This article identifies a role for the Arabidopsis thaliana Mediator complex subunit MED16 in bridging between specific transcription activators and the transcriptional machinery in the activation of salicylic acid–responsive defenses, including defense gene transcription, pathogen resistance, and systemic acquired resistance. MED16 also participates in ethylene- and jasmonic acid–induced defenses.
Abstract
Systemic acquired resistance (SAR) is a long-lasting plant immunity against a broad spectrum of pathogens. Biological induction of SAR requires the signal molecule salicylic acid (SA) and involves profound transcriptional changes that are largely controlled by the transcription coactivator NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1). However, it is unclear how SAR signals are transduced from the NPR1 signaling node to the general transcription machinery. Here, we report that the Arabidopsis thaliana Mediator subunit16 (MED16) is an essential positive regulator of SAR. Mutations in MED16 reduced NPR1 protein levels and completely compromised biological induction of SAR. These mutations also significantly suppressed SA-induced defense responses, altered the transcriptional changes induced by the avirulent bacterial pathogen Pseudomonas syringae pv tomato (Pst) DC3000/avrRpt2, and rendered plants susceptible to both Pst DC3000/avrRpt2 and Pst DC3000. In addition, mutations in MED16 blocked the induction of several jasmonic acid (JA)/ethylene (ET)–responsive genes and compromised resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. The Mediator complex acts as a bridge between specific transcriptional activators and the RNA polymerase II transcription machinery; therefore, our data suggest that MED16 may be a signaling component in the gap between the NPR1 signaling node and the general transcription machinery and may relay signals from both the SA and the JA/ET pathways.
INTRODUCTION
Plants, like animals, have evolved sophisticated innate immune systems to protect themselves against microbial invasion and colonization (Jones and Takemoto, 2004). Systemic acquired resistance (SAR) is a plant-specific immunity that develops throughout a plant after localized foliar infection by a pathogen (Ryals et al., 1996; Durrant and Dong, 2004). SAR provides long-lasting protection against subsequent infections by a broad spectrum of pathogens; therefore, the identification of key signaling components of SAR has been one of the major focuses in the field.
The signal molecule salicylic acid (SA) and the transcription coactivator NONEXPRESSOR OF PATHOGENESIS-RELATED (PR) GENES1/NON-INDUCIBLE IMMUNITY1/SA-INSENSITIVE1 (NPR1/NIM1/SAI1) have been identified as the key regulators of SAR (Gaffney et al., 1993; Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). Exogenous application of SA induces NPR1-dependent gene transcription and disease resistance, whereas mutations in NPR1 block SA- and pathogen-induced transcriptional reprogramming and compromise both basal immunity and SAR (Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997). NPR1 is thus a master transcriptional regulator functioning downstream of SA. NPR1 contains two protein–protein interaction domains (an ankyrin-repeat and a Broad-complex, Tramtrack, and Bric-a-brac/Pox virus and Zinc domain) and interacts with seven TGA transcription factors and three structurally related NIM1-INTERACTING (NIMIN) proteins (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000; Weigel et al., 2001). On the PR1 gene promoter, interaction of NPR1 with TGA transcription factors helps recruit SUPPRESSOR OF SNI1, 2 (SSN2) to counteract SUPPRESSOR OF NPR1, INDUCIBLE1 (SNI1) repression (Li et al., 1999; Song et al., 2011). The NIMIN proteins appear to negatively regulate SA/NPR1 signaling, preventing detrimental hyperactivation of immune responses (Weigel et al., 2005). The NPR1 signaling node thus plays a critical role in transducing the SAR signal. However, it is not known how the SAR signal is relayed from the NPR1 signaling node to the general transcription machinery.
SA-mediated immunity is generally effective against biotrophs, whereas jasmonic acid (JA)/ethylene (ET)-mediated signaling is central for resistance against necrotrophs (Glazebrook, 2005). SA and JA signaling mostly antagonize each other, although they may function synergistically under certain conditions (Pena-Cortés et al., 1993; Doares et al., 1995; Schenk et al., 2000; van Wees et al., 2000; Spoel et al., 2003; Mur et al., 2006). Proteins regulating SA-JA/ET crosstalk have been identified in Arabidopsis thaliana (Petersen et al., 2000; Kachroo et al., 2001; Li et al., 2004; Brodersen et al., 2006; Ndamukong et al., 2007). The SA signal transducer NPR1 acts as a crucial modulator in SA-mediated suppression of JA signaling and regulates SA-mediated expression of several genes encoding transcription (co)factors that suppress JA-dependent gene expression (Spoel et al., 2003; Li et al., 2004; Ndamukong et al., 2007). Whereas the NPR1-interacting TGA transcription factors participate in both SA- and JA/ET-induced defense signaling, their positive role in JA/ET signaling seems to be abolished in the presence of SA (Ndamukong et al., 2007; Zander et al., 2010). Crosstalk between SA and JA/ET signaling is believed to ensure efficient prioritization of immune responses against biotrophs and necrotrophs (Spoel et al., 2007); however, molecular mechanisms underlying the crosstalk remain elusive.
In eukaryotic cells, RNA polymerase II (RNAPII) catalyzes the transcription of all protein-encoding genes (Woychik and Hampsey, 2002). The transcription process is regulated by a collection of countless transcriptional regulatory proteins (Kadonaga, 2004). A multiprotein complex named Mediator has attracted considerable attention in recent years, because of its essential role in transcription (Kim et al., 1994; Kornberg, 2005; Takagi and Kornberg, 2006; Conaway and Conaway, 2011a). Mediator exists in the cell in multiple functionally distinct forms and serves as either a transcriptional activator or a repressor, depending on its associated protein components (Conaway and Conaway, 2011b). The Mediator core is composed of more than 20 subunits organized into three modules named head, middle, and tail (Guglielmi et al., 2004; Chadick and Asturias, 2005). The Mediator core can associate with the RNAPII complex to form the holoenzyme, which stimulates basal transcription and supports activation of transcription by specific transcription activators (Mittler et al., 2001; Baek et al., 2002; Zhu et al., 2006; Ansari et al., 2009). By interacting with a particular transcription activator or a class of transcription activators, individual Mediator subunits relay diverse signals to the general transcription machinery leading to pathway-specific gene transcription (Balamotis et al., 2009; Kagey et al., 2010; Takahashi et al., 2011). The Mediator core can also interact with a kinase module, which precludes its binding to the RNAPII complex, resulting in transcriptional repression (Holstege et al., 1998; Akoulitchev et al., 2000; Knuesel et al., 2009). The functionally distinct forms of Mediator thus provide various regulatory mechanisms to fine-tune gene-specific and pathway-specific transcriptional reprogramming (Balamotis et al., 2009).
The Mediator complex is highly conserved in eukaryotes ranging from yeast to humans (Boube et al., 2002; Bourbon, 2008). The Arabidopsis Mediator complex, recently purified to near homogeneity, contains 21 conserved and six putative Arabidopsis-specific Mediator subunits (Bäckström et al., 2007). Mediator functions as a bridge between the RNAPII complex and specific transcription activators; therefore, it is expected that individual Mediator subunits interact with and receive signals from a subset of the ∼1500 transcription factors encoded by the Arabidopsis genome (Riechmann et al., 2000). Indeed, several Arabidopsis Mediator subunits have been implicated in specific signaling pathways. Arabidopsis Mediator subunit25 (MED25)/PHYTOCHROME AND FLOWERING TIME1 (PFT1) and MED14/STRUWWELPETER (SWP) were first identified as key regulators of flowering and cell proliferation, respectively (Autran et al., 2002; Cerdán and Chory, 2003). Recently, MED25/PFT1 and two other Arabidopsis Mediator subunits, MED8 and MED21, were found to regulate JA-dependent defense responses (Dhawan et al., 2009; Kidd et al., 2009). In addition, Arabidopsis MED17, MED18, and MED20a have been shown to play a role in small and long noncoding RNA production (Kim et al., 2011).
MED16 is required for lipopolysaccharide-induced gene expression in Drosophila melanogaster (Kim et al., 2004). MED16 appears to be a specific binding partner of the differentiation-inducing factor, relaying the activation signal from the differentiation-inducing factor to the basal transcription machinery during lipopolysaccharide-induced innate immune response. In plants, Arabidopsis MED16 was first reported as a positive regulator of acclimation to freezing temperatures after exposure to low temperatures and was named SENSITIVE TO FREEZING6 (SFR6) (Knight et al., 2009). Knight et al. speculated that MED16/SFR6 might modulate the activity of C-box binding factor (CBF) transcription factors or help recruit CBF proteins into the nucleus in regulating cold on-regulated (COR) gene expression. It is likely that MED16/SFR6 is the target in Mediator for CBF-dependent pathway activation of COR gene transcription.
In a genetic screen for mutants insensitive to exogenous NAD+ (ien) (Zhang and Mou, 2009), we identified the ien1/med16-1 mutant, in which exogenous NAD+-induced PR1 gene expression was significantly inhibited. Characterization of ien1/med16/sfr6 in this current study revealed that MED16 is not only a key positive regulator of SAR, but also a convergence point of SA- and JA/ET-mediated defense pathways.
