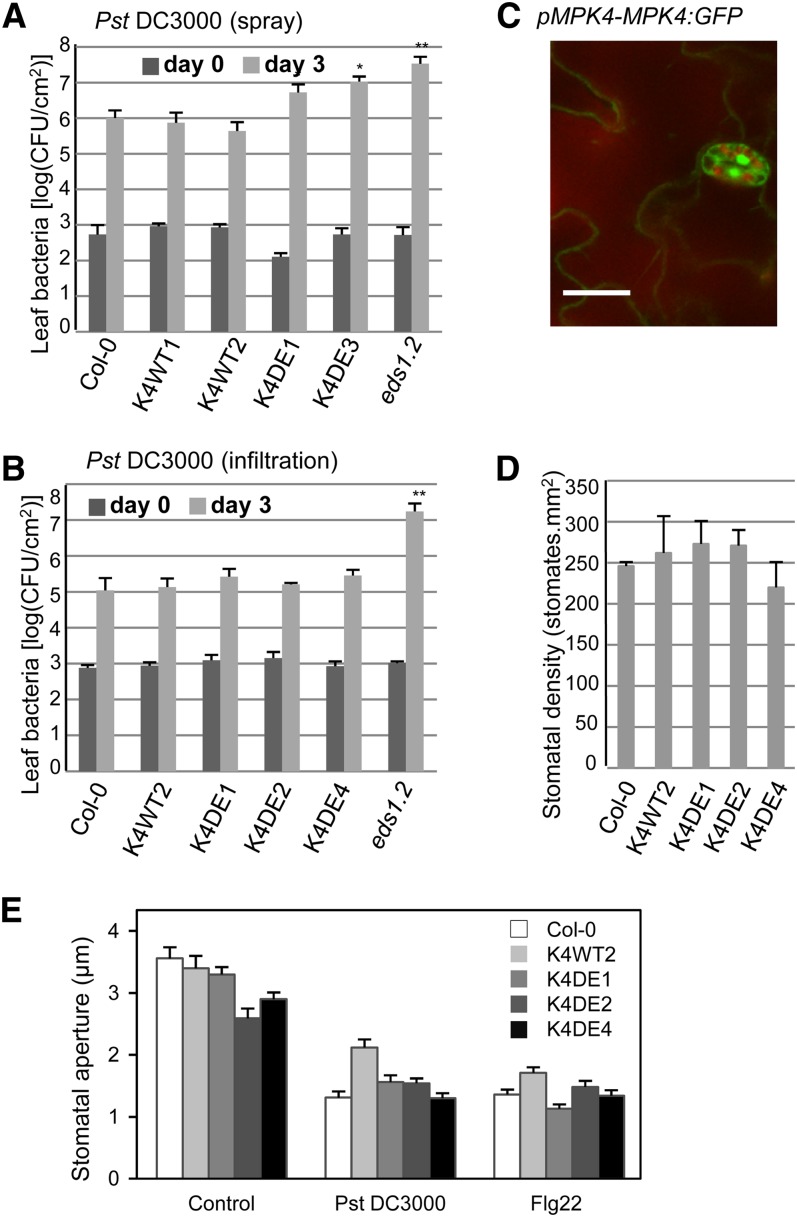
mpk4 mutant plants display enhanced resistance to the bacterial pathogen P. syringae, suggesting that MPK4 is a negative regulator of plant defenses to biotrophic pathogens (Petersen et al., 2000). According to this model, we reasoned that K4DE lines would be more susceptible to P. syringae infection. K4DE lines were first analyzed for their response to spray inoculation with virulent P. syringae pv tomato strain DC3000 (Pst DC3000). Bacterial titers at 3 d after inoculation were similar in Columbia-0 (Col-0) and K4WT leaves, but increased bacterial numbers were recovered from leaves of K4DE plants (Figure 4A), indicating that the higher MPK4 activity in K4DE lines compromises disease resistance. In order to distinguish if MPK4 functions in stomatal and postinvasive defenses, we quantified Pst DC3000 growth after inoculation through syringe infiltration. In this assay, the K4DE lines showed similar resistance levels as those observed in Col-0 and K4WT plants (Figure 4B), suggesting that MPK4 may play a role in stomatal-based defenses to bacterial entry into the apoplast rather than in postinvasive immunity. In agreement with this finding, mpk4 stable transgenic plants expressing functional MPK4-GFP (for green fluorescent protein) fusion under its own promoter displayed strong fluorescence in guard cells (Figure 4C).
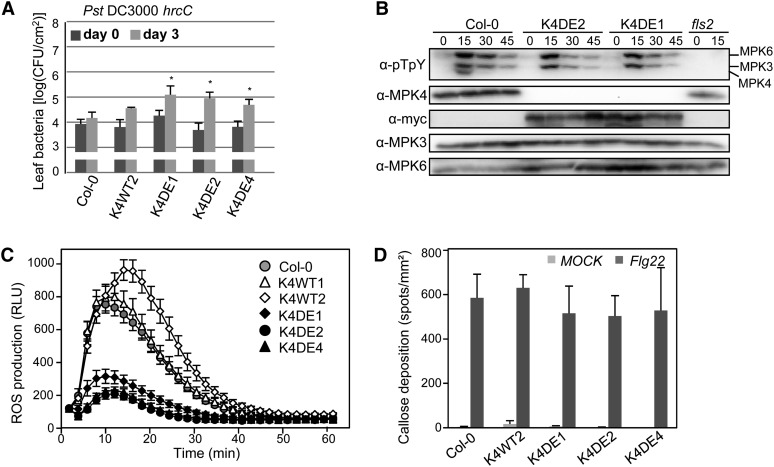
Figure 4.
