Abstract
Background
Many miR genes are located within or around CpG islands. It is unclear whether methylation of these CpG islands represses miR transcription regularly. The aims of this study are to characterize gastric carcinoma (GC)-related methylation of miR CpG islands and its relationship with miRNA expression.
Methods
Methylation status of 9 representative miR CpG islands in a panel of cell lines and human gastric samples (including 13 normal biopsies, 38 gastritis biopsies, 112 pairs of GCs and their surgical margin samples) was analyzed by bisulfite-DHPLC and sequencing. Mature miRNA levels were determined with quantitative RT-PCR. Relationships between miR methylation, transcription, GC development, and clinicopathological characteristics were statistically analyzed.
Results
Methylation frequency of 5 miR CpG islands (miR-9-1, miR-9-3, miR-137, miR-34b, and miR-210) gradually increased while the proportion of methylated miR-200b gradually decreased during gastric carcinogenesis (Ps < 0.01). More miR-9-1 methylation was detected in 62%-64% of the GC samples and 4% of the normal or gastritis samples (18/28 versus 2/48; Odds ratio, 41.4; P < 0.01). miR-210 methylation showed high correlation with H. pylori infection. miR-375, miR-203, and miR-193b methylation might be host adaptation to the development of GCs. Methylation of these miR CpG islands was consistently shown to significantly decrease the corresponding miRNA levels presented in human cell lines. The inverse relationship was also observed for miR-9-1, miR-9-3, miR-137, and miR-200b in gastric samples. Among 112 GC patients, miR-9-1 methylation was an independent favourable predictor of overall survival of GC patients in both univariate and multivariate analysis (P < 0.02).
Conclusions
In conclusion, alteration of methylation status of 6 of 9 tested miR CpG islands was characterized in gastric carcinogenesis. miR-210 methylation correlated with H. pylori infection. miR-9-1 methylation may be a GC-specific event. Methylation of miR CpG islands may significantly down-regulate their transcription regularly.
Background
miRNA are an abundant class of small non-coding RNAs that mainly regulate gene expression at the post-transcriptional level. They play critical roles in the renewal and differentiation of stem cells and help maintain cell lineages. Previous research has shown that in cancer several of the miR genes such as miR-200b/200a/429, miR-21, miR-30b, miR-30d, miR-31, and miR-423 are upregulated, while other miR genes such as miR-143 and miR-145 are downregulated [1-3]. Evidence suggests that changes in miRNA expression occur frequently in many cancers and these variations either contribute to carcinogenesis or reflect the development and progression of cancers.
There are a number of pathways that may affect mature miRNA levels in cells and tissues, such as gene amplification or deletion, transcriptional upregulation or downregulation, post-transcriptional processing, and miRNA degradation [4-7]. It is well known that some intragenic miR genes, such as miR-218-2, are coordinately transcribed with their host genes through co-regulation mechanisms [8]. However, many miR genes are extragenic and a certain proportion of intragenic miR genes such as miR-9-1 are transcribed in a host gene-independent pattern [9]. Because the exact promoter region of most miR genes are not characterized, especially with regard to the extragenic miR genes, the exact regulatory mechanisms of miR transcription are far from clear.
Methylation or hypermethylation of CpG islands in the region of transcription starting sites (TSS) is generally recognized to repress gene transcription epigenetically. Unlike protein-coding genes that may span multiple CpG islands, the miR genes may be shorter than a CpG island, and in some cases, multiple miR genes (i.e., a miR gene cluster) may be located within or flanking a single CpG island (Additional file 1: Table S1). Aberrant methylation of CpG islands associated with miR genes, such as let-7a-3 and miR-34a, is frequently observed in many cancers [10,11]. It has been suggested that methylation of the CpG islands that are associated with miR genes (i.e. miR-203, miR-152, miR-124-1, miR-34b/c, miR-129-2, miR-9-1, miR-130b, miR-124-2, and miR-181c) might inversely correlate with their expression levels [12-17]. However, whether or not transcription of miR genes is regularly affected by the methylation status of miR CpG islands has not been systemically studied.