RESULTS
An Arabidopsis Mutant Insensitive to Exogenous NAD+ Exhibits Increased Susceptibility to the Avirulent Bacterial Pathogen Pseudomonas syringae pv tomato DC3000/avrRpt2
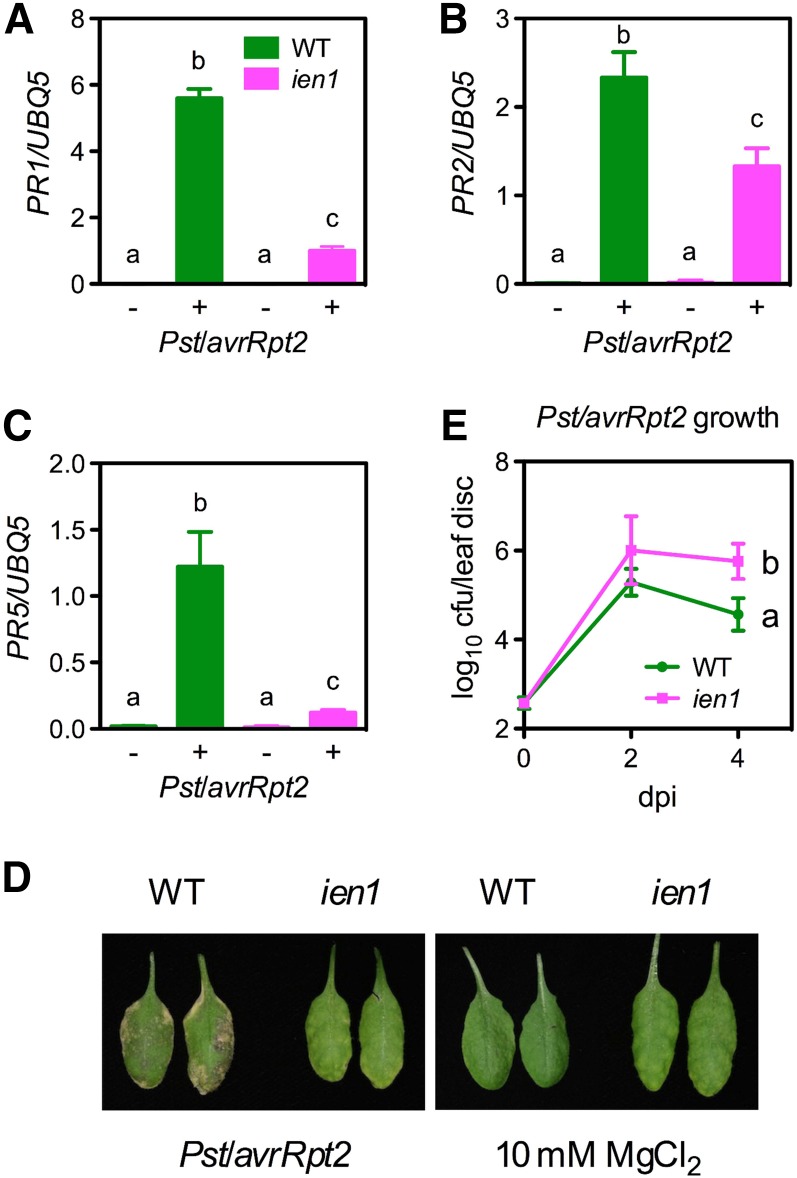
We previously reported that exogenous NAD(P) induces PR gene expression and disease resistance (Zhang and Mou, 2009), suggesting that extracellular NAD(P) [eNAD(P)] is likely an elicitor of plant immune responses. To identify new components in the eNAD(P)-activated signaling pathway, we performed a genetic screen designed to isolate ien mutants. Seeds of a PR1:luciferase transgenic line were mutagenized with ethyl methanesulfonate, and the M2 seedlings grown on one-half–strength Murashige and Skoog (MS) medium were treated with 5 mM of exogenous NAD+ and screened. One mutant, ien1, exhibited decreased PR1 expression after NAD+ treatment (Figures 1A and 1B). NAD(P) leaking into the extracellular compartment during pathogen infection might contribute to defense gene induction (Zhang and Mou, 2009); therefore, we asked whether the ien1 mutation affects pathogen-induced defense gene expression. To this end, we inoculated ien1 plants with the avirulent bacterial pathogen Pseudomonas syringae pv tomato (Pst) DC3000/avrRpt2. After 24 h, the inoculated leaves were collected for PR gene analysis. Compared with the wild type, Pst DC3000/avrRpt2–induced expression of PR1, PR2, and PR5 was drastically decreased in the ien1 plants (Figures 2A to 2C). Interestingly, Pst DC3000/avrRpt2–induced lesion formation, which is a visual defense phenotype (Dangl et al., 1996), was alleviated in ien1 (Figure 2D). Consistently, Pst DC3000/avrRpt2 grew significantly more in ien1 than in the wild type (Figure 2E). Thus, IEN1 is a positive regulator of Pst DC3000/avrRpt2–induced defense responses.
Figure 1.
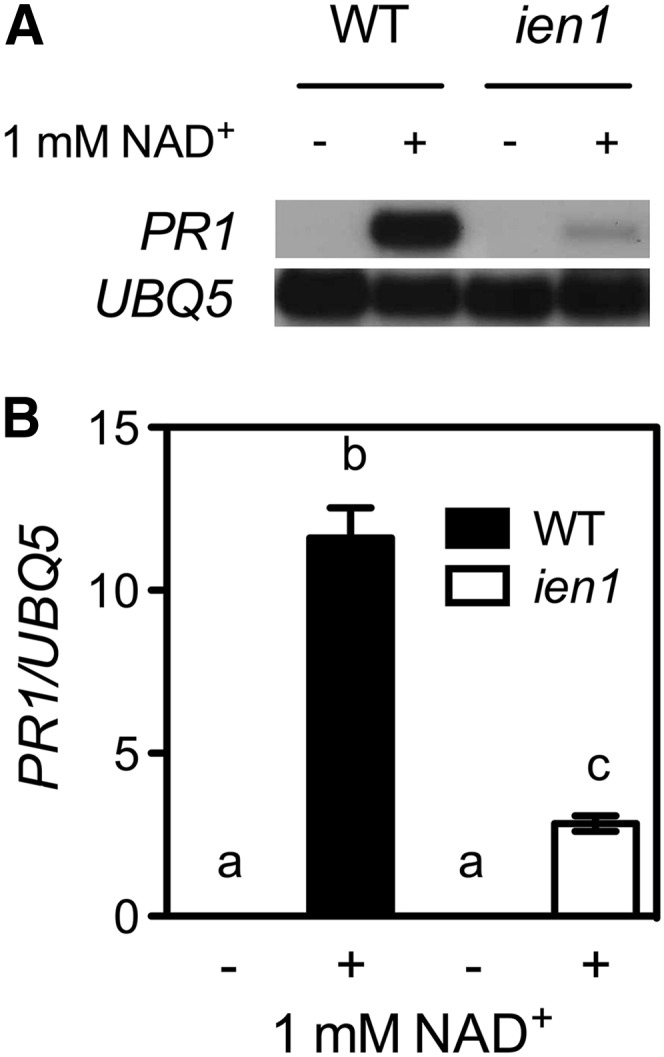
Isolation of the ien1 Mutant.
(A) Exogenous NAD+-induced PR1 gene expression in ien1 and wild-type (WT) seedlings. Seedlings grown on one-half–strength MS medium were treated with sprays of 1 mM of NAD+ solution. Total RNA was extracted from plant tissues collected 24 h later and subjected to RNA gel blot analysis. The UBQ5 gene was used as a loading control.
(B) Exogenous NAD+-induced PR1 gene expression in ien1 and wild-type soil-grown plants. Leaves of 4-week-old soil-grown plants were infiltrated with 1 mM of NAD+ solution. Total RNA was extracted from leaf tissues collected 24 h later and subjected to real-time qPCR analysis. Expression was normalized against constitutively expressed UBQ5. Data represent the mean of three independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test).
Figure 2.
Defense Phenotypes of the ien1 Mutant.
(A) to (C) Pst DC3000/avrRpt2–induced PR1 (A), PR2 (B), and PR5 (C) gene expression in ien1 and wild-type (WT) plants. Four-week-old soil-grown plants were inoculated with Pst DC3000/avrRpt2 (OD600 = 0.001). Total RNA was extracted from the inoculated leaves collected 24 h later and subjected to real-time qPCR analysis. Data represent the mean of three independent samples with sd.
(D) Visual phenotype of Pst DC3000/avrRpt2–infected ien1 and wild-type leaves. Four-week-old soil-grown plants were inoculated with Pst DC3000/avrRpt2 (OD600 = 0.001) or infiltrated with 10 mM of MgCl2 as a control. Photos were taken 4 d after inoculation.
(E) Growth of Pst DC3000/avrRpt2 in ien1 and wild-type plants. Four-week-old soil-grown plants were inoculated with Pst DC3000/avrRpt2 (OD600 = 0.001). The in planta bacterial titers were determined immediately and 2 and 4 days postinoculation (dpi). Data represent the mean of eight independent samples with sd. Different letters above the bars in (A), (B), and (C) or on the right side of the lines in (E) indicate significant differences (P < 0.05, Student’s t test). All experiments were repeated three times with similar results. cfu, colony-forming units.
IEN1 Encodes MED16
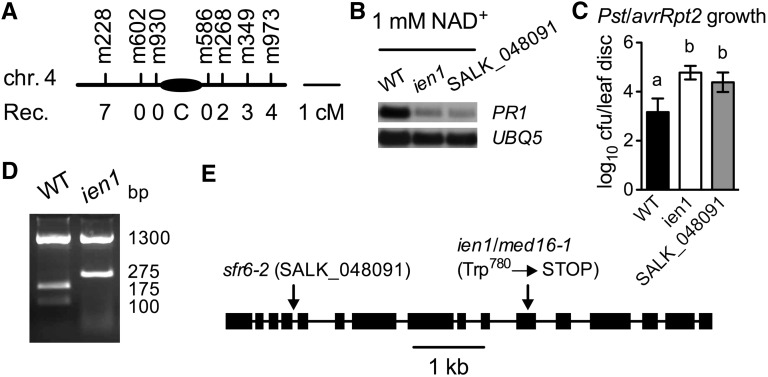
Morphologically, the ien1 plants were a paler shade of green than the wild type (see Supplemental Figure 1 online). This phenotype cosegregates with the ien1 defense phenotype and was therefore used to map the IEN1 locus. We crossed homozygous ien1, which is in the Columbia background, to the polymorphic ecotype Landsberg erecta. Linkage analysis of 118 F2 plants with ien1 morphology placed the IEN1 locus between the molecular markers CIW5 and CIW6 on chromosome 4. Recombination analysis of 1214 F2 mutant plants was only able to locate the IEN1 locus in an interval of ∼3.1 Mbp between markers m228 and m268, because of the low frequency of recombination in the centromeric region (Figure 3A). This low frequency of recombination was also encountered when the sfr6-1 mutation was cloned in the same interval (Knight et al., 2009). Similar to ien1, sfr6 mutants are also a paler shade of green than the wild type, indicating that ien1 might be allelic to sfr6. To test this, we obtained the T-DNA insertion line SALK_048091 (sfr6-2) carrying a T-DNA insertion in At4g04920 from the ABRC and crossed it with ien1. All F1 plants were paler than the wild type and similar to the parental ien1 and sfr6 plants (see Supplemental Figure 1 online). Moreover, SALK_048091 showed significantly decreased induction of PR1 by exogenous NAD+ and enhanced susceptibility to Pst DC3000/avrRpt2 (Figures 3B and 3C). These results indicate that ien1 is allelic to sfr6. To further confirm that the ien1 mutant phenotypes were caused by a mutation in SFR6, we attempted to complement the ien1 mutant with the full-length SFR6 genomic coding sequence driven by the 35S constitutive promoter. The transgenic plants exhibited wild-type morphology, strong induction of PR1 gene by NAD+, and wild-type levels of resistance to Pst DC3000/avrRpt2 (see Supplemental Figure 2 online), supporting that IEN1 is SFR6. To identify the ien1 mutation, a set of overlapping PCR fragments covering At4g04920 was amplified from ien1 and sequenced. A single base substitution of A for G was detected at the nucleotide 4396 (counting from the A in the ATG start codon). This mutation introduced a stop codon in the 11th exon (Figure 3E). We then successfully developed a cleaved amplified polymorphic sequence (CAPS) marker based on the mutation to genetically distinguish the ien1 mutant from the wild type (Figure 3D). At4g04920 was recently shown to encode Arabidopsis MED16; therefore, ien1 was renamed med16-1 (Bourbon et al., 2004; Bäckström et al., 2007).