Defense Responses of Arabidopsis mpk4-2 Mutants Complemented with CA-MPK4 Loci to Pst DC3000.
(A) and (B) Growth of Pst DC3000 after spray inoculation (A) or leaf infiltration (B) in the indicated genotypes. Bacterial titers were determined 2 h (day 0, dark-gray bars) and 3 d (light-gray bars) after inoculation. Bars are average of four replicates, and error bars show sd. Results are representative of three independent experiments. *P < 0.05 and **P < 0.01.
(C) Microscopy localization of a GFP-tagged MPK4 expressed from its own locus. Bar = 20 µm.
(D) Stomatal densities assessed in wild-type and three K4DE lines. Data are mean ± se (n = 6).
(E) Pst DC3000–induced (108 cfu/mL) and flg22-induced (5 µM) stomatal closure in wild-type and three K4DE lines. Data are mean ± se (n > 60).
We first investigated whether MPK4 could function during epidermal differentiation to control stomatal number; however, when compared with wild-type lines, no differences in stomatal densities were observed in K4DE lines (Figure 4D). Alternatively, MPK4 could be involved in stomatal aperture modulation in response to pathogens. To test this possibility, we therefore monitored the stomatal closure response of wild-type and K4DE lines after Pst DC3000 and flg22 treatments (Figure 4E; see Supplemental Figure 3 online). Stomates in the three K4DE lines tested opened in response to light and closed in response to 108 colony-forming units (cfu)/mL Pst DC3000 and 5 µM flg22 in a similar fashion as Col-0 plants. These results suggest that MPK4 function in plant immunity is not related to the regulation of stomatal closure in response to pathogen recognition.
CA-MPK4 Lines Are More Susceptible to a Type III Secretion-Defective Mutant of Pst DC3000 and Compromised in flg22-Induced ROS Production
To further understand the role of MPK4 in plant immunity, we tested the contribution of MPK4 to PTI by testing the CA-MPK4 lines for the activation of various PTI hallmarks. As MPK3 and MPK6 are known to be important regulators of PTI, we tested whether CA-MPK4 would affect the flg22-induced activation of the other MAPKs using an anti-phospho-ERK1 antibody that detects the active forms of wild-type MAPKs. The activation of MPK3 and MPK6 was similar in the wild type and the three K4DE lines, showing maximal activation at 15 min (Figure 5B). This result indicates that the CA mutations in MPK4 do not affect PAMP-dependent activation of other MAPK signaling pathways and suggests that CA-MPK4 lines can be used to define events specifically controlled by MPK4. The PAMP-induced RbohD-dependent oxidative burst is one of the fastest known PTI-related responses, occurring within minutes after PAMP treatment (Zhang et al., 2007; Ranf et al., 2011). To determine whether MPK4 has a role in regulating PAMP-induced ROS production, we measured the flg22-induced oxidative burst in leaf disks of adult plants. Col-0 and K4WT plants responded to flg22 treatment with a similar peak of ROS production, whereas K4DE lines exhibited a strong reduction of the oxidative burst (Figure 5C), indicating that MPK4 activity negatively regulates PAMP-induced ROS production. Callose deposition is another defense response that occurs after PAMP detection (Gómez-Gómez et al., 1999). In resting conditions, none of the lines showed any callose depositions in leaves, but infiltration of flg22 into the leaves triggered strong callose deposition appearing as aniline blue–stained spots (Figure 5C). When compared with Col-0 and K4WT, K4DE lines showed no statistical difference in flg22-induced callose deposition. Given that mpk4 mutant plants display constitutive ROS and callose accumulation (Gao et al., 2008), the analysis of CA-MPK4 lines allows discrimination between responses regulated by MPK4 activity and secondary effects of the mpk4 mutation.
Figure 5.
Characterization of PTI Responses in CA-MPK4 Lines.
(A) Growth of Pst DC3000 hrcC on the indicated genotypes. Bacterial titers were determined 2 h (day 0, dark-gray bars) and 3 d (light-gray bars) after spray inoculation. Bars are average of four replicates, and error bars show sd. Results are representative of three independent experiments. *P < 0.05
(B) Flg22-induced activation of MAPKs in Col-0 and two CA-MPK4 lines at the indicated time points, using anti-pTEpY immunoblot.
(C) Oxidative burst response in wild-type and CA lines in response to 0.5 µM flg22. Results are means ± se (n > 20) of a representative experiment among four. P value < 10−7 at 11 min for all K4DE lines. RLU, relative light units.
(D) Callose production in 4- to 5-week-old plants of the indicated genotypes infiltrated with half-strength MS or half-strength MS supplemented with 300 nM flg22. Data are means ± se of four independent experiments except for K4DE4 line with two experiments (n > 10 leaves).
To further assess the role of MPK4 in PTI, we used the type III secretion mutant Pst DC3000 hrcC, which is impaired in effector delivery and therefore induces mainly PTI (Tsuda et al., 2008). Bacterial quantification after spray inoculation showed that K4DE lines are significantly more susceptible to Pst DC3000 hrcC than Col-0 and K4WT lines (Figure 5A). These results indicate that even if MPK4 activity does not control all PAMP-induced responses, MPK4 performs an important role in PTI.