It is well known that abnormal methylation or demethylation of CpG islands in a small proportion (<1%) of the cell population can be sensitively detected in cellular heterozygous tissue samples. This demonstrates the advantage of methylation analysis over alterations of gene expression at the RNA and protein levels that can only be detected when such a changes is present in a large proportion of a cell population in a sample [18]. Our bioinformatic analysis shows 50, 9, and 70 of 721 human miR genes in the miRbase (Release 14.0) are located, respectively, within, flanking, and near CpG islands (collectively we will refer to these as miR CpG islands; Additional file 1: Table S1). We hypothesize that aberrant methylation of miR CpG islands may occur during development and progression of cancers. Therefore they could be used as candidate genes not only for prediction of cancer prognosis, but also for investigation of the methylation-expression association in vivo. Thus, CpG islands of 9 disease-related miR genes, including 5 extragenic miR genes or gene clusters (miR-9-3, miR-137, miR-200b/200a/429, miR-203, and miR-375) and 4 intragenic genes or gene clusters (miR-9-1, miR-34b/c, miR-193b/365-1, and miR-210), were selected as the representative genes in the present study (Additional file 1: Table S1). The methylation-expression association for 3 of these miR genes has not been previously established (Additional file 1: Table S2) [12,14,19-27]. We initially screened for gastric carcinoma (GC)- or host-related aberrant miR methylation and then investigated the methylation-expression association in vitro and in vivo. Associations between clinicopathological features of GC patients and methylation of these miR CpG islands were also analyzed.
Methods
Cell line sources and cell culture
Source information of used cell lines used in this study: RKO cell line, provided by Dr. Guoren Deng at University of California San Francisco; SW480 and HCT116 were provided by Dr. Yuanjia Chen at Peking Union Medical College Hospital; MKN74 and 293 T provided by Tokyo Medical and Dental University; PC-3 was purchased from the Cell Line Bank at the Chinese Academy of Medical Science; HL60 and KG1A were obtained from the Hematology Department of Peking University First Hospital; Du145 was obtained from Hanmi Pharmacy Company; Siha was provided by Peking University People’s Hospital. HepG2 was provided by Dr. Qingyun Zhang, Calu3 and A549 by Dr. Zhiqian Zhang, H1299 and AGS by Dr. Chengchao Shou, MKN45 by Dr Youyong Lv, and other cell lines (SGC7901, BGC823, MGC803, HeLa and GES-1) by Dr. Yang Ke, all at Peking University Cancer Hospital/Institute. These cell lines were cultured at 37°C in 5% CO2, using various culture media. MKN45, MKN74, SGC7901, BGC823, MGC803, HL60, KG1A, A549, H1299, GES-1, HepG2, 293 T, Du145, and RKO were cultured in 90% RPMI-1640 and 10% FBS. PC-3 and AGS were cultured in 90% F-12 and 10% FBS. Calu3, HeLa and Siha were cultured in 90% DMEM and 10% FBS. SW480 and HCT116 were cultured in 90% DMEM:RPMI-1640 (1:1) and 10% FBS.
Patients and tissue samples
Surgical primary GC samples and their paired non-cancerous surgical margin (SM) samples were collected from 112 inpatients (average age 59.2 years [range, 32–79]; 80 males and 32 females; 78 non-cardiac GCs and 34 cardiac GCs; 40 GCs at pTNM stage I ~ II and 59 at the stage III ~ IV) at Peking University Cancer Hospital. Follow-up data for all patients was collected for at least five years. All clinical samples, as well as histopathological and followup information for each case were obtained according to approved institutional guidelines. Gastric biopsies from 13 healthy subjects and 38 gastritis outpatients collected from the same hospital were used as non-cancer patient controls. Before bisulfite modification each patient’s gastric genomic DNA sample was analyzed for the presence of H. pylori-specific 23 S rDNA by a PCR assay as described previously [28]. The Institutional Review Boards of Peking University Cancer Hospital and Institute approved the study (#2011041207), and all patients gave written informed consent.