Figure 3.
Map-Based Cloning of ien1.
(A) A schematic diagram of the map-based cloning process. A total of 118 F2 progeny homozygous for ien1 were used to determine the approximate position of the ien1 mutation using bulked segregant analysis. The ien1 mutation was linked to the markers CIW5 and CIW6 on chromosome 4. Out of a total mapping population of 1214 plants homozygous for ien1, seven were heterozygous at m228, and two were heterozygous at m268. The heterozygotes found by these two markers were mutually exclusive. No heterozygotes were found at m602, m930, and m586. cM, centimorgan; Rec., recombination.
(B) Exogenous NAD+-induced PR1 gene expression in ien1, SALK_048091 (sfr6-2), and wild-type (WT) plants. Leaves on 4-week-old soil-grown plants were infiltrated with 1 mM of NAD+ solution. Total RNA was extracted from leaf tissues collected 24 h later and subjected to RNA gel blot analysis. The UBQ5 gene was used as a loading control.
(C) Growth of Pst DC3000/avrRpt2 in ien1, SALK_048091, and wild-type plants. Four-week-old soil-grown plants were inoculated with Pst DC3000/avrRpt2 (OD600 = 0.001). The in planta bacterial titers were determined immediately and 4 d after inoculation. Data represent the mean of eight independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test). The experiment was repeated three times with similar results. cfu, colony-forming units.
(D) A CAPS marker generated based on the ien1 mutation. The PCR products amplified from ien1 and wild-type genomic DNA were digested with NcoI and separated on an agarose gel.
(E) Structure of the IEN1/MED16 gene (At4g04920), the ien1 mutation, and the insertion site of the T-DNA insertion line SALK_048091. Boxes denote the translated regions and lines between boxes denote introns.
MED16 Suppresses SAR-Negative Regulators and Promotes SAR-Positive Regulators
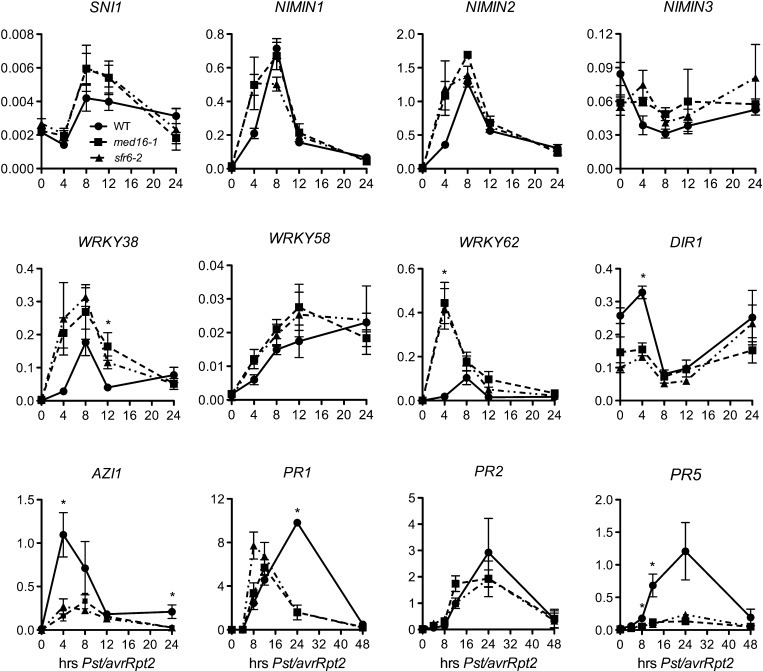
Mediator is a transcription coregulator, and mutations in the MED16 gene compromise resistance to Pst DC3000/avrRpt2, suggesting that the med16/sfr6 mutation may alter Pst DC3000/avrRpt2–induced gene expression. To identify potential candidate genes that are regulated by MED16, we performed a microarray experiment to compare Pst DC3000/avrRpt2–induced transcriptome changes in med16-1 and the wild type, and then examined genes that showed a twofold or larger difference in their expression levels between med16-1 and the wild type (National Center for Biotechnology Information Gene Expression Omnibus series number GSE38999). As shown in Supplemental Table 1 online, after Pst DC3000/avrRpt2 infection, a large number of defense genes, including ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5)/SA INDUCTION DEFICIENT1 (SID1), AVRPPH3 SUSCEPTIBLE3 (PBS3)/HOPW1-1-INTERACTING3 (WIN3)/GH3-LIKE DEFENSE GENE1 (GDG1), AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1), FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1), ACCELERATED CELL DEATH6 (ACD6), AZELAIC ACID INDUCED1 (AZI1), and SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 (SFD1)/GLY1, and many NPR1 target genes, such as PR1, PR2, PR5, VACUOLAR SORTING RECEPTOR6 (VSR6), WRKY30, WRKY53, WRKY54, WRKY59, WRKY66, and WRKY70, were potentially either upregulated or downregulated in med16-1. Interestingly, a group of SAR-negative regulators, including SNI1, NIMIN1, NIMIN2, NIMIN3, WRKY38, WRKY58, and WRKY62, were potentially all upregulated in med16-1 (see Supplemental Table 1 online). The numbers of differentially expressed genes were based on comparisons of single arrays with no replication; therefore, statistical significance could not be assessed. However, this microarray data provided a set of candidate genes for further analysis. To confirm and extend the microarray results for the selected candidate genes, we used real-time quantitative PCR (qPCR) to monitor the induction kinetics of SNI1, NIMIN1, NIMIN2, NIMIN3, WRKY38, WRKY58, WRKY62, DEFECTIVE IN INDUCED RESISTANCE1 (DIR1), AZI1, PR1, PR2, and PR5 in med16-1, sfr6-2, and the wild type after Pst DC3000/avrRpt2 infection. Consistent with the microarray results, the induction of two SAR-negative regulators (WRKY38 and WRKY62) in med16/sfr6 was faster and stronger than in the wild type (Kim et al., 2008), whereas the induction of the SAR-positive regulator AZI1 in med16/sfr6 was inhibited (Figure 4) (Jung et al., 2009). In addition, the expression of the SAR-positive regulator DIR1 was decreased in med16/sfr6 plants (Maldonado et al., 2002). As a late response gene, PR1 was upregulated at 8 and 16 h after infection, but its expression was significantly decreased at 24 h in med16/sfr6 mutants (Figure 4). Interestingly, PR5 was only marginally induced in med16/sfr6 (Figure 4). Taken together, these results indicate that MED16 may regulate plant immune responses by suppressing SAR-negative regulators and promoting SAR-positive regulators.
Figure 4.
Pst DC3000/avrRpt2–Induced Kinetic Expression of 12 Defense Genes in med16/sfr6 Mutants.
Plants were inoculated with the avirulent bacterial pathogen Pst DC3000/avrRpt2 (OD600 = 0.001). Leaf tissues were collected at the indicated time points. Total RNA was extracted from the inoculated leaves and analyzed for the expression of indicated genes using real-time qPCR. Expression was normalized against constitutively expressed UBQ5. Data represent the mean of three independent samples with sd. An asterisk (*) indicates that the expression level of the gene in the wild type (WT) was either significantly lower (WRKY38 and WRKY62) or significantly higher (DIR1, AZI1, PR1, and PR5) than in both med16-1 and sfr6-2 (P < 0.05, Student’s t test). The experiment was repeated with similar results.
MED16 Functions Downstream of SA
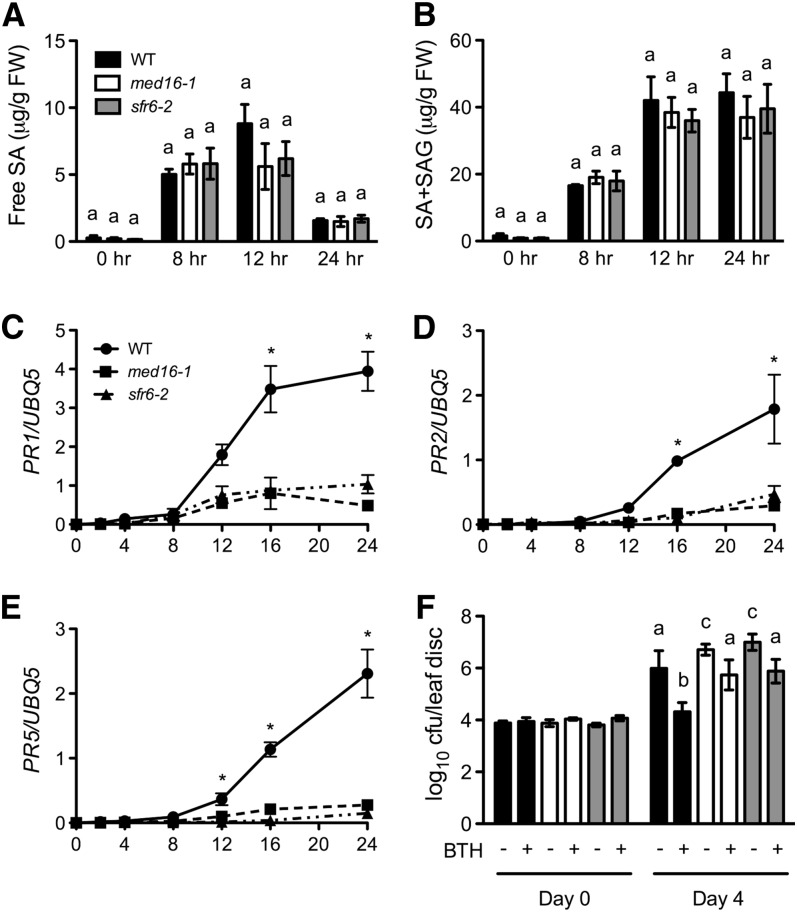
To test whether pathogen-induced SA accumulation is altered in med16/sfr6 plants, we measured SA levels in Pst DC3000/avrRpt2–infected med16/sfr6 and wild-type plants. As shown in Figures 5A and 5B, although free SA levels in the med16/sfr6 mutant plants at 12 h after Pst DC3000/avrRpt2 infection were lower than in the wild type, they were not significantly different from those in the wild type, suggesting that MED16 is not a major contributor to SA biosynthesis. The med16/sfr6 mutation inhibits Pst DC3000/avrRpt2–induced PR gene expression; therefore, MED16 may function downstream of SA to regulate immune responses. To test this, PR gene expression in med16/sfr6 plants treated with soil drenches and foliar sprays of the SA biologically active analog benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH) was examined. We used BTH instead of SA, because BTH is a stronger inducer of defense genes than SA (Friedrich et al., 1996). In agreement with an earlier report (Wathugala et al., 2012), BTH-induced expression of PR1, PR2, and PR5 was dramatically decreased in the med16/sfr6 mutant plants (Figures 5C to 5E). Thus, MED16 plays a role downstream of SA in regulating defense gene expression, and mutations in MED16 significantly compromise SA responsiveness.