CA-MPK4 Affects Pathogen Resistance Mediated by TIR-NB-LRR, but Not CC-NB-LRR, Receptors
To investigate the function of MPK4 during ETI, we challenged K4DE lines with the avirulent strain Pst DC3000 expressing the effector AvrRps4. In Arabidopsis, AvrRps4 triggers ETI through recognition mediated by the TIR-NB-LRR RPS4 (Gassmann et al., 1999; Heidrich et al., 2011). Disease symptoms as well as bacterial growth were more pronounced on leaves of the K4DE lines at 3 d after inoculation (Figures 6A and 6B). As expected, eds1-2 mutant plants showed hypersusceptibility to Pst DC3000 AvrRps4 (García et al., 2010). Since the mpk4 resistance phenotype is linked to the constitutive accumulation of SA, we also assessed if the K4DE lines behave in an opposite way and accumulate less SA than K4WT lines in response to pathogen treatment. Col-0 and the K4WT lines accumulated similar amounts of SA at 24 h after spray inoculation with Pst DC3000 AvrRps4, whereby the K4DE lines showed lower pathogen-induced SA accumulation (Figure 6C). This result suggests that the reduced SA accumulation in K4DE plants could contribute to the reduced resistance of K4DE lines to P. syringae.
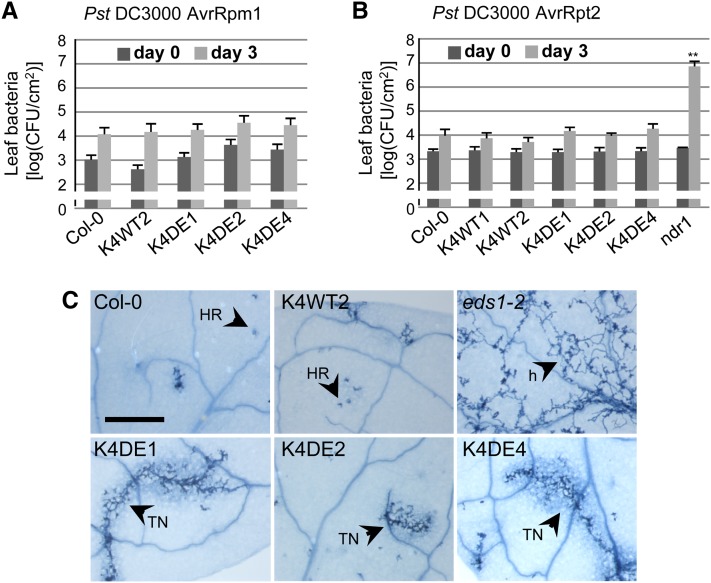
Figure 6.
Characterization of RPS4-Mediated ETI in CA-MPK4 Lines.
(A) Growth of Pst DC3000 AvrRps4 in the indicated genotypes. Bacterial titers were determined 2 h (day 0, dark-gray bars) and 3 d (light-gray bars) after spray inoculation. Bars are average of at least four replicates, and error bars show sd. *P value < 0.05 and **P value < 0.01. Similar results were obtained in at least three independent experiments.
(B) Disease symptoms observed on K4WT and K4DE lines 3 d after inoculation with Pst DC3000 AvrRps4.
(C) SA content before (T0) and 24 h after spray inoculation with Pst DC3000 AvrRps4 or after mock treatment with 10 mM MgCl2. Bars represent the average of three independent biological replicates. After Pst DC3000 AvrRps4 treatment, the SA content of Col-0 and wild-type lines taken together was 8.2 ± 1.0 ng⋅mg−1 fresh weight (FW) (n = 9) and of K4DE lines was 5.1 ± 1.0 ng⋅mg−1 fresh weight (n = 9) (the difference is statistically significant; P value < 0.05).
To further investigate the role of MPK4 in ETI, we analyzed the resistance of CA-MPK4 lines to strains of Pst DC3000 expressing the effectors AvrRpm1 or AvrRpt2. In Arabidopsis, AvrRpm1 and AvrRpt2 trigger ETI following recognition by the CC-NB-LRR–type receptors RPM1 and RPS2, respectively (Kunkel et al., 1993; Boyes et al., 1998). Interestingly, K4DE lines exhibited complete resistance against Pst DC3000 AvrRpm1 and Pst DC3000 AvrRpt2 similar to Col-0 and K4WT plants (Figures 7A and 7B).
Figure 7.
ETI Responses in CA-MPK4 Lines.
(A) and (B) Growth of Pst DC3000 AvrRpm1 (A) and Pst DC3000 AvrRpt2 (B) in the indicated genotypes. Bacterial titers were determined 2 h (day 0, dark-gray bars) and 3 d (light-gray bars) post spray inoculation. Bars are average of at least four replicates, and error bars show sd. **P value < 0.01. Similar results were obtained in at least three independent experiments.
(C) Growth of H. arabidopsidis Emwa1 and cell death structures in leaves of the indicated genotypes. Leaves were stained with lactophenol trypan blue 6 d after inoculation. h, hyphae; HR, hypersensitive response; TN, trailing necrosis. Bar = 500 nm.
[See online article for color version of this figure.]
These data suggest that the function of MPK4 in ETI may be limited to resistance mediated by TIR-NB-LRR receptors. To assess this possibility, we examined the resistance conferred by the TIR-NB-LRR RPP4 to the oomycete Hyaloperonospora arabidopsidis by trypan blue staining of leaves for pathogen structures and plant cell death (van der Biezen et al., 2002). Leaves of Col-0 and K4WT displayed the hypersensitive response–associated localized cell death, but the K4DE lines showed reduced RPP4-mediated resistance as manifested by the occurrence of a trailing necrosis and limited hyphal growth (Figure 7C). As expected, eds1-2 plants displayed complete susceptibility visible by the free hyphal growth and pathogen sporulation. These results suggest that MPK4 functions broadly during TIR-NB-LRR resistance signaling but with a stronger impact on the resistance conferred by certain TIR-NB-LRRs.
DISCUSSION
CA Mutations Are a Useful Tool to Activate MAPKs
In this study, we identified mutations that increase MAPK activity independently of MAP2K activation. Using a genetic screen in yeast and Arabidopsis MPK6 as a model, we identified three sets of mutations that triggered constitutive MPK6 activity. Furthermore, introducing the corresponding mutations in MPK3 and MPK4 resulted in constitutive kinase activity. These results suggest that the mutations identified provide a strategy to render MAPKs constitutively active.