DNA extraction and bisulfite modification
Cancer cell line and tissue sample genomic DNA (1.8 μg) was isolated using phenol/chloroform extraction [29]. The unmethylated cytosine residues in the DNA samples were converted to uracil residues (becoming thymidine residues in PCR products) by the addition of 5 M sodium bisulfite [30]. The Wizard® DNA Clean-Up System Kit (Promega) was used to purify the bisulfite-treated DNA before PCR amplification.
PCR amplification and quantification of miR CpG island methylation by DHPLC
CpG-free universal primer sets were used to amplify miR CpG islands by hot-start PCR polymerase (Additional file 1: Figure S1 and Additional file 1: Table S3). Sequences of CpG islands embedded or flanked these related miR genes were used to design the primers. The PCR products were then analyzed quantitatively by DHPLC on the WAVE® DNA Fragment Analysis System [31]. Elution profiles of miR-9-3, miR-200b, and miR-203 methylation was analyzed with an ultraviolet detector; other miR gene methylation was detected with the post column HSX-3500 Accessory (Transgenomic, Inc., Omaha, USA) and a high–sensitivity fluorescence (FL) detector (excitation at 450 nm, emission at 520 nm) [32]. Methylated and unmethylated miR gene PCR products were separated by a DNASep® analytical column (Transgenomic) at the corresponding partial denaturing temperature (Additional file 1: Table S3 and Figure S2-10). The peak areas corresponding to the methylated and unmethylated PCR products were used to calculate the proportion of methylated miR CpG island [the proportion of methylated copies = methylation-peak area/ total peak area] as previously described [33]. M.SssI-methylated genomic DNA from blood samples was used as a positive control.
Bisulfite clone-sequencing
Fresh PCR products of miR CpG islands amplified with the CpG-free universal primer sets were cloned with the pGEM-T Easy kit (Promega, Madison, USA) and sequenced with an Applied Biosystems 3730xl DNA Analyzer at SinoGeneMox Company (Beijing, China).
Extraction of RNA and detection of mature miRNA level with quantitative RT-PCR assays
Total 50 ng RNA was extracted from fresh tissue samples or cell lines using the TRIzol reagent (Life Technologies, Carlsbad, USA) according to the manufacturer’s protocol. Corresponding cDNA samples were synthesized using the TaqMan® MicroRNA Reverse Transcription Kit (Life Technologies) with miR-specific stem-loop inverse transcription (RT) primers (specific for miR-375 #RT000564, miR-34b #RT000427, miR-137 #RT000593, and miR-9 #RT000583). The RT conditions used were 16°C for 30 min ➔ 42°C for 30 min ➔ 85°C for 5 min. The miRNA levels were then analyzed using a TaqMan Gene Expression Master Mix kit (Life Technologies) with the corresponding probe and primers (Life Technologies, miR-375 #TM000564, miR-34b #TM000427, miR-137 #TM000593, and miR-9 #TM000583). U6 (Life Technologies, #RT001093 and #TM001093) was used as the internal reference. The PCR cycling conditions were 95°C for 10min ➔ followed by 40 cycles of 95°C for 20 sec ➔ 60°C for 1 min. Expression levels of miR-200b and miR-210 were determined using a standard polyA RT-PCR assay. Sequences of the RT adaptor primer, the universal inverse primer and the U6 primer in the regular polyA RT-PCR assay are shown in Additional file 1: Table S5. RT conditions were 55°C for 5 min ➔ followed by 25°C for 10 min ➔ 42°C for 1 hr ➔ and 70°C for 5 min. The PCR cycling conditions were 95°C for10min ➔ followed by 40 cycles of 95°C for 20 sec ➔ and 61°C for 1 min.