Figure 5.
Pst DC3000/avrRpt2–Induced SA Accumulation and BTH-Induced Defense Responses in med16/sfr6 Mutants.
(A) and (B) Free SA (A) and total (B) levels in Pst DC3000/avrRpt2–infected med16/sfr6 and wild-type (WT) plants. Four-week-old soil-grown plants were inoculated with Pst DC3000/avrRpt2 (OD600 = 0.002). The inoculated leaves were collected at the indicated time points for SA measurement. Data represent the mean of four independent samples with sd. FW, fresh weight; SAG, SA-2-O-β-d-glucoside.
(C) to (E) BTH-induced PR1 (C), PR2 (D), and PR5 (E) gene expression in med16/sfr6 and wild-type plants. Four-week-old soil-grown plants were treated with soil drenches plus foliar sprays of 0.3 mM of BTH solution. Leaf tissues were collected at the indicated time points and subjected to total RNA extraction and real-time qPCR analysis. Data represent the mean of three independent samples with sd.
(F) BTH-induced resistance to Pst DC3000 in med16/sfr6 and wild-type plants. Four-week-old soil-grown plants were treated with soil drenches plus foliar sprays of 0.3 mM of BTH solution (+BTH) or water (−BTH). After 24 h, the plants were inoculated with Pst DC3000 (OD600 = 0.001). The in planta bacterial titers were determined immediately and 4 d after inoculation. Data represent the mean of eight independent samples with sd. Different letters above the bars in (A), (B), and (F) indicate significant differences (P < 0.05, Student’s t test), and an asterisk (*) in (C), (D), and (E) indicates that the expression level of the gene in the wild type was significantly higher than in both med16-1 and sfr6-2 (P < 0.05, Student’s t test). Note that the comparison was made separately among the wild type, med16-1, and sfr6-2 for each time point. All experiments were repeated three times with similar results. cfu, colony-forming units.
We also examined BTH-induced pathogen resistance in med16/sfr6 plants. The growth of the virulent bacterial pathogen Pst DC3000 in BTH-treated med16/sfr6 plants was significantly higher than in BTH-treated wild-type plants (Figure 5F). This result is consistent with the conclusion that MED16 functions downstream of SA as a positive regulator of plant immune responses.
MED16 Plays a Positive Role in Basal Immunity
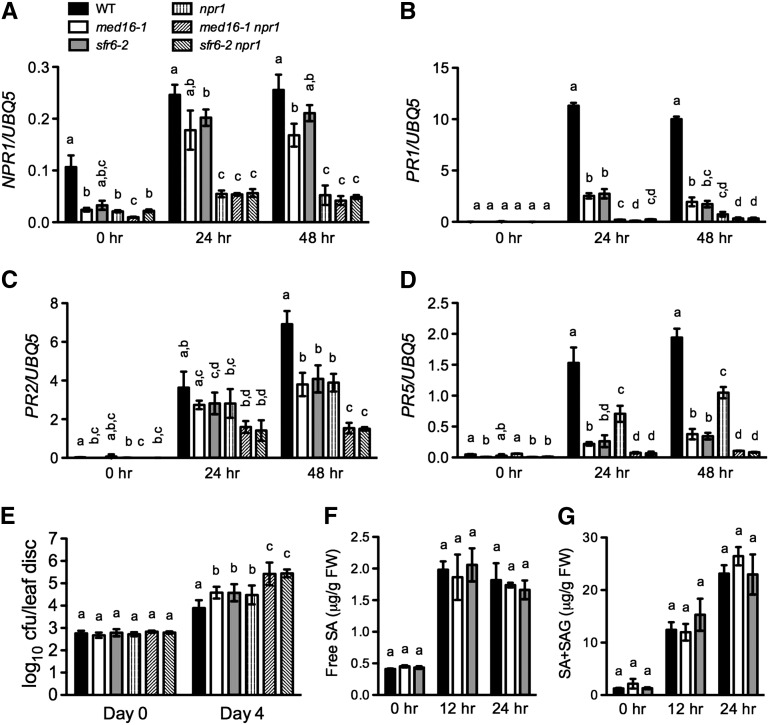
Because med16 influences the expression of many NPR1 target genes (see Supplemental Table 1 online), Arabidopsis MED16 may positively contribute to basal immunity like NPR1 does (Wathugala et al., 2012). To compare the roles of MED16 and NPR1 in basal defense responses, we examined the expression of NPR1, PR1, PR2, and PR5 in med16-1, sfr6-2, npr1, med16-1 npr1, sfr6-2 npr1, and wild-type plants during the infection of the virulent bacterial pathogen Pst DC3000. The background expression levels of NPR1 were lower in med16/sfr6 than in the wild type, but after Pst DC3000 infection, NPR1 was induced to a level just slightly lower than that in the wild type (Figure 6A). Note that the expression levels of NPR1 were significantly decreased in the npr1-3 mutant, probably because of mRNA decay caused by the premature stop codon in the mutant (Cao et al., 1997; Chang et al., 2007). Compared with the wild type, both med16/sfr6 and npr1 compromised Pst DC3000–induced PR gene expression (Figures 6B to 6D), which is consistent with the previous report (Wathugala et al., 2012). The med16/sfr6 mutations showed a stronger effect on PR5, whereas npr1 had a stronger effect on PR1, and med16/sfr6 and npr1 displayed a comparable effect on the expression of PR2. In the med16/sfr6 npr1 double mutants, expression of PR2 and PR5 was further decreased to a level lower than that in either single mutant (Figures 6B to 6D). These results suggest that MED16, like NPR1, plays a positive role in Pst DC3000–induced PR gene expression.
Figure 6.
Genetic Relationship between med16/sfr6 and npr1.
(A) to (D) Pst DC3000–induced expression of NPR1 (A), PR1 (B), PR2 (C), and PR5 (D) in med16/sfr6, npr1, med16/sfr6 npr1, and wild-type (WT) plants. Plants were inoculated with Pst DC3000 (OD600 = 0.001). Total RNA was extracted from the inoculated leaves collected at the indicated time points and analyzed for the expression of NPR1 using real-time qPCR. Expression was normalized against constitutively expressed UBQ5. Data represent the mean of three independent samples with sd.
(E) Growth of Pst DC3000 in med16/sfr6, npr1, med16/sfr6 npr1, and wild-type plants. Leaves of 4-week-old plants were inoculated with Pst DC3000 (OD600 = 0.0001). The in planta bacterial titers were determined immediately and 4 d after inoculation. Data represent the mean of eight independent samples with sd. cfu, colony-forming units.
(F) and (G) Free SA (F) and total (G) levels in Pst DC3000–infected med16/sfr6 and wild-type plants. Plants were inoculated with Pst DC3000 (OD600 = 0.001). The inoculated leaves were collected at the indicated time points for SA measurement. Data represent the mean of four independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test). The comparison was made separately among the wild type, med16-1, sfr6-2, npr1, med16-1 npr1, and sfr6-2 npr1 for each time point. All experiments were repeated with similar results. FW, fresh weight; SAG, SA-2-O-β-d-glucoside.
We also tested Pst DC3000 growth in med16/sfr6, npr1, and the double mutant med16/sfr6 npr1. Pst DC3000 grew significantly more in med16/sfr6 and npr1 than in the wild type, and the growth of Pst DC3000 in med16/sfr6 and npr1 was comparable, suggesting that MED16 also plays a role in basal resistance (Figure 6E). Interestingly, Pst DC3000 growth increased about sevenfold in the med16/sfr6 npr1 double mutant plants compared with that in either single mutant (Figure 6E). Because med16/sfr6 did not significantly affect Pst DC3000–induced SA accumulation (Figures 6F and 6G), these results support the conclusion that MED16 functions downstream of SA in plant immunity (Wathugala et al., 2012).
MED16 Is Required for the Establishment of SAR
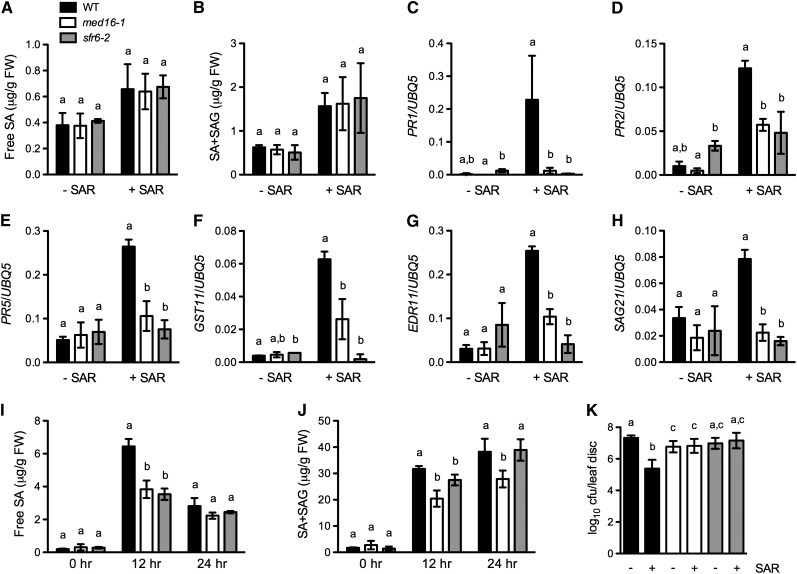
Mutations in the MED16 gene compromise SA responsiveness; therefore, MED16 may play a role in SAR. To test this hypothesis, we analyzed the biological induction of SAR in med16/sfr6 plants. Three lower leaves on each plant were infiltrated with either 10 mM of MgCl2 (mock treatment) or P. syringae pv maculicola (Psm) ES4326 (SAR treatment). After 2 d, we measured SAR treatment–induced SA accumulation and gene expression in the upper, untreated systemic leaves. As shown in Figures 7A and 7B, free SA and total SA levels in the systemic leaves of the med16/sfr6 plants were comparable with those in the wild type. However, the expression of six genes (PR1, PR2, PR5, GST11, EDR11, and SAG21), which are induced in systemic leaves during SAR (Maleck et al., 2000), was significantly decreased in the med16/sfr6 plants (Figures 7C to 7H), suggesting that MED16 is required for SAR treatment–induced defense gene expression in systemic leaves.
