Abstract
Monogenic neurodevelopmental disorders provide key insights into the pathogenesis of disease and help us understand how specific genes control the development of the human brain. Timothy syndrome is caused by a missense mutation in the L-type calcium channel Cav1.2 that is associated with developmental delay and autism 1. We generated cortical neuronal precursor cells and neurons from induced pluripotent stem cells derived from individuals with Timothy syndrome. Cells from these individuals have defects in calcium (Ca2+) signaling and activity-dependent gene expression. They also show abnormalities in differentiation, including decreased expression of genes that are expressed in lower cortical layers and in callosal projection neurons. In addition, neurons derived from individuals with Timothy syndrome show abnormal expression of tyrosine hydroxylase and increased production of norepinephrine and dopamine. This phenotype can be reversed by treatment with roscovitine, a cyclin-dependent kinase inhibitor and atypical L-type–channel blocker 2, 3, 4. These findings provide strong evidence that Cav1.2 regulates the differentiation of cortical neurons in humans and offer new insights into the causes of autism in individuals with Timothy syndrome.
Human genetic studies have implicated voltage-gated calcium channels, in particular the L-type channel CaV1.2, in the development of psychiatric diseases such as autism5, bipolar disorder6 and schizophrenia7. Although calcium influx through these channels is important for a variety of neuronal processes including regulation of gene expression8, the cellular defects caused by mutations in these channels and how these defects lead to psychiatric symptoms is unknown. TS is caused by a point mutation in an alternatively spliced exon of CACNA1C, the gene that encodes the α1 subunit of Cav1.21. This mutation leads to decreased calcium- and voltage-dependent inactivation of the channel1,9. TS patients suffer from cardiac arrhythmia, hypoglycemia and global developmental delay. Over 60% of TS patients also fulfill the criteria for an autism spectrum disorder (ASD)1 making TS one of the most penetrant monogenic forms of autism.
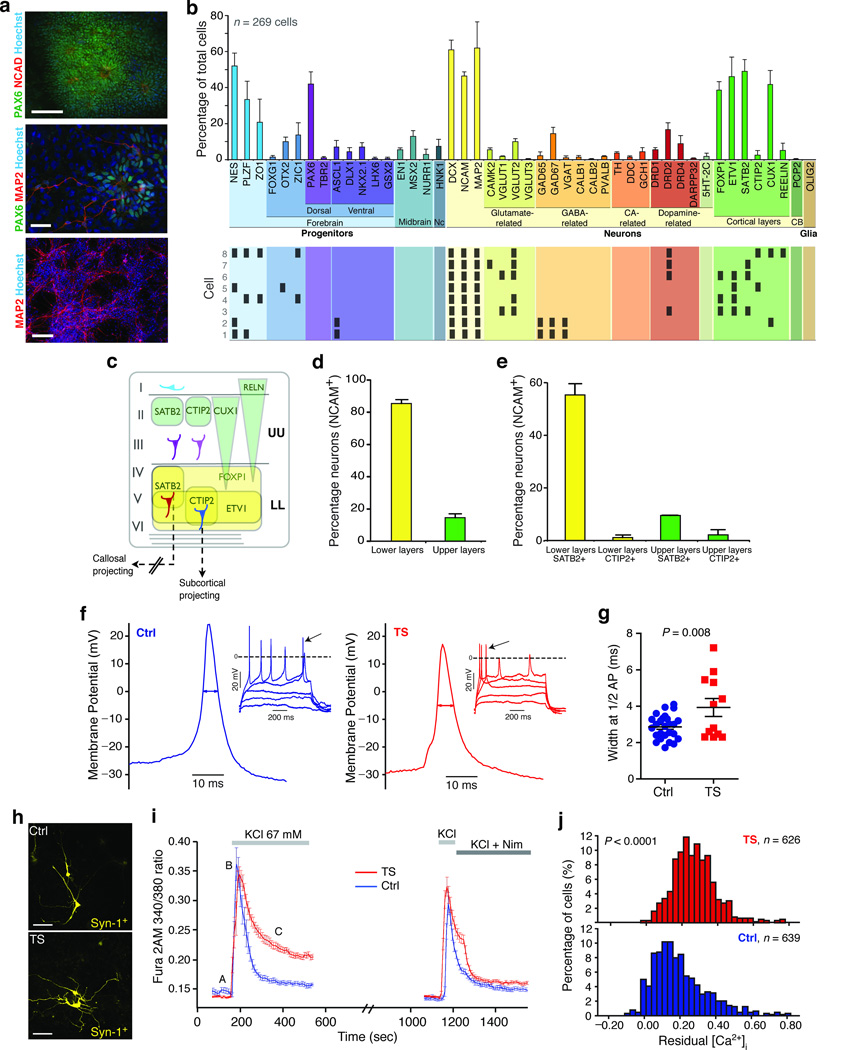
To determine the cellular consequences of the TS mutation, we used somatic cell reprogramming10,11 to generate iPSCs from individuals with TS (see Supplementary Figs. 1, 2, Methods and reference12 for characterization, and Supplementary Table 1 for a list of lines). We differentiated the iPSC lines into NPCs and neurons using conditions that favor the generation of cortical neurons13–15 (Fig. 1, Supplementary Fig. 3, and Methods for details on differentiation). To identify the types of cells in these cultures, we used Fluidigm Dynamic Arrays16 to measure the expression of region and cell-type specific marker genes in single cells (Fig. 1b; see Supplementary Table 2). The reliability and accuracy of this method for measuring single cell gene expression was verified in a variety of ways (see17, Supplementary Figs. 4, 5 and Methods). Overall, 64.9% of the cells at day 45 of differentiation in vitro expressed a neuronal marker (MAP2 or NCAM), and 76% of these cells were positive for the early neuronal marker Doublecortin (DCX)18. Individual neurons expressed combinations of genes that could be used to determine their neurotransmitter identity and cortical layer specificity (Fig. 1b lower). A substantial number of neurons expressed excitatory markers like VGLUT1 and VGLUT2 along with the dopamine receptor DRD2, whereas others expressed the inhibitory markers GAD65, GAD67 and VGAT.