The MPK6 residue Tyr-144 is located in the ATP binding pocket and is homologous to the well-described Gatekeeper residue of the mammalian MAPK ERK2 (Emrick et al., 2006). Mutation of this residue is known to allow ERK2 intramolecular autophosphorylation and, therefore, its activation in a MAP2K-independent manner. MPK6 Y144C or Y144F mutations might have a similar effect. Despite the fact that some activating mutations were identified in the activation loop of human or yeast MAPKs (Bell et al., 2001; Diskin et al., 2004), the effect of the combination of the MPK6 mutations D218G and E222A is novel. Interestingly, these mutations change two acidic amino acid residues for neutral ones and therefore should have an opposite effect compared with the phosphorylation of the TEY motif that leads to MAPK activation. A plausible explanation is that these mutations change the activation loop flexibility, resulting in higher autophosphorylation and kinase activation.
Since we had no clear evidence how these mutations enhance MAPK catalytic activity, we were concerned of their proper functioning. Although we cannot exclude that some interactions with unidentified targets or regulators might be affected, CA mutations did not change the MAPK binding specificities for the known activating MAP2Ks or substrates we tested. Additionally, the use of a semidegenerate peptide array allowed us to confirm that CA-MPK3, 4, and 6 recognize the typical phosphorylation motif for MAPKs (Hutti et al., 2004). Finally, the fact that both MPK4Y124C and MPK4D198G/E202A complemented the developmental defects of mpk4-2 knockout mutant plants proves that the CA-MPK4 variants retain full wild-type kinase specificity. Interestingly, only the MPK4D198G/E202A lines showed an increase in MPK4 kinase activity in planta. This result suggests that the MAPK phosphatases dephosphorylate the activation loop of MPK4Y124C but not of MPK4D198G/E202A and indicates that not all CA mutations leading to enhanced activity in heterologous systems work similarly in planta. Strikingly, the MPK4D198G/E202A construct also suppressed the severe dwarf phenotype of mekk1 plants, which are defective in the upstream activation of MPK4. This result provides clear evidence that MPK4D198G/E202A retains activation-independent activity in planta and that a certain activity level of the MEKK1-MKK1/2-MPK4 pathway is required to ensure normal growth homeostasis. Although it cannot be excluded that CA-MAPKs in planta may have consequences distinct from a transient activation, they clearly provide an important tool to analyze MAPK pathways.
MPK4 Regulates PAMP-Induced ROS Production
To provide a proof-of-concept that the CA strategy can be used to understand the physiological role of MAPKs, CA-MPK4 was studied in planta. MPK3, MPK4, MPK6, and MPK11 are activated by PAMPs (Asai et al., 2002; Droillard et al., 2004; Bethke et al., 2012). MPK3 and MPK6 have been shown to be positive regulators of PTI (Asai et al., 2002), but the strong growth defect of mpk4 plants complicates the examination of its function. Taking advantage of our CA mutants, we demonstrated that MPK4 plays an important role in PTI as the CA-MPK4 lines are compromised in resistance to the Pst DC3000 hrcC mutant strain. Interestingly, flg22-induced ROS production is strongly reduced in the K4DE lines, showing that MPK4 activity negatively regulates ROS production. The NADPH oxidase RbohD has been shown to be the key producer of ROS upon PAMP induction (Torres et al., 2002; Zhang et al., 2007) and so far only CDPKs have been shown to regulate the enzymatic activity of RbohD through phosphorylation (Boudsocq et al., 2010). Future research will have to unravel whether the NADPH oxidase is directly or indirectly regulated by MPK4. Whatever the exact mechanism, our results also explain the constitutive ROS accumulation in mpk4 mutant plants, which seems to be the consequence of a lack of MPK4 regulation, resulting in increased NADPH oxidase activity (Nakagami et al., 2006).
Although ROS has been proposed to be a positive signal in flg22-induced callose deposition (Zhang et al., 2007; Daudi et al., 2012), we only observed a slight and statistically nonsignificant reduction in flg22-induced callose deposition in CA-MPK4 plants. One explanation is that the reduced flg22-triggered oxidative burst observed in K4DE lines is sufficient to allow normal callose deposition. Alternatively, Daudi et al. (2012) found only a partial reduction of callose deposition in the rbohD knockout mutant, suggesting a certain degree of RbohD-independent callose deposition. In summary, our results show that even if MPK4 activity has an important impact on resistance to a PTI-inducing Pst strain, we found that MPK4 activity controls only certain aspects of PTI signaling. The other PTI-related responses might be controlled by other MAP kinases like MPK3, MPK6, and MPK11, which are also rapidly activated upon PAMP perception (Asai et al., 2002; Bethke et al., 2012) or alternatively by MAPK-independent pathways (Boudsocq et al., 2010). The fact that we did not detect MPK11 activation in the CA-MPK4 plants could suggest a misregulation of this specific branch.