Figure 7.
SAR Induction in med16/sfr6 Mutants.
(A) and (B) Free SA (A) and total SA (B) levels in systemic leaves of med16/sfr6 and wild-type (WT) plants. Three lower leaves on each plant were inoculated with Psm ES4326 (OD600 = 0.002) (+SAR) or mock-treated with 10 mM of MgCl2 (−SAR). The upper uninfected/untreated systemic leaves were collected 48 h later for SA measurement. Data represent the mean of three independent samples with sd. FW, fresh weight; SAG, SA-2-O-β-d-glucoside.
(C) to (H) Expression of PR1, PR2, PR5, GST11, EDR11, and SAG21 in systemic leaves of med16/sfr6 and wild-type plants. Three lower leaves on each plant were inoculated with Psm ES4326 (OD600 = 0.002) (+SAR) or mock-treated with 10 mM of MgCl2 (−SAR). After 2 d, total RNA was extracted from the upper uninfected/untreated systemic leaves and analyzed for the expression of indicated genes using real-time qPCR. Expression was normalized against constitutively expressed UBQ5. Data represent the mean of three independent samples with sd.
(I) and (J) Free SA (I) and total SA (J) levels in challenge-inoculated systemic leaves of med16/sfr6 and wild-type plants. Three lower leaves on each plant were inoculated with Psm ES4326 (OD600 = 0.002). After 2 d, the upper uninfected systemic leaves were challenge-inoculated with Psm ES4326 (OD600 = 0.001). Leaf samples were collected at the indicated time points for SA measurement. Data represent the mean of three independent samples with sd.
(K) SAR-mediated resistance in med16/sfr6 and wild-type plants. Three lower leaves on each plant were inoculated with Psm ES4326 (OD600 = 0.002) (+SAR) or mock-treated with 10 mM of MgCl2 (−SAR). After 2 d, two upper uninfected/untreated leaves were challenge-inoculated with Psm ES4326 (OD600 = 0.001). The in planta bacterial titers were determined immediately and 3 d after challenge inoculation. Data represent the mean of eight independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test). The comparison was made separately among the wild type, med16-1, and sfr6-2 for each time point or treatment. All the experiments were repeated with similar results. cfu, colony-forming units.
To test whether MED16 plays a role in the execution of SAR on secondary infection, we challenge-inoculated the upper, untreated systemic leaves with Psm ES4326 2 d after the primary infection and monitored SA accumulation. At 12 h after the challenge inoculation, med16/sfr6 plants accumulated significantly less free SA and total SA than wild-type plants (Figures 7I and 7J), indicating that MED16 is required for full accumulation of SA in systemic leaves after challenge inoculation. To test whether SAR induction in med16/sfr6 plants is effective for limiting pathogen growth, bacterial titers were determined 3 d after the challenge inoculation. As shown in Figure 7K, SAR activation induced strong resistance in the wild-type plants. By contrast, SAR treatment did not induce any resistance in the systemic leaves of med16/sfr6 plants, as illustrated by the similar levels of Psm ES4326 growth in both the mock-treated and the SAR-treated med16/sfr6 plants. These results demonstrate that MED16 is a key positive regulator of SAR.
MED16 Functions Downstream of JA and ET and Is Required for Resistance to Necrotrophic Fungal Pathogens
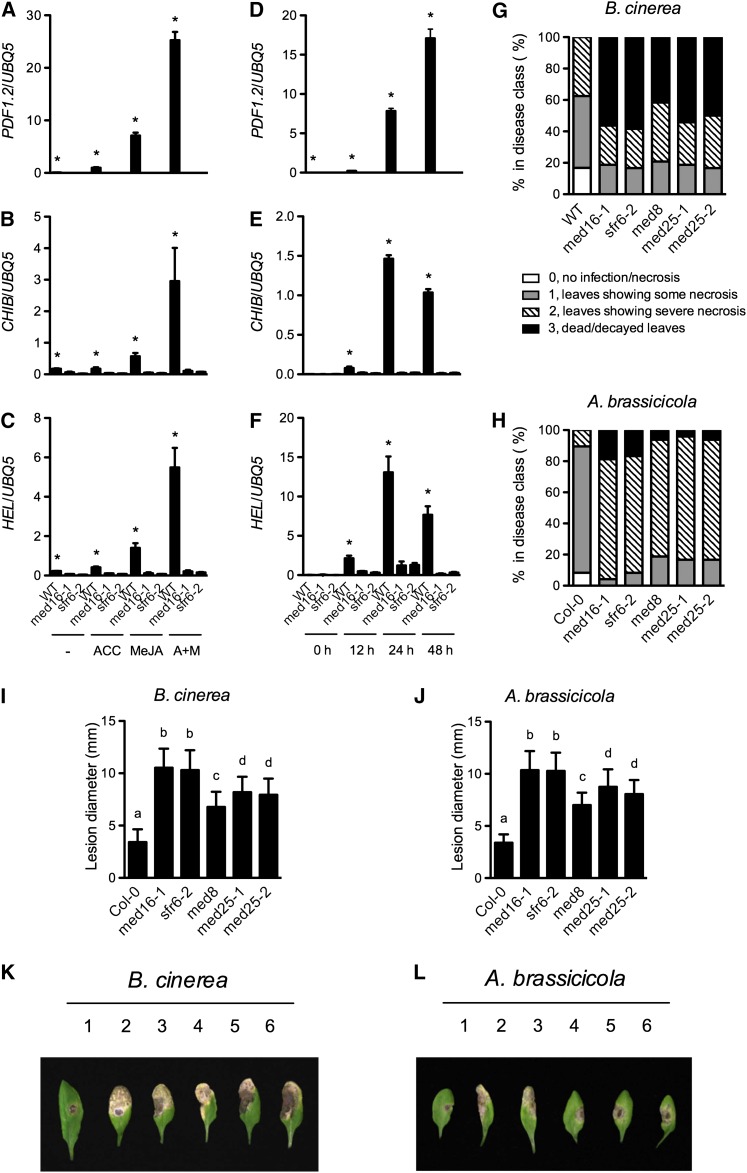
Arabidopsis MED8 and MED25 have been implicated in JA-mediated defense against necrotrophic pathogens in Arabidopsis (Kidd et al., 2009). Induction of the JA/ET-responsive gene PDF1.2 by methyl jasmonate (MeJA) is inhibited in med25/pft1 mutants and further decreased in the med8 med25 double mutant. Both med8 and med25 are more susceptible to the leaf-infecting necrotrophic pathogens Botrytis cinerea and Alternaria brassicicola (Kidd et al., 2009). We reasoned that MED16 might also play a role in JA/ET-mediated antimicrobial signaling. To this end, we analyzed the induction of several JA/ET-responsive genes by 1-aminocyclopropane-1-carboxylic acid (ACC; the immediate precursor of ET), MeJA, or a combination of ACC and MeJA. Consistent with the previous reports (Penninckx et al., 1998; Norman-Setterblad et al., 2000), the JA/ET-responsive genes PDF1.2, CHIB/PR3, and HEL/PR4 were induced by both ACC and MeJA, and the induction was synergistically enhanced by the combination of ACC and MeJA (Figures 8A to 8C). Interestingly, the induction of PDF1.2 and CHIB by ACC, MeJA, and their combination was almost completely blocked in med16, (Wathugala et al., 2012) and the induction of HEL was also significantly suppressed. To test whether MED16 is required for the induction of PDF1.2, CHIB, and HEL during pathogen infection, we inoculated med16-1, sfr6-2, and wild-type plants with B. cinerea, a necrotrophic pathogen activating JA and ET responses. As shown in Figures 8D to 8F, whereas all three genes were significantly induced by B. cinerea in the wild-type plants, the induction of PDF1.2 and CHIB was completely blocked, and that of HEL was also dramatically decreased in the med16/sfr6 plants. Taken together, these results indicate that MED16 is required for JA/ET-mediated defense gene expression.
Figure 8.
Induction of JA/ET-Responsive Genes and Resistance to Necrotrophic Pathogens in med16/sfr6 Mutants.
(A) to (C) ACC- and MeJA-induced expression of PDF1.2, CHIB, and HEL in med16/sfr6 and wild-type (WT) plants. Ten-d-old seedlings grown on one-half–strength MS medium were transplanted onto one-half–strength MS medium (−) or one-half–strength MS medium supplemented with 0.1 mM of ACC, 0.1 mM of MeJA, or both (A+M). Total RNA was extracted from plant tissues except roots collected 24 h later and subjected to real-time qPCR analysis. The UBQ5 gene was used as a loading control.
(D) to (F) B. cinerea–induced expression of PDF1.2, CHIB, and HEL in med16/sfr6 and wild-type plants. Four-week-old soil-grown plants were inoculated with B. cinerea spores, and the inoculated leaves were collected at the indicated time points and analyzed as in (A).
(G) and (H) Growth of B. cinerea (G) or A. brassicicola (H) on med16/sfr6, med8, med25, and wild-type plants. Four-week-old soil-grown plants were inoculated with B. cinerea or A. brassicicola spores, and the inoculated leaves were scored 4 d later and classified according to the disease symptoms. A total of 64 leaves on 16 plants were scored for each genotype. Col-0, ecotype Columbia.
(I) and (J) Size of the necrotic lesions formed on B. cinerea–infected (I) or A. brassicicola–infected (J) med16/sfr6, med8, med25, and wild-type plants. Data represent the mean of lesion sizes on 36 leaves with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test).
(K) and (L) Symptoms on rosette leaves of 4-week-old soil-grown med16, med8, med25, and wild-type plants inoculated with B. cinerea (K) or A. brassicicola (L) spores. Photos were taken 4 d after inoculation. 1: the wild type; 2: med16-1; 3: sfr6-2; 4: med8; 5: med25-1; 6: med25-2. An asterisk (*) in (A) to (F) indicates that the expression level of the gene in the wild type was significantly higher than in both med16-1 and sfr6-2 (P < 0.05, Student’s t test). The comparison was made separately between the wild type and med16-1 or sfr6-2 for each time point or treatment. All experiments were repeated with similar results.