Figure 1. Characterization of iPSC-derived NPCs and neurons.
(a) Immunostaining of neural rosettes (upper: PAX6, N-Cadherin, Hoescht; scale bar is 100 µm), neurons (MAP2) and progenitors (PAX6) after plating of the neurospheres (middle, scale bar 100 µm) and cultures at day 43 of differentiation (lower: MAP2, Hoescht; scale bar is 200 µm). The middle image is from a Timothy syndrome–derived culture, and the other two are from cultures derived from control individuals. (b) Single cell gene expression analysis (Fluidigm) of the population of cells at 45 days of differentiation. Shown are the proportion of cells that express a cell-specific marker (mean ± s.e.m.; n = 269 cells from three control iPSC lines). The lower panel shows gene expression profiles of single neurons. (c) Scheme illustrating marker gene expression in upper and lower layers of the human cortex. (d) Fraction of neurons (NCAM+) expressing lower (FOXP1+/ETV1+) or upper layer (FOXP1–/ETV1–) cortical markers as assessed by Fluidigm (mean ± s.e.m; n = 116 cells from three controls iPSC lines). (e) Fraction of subpopulations of upper and lower layer neurons (NCAM+) expressing CTIP2 or SATB2. (f) Representative current clamp recordings (holding potential −65 mV; 1s current pulses, ΔIinj = 5–10 pA) from TS and control derived neurons. (g) APs recorded from TS neurons were significantly wider. (h) Representative images of control and TS neurons expressing YFP under the control of the Synapsin-1 promoter (scale bars are 50 µm). (i) Average [Ca2+]i measurements in Synapsin-1 expressing neurons depolarized twice with 67 mM KCl and treated with 5 µM nimodipine (TS; n = 10 neurons; Ctrl; n = 9 neurons). (j) Histogram of residual [Ca2+]i ([C–A]/[B–A]) in neurons (three TS lines, three control lines, t-test, P < 0.0001; see also Supplementary Fig. 8c).
The expression of FOXP1, ETV1, SATB2, CTIP2, CUX1 and RELN was used to define cortical layer identity, based on immunohistochemical analysis of human brains19,20 (Fig. 1c). Ninety-one percent of the NCAM+ neurons expressed at least one cortical layer marker. The expression of ETV1 and FOXP1 was used to indicate a lower cortical layer identity, while the expression of CUX1, SATB2, CTIP2 and RELN in the absence of ETV1 and FOXP1 was used to define upper layer neurons. Approximately 85% of cortical neurons could be classified as lower layer neurons while the remaining 15% expressed markers of upper cortical layers (Fig. 1d). Among the lower layer neurons we could identify two distinct subpopulations (Fig. 1e): cells expressing CTIP2, which defines a population of subcortical projection neurons, and cells expressing SATB2, which defines neurons that project to distant cortical regions via the corpus callosum21–23.
A notable finding from this analysis was the remarkable reproducibility of the neuronal differentiation protocol across multiple iPSC lines and individuals (average standard deviation for the proportion of cells expressing a particular marker was 4.47%), indicating that single cell analysis of gene expression is reproducible and can be used to identify defects in neuronal differentiation.
The ability to generate well-defined populations of NPCs and neurons from iPSCs prompted us to ask whether we could identify cellular phenotypes associated with TS. We examined the proliferation (Supplementary Fig. 6a) and migration of NPCs (Supplementary Fig. 6b,c) and the total number of neurons generated (Supplementary Fig. 5), but found no differences between controls and TS cultures. We next used patch clamp recording and calcium imaging to assess the physiological properties of the neurons generated from iPSC lines (Fig. 1f). We found that 57% of the iPSC-derived cells fired mature action potentials (APs) (Supplementary Fig. 7). Comparison of TS and control neurons did not reveal any significant differences in AP threshold or amplitude, resting membrane potential, input resistance or capacitance (Supplementary Table 3). However, the APs of TS neurons were approximately 37% wider at the midpoint than those of controls (TS: 3.92 ± 0.49 versus Ctrl: 2.86 ± 0.12 ms, P = 0.008), which is consistent with a loss of channel inactivation (Fig. 1f,g) and is similar to the defect we observed in TS-derived cardiomyocytes12.
We next examined intracellular calcium ([Ca2+]i) signals in TS and control NPCs and neurons using Fura-2 and time-lapse video microscopy. Because the cultures contain a mixed population of neurons and NPCs, we first measured [Ca2+]i in mature neurons expressing a YFP reporter gene under the control of the Synapsin-1 promoter (Fig. 1h). In TS cells we observed a significant increase in the sustained [Ca2+]i rise following depolarization that was abolished by treatment with nimodipine (Fig. 1i). This increased [Ca2+]i rise in TS neurons was observed in neurons derived from multiple lines and multiple independent differentiations (Supplementary Fig. 8a,b, Fig. 1j). Similarly, increased [Ca2+]i elevations were observed in TS-derived NPCs (Supplementary Fig. 8c,d). Taken together, these results provide strong evidence that NPCs and neurons derived from TS individuals have defects in AP firing and [Ca2+]I signaling.