The activation of the MPK4 pathway by PAMPs suggests that certain PAMP-activated processes might play a strategic role to repress defenses in favor of the pathogen or to reduce detrimental effects for the plant. The hypothesis that the activation of the MPK4 pathway benefits the pathogen is also corroborated by the finding that the P. syringae effector avrB directly activates MPK4 and thereby increases Arabidopsis susceptibility to Pst DC3000 infection (Cui et al., 2010). Overall, these results contradict the common belief that PAMP-induced signaling enhances resistance against microbial infection and show that certain PAMP-induced immune signaling events are aimed at dampening plant immunity.
mpk4 knockout mutant plants show constitutive activation of SA-mediated defenses (Petersen et al., 2000). Consistent with this, our data show that MPK4 activity inhibits basal defenses to virulent Pst DC3000 and pathogen-induced SA accumulation. Interestingly, CA-MPK4 lines were not compromised in resistance when bacteria were directly injected into the leaf apoplast, indicating that MPK4 activity could inhibit an early layer of plant immunity controlling bacterial entry. Previous reports have postulated a role for MAPK signaling in controlling stomatal aperture. The MAPK phosphatase PP2C5, which modulates abscisic acid–dependent MPK4 activation, positively regulates stomatal aperture (Brock et al., 2010). Furthermore, the MPK4 homologs in Nicotiana attenuata and Nicotiana tabacum were proposed to positively regulate stomatal closure, as MPK4 gene silencing led to increased stomatal aperture (Gomi et al., 2005; Hettenhausen et al., 2012). Our results show that CA-MPK4 lines open and close stomates in response to PAMPs or bacteria similarly to wild-type plants. Therefore, Arabidopsis MPK4 may play a role in plant immunity, which is not related to stomatal closure or at least not measurably for the time points and conditions analyzed.
MPK4 Plays a Role in ETI
CA-MPK4 lines were compromised in resistance conditioned by the TIR-NB-LRR receptors RPS4 to P. syringae and, to a lesser extent, RPP4-mediated resistance to H. arabidopsidis, proving that MPK4 functions in ETI. It is intriguing that the CA-MPK4 lines retained normal resistance to Pst DC3000 AvrRpm1 and AvrRpt2 recognized by two different CC-NB-LRR receptors. Interestingly, the immune regulator EDS1, which genetically interacts with MPK4, is specifically required for resistance conferred by TIR-NB-LRR receptors during ETI (Aarts et al., 1998; Brodersen et al., 2006). An interesting possibility could be that MPK4 might negatively regulate an EDS1-dependent plant immunity branch downstream of TIR-NB-LRR receptors, and further investigations will be necessary to clarify the role of MPK4 in ETI.
Recently, a piece of the puzzle was clarified by the analysis of the Arabidopsis mutant summ2, which carries a mutation in a CC-NB-LRR receptor and restores normal growth to mutants of the MEKK1-MKK1/MKK2-MPK4 cascade (Zhang et al., 2012). The authors hypothesized that this cascade, which positively regulates disease resistance, also functions to detect microbial effectors injected to suppress immune responses. These results confirm the modulation of ETI by early PAMP signaling events and that PTI and ETI are not independent as often presented in simplified models, and further studies will be necessary to clarify the role of MPK4 in ETI.
In conclusion, we identified a strategy to generate CA-MAPKs. Our data show that the use of CA-MAPKs can help to provide answers that go beyond classical genetic analysis with loss-of-function or knockout mutants. This strategy should help to unravel the multiple functions that MAPKs play in various pathways, including defense, growth, cell cycle, and development.
METHODS
Plant Growth Conditions and Genotyping
Arabidopsis thaliana Col-0, Col-0 eds1-2 (Bartsch et al., 2006), mekk1-1 (Salk_052557), and mpk4-2 (Salk_056245) derived lines were grown either on soil or on plates (half-strength Murashige and Skoog [MS], 1% Suc, 1.2% agar, and 0.5% MES, pH adjusted to 5.7 with KOH) in long-day conditions. For pathogen assays and callose deposition, plants were grown for 4 to 5 weeks in Percival growth cabinets in short days at 22°C and 70% humidity.
Gateway Cloning and Point Mutations
PCR reactions used iProof enzyme (Bio-Rad).
Gene ORFs were amplified from Col-0 cDNA and recombined in pDNR207 using Gateway technology (Invitrogen). The primers are described in Supplemental Table 2 online, and the protocol was kindly provided by Lurin and coworkers (http://www-urgv.versailles.inra.fr/atome/protocols.htm). Clones with and without the stop codon were amplified, allowing N- and C-terminal protein fusions. Clones were systematically sequenced. For functional studies, ORFs were recombined using the LR enzyme mix following the manufacturer’s indications (Invitrogen) in devoted plasmids (see Supplemental Table 3 online).
When necessary, mutations were introduces in ORFs using a site-directed mutagenesis classical protocol (Papworth et al., 1996). Briefly, complementary primers carrying a mutation(s) were used in a 20-cycle PCR reaction on the appropriate vector substrate with a long extension time (>5 min) and the iProof enzyme (primer sequences in Supplemental Table 2 online). Mixture was digested by DpnI (NEB Biolabs) and directly transformed into Escherichia coli DH5α cells. Candidate clones were resequenced for the part of interest.
Genetic Screen in Yeast
To perform the genetic screen in yeast, MPK6 ORFs were LR recombined in pDR195gtw to generate pDR195gtw-MPK6. The PCR fragment of randomly mutated MPK6 ORF was produced using the Mutazyme II kit (Stratagene), with pDR195gtw-MPK6 as substrate and DR_F/DR_R primers (see Supplemental Table 2 online). pDR195 was linearized using BamHI and cotransformed with MPK6 randomly mutated PCR fragments into hog1Δpbs2Δ yeasts using a classical lithium/polyethylene glycol/heat shock method (Schiestl and Gietz, 1989). Yeast cells having reconstructed the pDR195gtw-MPK6 plasmids were selected on synthetic complete (SC; 0.17% yeast nitrogen base without carbohydrate and amino acids, 0.5% NH4SO4, 2% d-Glc, and 2% BactoAgar) medium lacking uracil (SC + 0.2% dropout minus uracil (US Biological), pH 5.6 adjusted with NaOH). Colonies were replicated on yeast extract peptone dextrose medium supplemented with 2% BactoAgar and 400 to 600 mM NaCl.