Statistical analysis
SPSS 16.0 Trend-test and Pearson’s Chi-square test were used to analyze the miR methylation frequency difference between normal biopsies, gastritis lesions, GC and SM samples. Kruskal-Wallis H-test and One-Way ANOVA were used to analyze the miR methylation proportion differences between normal biopsies, gastritis lesions, GC, and SM samples. Fisher’s exact test, Pearson’s Chi-square test, and Trend-test were used to analyze the association between miR methylation positive rates and the clinicopathological features. The Mann–Whitney U-test and Student’s t-test were used to analyze the association between the proportion of methylated miR alleles and the clinicopathological features. Kaplan-Meier and Cox-Proportional Hazards methods were used for univariate and multivariate analysis to compare overall survival of GC patients with differences in methylation status of miR CpG islands. All statistical tests were two-sided, and P < 0.05 was considered statistically significant.
Results
Characterization of methylation or demethylation of 6 miR CpG islands related to the development of GCs
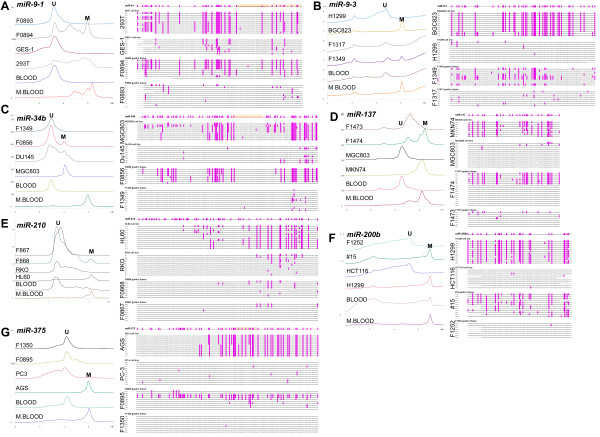
We amplified bisulfite-treated templates of 9 representative miR CpG islands with CpG-free primers and developed 9 DHPLC assays to analyze the methylation status of the CpG islands in PCR products of 4 intragenic miR genes (miR-9-1, miR-34b, miR-193b, miR-210) and 5 extragenic miR genes (miR-9-3, miR-137, miR-200b, miR-203 and miR-375), respectively (Figure 1, Additional file 1: Table S3, and Additional file 1: Figure S1–S10). These DHPLC assays revealed that the methylation positive rate of 5 miR CpG islands (miR-9-1, miR-9-3, miR-34b, miR-137, and miR-210) was significantly increased concurrently with the severity of pathological changes in the stomach. These findings strongly suggest that methylation of these miR CpG islands is related to the development of GCs (trend-test, miR-9-1, P < 0.001; miR-9-3, P < 0.001; miR-34b, P = 0.008; miR-137, P < 0.001; miR-210, P = 0.001; Table 1). miR-9-1 methylation was detected in 18 of 28 GCs (sensitivity, 64%), but only in 2 of 48 normal or gastritis biopsies showed methylation (specificity, 96%). Although methylation of miR-200b was detected in almost all gastric tissues samples, the proportion of methylated miR-200b in normal or gastritis tissues (46% ~ 100%) was significantly higher than that in both SM and GC samples (41% ~ 47%) (Mann–Whitney U-test, gastritis biopsies versus SMs, P = 0.001; Table 1). This suggests that miR-200b demethylation was a GC, patient-specific event. Bisulfite sequencing of these miR CpG islands confirmed the data obtained from DHPLC analysis (Figure 1).
Figure 1.
DHPLC chromatograms and bisulfite sequencing of 7 miR CpG islands in representative cell lines and gastric tissue samples. Both human peripheral blood DNA (BLOOD) and its M.SssI-methylated products (M.BLOOD) were used as controls. All CpG sites in the amplicon of each miR CpG island are also listed above the results of bisulfite sequencing. Each row represents one clone; each pink bar represents a methylated CpG site. The corresponding chromatogram of each sequenced sample (right column) is shown (left column).