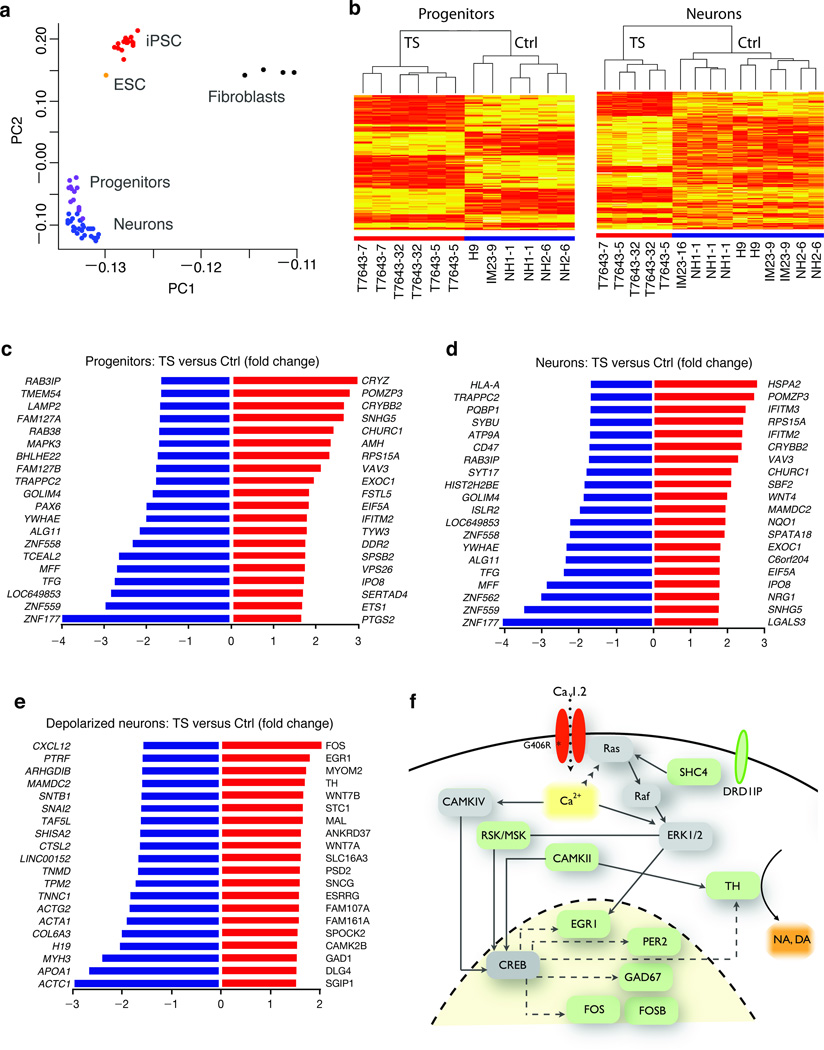
CaV1.2 plays an important role in regulating activity-dependent gene expression in the nervous system8. We therefore used Illumina microarrays to compare the gene expression profile of TS and control NPCs and neurons (Fig. 2a–d). Hierarchical clustering based on differentially expressed genes showed that TS-derived cells clustered separately from controls. The expression levels of 211 genes in neurons (126 upregulated, 85 downregulated) and 136 genes in NPCs (58 upregulated, 78 downregulated) were significantly altered in TS cells (Fig. 2c,d and Supplementary Table 4). Of the genes that were altered in TS neurons, 11 have been previously implicated in either ASD or intellectual disability (ID)24,25 (Supplementary Table 5).
Figure 2. Characterization of NPCs and neurons derived from TS individuals and controls by genome-wide microarrays.
(a) Principal component analysis of whole-genome gene expression profiles for fibroblasts, iPSC, embryonic stem cells (ESC), NPCs and neurons showing clustering of cell types based on the first two principal components (PC1 and PC2). (b) Heatmaps depicting expression levels of genes differentially expressed between TS and control NPCs and neurons. Each column represents an independent differentiation of an iPSC line. Genes that are highly expressed in TS cells relative to controls are shown in red. Dendrograms show hierarchical clustering of samples based on differentially expressed genes. (c) and (d) List of top 20 genes showing the highest expression differences between TS and control cells (c, progenitors; d, neurons). (e) Differentially expressed genes in TS neurons relative to control neurons after electrical stimulation. (f) Scheme illustrating interactions between a subset of calcium regulated genes upregulated in TS (shown in green).
We also identified 223 genes (135 upregulated, 88 downregulated) that were altered in TS relative to control neurons upon depolarization. A number of the genes that were altered in TS cells are linked to Ca2+-dependent regulation of the transcription factor CREB (Fig. 2e,f) including RSK/MSK and CAMKII. Others such as EGR1, FOS, FOSB, GAD67 and TH are downstream targets of CREB. In addition to TH, which is the rate-limiting enzyme in the production of dopamine and norepinephrine, DRD1IP (Calcyon), another gene involved in dopamine signaling, was also upregulated in TS neurons. These results suggest that the TS mutation leads to misregulation of Ca2+-dependent gene expression and perturbs catecholamine signaling.