To determine whether MED16 plays a role in resistance to necrotrophic pathogens, we inoculated med16-1, sfr6-2, and wild-type plants with B. cinerea and A. brassicicola. The med8, med25-1, and med25-2 plants were included as controls in the experiments. Results showed that all five mutants exhibited enhanced susceptibility to both B. cinerea and A. brassicicola (Figures 8G to 8L). In the experiment for B. cinerea infection, 0% and ∼17% of the inoculated leaves from med16/sfr6 and wild-type plants, respectively, showed no necrosis, whereas ∼56% and 0% of the inoculated leaves from med16/sfr6 and wild-type plants, respectively, were dead or decayed (Figure 8G). The average lesion sizes (diameter) on med16/sfr6 plants were ∼10.4 and ∼3.4 mm, respectively (Figure 8I). BTH treatment made both med16/sfr6 and wild-type plants more susceptible to B. cinerea infection, as indicated by the increased lesion sizes (see Supplemental Figure 3 online). In the test for A. brassicicola infection, 0% and ∼8% of the inoculated leaves from med16/sfr6 and wild-type plants, respectively, showed no necrosis, whereas ∼17% and 0% of the inoculated leaves from med16/sfr6 and wild-type plants, respectively, were dead or decayed (Figure 8H). The average lesion sizes on med16/sfr6 plants were ∼10.3 and ∼3.3 mm, respectively (Figure 8J). These results together demonstrate that MED16 is another Mediator subunit playing an important role in basal resistance against necrotrophic fungal pathogens.
MED16 Modulates NPR1 Protein Accumulation
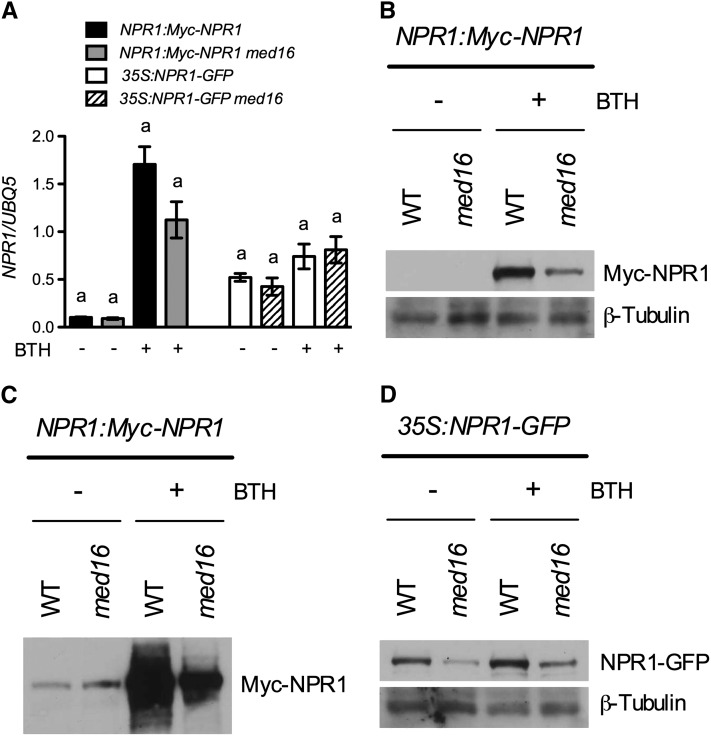
To explore the possible mechanism of action of MED16 in SAR, we first examined whether BTH treatment alters the subcellular localization of the MED16 protein. A green fluorescent protein (GFP)-MED16 fusion has been shown to be localized predominantly in the nucleus (Knight et al., 2009). BTH treatment did not change the subcellular localization of the GFP-MED16 fusion and a MED16-GFP fusion (see Supplemental Figure 4 online). Both MED16 and NPR1 positively contribute to SAR; therefore, MED16 might transcriptionally and/or posttranscriptionally regulate NPR1. To test this possibility, we generated transgenic npr1-3 plants expressing a Myc-NPR1 fusion driven by its endogenous promoter. The NPR1:Myc-NPR1 transgene complemented all of the npr1-3 mutant phenotypes, including reduced tolerance to SA toxicity, lack of inducible PR gene expression, and enhanced pathogen susceptibility (see Supplemental Figure 5 online). The transgene was crossed into the med16 npr1-3 double mutant. Pathogen infection does not induce NPR1 transcript accumulation in npr1-3 (Figure 6A); therefore, these plants allowed us to monitor the induction of the transgene. As shown in Figure 9A, whereas BTH treatment induced the expression of the transgene in both genetic backgrounds, the induction level was lower in NPR1:Myc-NPR1 med16 plants than in NPR1:Myc-NPR1 plants. Although we found that NPR1:Myc-NPR1 med16 plants accumulated less Myc-NPR1 protein than NPR1:Myc-NPR1 plants (Figures 9B and 9C), it is not clear whether the reduction in Myc-NPR1 protein levels was caused by the decreased transcript level. To unambiguously address whether MED16 posttranscriptionally regulates NPR1, we crossed the previously characterized 35S:NPR-GFP transgene into the med16 npr1-3 double mutant (Kinkema et al., 2000). The med16 mutation did not alter the expression of the transgene no matter whether the plants were treated with or without BTH (Figure 9A), which allowed us to determine whether med16 modulates NPR1-GFP nuclear localization and/or total protein accumulation. The mutation med16 did not block BTH-induced NPR1-GFP nuclear localization (see Supplemental Figure 6 online). However, total NPR1-GFP protein levels decreased dramatically in the med16 genetic background (Figure 9D), indicating that MED16 is required for NPR1 protein accumulation.
Figure 9.
NPR1 Protein Accumulation in med16 Plants.
(A) Expression levels of NPR1 in NPR1:Myc-NPR1, NPR1:Myc-NPR1 med16-1, 35S:NPR1-GFP, and 35S:NPR1-GFP med16-1 plants treated with or without BTH. Four-week-old soil-grown plants were treated with soil drenches plus foliar sprays of 0.3 mM of BTH solution or water. Leaf tissues were collected 24 h later and subjected to total RNA extraction and real-time qPCR analysis or protein analysis in (B) and (D). Data represent the mean of three independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test). The comparison was made separately between NPR1:Myc-NPR1 and NPR1:Myc-NPR1 med16-1 or between 35S:NPR1-GFP and 35S:NPR1-GFP med16-1 for each treatment.
(B) Myc-NPR1 protein levels in NPR1:Myc-NPR1 and NPR1:Myc-NPR1 med16-1 plants treated with or without BTH. Plants were treated as in (A). Total protein was analyzed by reducing SDS-PAGE and immunoblotting using an anti-Myc antibody. Detection of the constitutively expressed β-tubulin confirmed equal loading. WT, wild type.
(C) Myc-NPR1 protein levels in untreated NPR1:Myc-NPR1 and NPR1:Myc-NPR1 med16-1 plants. A longer exposure of the protein gel blot filter in (B) was used to detect the background protein levels.
(D) NPR1-GFP protein levels in 35:NPR1-GFP and 35:NPR1-GFP med16-1 plants treated with or without BTH. Total protein was analyzed by reducing SDS-PAGE and immunoblotting using an anti-GFP antibody. Detection of the constitutively expressed β-tubulin confirmed equal loading. All experiments were repeated three times with similar results.
DISCUSSION
The understanding that establishment of SAR involves profound transcriptional changes has been well documented (Maleck et al., 2000). Although the transcription coactivator NPR1 and its interacting TGA transcription factors have been shown to mediate these transcriptional changes (Cao et al., 1994; Zhang et al., 2003), it is unclear how SAR signals are transduced from the NPR1 signaling node to the RNAPII transcription machinery. In a genetic screen for Arabidopsis mutants insensitive to exogenous NAD+, which activates defense responses in plants, we identified MED16 as a key regulator functioning downstream of eNAD+. Results from characterization of the med16/sfr6 mutant plants suggest that MED16, a subunit in the tail module of the Arabidopsis Mediator complex, may be a signaling component in the gap between SAR-specific transcription activators and the general transcription machinery.
In animal cells, eNAD(P) is a well established signal molecule that activates intracellular signaling events, including immune responses (Billington et al., 2006). However, it is not clear whether plants and animals use similar mechanisms to process or perceive eNAD(P) and to transduce eNAD(P)-activated signals. The animal NAD(P)-metabolizing ectoenzyme CD38 converts eNAD(P) into the secondary messengers cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate, which in turn activates intracellular signaling (Ceni et al., 2003; Partida-Sánchez et al., 2003; Krebs et al., 2005). We recently showed that expression of the human CD38 compromises SAR in Arabidopsis (Zhang and Mou, 2012), suggesting that plants may use different mechanisms to perceive eNAD(P). Here we found that mutations in MED16 block exogenous NAD+-induced PR1 gene expression and compromise SAR (Figures 3B and 7K), indicating that MED16 is a downstream regulator that transduces eNAD+-activated signals to the RNAPII transcription machinery in Arabidopsis. MED16 and the Mediator complex are highly conserved in plants and animals (Bäckström et al., 2007); therefore, it may be possible that MED16 is also the Mediator subunit transferring eNAD(P)-activated signals to the transcriptional machinery in animal cells.
Biological induction of SAR was abolished completely in med16/sfr6 plants (Figure 7D), similar to that in npr1 and the tga6 tga2 tga5 triple knockout mutant (Cao et al., 1994; Zhang et al., 2003), suggesting that MED16, like NPR1 and the TGA factors (TGA2, TGA5, and TGA6), is an essential positive regulator of SAR. We found that expression of the SAR marker genes PR1, PR2, PR5, GST11, EDR11, and SAG21 was significantly decreased in med16/sfr6 (Figures 7C to 7H), indicating that MED16, like NPR1, is required for the transcriptional changes occurring in systemic leaves during the establishment of SAR (Maleck et al., 2000). Therefore, NPR1, the TGA factors, and MED16 may constitute a signaling pathway operating during biological induction of SAR.
In contrast with biological induction of SAR, induction of SAR by chemical inducers revealed differences among med16/sfr6, npr1, and tga6 tga2 tga5 plants. In npr1 plants, SA and its analog 2,6-dichloroisonicotinic acid did not induce PR gene expression and disease resistance (Cao et al., 1994). In tga6 tga2 tga5 plants, although one report showed that induction of PR1 by SA was only slightly delayed (Blanco et al., 2009), 2,6-dichloroisonicotinic acid did not induce resistance (Zhang et al., 2003). However, BTH induced low levels of PR gene expression and a considerable level of disease resistance in med16/sfr6 plants (Figures 5C to 5F) (Wathugala et al., 2012). These results suggest that MED16, NPR1, and the TGA factors may use different mechanisms in regulating SA-activated defense responses, and that BTH treatment activates both MED16-dependent and MED16-independent defense responses.
NPR1 and TGA factors are pathway-specific transcription activators, whereas MED16 is a subunit of Mediator, a protein complex in the general transcription machinery; therefore, MED16 likely functions downstream of NPR1 and the TGA factors to relay signals from the NPR1 signaling node to the general transcription machinery. Mediator is well known to transfer signals from pathway-specific transcription activators (Balamotis et al., 2009; Kagey et al., 2010; Takahashi et al., 2011); however, it is unclear whether Mediator also influences the homeostasis of the transcription activators. Here we show that NPR1 protein levels were decreased in med16 plants (Figures 9B and 9D), suggesting that MED16 may regulate SA responsiveness and basal immunity partially through modulating NPR1 protein accumulation (Figures 5C to 5F and 6A to 6E). Although the mechanism underlying the regulation is unclear, this result indicates that Mediator not only perceives signals from specific transcription activators but also actively regulates the homeostasis of the transcription activators. Whether NPR1 or TGA factors are physically associated with MED16 during biological induction of SAR needs to be addressed in future research.