Table 1.
Methylation status of miR CpG islands in gastric mucosa samples with different pathological changes in GC and non-cancerous control patients
| miR CpG islands | Group of gastric samples |
miR -Methylation |
||||||
|---|---|---|---|---|---|---|---|---|
|
Positive rate |
Proportion (%) of methylated miR in the miR methylation-positive samples |
|||||||
| Positive rate (%) | χ2-value | Trend-test (P-value) | Median[25% ~ 75%] | Mean±SD | t/F/χ 2-value | P-value | ||
|
miR-9-1 |
Normal |
1/13 (7.7) |
27.598 |
<0.001 a |
NA |
|
|
NA |
| |
Gastritis |
1/35 (2.9) |
|
|
NA |
|
|
|
| |
SM |
9/28 (32.1) |
|
|
17 [13–30] |
20 ± 4 |
t = −3.039 |
0.005 g |
| |
GC |
18/28 (64.3) |
|
|
29 [19–40] |
32 ± 4 |
|
|
|
miR-9-3 |
Normal |
6/13 (46.2) |
23.389 |
<0.001 a |
38 [31–59] |
43 ± 6 |
F = 1.639 |
0.189 h |
| |
Gastritis |
15/37 (40.5) |
|
|
43 [36–48] |
42 ± 2 |
|
|
| |
SM |
26/28 (92.9) |
|
|
38 [26–44] |
36 ± 2 |
|
|
| |
GC |
26/28 (92.9) |
|
|
37 [26–43] |
36 ± 2 |
|
|
|
miR-137 |
Normal |
5/13 (38.5) |
18.626 |
<0.001 a |
10 [4–41] |
|
χ2 = 8.065 |
0.045 d |
| |
Gastritis |
24/38 (63.2) |
|
|
19 [7–36] |
|
|
|
| |
SM |
25/26 (96.4) |
|
|
15 [7–40] |
21 ± 3g |
|
|
| |
GC |
24/26 (92.3) |
|
|
36 [20–51] |
37 ± 4 |
|
|
|
miR-34b |
Normal |
6/13 (46.2) |
6.947 |
0.008 a |
13 [7–19] |
|
χ2 = 0.658 |
0.883 d |
| |
Gastritis |
13/36 (36.1) |
|
|
11 [3–20] |
|
|
|
| |
SM |
22/28 (78.6) |
|
|
11 [4–18] |
|
|
|
| |
GC |
19/28 (67.9) |
|
|
10 [5–32] |
|
|
|
|
miR-200b |
Normal |
13/13 (100) |
0.486 |
0.922 f |
54 [52–83] |
|
χ2 = 17.883 |
<0.001 d |
| |
Gastritis |
35/36 (97.2) |
|
|
52 [46–100]e |
|
|
|
| |
SM |
27/28 (96.4) |
|
|
44 [40–47] |
|
|
|
| |
GC |
27/28 (96.4) |
|
|
48 [40–53] |
|
|
|
|
miR-210 |
Normal |
2/13 (15.4) |
11.908 |
0.001 a |
NA |
|
|
NA |
| |
Gastritis |
13/38 (34.2) |
|
|
7 [5–10] |
|
|
|
| |
SM |
22/27 (81.5) |
|
|
11 [5–22] |
|
|
|
| |
GC |
16/27 (59.3) |
|
|
9 [4–16] |
|
|
|
|
miR-193b |
Normal |
0/13 |
7.045 |
0.008 b |
NA |
|
|
NA |
| |
Gastritis |
2/37 (5.4) |
|
|
NA |
|
|
|
| |
SM |
7/28 (25.0) |
|
|
5 [4–14] |
|
|
|
| |
GC |
4/28 (14.3) |
|
|
5 [2–12] |
|
|
|
|
miR-203 |
Normal |
5/13 (38.5) |
5.617 |
0.018 b |
32 [29–75] |
|
χ2 = 2.957 |
0.398 d |
| |
Gastritis |
20/38 (52.6) |
|
|
31 [26–36] |
|
|
|
| |
SM |
21/28 (75.0) |
|
|
34 [30–36] |
|
|
|
| |
GC |
11/28 (39.3)c |
|
|
32 [29–35] |
|
|
|
|
miR-375 |
Normal |
0/13 |
12.266 |
<0.001 b |
NA |
|
|
NA |
| |
Gastritis |
13/38 (34.2) |
|
|
3 [3–4]e |
|
|
|
| |
SM |
16/28 (57.1) |
|
|
7 [4–14] |
|
|
|