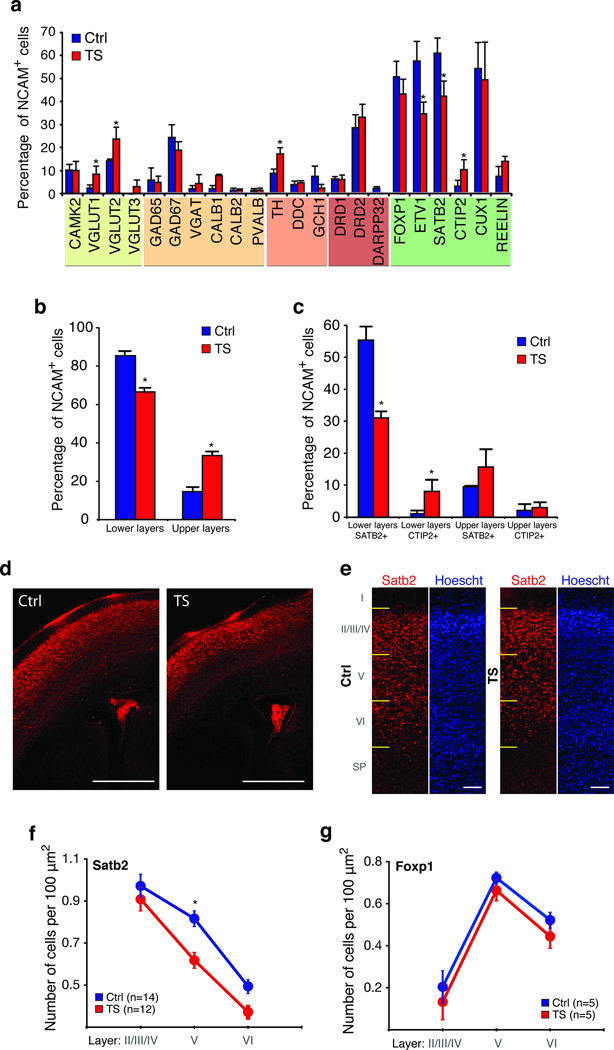
To determine whether the TS mutation leads to defects in neuronal differentiation, we used Fluidigm Arrays to study the identity of cells generated from individuals with TS versus controls (Fig. 3a). We found a significant decrease in the fraction of neurons expressing lower layer markers in TS relative to controls (TS: 66.7% versus Ctrl: 85.5%, P < 0.001; Fig. 3b), and an increase in the fraction of neurons expressing upper layer markers (TS: 33.3% versus Ctrl: 14.5%, P = 0.002; Fig. 3b). TS cells expressing lower layer markers cells (Fig. 3c) contained a significantly lower proportion of SATB2 expressing cells (TS: 31.0% versus Ctrl: 55.3%, P < 0.001) and an increase in CTIP2 expressing cells (TS: 8.0% versus Ctrl: 1.0%, P = 0.01). Because SATB2 is both necessary and sufficient for the formation of callosal projection neurons21,22, this finding is consistent with the notion that the TS mutation decreases the fraction of callosal projection neurons and increases the number of cells that project to subcortical structures.
Figure 3. Characterization of neuronal subpopulations differentiated from TS and control iPSCs.
(a) Single cell qPCR analysis (Fluidigm) showing the proportion (mean ± s.e.m., *P < 0.05, Chi-square test) of NCAM+ neurons expressing subtype-specific markers, for control (n = 125 cells, three lines) and TS cultures (n = 140 cells, three lines). (b) Fraction of neurons expressing upper and lower cortical layer makers in TS (n = 125 cells) and control cortical cultures (n = 116 cells; mean ± s.e.m. *P < 0.05, Chi-square test). (c) Fraction of neurons expressing SATB2 and CTIP2 and either lower or upper layer cortical markers in TS and control cultures (mean ± s.e.m. *P < 0.05, Chi-square test). (d) 50× image of Satb2 stained cortical sections from a control mouse and a mouse expressing the TS channel in the forebrain showing a decrease in Satb2+ cells. Scale bar is 500 µm. (e) Image of Satb2 stained cortical sections from control and TS mouse at P0.5 (scale bar is 50 µm). (f) Measurement of the number of Satb2+ cortical neurons in cortical sections from TS and control mice (mean ± s.e.m.; TS: n = 12 animals, Ctrl: n = 14 animals; two-way ANOVA, P = 0.001; asterisk denotes P < 0.05 for posthoc analysis) (g) Distribution of Foxp1+ neurons in cortical sections from TS and control mice (mean ± s.e.m.; n = 5 animals per group; two-way ANOVA, P > 0.05).
To validate these findings in vivo we also measured Satb2 expression in a transgenic mouse expressing the TS channel driven by the Foxg1 promoter in the forebrain (Supplementary Fig. 9). TS transgenic mice had a reduced number of Satb2 expressing cells that was most pronounced in lower layers (Fig. 3d–f), whereas Foxp1 expression (Fig. 3g) and the total number of NeuN expressing neurons in the cortex were unchanged. We therefore conclude that the TS mutation alters SATB2 expression both in vitro and in vivo. Interestingly, we did not observe an increase in the number of Ctip2 expressing cells in the TS channel-expressing mice. This could be either because of differences between in vivo and in vitro neuronal differentiation or because of species-specific differences in Ctip2 regulation. CTIP2 is in fact expressed more broadly in humans, labeling cells in the subventricular zone as well as in cortical layers II and V26.