To extract plasmids from yeast clones, pellets from 2-mL overnight cultures were resuspended in 200 μL buffer (100 mM NaCl, 10 mM Tris-HCl, pH 8, 1 mM EDTA, and 0.1% SDS). Glass beads (0.3 g) and 200 μL of phenol were added, followed by 3 min of vigorous shaking at room temperature. Phases were separated (15,000 rpm, 5 min) and a second phenol extraction was performed with the supernatant. DNA was precipitated from the supernatant with 30 μL of 3 M sodium acetate and 800 μL of 96% ethanol for 10 min at −20°C, followed by 20 min centrifugation at 4°C. The pelleted DNA was washed with 70% ethanol and air-dried for 1 h. DNA was resuspended in 50 μL of TE buffer (1 mM EDTA, 10 mM Tris/HCl, pH 8), and used to transform E. coli for amplification and sequencing.
Immunoprecipitated Kinase Assay from Yeast and Plant Samples
To measure MPK6 activity from yeast expressing wild-type pDR195gtw-MPK6 and mutants, the supernatants of 5 mL overnight cultures were resuspended in 300 μL yeast lacus buffer (25 mM Tris, pH 7.8, 75 mM NaCl, 10 mM MgCl2, 15 mM EGTA, 75 mM NaCl, 1 mM DTT, 1 mM NaF, 0.5 mM NaVO3, 15 mM B-glycerophosphate, 15 mM p-nitrophenylphosphate, 0.1% Tween 20, 0.5 mM phenylmethylsulfonyl fluoride, 5 µg/mL leupeptin, and 5 µg/mL aprotinin) and ground at 4°C using 0.3 mg glass beads (Sigma-Aldrich). Two hundred microliters of the supernatant cleared off beads and cell debris were used for the immunoprecipitation with anti-At-MPK6 antibody (polyclonal; Davids Biotechnologie) as previously described (Nakagami et al., 2004).
Production and Activity Assay of Recombinant MAPKs
Cultures (0.1 to 0.3 liters) were grown at 37°C until they reached an OD of 0.5 to 0.6. Protein expression was induced overnight at 25°C using 0.1 mM isopropyl β-d-1-thiogalactopyranoside. The cells were spun down and stored at −20°C. The frozen pellet was thawed on ice and gently resuspended in the lysis buffer (30 mM Tris-HCl, pH 7.5, 300 mM NaCl, 5 mM imidazole, 2 mg/mL lysozyme, and 0.1% Triton). After sonication, the lysate was centrifuged at 10,000g for 20 min at 4°C. The supernatant containing the soluble protein was incubated for 30 min with 2 mL of cobalt resin. The matrixes were washed three times (30 mM Tris-HCl, pH 7.5, 300 mM NaCl, and 5 mM imidazole) and elution from the columns performed with 500 μL 30 mM Tris-HCl, pH 7.5, 300 mM NaCl, 200 mM imidazole, and 10% glycerol. The protein solution was then dialyzed overnight (30 mM Tris-HCl, pH 7.5, and 20% glycerol), and the concentration was determined with Bradford assay and SDS-PAGE.
In vitro kinase assay activity with purified kinases (0.6 to 1 µg purified) and kinase assay on semidegenerate peptide arrays were performed as previously reported (Nakagami et al., 2004).
Construction of mpk4-2 Lines Expressing MPK4 Locus-Based Constructs
PC2 (see sequence in Supplemental Text 1 online, containing 9xc-Myc) and GFP tags were amplified with PC2_F/PC2_R and GFP_S/GFP_R primers (see Supplemental Table 2 online) from pC2n1 (kindly provided by H. Mireau) and pCAMBIA1302 vectors, respectively, cloned in pGEMTeasy (Promega), and sequenced. The BglII DNA fragment containing PC2 tag and the BamHI DNA fragment containing the GFP were then subcloned in the unique compatible BamHI site of pGREEN0229-MPK4L, previously described by Kosetsu et al. (2010), to generate pGREEN0229-MPK4L (locus)-PC2 and pGREEN0229-MPK4L-GFP, respectively. The constructs were confirmed by sequencing. Point mutations were introduced as described above to generate pGREEN0229-MPK4LY124C-PC2 and pGREEN0229-MPK4LD198G/E202A-PC2. These vectors were transformed in the Agrobacterium tumefaciens strain C58C1 carrying pSOUP (Hellens et al., 2000). The progeny of mpk4-2/MPK4 plants were transformed by floral dipping (Clough and Bent, 1998). In the progeny, Basta-resistant plants were genotyped to identify mpk4-2/mpk4-2 plants as previously described (Kosetsu et al., 2010).
Yeast Two-Hybrid Analysis
Qualitative yeast two-hybrid assays were performed using yeast strain MaV203 (Vidal et al., 1996) and Gateway vectors pDEST22 for GAL4-AD fusion and substrates and a modified pDEST32 (carrying a kanamycin-resistant gene) for GAL-BD fusion (Invitrogen). Cells were cotransformed with the two two-hybrid plasmids using a classical lithium/polyethylene glycol/heat shock method (Schiestl and Gietz, 1989) and selected on medium lacking Trp and Leu (SC with 0.2% dropout-T-L [US Biological] at 30°C for 2 d). Colonies were grown overnight in the same medium without agar and then diluted 200 times, and 5 μL were spotted on an SC plate (SC with 0.2% dropout-T-L-H [US Biological] and with indicated concentrations of 3-amino-1,2,4-triazole.