| GC | 8/28 (28.6) c | 17 [8–21] | ||||||
a,Trend test, among Normal, Gastritis, SM and GC samples; b,Trend-test, among Normal, Gastritis, and SM samples; c, Pearson’s Chi-square test, GC versus SM: miR-203, χ2 = 7.292, P = 0.007; miR-375, χ2 = 4.667, P = 0.031; d, Kruskal-Wallis H-test; e, Mann–Whitney U-test: Gastritis versus SM, miR-200b,U = 230.000, P = 0.001; Gastritis versus GC, miR-375, U = 46.000, P = 0.011; SM versus GC, miR-137, U = 170.000, P = 0.009; f, Pearson’s Chi-square test, among Normal, Gastritis, SM and GC samples; g. Paired t-test: SM versus GC, miR-137, t = −2.652, P = 0.014; h. One-way ANOVA; NA, not available.
Except for miR-9-1 and miR-137, the methylation positive rates or proportions of methylated miR-9-3, miR-34b, miR-210, and miR-200b in GCs were similar to those in SMs (Table 1). To validate if methylation of some of these miR genes is a field-effect happens simultaneously in both cancerous and non-cancerous tissues in the stomach due to the same exposure to environmental factors, we detected the methylation data in another subset of GC and SM samples from 84 patients and found that the positive rate and proportion of methylated miR-9-1 in GCs was still significantly higher than that in SMs (60.7% versus 32.1%; Pearson’s Chi-square test, P = 0.001; Sign rank test, P < 0.001; Additional file 1: Table S4, Subset-2). The average proportion of the methylated miR-137 was also significantly higher in GCs than SMs (Mean ± SD, 26 ± 2 versus 38 ± 2, Paired t-test, P < 0.001). As expected a significant difference in the positive rate or the proportion of methylated miR-9-3, miR-34b, miR-210, and miR-200b was not observed between GC and SM samples. These results confirmed that miR-9-1 and miR-137 methylation was a tumor-specific event and that miR-9-3, miR-34b, and miR-210 methylation, as well as miR-200b demethylation, was a field-effect that occurred during gastric carcinogenesis.
Furthermore, the positive rate of methylated miR-203 and miR-375 gradually increased from normal to gastritis to SM samples, but significantly decreased in GCs compared with SMs (Pearson Chi-square test, miR-203, P = 0.007; miR-375, P = 0.031; Table 1). In the Subset-2 samples, we also analyzed miR-375 methylation and found more miR-375 methylation in SMs than in GCs again (Pearson Chi-square test, P = 0.034; Additional file 1: Table S4). These data imply that miR-375 (and miR-203) methylation is not GC-specific and might be one kind of host adaptation in the non-malignant tissues to the development of GCs.