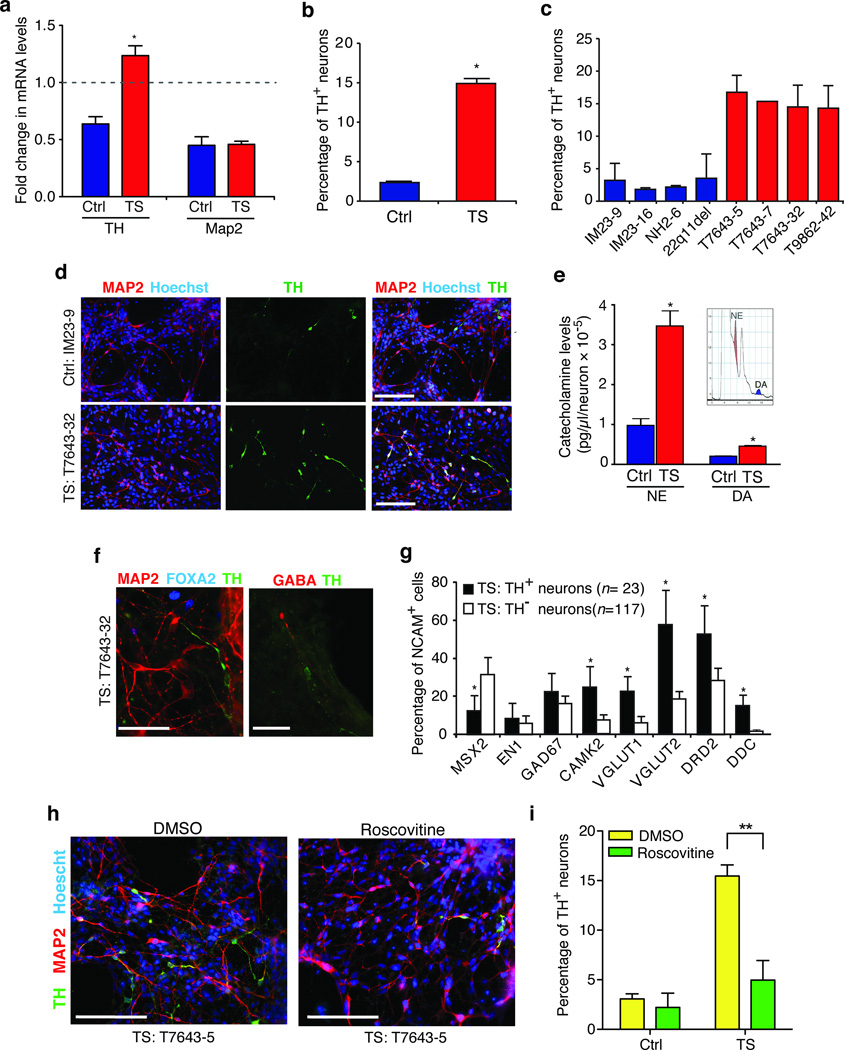
We also observed a significant increase in the fraction of cells that expressed TH in TS-derived neuronal cultures (TS: 16.4% versus Ctrl: 8.0%, P = 0.03; Fig. 3a). This agrees with our earlier finding that genes involved in catecholamine synthesis are misregulated in TS. To determine whether the TS mutation alters the regulation of TH, we measured TH mRNA in TS and control neurons following electrical activation (Fig. 4a). After nine hours of stimulation, TH was downregulated in control neurons but upregulated in TS neurons, indicating that the TS mutation prevents down-regulation of TH in response to prolonged electrical activity. To provide evidence that this increase in TH mRNA has functional consequences we measured expression of TH protein using a TH specific antibody. We found that TS neuronal cultures contained 6.3 times more TH+ neurons (TH+/MAP2+ cells) than control cultures (Fig. 4b–d; TS: 15.03% ± 0.92%, n = 9 differentiations; Ctrl: 2.52% ± 0.62%, n = 7 differentiations; t-test, P < 0.001 or than cultures generated from an individual with 22q11.2 deletion syndrome, another neurodevelopmental disorder (Fig. 4c). To determine whether this change in TH protein caused an increase in the production of catecholamines, we used high-pressure liquid chromatography (HPLC) to measure the level of norepinephrine and dopamine in the media collected from neuronal cultures. We found that the TS neurons secreted 3.5 times more norepinephrine (TS: 34.7 ± 4.02 × 10−5 versus Ctrl: 9.79 ± 1.83 × 10−5 pg µl−1 neuron−1, t-test, P = 0.004) and 2.3 times more dopamine (TS: 4.56 ± 0.36 × 10−5 versus Ctrl: 2.05 ± 0.24 × 10−5 pg µl−1 neuron−1, t-test, P = 0.001) than control lines, strongly suggesting that the TS mutation leads to increased TH expression and to an excess secretion of catecholamines (Fig. 4e).
Figure 4. Abnormal expression of TH in neurons from TS individuals.
(a) Fold changes in TH and MAP2 mRNA levels in TS and control neurons after nine hours of depolarization with 67 mM KCl. (b) TS neuronal cultures contain an excess number of TH+ neurons (*P < 0.001, t-test). (c) The proportion of TH+ neurons (TH+/MAP2+) is shown for three control lines from two healthy individuals (IM23-9, IM23-16, NH2-6), one line derived from a individual with 22q11.2 deletion syndrome, and four TS lines from two individuals (T7643-5, T7643-7, T7643-32, T9862-42). (d) Representative images of neurons stained with TH and MAP2 specific antibodies and Hoechst 33258 (scale bars are 200 µm). (e) Norepinephrine (NE, *P = 0.004) and dopamine (DA, *P = 0.001) levels are elevated in media collected from TS neuronal cultures relative to controls. Inset shows an HPLC chromatogram with the peaks for NE and DA. (f) TH+ neurons (green) in TS cultures (T7643-32 line is shown) are not immunostained by antibodies to FOXA2 (left, blue) or GABA (right, red). Scale bar is 50 µm. (g) Fraction of TH+ or TH− neurons from TS individuals that co-express other neuronal marker genes (mean ± s.e.m.,* P < 0.05, Chi-square test). (h) Cultures of TS neurons treated with roscovitine (15 µM at day 39 and 10 µM at day 41) or DMSO and stained with TH and MAP2 specific antibodies at day 43 of differentiation. Scale bar is 200 µm. (i) Proportion of TH+ neurons (TH+/MAP2+) in control and TS cultures after treatment with roscovitine or DMSO (mean ± s.e.m., two-way ANOVA, **P < 0.001).