NPR1 negatively regulates SA biosynthesis by suppressing the SA biosynthesis gene ICS1/SID2 (Nawrath and Métraux, 1999; Wildermuth et al., 2001). In npr1 plants, ICS1 transcripts and SA accumulate to much higher levels than in the wild type. We found that, on pathogen infection, SA levels in med16/sfr6 and wild-type plants were not significantly different (Figures 5A, 5B, 6F, and 6G), suggesting that MED16 may not participate in the NPR1-mediated feedback inhibition of SA biosynthesis. High levels of SA cause toxicity to plants; therefore, both npr1 and tga6 tga2 tga5 exhibited hypersensitivity to high concentrations of SA (Kinkema et al., 2000; Zhang et al., 2003). However, med16/sfr6 seedlings were as tolerant as the wild type to SA toxicity, suggesting that MED16 is not involved in NPR1- and TGA factor–mediated SA tolerance. These MED16-independent pathways mediated by NPR1 and the TGA factors also deserve future investigation.
Mutations in the MED16 gene suppressed JA/ET- and the necrotrophic pathogen B. cinerea–induced expression of several JA/ET-responsive genes (Figures 8A to 8F) (Wathugala et al., 2012), and med16/sfr6 mutants exhibited enhanced susceptibility to B. cinerea and A. brassicicola (Figures 8G to 8L), suggesting that MED16 functions downstream of JA/ET in defense signaling pathway. Recently, Arabidopsis MED8, MED21, and MED25 have been shown to regulate resistance to necrotrophic pathogens (Dhawan et al., 2009; Kidd et al., 2009). MED25 seems to be required for both background and JA-induced expression of JA-responsive genes, whereas MED8 seems to play a minor role in regulating PDF1.2 expression. The effect of the med8 mutation on JA-induced expression of PDF1.2 could only be readily detected in the med8 med25 double mutant (Kidd et al., 2009). Although MED21 RNA interference lines showed increased susceptibility to B. cinerea and A. brassicicola (Dhawan et al., 2009), its function in regulating defense gene expression has not been reported. These results together indicate that Mediator is a pivotal player in plant immunity, with four subunits having been shown to be required for resistance to necrotrophic fungal pathogens in Arabidopsis.
Our data show that MED16 is required for induction of both SA- and JA/ET-responsive genes, indicating that MED16 is a positive regulator of the SA and the JA/ET signaling pathway. Although Kidd et al. (2009) reported that the med25 mutation also decreased SA-induced defense gene expression, we did not detect any enhanced susceptibility to the biotrophic bacterial pathogen Pst DC3000 in med25 mutant plants (see Supplemental Figure 7A online). Consistent with this, biological induction of SAR was not significantly altered in med25 plants (see Supplemental Figure 7B online). The med8 mutant displayed enhanced susceptibility to Pst DC3000 (see Supplemental Figure 7A online) but did not show significant defects in biological induction of SAR (see Supplemental Figure 7B online). Therefore, the MED16 Mediator subunit is required for both biological induction of SAR and JA/ET-mediated resistance to necrotrophs.
It is generally accepted that SA and JA signaling pathways antagonize each other (Kunkel and Brooks, 2002; Pieterse et al., 2009). In recent years, a number of proteins regulating SA-JA/ET crosstalk have been identified in Arabidopsis (Petersen et al., 2000; Kachroo et al., 2001; Spoel et al., 2003; Li et al., 2004; Brodersen et al., 2006; Ndamukong et al., 2007). Whereas most of these regulators, including NPR1, inversely regulate SA and JA signaling pathways, the TGA factors seem to positively contribute to both SA and JA/ET signaling (Zander et al., 2010). The TGA factors are required for antagonizing the negative effect of MYC2/JIN1 (a negative regulator of PDF1.2) in the JA/ET pathway, and this positive function might be abolished in the presence of SA (Zander et al., 2010). Interestingly, MED16 suppresses several negative regulators of the SA signaling pathway (Figure 4). Whether this function of MED16 is involved in crosstalk between SA and JA/ET is currently unknown. MED16 positively regulates both SA and JA/ET signaling; therefore, it might function as a convergence point in Mediator conveying signals from both SA and JA/ET pathways.
The multiprotein Mediator complex functions as a bridge between specific transcription activators and the general transcription machinery. Individual Mediator subunits interact with pathway-specific activators to coordinate and transfer pathway-specific signals to the transcription machinery. Arabidopsis MED21 has been shown to interact with a RING E3 ligase histone monoubiquitination1 (HUB1), which positively regulates resistance to necrotrophs (Dhawan et al., 2009). Although several other Mediator subunits, including MED8, MED16, and MED25, have been implicated in SAR, basal resistance, and/or resistance to necrotrophs, their interacting partners in defense signaling pathways are unknown. It is also unknown whether the remaining subunits function in plant immunity. Future research focusing on these questions would shed new light on the molecular mechanisms by which Mediator regulates gene transcription in plant immune responses.
METHODS
Plant Materials and Growth Conditions
The wild-type Columbia ecotype of Arabidopsis thaliana was used. Transgenic plants and homozygous T-DNA insertion lines were generated or identified as described in the Supplemental Methods 1 online using gene-specific primers (see Supplemental Table 2 online). The med16/sfr6 npr1 double mutants were generated by crossing med16/sfr6 with npr1-3 and identified in the segregating F2 populations based on their morphology and confirmed by the med16-1 CAPS marker (see Supplemental Table 3 online) or by PCR using the primers flanking the T-DNA insertion in sfr6-2, and the npr1-3 mutation was confirmed by a derived CAPS marker (DeFraia et al., 2010). Arabidopsis seeds were sown on autoclaved soil (Metro-Mix 200; Grace-Sierra) and vernalized at 4°C for 3 d. Plants were germinated and grown at 23 to 25°C under a 16-h light/8-h dark regime.
Pathogen Infection
Inoculation of plants with Pseudomonas syringae pv tomato (Pst) DC3000/avrRpt2 and Pst DC3000 was performed by pressure-infiltration with a 1-mL needleless syringe as described previously (Clarke et al., 1998).
For Botrytis cinerea and Alternaria brassicicola inoculation, pathogens were grown on BD Difco Potato Dextrose Agar (Becton, Dickinson and Company) for ∼10 d at 25°C. Spores were harvested, resuspended in BD Difco Potato Dextrose Broth (Becton, Dickinson and Company) at a density of 1 to 5 × 105 spores/mL, and incubated for 2 h before inoculation. Then, 5-μL spore suspensions were dropped on the adaxial surface of rosette leaves, where the leaves were gently wounded with a needle. Symptoms were monitored for 3 to 4 d, and infection ratings from 0 to 3 were assigned to the inoculated leaves (0, no infection/necrosis; 1, leaves showing some necrosis; 2, leaves showing severe necrosis; 3, dead/decayed leaves). In each experiment, 16 to 20 plants per genotype were inoculated, and three to four leaves from each plant were scored after 4 d for symptom development.
SA Measurement
SA measurement was done by HPLC as described by Verberne et al. (2002).
Chemical Treatment
Exogenous NAD+ and BTH treatment were performed as previously described (Zhang and Mou, 2009). For ACC and MeJA treatment, 10-d-old seedlings grown on one-half–strength MS medium were transplanted onto one-half–strength MS medium supplemented with either 0.1 mM of ACC, 0.1 mM of MeJA, or both. Seedlings for the negative control were also transplanted onto one-half–strength MS medium. Plant tissues except roots were collected and subjected to total RNA extraction.
RNA and Protein Analysis
RNA extraction and RNA gel blot analysis were performed as described by Cao et al. (1997). Reverse transcription and real-time qPCR were performed as previously described using gene-specific primers (see Supplemental Table 4 online; DeFraia et al., 2010). The relative quantity of a gene is expressed in relation to ubiquitin5 (UBQ5) using the equation 2^(Ct[UBQ5] − Ct[GENE]), where 2 represents perfect PCR efficiency and Ct stands for cycle threshold. Total protein extraction and protein gel blot analysis were conducted as described by Mou et al. (2003).
Microarray Analysis
Three biological replicates with leaves from eight plants per sample were collected at 0, 4, 8, and 24 h after inoculation. RNA concentration was determined on a NanoDrop Spectrophotometer (Thermofisher Scientific), and sample quality was assessed using the 2100 Bioanalyzer (Agilent Technologies). Equal quantities of RNA from the three biological replicates were pooled. cDNA was synthesized from 200 ng of total RNA and used as a template for in vitro transcription in the presence of T7 RNA Polymerase and cyanine-labeled CTPs using the Quick Amp Labeling kit (Agilent Technologies) according to the manufacturer’s protocol. The amplified, labeled complementary RNA was purified using the RNeasy Mini kit (Qiagen). For each array, 1.65 μg of Cy 3–labeled complementary RNA was fragmented and hybridized with rotation at 65°C for 17 h. Samples were hybridized to Arabidopsis 4 × 44k arrays (Agilent Technologies). The arrays were washed according to the manufacturer’s protocol and then scanned on a G2505B scanner (Agilent Technologies). Data were extracted using Feature Extraction 10.1.1.1 software (Agilent Technologies). Microarray experiments were performed in the microarray core laboratory of the Interdisciplinary Center for Biotechnology Research at the University of Florida.
Before comparative analysis, the individual signal intensity values obtained from the eight microarray probes were log transformed (using 2 as the base) and normalized to ensure that meaningful biological comparisons can be made. More specifically, we first estimated the lower quartile, median, and upper quartile values by pooling all samples and then scaling and shifting the individual log-transformed signal intensities using the estimated quartile values as the references. As shown in Supplemental Figure 8 online, the normalized signal intensity data sets have similar median intensities and dynamic ranges. After normalization, a probe-by-probe comparison was performed between different time points of the same genotype using the 0-h sample as the reference and between med16-1 and the wild type at the same time point. In each comparison, fold change values were computed for each gene. The gene expression fold changes were computed based on the normalized log-transformed signal intensity data. The comparison results were further explored to obtain numbers of overlapped genes between med16-1 and the wild type.