miR methylation and H. Pylori infection
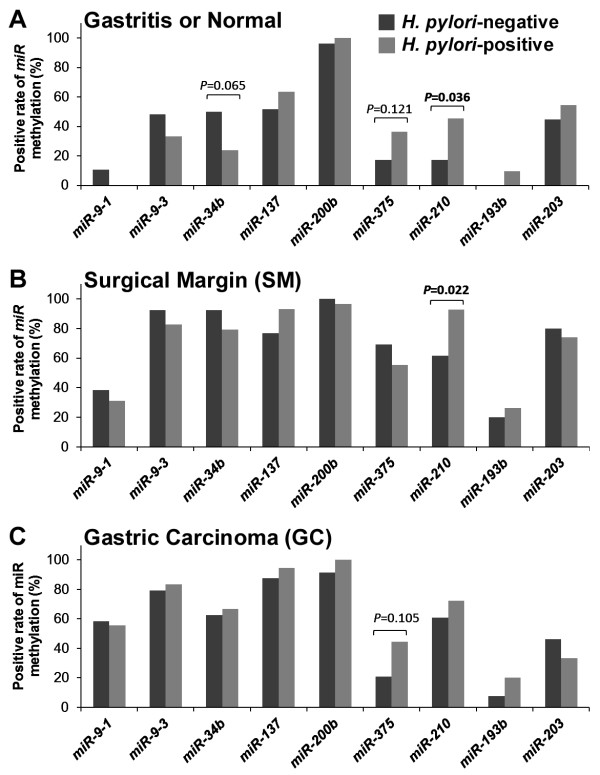
To determine if H. pylori infection plays a role in miR methylation, we looked for H. pylori-specific 23 S rDNA in gastric genomic DNA samples by PCR and found that the H. pylori-positive rate increased concurrently with the severity of pathological changes [15.4% (2 of 13) of normal gastric biopsies, 52.6% (20 of 38) of gastritis lesions, and 69.0% (29 of 42) SM biopsies; trend-test, P = 0.001] and decreased in GC samples (18 of 42 = 42.9%; GCs versus SMs, Pearson Chi-square test, P = 0.016; Additional file 1: Figure S11). H. pylori-positive gastritis/normal and SM samples showed significantly higher methylation of miR-210 methylation than H. pylori-negative samples (P = 0.036 and 0.022, respectively; Figure 2A and B).
Figure 2.
Comparison of miR methylation in various gastric mucosa samples with and without existence of H. pylori. (A) Normal or gastritis biopsies from control outpatients without malignant disease; (B and C) Surgical margin and tumor samples from patients with gastric carcinoma, respectively; H. pylori-specific 23 S rDNA was detected by PCR.
Inversed relationship between miR methylation of CpG islands and their corresponding expression levels
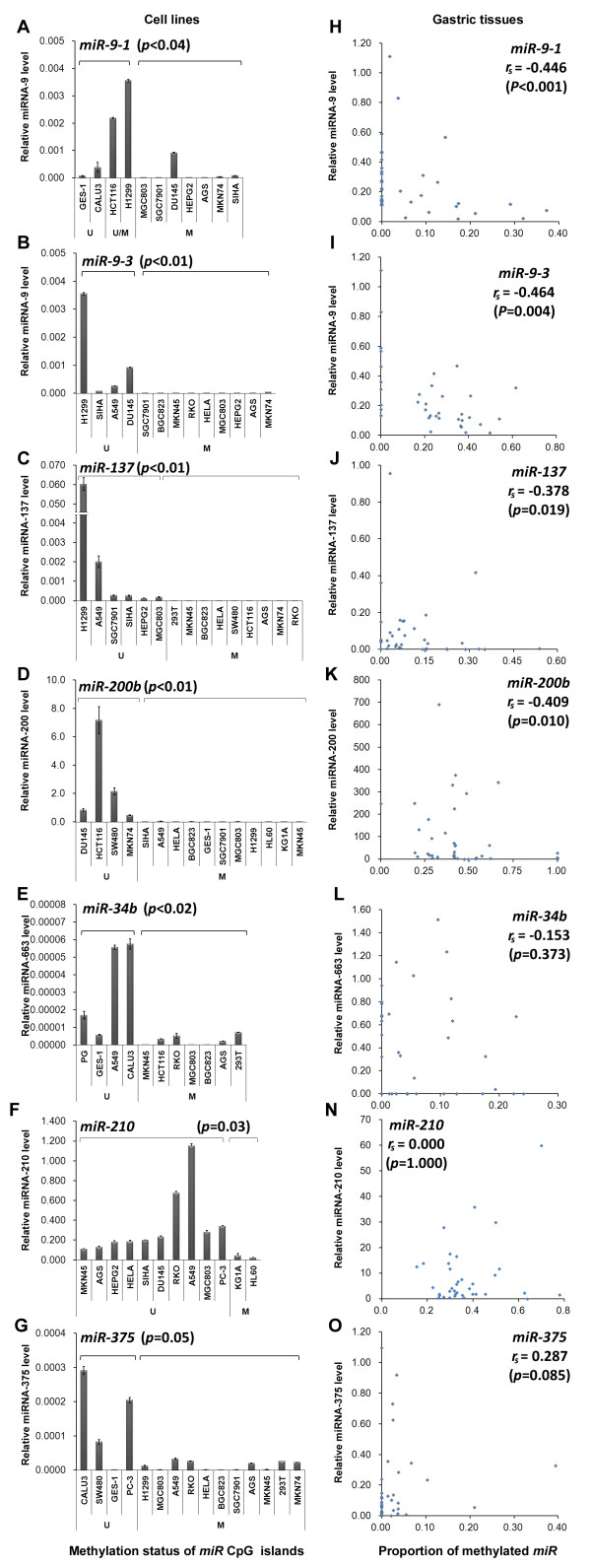
To investigate the relationship between the above aberrant miR methylation and the transcription of the corresponding miR gene, we quantified the mature miRNA levels of miR-9-1, miR-9-3, miR-34b, miR-137, miR-210, miR-200b, (and miR-375), whose methylation status is related to the development of GC (and GC host adaptation) as described above, in a set of human cell lines with different methylation status of miR CpG islands. The methylation status of each miR CpG island in these cell lines was determined by DHPLC as