We next investigated the cellular identity of neurons that produce TH in TS and control cultures. In both control and TS cultures, TH expressing neurons were not stained with antibodies to FOXA2 or EN1, markers of midbrain neurons, or GABA, a marker for dopaminergic olfactory neurons in the forebrain (Fig. 4f). Using Fluidigm chip analysis we found that TH expression was not confined to any specific class of neurons, although these cells were more likely to co-express excitatory markers and dopamine-related genes (Fig. 4g). This suggests that the TS channel does not promote a catecholaminergic cell fate, but instead increases the expression of TH in a variety of cortical cell types. Interestingly, we did not observe an increase in TH staining in the cortex of transgenic mice expressing the TS channel. This likely reflects the low homology between the promoter regions of the mouse and human TH genes and differences in gene regulation27,28.
Finally, to determine if the increase in TH expression in TS neurons was reversible and a result of L-type channel activity, we treated 39 day-old neurons from TS individuals with L-type channel blockers. The conventional L-type calcium channel blocker nimodipine failed to reverse the excess expression of TH in these neurons. Earlier studies in cardiomyocytes12 from TS individuals indicated that roscovitine2,3,29, a cyclin-dependent kinase blocker that also increases L-type channel inactivation, can reduce the prolongation of the cardiac AP in cells from TS individuals. Treatment with roscovitine caused a 68% reduction in the proportion of TH+ neurons (Fig. 4h,i; two-way ANOVA, P < 0.01, n = 3,975 neurons from three TS lines and n = 2,679 from three control lines) without affecting the fraction of MAP2 expressing cells. This result suggests that the increase in TH expression probably results from lack of inactivation of the TS channels and, further, that restoring channel inactivation in mature neurons can decrease the abnormal expression of TH in patient cells.
In this study we show that neurons from patient-derived iPSCs can be used to identify cellular phenotypes associated with a neurodevelopmental disorder. These findings provide insight both into the function of Cav1.2 in the developing human brain and its role in the pathogenesis of psychiatric diseases. We report an increase in the amplitude of Ca2+ elevations in TS derived NPCs and neurons indicating that loss of inactivation in a single splice variant of Cav1.2 can have profound effects on neuronal signaling. A consequence of this defect was a change in activity-dependent gene expression and an increase in cells producing norepinephrine and dopamine, consistent with previous experiments showing that dopaminergic specification is activity dependent39. The TS mutation also caused a decrease in neurons expressing SATB2, a marker for callosal projection neurons. This finding is unexpected because Cav1.2 has not been previously linked to the specification of these cells. The increase in SATB2 expressing cells was recapitulated in the cortex of a mouse expressing the TS channel. However, this mouse did not show an increase in TH expression probably reflecting species-specific differences in gene regulation and the structure of the TH promoter27,28.
A key question is whether these cellular defects help to explain developmental delay and ASD in individuals with TS. The reduction in cortical projecting neurons in TS is consistent with the emerging view that ASDs arise from defects in connectivity between cortical areas30,31, and agrees with studies that show a decreased size of the corpus callosum in ASD32. Ectopic production of TH and a subsequent increase in catecholamine synthesis agrees with findings from valproic-acid based models of ASD33 and postmortem studies of patients with schizophrenia34. As catecholamines play an important role in sensory gating and in social behavior, an increase in their synthesis might be important in the pathophysiology of ASDs.
ONLINE MATERIALS AND METHODS
iPSC maintenance
Human iPSCs and the embryonic H9 line were cultured on irradiated DR4 mouse embryonic fibroblast feeders.
Neural differentiation protocol
Neural differentiation was carried out using a modified version of a previously described protocol13. Briefly, the iPSCs were suspended to generate embryoid bodies (EBs) and plated to produce neural rosettes (Fig. 1a upper). The rosettes were mechanically isolated and expanded as neurospheres, and then either purified by fluorescence activated cell sorting (FACS) using the forebrain progenitor marker FORSE-135,36 or plated for differentiation into neurons. Seven days after plating of the neurospheres, MAP2ab positive cells were observed migrating away from radial clusters of undifferentiated precursors expressing the dorsal forebrain marker PAX6 (Fig. 1a middle). After 43 of differentiation in vitro days significant numbers of cells expressing the cortical marker DCX (Supplementary Fig. 3) and the mature form of MAP2 were observed in the cultures (Fig. 1a lower). Five independent differentiations were performed with four control lines, four TS lines, one 22q11 deletion syndrome line, and one human ESC line (H9). Each line was differentiated at least twice. In addition, two more control lines and one TS line were used for the rescue experiment (see also Supplementary Table 1). R-roscovitine (Sigma-Aldrich, R7772) was dissolved in DMSO. Neuronal cultures were treated with roscovitine or with the same volume of DMSO at day 39 and day 41. Analyses of neuronal cultures were performed at day 43 of differentiation.