Flagellin Stress Assays
For kinase activity measurement, 2-week-old seedlings grown in liquid MS plates were used. flg22 peptide solution (1 µM) was added to media and gently mixed, and the plants were incubated for 15, 30, or 45 min. Stress was stopped by liquid nitrogen freezing. For each treatment, at least 10 plants were used. For radioactive kinase assay, immunoprecipitation of MPK4 was done as previously described (Bögre et al., 1999; Cardinale et al., 2002) using anti-c-Myc antibody. For direct immunoblotting, samples were ground using a homogenizer, proteins were extracted in 2 volumes of Laemmli buffer two times, boiled for 5 min, and debris pelleted. Anti-MPK3 and anti-MPK4 antibodies were raised against MIYQEAIALNPTY and ELIYRETVKFNPQDSV epitopes, respectively. Anti-pTEpY (4370s; Ozyme), anti-c-Myc (C3956; Sigma-Aldrich), and anti-MPK6 (Davis) were purchased.
ROS detection using leaf discs from long-day-grown plants was performed as described previously (Gómez-Gómez et al., 1999). ROS production was assessed using a TECAN infinite M200 spectrophotometer
The callose assay was performed following a protocol described previously (Daudi et al., 2012) on 4- to 5-week-old plants grown under short days. At least three independent plants were used in each biologically independent experiment, where four rosette leaves were sampled from each plant. Four biologically independent experiments were performed. Callose deposition was visualized with a Leica M216F fluorescence microscope at ×100 magnification and with a Leica 10447354 4′,6-diamidino-2-phenylindole filter. Callose experiments were scored blind.
Stomatal Bioassays
Leaves from 3- to 4-week old plants were used for stomatal aperture measurements. Epidermal strips were prepared from the abaxial epidermis as previously described (Merlot et al., 2007) and incubated in 10 mM MES-Tris, pH 6.0, 30 mM KCl, and 1 mM CaCl2 for 2 h under light (300 µE⋅m−2⋅s−1) to promote ostiole opening. To induce closure, flg22 (at indicated concentrations) or Pst DC3000 (final concentration 108 cfu/mL) was directly diluted in the buffer in contact with epidermal peels. Control and flg22-treated peels were incubated under light for 2.5 h, whereas Pst DC3000–treated peels were incubated under light for 1 h. Stomatal aperture measurements were performed with a light microscope (Optiphot-2; Nikon) fitted with a Lucida camera and a digitizing table linked to a personal computer. Sixty aperture widths were measured blind for each genotype.
Pathogen Assays and SA Measurements
Spray and injection infections with Pst DC3000, Pst DC3000 hrcC mutant, Pst DC3000 AvrRps4, Pst DC3000 AvrRpm1, and Pst DC3000 AvrRpt2 and determination of bacterial titers were performed on 4- to 5-week-old plants as previously described (Bartsch et al., 2006; García et al., 2010). Spray inoculations were performed with bacterial suspensions at 108 cfu/mL in 10 mM MgCl2 with 0.04% Silwet L-77. Infiltration experiments were performed with bacterial suspensions at 105 cfu/mL in 10 mM MgCl2. At least four plants of each genotype were used per experiment ,and the experiments repeated at least three times. Bacterial numbers in mutant or transgenic lines were compared with Col-0 using a two-tailed Student's t test.
Hyaloperonospora arabidopsidis isolate Emwa1 was inoculated onto 2-week-old seedlings at 5 × 104 spores/mL as previously described (García et al., 2010). To assess plant cell death and H. arabidopsidis infection structures, trypan blue staining of the leaves was performed 6 d after inoculation. Experiments were repeated three times with similar results.
SA contents were determined after bacterial spray inoculation as previously described (Rozhon et al., 2005) except that 10 µM EDTA was added to the HPLC eluent.
Microscopy
GFP signals due to MPK4-GFP fusion were monitored with a confocal microscope (TCS SP2-AOBS; Leica).
Protein Structure
MPK6 structure was predicted from the crystallized structure of ERK2 (PDB-ID: 2erkA) using SWISS-MODEL (http://swissmodel.expasy.org) and visualized using SWISS-pdbviewer.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g45640 (MPK3), At4g01370 (MPK4), At2g43790 (MPK6), At4g26070 (MKK1), At4g29810 (MKK2), At1g51660 (MKK4), At4g08500 (MeKK1), At1g43700.1 (VIP1), At3g18690.1 (MKS1), and At5g61600.1 (ERF104).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Local Sequence Alignment of 20 Arabidopsis MAPKs.
Supplemental Figure 2. Substrate Preference of Some MAPKs.
Supplemental Figure 3. Stomatal Closure Induced by flg22 at Indicated Concentrations in Col-0, K4WT2, and K4DE2 Lines.
Supplemental Table 1. List of MPK6 Mutations Identified in the Yeast Complementation Screen.
Supplemental Table 2. Pairs of Primers Used in This Work.
Supplemental Table 3. Plasmids Used in This Study.
Supplemental Text 1. DNA Sequence of the PC2 Tag.
Supplementary Material
Acknowledgments
We thank Markus Teige, Hakim Mireau, and Sébastien Thomine who kindly provided the pbs2Δhog1Δ yeast strain, pPC2, and the pDR195gtw vectors, respectively, Sylvain Merlot and Florina Vlad for the peptide array technology, Elodie Hudik for establishing screening protocol, Marie Garmier for providing H. arabidopsidis isolate Emwa1, and the Unité de Recherche en Génomique Végétale laboratory for daily help on experiments. This work was supported by the Agence Nationale de la Recherche Program and French Ministry of Research PhD fellowship to S.B.
AUTHOR CONTRIBUTIONS
S.B., H.H., and J.C. designed the research. S.B., A.V.G., N.F.F., W.R., S.P., N.L., J.-L.M., and J.C. performed research. J.L. contributed analytic tools. S.B., A.V.G., H.H., and J.C. wrote the article.