Statistical Methods
All statistical analyses were performed with the data analysis tools (Student’s t test: Two Sample Assuming Unequal Variances) in Microsoft Excel (Microsoft Office 2004 for Macintosh).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MED16 (At4g04920); NPR1 (At1g64280); SNI1 (At4g18470); NIMIN1 (At1g02450); NIMIN2 (At3g25882); NIMIN3 (At1g09415); WRKY38 (At5g22570); WRKY58 (At3g01080); WRKY62 (At5g01900); DIR1 (At5g48485); AZI1 (At4g12470); PR1 (At2g14610); PR2 (At3g57260); PR5 (At1g75040); GST11 (At1g02920); EDR11 (At1g02930); SAG21 (At4g02380); PDF1.2 (At5g44420); CHIB (At3g12500); HEL (At3g04720).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphology of ien1, SALK_048091 (sfr6-2), and Their F1 Progeny.
Supplemental Figure 2. Genetic Complementation of the ien1 Mutant.
Supplemental Figure 3. Effect of BTH Treatment on Resistance to B. cinerea in med16/sfr6 Mutants.
Supplemental Figure 4. Effect of BTH Treatment on the Subcellular Localization of the MED16 Protein.
Supplemental Figure 5. Characterization of the NPR1:Myc-NPR1 Transgene.
Supplemental Figure 6. Subcellular Localization of NPR1-GFP in med16 Plants.
Supplemental Figure 7. Basal Resistance and SAR Induction in med8 and med25 Mutants.
Supplemental Figure 8. Normalization of the Microarray Data Sets Obtained from the Eight Microarray Probes.
Supplemental Table 1. Defense Genes Differentially Expressed between med16-1 and the Wild Type during Pst DC3000/avrRpt2 Infection.
Supplemental Table 2. Primers for Identification of Homozygous T-DNA Insertion Lines.
Supplemental Table 3. The CAPS Marker for the med16-1 Mutation.
Supplemental Table 4. Primers Used for qPCR in This Study.
Supplemental Methods 1. Supplemental Methods for the Supplemental Data.
Supplementary Material
Acknowledgments
We thank Jeffrey A. Rollins (University of Florida) for providing the fungal pathogens B. cinerea and A. brassicicola, Sixue Chen (University of Florida) for access to the HPLC equipment, Heather Knight (Durham University, United Kingdom) for 35:GFP-MED16 and 35S:MED16-GFP seeds, and the Interdisciplinary Center for Biotechnology Research at the University of Florida for microarray experiment and data analysis. We also thank Jeffrey A. Rollins and Chuanfu An for critical comments on the article. This study was supported by a grant from the National Science Foundation (IOS-0842716) awarded to Z.M.
AUTHOR CONTRIBUTIONS
X.Z, C.W., and Z.M. designed the research; X.Z. and C.W. performed research and analyzed data; Y.Z. performed the microarray experiment; Y.S. analyzed the microarray data; Z.M. wrote the article.
Glossary
- SAR
systemic acquired resistance
- SA
salicylic acid
- Pst
Pseudomonas syringae pv tomato
- JA
jasmonic acid
- ET
ethylene
- RNAPII
RNA polymerase II
- eNAD(P)
extracellular NAD(P)
- MS
Murashige and Skoog
- CAPS
cleaved amplified polymorphic sequence
- qPCR
quantitative PCR
- BTH
benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester
- MeJA
methyl jasmonate
- ACC
1-aminocyclopropane-1-carboxylic acid
- Psm
Pseudomonas syringae pv maculicola
References
- Akoulitchev S., Chuikov S., Reinberg D. (2000). TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407: 102–106 [DOI] [PubMed] [Google Scholar]
- Ansari S.A., He Q., Morse R.H. (2009). Mediator complex association with constitutively transcribed genes in yeast. Proc. Natl. Acad. Sci. USA 106: 16734–16739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D., Jonak C., Belcram K., Beemster G.T., Kronenberger J., Grandjean O., Inzé D., Traas J. (2002). Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Baek H.J., Malik S., Qin J., Roeder R.G. (2002). Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol. 22: 2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamotis M.A., Pennella M.A., Stevens J.L., Wasylyk B., Belmont A.S., Berk A.J. (2009). Complexity in transcription control at the activation domain-mediator interface. Sci. Signal. 2: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington R.A., Bruzzone S., De Flora A., Genazzani A.A., Koch-Nolte F., Ziegler M., Zocchi E. (2006). Emerging functions of extracellular pyridine nucleotides. Mol. Med. 12: 324–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F., Salinas P., Cecchini N.M., Jordana X., Van Hummelen P., Alvarez M.E., Holuigue L. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Boube M., Joulia L., Cribbs D.L., Bourbon H.M. (2002). Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151 [DOI] [PubMed] [Google Scholar]
- Bourbon H.M. (2008). Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H.M., et al. (2004). A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14: 553–557 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M.A., Shokat K.M., Rietz S., Parker J., Mundy J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Ceni C., Pochon N., Brun V., Muller-Steffner H., Andrieux A., Grunwald D., Schuber F., De Waard M., Lund F., Villaz M., Moutin M.J. (2003). CD38-dependent ADP-ribosyl cyclase activity in developing and adult mouse brain. Biochem. J. 370: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán P.D., Chory J. (2003). Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chadick J.Z., Asturias F.J. (2005). Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30: 264–271 [DOI] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Clarke J.D., Liu Y., Klessig D.F., Dong X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R.C., Conaway J.W. (2011a). Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 22: 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R.C., Conaway J.W. (2011b). Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Dietrich R.A., Richberg M.H. (1996). Death don't have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFraia C.T., Zhang X., Mou Z. (2010). Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 64: 511–523 [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Friedrich L., Ryals J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92: 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten Scheid O., Mengiste T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doares S.H., Narvaez-Vasquez J., Conconi A., Ryan C.A. (1995). Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 108: 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W.E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Friedrich L., et al. (1996). A benzothiadiazole induces systemic acquired resistance in tobacco. Plant J. 10: 61–70 [DOI] [PubMed] [Google Scholar]
- Gaffney T., Friedrich L., Vernooij B., Negrotto D., Nye G., Uknes S., Ward E., Kessmann H., Ryals J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H.M., Holstege F.C., Werner M. (2004). A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. (1998). Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Jones D.A., Takemoto D. (2004). Plant innate immunity - Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16: 48–62 [DOI] [PubMed] [Google Scholar]
- Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. (2009). Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kachroo P., Shanklin J., Shah J., Whittle E.J., Klessig D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257 [DOI] [PubMed] [Google Scholar]
- Kagey M.H., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Kwon Y.J., Kim J.M., Song Y.H., Kim S.N., Kim Y.J. (2004). MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc. Natl. Acad. Sci. USA 101: 12153–12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Björklund S., Li Y., Sayre M.H., Kornberg R.D. (1994). A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608 [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Mugford S.G., Ülker B., Gao D., Thorlby G., Knight M.R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Knuesel M.T., Meyer K.D., Bernecky C., Taatjes D.J. (2009). The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Krebs C., Adriouch S., Braasch F., Koestner W., Leiter E.H., Seman M., Lund F.E., Oppenheimer N., Haag F., Koch-Nolte F. (2005). CD38 controls ADP-ribosyltransferase-2-catalyzed ADP-ribosylation of T cell surface proteins. J. Immunol. 174: 3298–3305 [DOI] [PubMed] [Google Scholar]
- Kunkel B.N., Brooks D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Clarke J.D., Li Y., Dong X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339 [DOI] [PubMed] [Google Scholar]
- Maldonado A.M., Doerner P., Dixon R.A., Lamb C.J., Cameron R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K.A., Dangl J.L., Dietrich R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Mittler G., Kremmer E., Timmers H.T., Meisterernst M. (2001). Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Métraux J.-P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A.A., Thurow C., Fode B., Zander M., Weigel R., Gatz C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Norman-Setterblad C., Vidal S., Palva E.T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact. 13: 430–438 [DOI] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Partida-Sánchez S., Randall T.D., Lund F.E. (2003). Innate immunity is regulated by CD38, an ecto-enzyme with ADP-ribosyl cyclase activity. Microbes Infect. 5: 49–58 [DOI] [PubMed] [Google Scholar]
- Pena-Cortés H., Albrecht T., Prat S., Weiler E.W., Willmitzer L. (1993). Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 191: 123–128 [Google Scholar]
- Penninckx I.A.M.A., Thomma B.P.H.J., Buchala A., Métraux J.-P., Broekaert W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., Leon-Reyes A., Van der Ent S., Van Wees S.C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ryals J., Weymann K., Lawton K., Friedrich L., Ellis D., Steiner H.-Y., Johnson J., Delaney T.P., Jesse T., Vos P., Uknes S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I κ B. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J.A., Neuenschwander U.H., Willits M.G., Molina A., Steiner H.-Y., Hunt M.D. (1996). Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P.M., Kazan K., Wilson I., Anderson J.P., Richmond T., Somerville S.C., Manners J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Song J., Durrant W.E., Wang S., Yan S., Tan E.H., Dong X. (2011). DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 9: 115–124 [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y., Kornberg R.D. (2006). Mediator as a general transcription factor. J. Biol. Chem. 281: 80–89 [DOI] [PubMed] [Google Scholar]
- Takahashi H., et al. (2011). Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees S.C., de Swart E.A., van Pelt J.A., van Loon L.C., Pieterse C.M. (2000). Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 8711–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberne M.C., Brouwer N., Delbianco F., Linthorst H.J., Bol J.F., Verpoorte R. (2002). Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochem. Anal. 13: 45–50 [DOI] [PubMed] [Google Scholar]
- Wathugala D.L., Hemsley P.A., Moffat C.S., Cremelie P., Knight M.R., Knight H. (2012). The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195: 217–230 [DOI] [PubMed] [Google Scholar]
- Weigel R.R., Bäuscher C., Pfitzner A.J.P., Pfitzner U.M. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46: 143–160 [DOI] [PubMed] [Google Scholar]
- Weigel R.R., Pfitzner U.M., Gatz C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Woychik N.A., Hampsey M. (2002). The RNA polymerase II machinery: Structure illuminates function. Cell 108: 453–463 [DOI] [PubMed] [Google Scholar]
- Zander M., La Camera S., Lamotte O., Métraux J.P., Gatz C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61: 200–210 [DOI] [PubMed] [Google Scholar]
- Zhang X., Mou Z. (2009). Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J. 57: 302–312 [DOI] [PubMed] [Google Scholar]
- Zhang X., Mou Z. (2012). Expression of the human NAD(P)-metabolizing ectoenzyme CD38 compromises systemic acquired resistance in Arabidopsis. Mol. Plant Microbe Interact. 25: 1209–1218 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M.J., Lassner M., Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-M., Trifa Y., Silva H., Pontier D., Lam E., Shah J., Klessig D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Zhu X., Wirén M., Sinha I., Rasmussen N.N., Linder T., Holmberg S., Ekwall K., Gustafsson C.M. (2006). Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell 22: 169–178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.