Calcium Imaging
NPCs (at passage two after plating of the neurospheres) or neuronal cultures at day 43 of differentiation, were loaded with 1 µM Fura-2 acetoxymethyl ester (Invitrogen) for 30 min at 37°C in Neurobasal/B27 media, washed with Tyrode's solution, and placed in a perfusion chamber on the stage of an inverted fluorescence microscope (TE2000U; Nikon). Cells were stimulated with high KCl Tyrode's solution (67 mM NaCl, 67 mM KCl for neurons and 100 mM for NPCs, 2 mM CaCl2, 1 mM MgCl2, 30 mM glucose, and 25 mM Hepes, pH 7.4) without or with nimodipine (final concentration 5 µM). Imaging was performed at room temperature on an epifluorescence microscope equipped with an excitation filter wheel and an automated stage. Openlab software (PerkinElmer) software was used to collect and quantify time-lapse excitation ratio images. Fluorescence images were analyzed using IGOR Pro software (WaveMetrics) software.
Single cell qPCR
Neuronal cultures at day 45 of differentiation were rinsed with HBSS and incubated with Accutase (StemCell Technologies). After one wash with fresh Neurobasal medium (Invitrogen), cells were resuspended in Neurobasal/B27 medium containing 1µg ml−1 propidium-iodide (Molecular Probes) and filtered through a 40 µm nylon cell strainer (BD Biosciences). Clone sorting in 96-well qPCR plates (Eppendorf) was performed at the Stanford Shared FACS Facility on a BD Influx cell sorter. Cells were sorted into 10 µl pre-amplification mix containing 40 nM of all primers for the 96 genes of interest, and the following components of the CellsDirect One-Step qRT-PCR Kit (Invitrogen): 2× Reaction Mix, SuperScript III RT/Platinum Taq Mix. After sorting, samples were reverse transcribed and pre-amplified for 18 cycles. Pre-amplified samples were diluted (2×) with TE buffer and stored at −20°C. Sample and assay (primer pairs) preparation for 96.96 Fluidigm Dynamic arrays was done according to the manufacturer’s recommendation. Briefly, the sample was mixed with 20× DNA Binding Dye Sample Loading Reagent (Fluidigm Corp.), 20× EvaGreen (Biotium) and TaqMan® Gene Expression Master Mix (Applied Biosystems). Assays were mixed with 2× Assay loading reagent (Fluidigm Corp.) and TE to a final concentration of 5 µM. The 96.96 Fluidigm Dynamic Arrays (Fluidigm Corp.) were primed and loaded on an IFC Controller HX (Fluidigm Corp.) and qPCR experiments were run on a Biomark System for Genetic Anaylsis (Fluidigm Corp.). See Supplementary Methods for details on analysis.
Microarrays
RNA from fibroblasts, iPSCs, NPCs, neurons at rest or neurons stimulated with KCl (day 43 of differentiation), was isolated using the RNeasy Mini kit and the RNase-Free DNase set (Qiagen). Total RNA (200 ng) was amplified, labeled, and hybridized on Illumina HumanRef-8 v3 Expression BeadChips (Illumina Inc, San Diego, CA) according to the manufacturer’s protocol. Data analysis was performed using the Lumi R37 and Bioconductor (www.bioconductor.org) packages, as we have previously shown38. Briefly, absolute expression values were log2 transformed and normalized using quantile normalization. Data quality control measures included inter-array Pearson correlation, clustering based on most variable genes (coefficient of variance > 0.05), and detection of outlier arrays, as well as probe detection (P values < 0.05). Differential expression analysis was performed using Significance analysis of Microarrays (http://www-stat.stanford.edu/~tibs/SAM/). The statistical criteria for differential expression were FDR < 0.05 and fold change > 1.3. Microarray data is available at GEO (accession code GSE25542).
Supplementary Material
Acknowledgements
We thank K. Timothy and the individuals with Timothy syndrome who participated in this study; E. Nigh for editing of the manuscript; U. Francke for karyotyping; A. Cherry and D. Bangs for help with fibroblast cultures; G. Panagiotakos and C. Young-Park for insightful discussions, and A. Krawisz, R. Schwemberger, D. Fu and R. Shu for help with data analysis. Antibodies to FORSE-1 were developed by P.H. Patterson and were obtained from the Developmental Studies Hybridoma Bank (University of Iowa). Financial support was provided by a US National Institutes of Health Director’s Pioneer Award, and by grants to R.E.D. from the US National Institute of Mental Health, the California Institute for Regenerative Medicine and the Simons Foundation for Autism Research. S.P.P. was supported by awards from the International Brain Research Organization Outstanding Research Fellowship and the Tashia and John Morgridge Endowed Fellowship, M.Y. by a Japan Society of the Promotion for Science Postdoctoral Fellowship for Research Abroad and an American Heart Association Western States postdoctoral fellowship, T.P. by a Swiss National Science Foundation Postdoctoral Fellowship and A.S. by a California Institute for Regenerative Medicine Postdoctoral Fellowship. We are also grateful for funding from B. and F. Horowitz, M. McCafferey, B. and J. Packard, P. Kwan and K. Wang and the Flora foundation.