Glossary
- MAPK
mitogen-activated protein kinase
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- NB
nucleotide binding
- LRR
Leu-rich repeat
- ETI
effector-triggered immunity
- SA
salicylic acid
- ROS
reactive oxygen species
- CA
constitutively active
- TIR
Toll Interleukin-1 Receptor
- CC
coiled-coil
- MAP2K
to be defined
- Pst DC3000
Pseudomonas syringae pv tomato strain DC3000
- Col-0
Columbia-0
- GFP
green fluorescent protein
- cfu
colony-forming unit
- MS
Murashige and Skoog
- ORF
open reading frame
- SC
synthetic complete
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113 [DOI] [PubMed] [Google Scholar]
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana Nature 408: 796–815. [DOI] [PubMed]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Komis G., Müller J., Menzel D., Samaj J. (2010). Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22: 755–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M., Capone R., Pashtan I., Levitzki A., Engelberg D. (2001). Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J. Biol. Chem. 276: 25351–25358 [DOI] [PubMed] [Google Scholar]
- Bethke G., Pecher P., Eschen-Lippold L., Tsuda K., Katagiri F., Glazebrook J., Scheel D., Lee J. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25: 471–480 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L., Calderini O., Binarova P., Mattauch M., Till S., Kiegerl S., Jonak C., Pollaschek C., Barker P., Huskisson N.S., Hirt H., Heberle-Bors E. (1999). A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell 11: 101–113 [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes D.C., Nam J., Dangl J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A.K., Willmann R., Kolb D., Grefen L., Lajunen H.M., Bethke G., Lee J., Nürnberger T., Gust A.A. (2010). The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 153: 1098–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M.A., Shokat K.M., Rietz S., Parker J., Mundy J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Cardinale F., Meskiene I., Ouaked F., Hirt H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14: 703–711 [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A., Cheng Z., O’Brien J.A., Mammarella N., Khan S., Ausubel F.M., Bolwell G.P. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R., Askari N., Capone R., Engelberg D., Livnah O. (2004). Active mutants of the human p38alpha mitogen-activated protein kinase. J. Biol. Chem. 279: 47040–47049 [DOI] [PubMed] [Google Scholar]
- Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. (2007). Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318: 453–456 [DOI] [PubMed] [Google Scholar]
- Droillard M.J., Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: Activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett. 574: 42–48 [DOI] [PubMed] [Google Scholar]
- Emrick M.A., Lee T., Starkey P.J., Mumby M.C., Resing K.A., Ahn N.G. (2006). The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc. Natl. Acad. Sci. USA 103: 18101–18106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- García A.V., Blanvillain-Baufumé S., Huibers R.P., Wiermer M., Li G., Gobbato E., Rietz S., Parker J.E. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Hinsch M.E., Staskawicz B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Felix G., Boller T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Gomi K., Ogawa D., Katou S., Kamada H., Nakajima N., Saji H., Soyano T., Sasabe M., Machida Y., Mitsuhara I., Ohashi Y., Seo S. (2005). A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol. 46: 1902–1914 [DOI] [PubMed] [Google Scholar]
- Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L., Parker J.E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hettenhausen C., Baldwin I.T., Wu J. (2012). Silencing MPK4 in Nicotiana attenuata enhances photosynthesis and seed production but compromises abscisic acid-induced stomatal closure and guard cell-mediated resistance to Pseudomonas syringae pv tomato DC3000. Plant Physiol. 158: 759–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti J.E., Jarrell E.T., Chang J.D., Abbott D.W., Storz P., Toker A., Cantley L.C., Turk B.E. (2004). A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1: 27–29 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Irie K., Morris P., Giraudat J., Matsumoto K., Shinozaki K. (1998). Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 253: 532–543 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kosetsu K., Matsunaga S., Nakagami H., Colcombet J., Sasabe M., Soyano T., Takahashi Y., Hirt H., Machida Y. (2010). The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B.N., Bent A.F., Dahlbeck D., Innes R.W., Staskawicz B.J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Müller A., Giraudat J., Leung J. (2007). Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardóczy V., Teige M., Koncz C., Peck S., Bögre L. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Kiegerl S., Hirt H. (2004). OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J. Biol. Chem. 279: 26959–26966 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Papworth C., Bauer J.C., Braman J., Wright D.A. (1996). QuikChange site-directed mutagenesis. Strategies 9: 3–4 [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S., Eschen-Lippold L., Pecher P., Lee J., Scheel D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Rozhon W., Petutschnig E., Wrzaczek M., Jonak C. (2005). Quantification of free and total salicylic acid in plants by solid-phase extraction and isocratic high-performance anion-exchange chromatography. Anal. Bioanal. Chem. 382: 1620–1627 [DOI] [PubMed] [Google Scholar]
- Schiestl R.H., Gietz R.D. (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Stulemeijer I.J., Stratmann J.W., Joosten M.H. (2007). Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 144: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Glazebrook J., Katagiri F. (2008). The interplay between MAMP and SA signaling. Plant Signal. Behav. 3: 359–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen E.A., Freddie C.T., Kahn K., Parker J.E., Jones J.D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29: 439–451 [DOI] [PubMed] [Google Scholar]
- Vidal M., Braun P., Chen E., Boeke J.D., Harlow E. (1996). Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc. Natl. Acad. Sci. USA 93: 10321–10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Turk B.E., Peynot P., Leung J., Merlot S. (2008). A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55: 104–117 [DOI] [PubMed] [Google Scholar]
- Wang D., Harper J.F., Gribskov M. (2003). Systematic trans-genomic comparison of protein kinases between Arabidopsis and Saccharomyces cerevisiae. Plant Physiol. 132: 2152–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Chen J.G., Ellis B.E. (2011). AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 67: 895–906 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.