Footnotes
Author Contributions
R.E.D. and S.P.P. designed the experiments and wrote the manuscript. S.P.P. generated iPSC lines, differentiated the iPSC lines into neurons, performed the calcium imaging and immunocytochemistry studies and contributed to the mutant mouse characterization. T.P. designed and analyzed the Fluidigm microarray studies. M.Y. generated and characterized the iPSC lines, and generated and characterized the mutant mice. I.V. and D.H.G. performed and analyzed the microarray gene expression experiments. A.S. derived neurons and designed and performed the electrophysiological experiments. A.M.P. performed the karyotyping and immunocytochemistry. S.C. and N.S. performed and analyzed catecholamine concentrations by HPLC. B.C. and T.D.P. contributed to the Fluidigm studies. J.A.B. and J.H. recruited and characterized the subjects.
REFERENCES
- 1.Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Yarotskyy V, et al. Roscovitine binds to novel L-channel (CaV1.2) sites that separately affect activation and inactivation. J Biol Chem. 2010;285:43–53. doi: 10.1074/jbc.M109.076448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yarotskyy V, Elmslie KS. Roscovitine, a cyclin-dependent kinase inhibitor, affects several gating mechanisms to inhibit cardiac L-type (Ca(V)1.2) calcium channels. Br J Pharmacol. 2007;152:386–395. doi: 10.1038/sj.bjp.0707414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. Phospholemman modulates the gating of cardiac L-type calcium channels. Biophys J. 2010;98:1149–1159. doi: 10.1016/j.bpj.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moskvina V, et al. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry. 2009;14:252–260. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyegaard M, et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry. 2010;15:119–121. doi: 10.1038/mp.2009.69. [DOI] [PubMed] [Google Scholar]
- 8.Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- 9.Barrett CF, Tsien RW. The Timothy syndrome mutation differentially affects voltage- and calcium-dependent inactivation of CaV1.2 L-type calcium channels. Proc Natl Acad Sci U S A. 2008;105:2157–2162. doi: 10.1073/pnas.0710501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Yazawa M, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 14.Pankratz MT, et al. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li XJ, et al. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flatz L, et al. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1013084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J, Mizuguchi M, Itoh M, Takashima S. Immunohistochemical expression of doublecortin in the human cerebrum: comparison of normal development and neuronal migration disorders. Brain Res. 2000;863:225–232. doi: 10.1016/s0006-8993(00)02099-0. [DOI] [PubMed] [Google Scholar]
- 19.Garbelli R, et al. Layer-specific genes reveal a rudimentary laminar pattern in human nodular heterotopia. Neurology. 2009;73:746–753. doi: 10.1212/WNL.0b013e3181af3397. [DOI] [PubMed] [Google Scholar]
- 20.Saito T, et al. Neocortical Layer Formation of Human Developing Brains and Lissencephalies: Consideration of Layer-Specific Marker Expression. Cereb Cortex. 2010 doi: 10.1093/cercor/bhq125. [DOI] [PubMed] [Google Scholar]
- 21.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 23.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garbett K, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ip BK, Bayatti N, Howard NJ, Lindsay S, Clowry GJ. The corticofugal neuron-associated genes ROBO1, SRGAP1, and CTIP2 exhibit an anterior to posterior gradient of expression in early fetal human neocortex development. Cereb Cortex. 2011;21:1395–1407. doi: 10.1093/cercor/bhq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano G, Suon S, Jin H, Donaldson AE, Iacovitti L. Characterization of five evolutionary conserved regions of the human tyrosine hydroxylase (TH) promoter: implications for the engineering of a human TH minimal promoter assembled in a self-inactivating lentiviral vector system. J Cell Physiol. 2005;204:666–677. doi: 10.1002/jcp.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghanti MA, et al. Species-specific distributions of tyrosine hydroxylase-immunoreactive neurons in the prefrontal cortex of anthropoid primates. Neuroscience. 2009;158:1551–1559. doi: 10.1016/j.neuroscience.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarotskyy V, Gao G, Peterson BZ, Elmslie KS. The Timothy syndrome mutation of cardiac CaV1.2 (L-type) channels: multiple altered gating mechanisms and pharmacological restoration of inactivation. J Physiol. 2009;587:551–565. doi: 10.1113/jphysiol.2008.161737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barttfeld P, et al. A big-world network in ASD: Dynamical connectivity analysis reflects a deficit in long-range connections and an excess of short-range connections. Neuropsychologia. 2011;49:254–263. doi: 10.1016/j.neuropsychologia.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Casanova MF, et al. Reduced gyral window and corpus callosum size in autism: possible macroscopic correlates of a minicolumnopathy. J Autism Dev Disord. 2009;39:751–764. doi: 10.1007/s10803-008-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Souza A, Onem E, Patel P, La Gamma EF, Nankova BB. Valproic acid regulates catecholaminergic pathways by concentration-dependent threshold effects on TH mRNA synthesis and degradation. Brain Res. 2009;1247:1–10. doi: 10.1016/j.brainres.2008.09.088. [DOI] [PubMed] [Google Scholar]
- 34.Toru M, Nishikawa T, Mataga N, Takashima M. Dopamine metabolism increases in post-mortem schizophrenic basal ganglia. J Neural Transm. 1982;54:181–191. doi: 10.1007/BF01254928. [DOI] [PubMed] [Google Scholar]
- 35.Elkabetz Y, et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tole S, Patterson PH. Regionalization of the developing forebrain: a comparison of FORSE-1, Dlx-2, and BF-1. J Neurosci. 1995;15:970–980. doi: 10.1523/JNEUROSCI.15-02-00970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 38.Coppola G, et al. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol. 2008;64:92–96. doi: 10.1002/ana.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dulcis D, Spitzer NC. Illumination controls differentiation of dopamine neurons regulating behaviour. Nature. 2008;456:195–201. doi: 10.1038/nature07569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.