illustrated in Additional file 1: Figure S2–S10. Results of quantitative RT-PCR assays showed that in the tested cell lines containing methylated miR alleles the mature miRNA levels of all 6 GC-related miR genes and one host adaptation miR gene were significantly lower than those detected in the cell lines containing unmethylated miR alleles (Figure 3A-G).
Figure 3.
The relationship between expression level and methylation status of miR genes in human cell lines and gastric tissue samples. (A-G) The correlation analysis between methylation proportion of miRNA CpG islands and their mature miRNA expression levels in the target CpG island methylated and unmethylated human cell lines; (H-P) Expression of miRNAs in 20 paired, fresh gastric carcinoma samples; (U) Target CpG island unmethylated cell lines; (M) Target CpG island methylated cell lines; (U/M) Target CpG island partially methylated cell lines.
We further analyzed the miRNA levels of 20 pairs of fresh GC and SM samples and found a significantly higher expression level of miRNA-200b, miRNA-375, and miRNA-210 in SMs than in GCs (Paired t-test: Ps≤0.030; Additional file 1: Figure S12). The miRNA-137 levels in SMs were also higher than those in GCs, but was not statistically significant (P = 0.059). The expression levels of miRNA-9 and miRNA-34b were similar between SMs and GCs. Most importantly, an inverse relationship between miR methylation and the corresponding expression level was observed for miR-9-1, miR-9-3, miR-137, and miR-200b in these gastric tissue samples (Spearman’s Rank Correlation analysis, miR-9-1, rs = −0.533, P = 0.001; miR-9-3, rs = −0.464, P = 0.004; miR-137, rs = −0.378, P = 0.019; miR-200b, rs = −0.409, P = 0.010; Figure 3H-K). A weak inverse methylation-expression relationship was also found for miR-375 in these tissue samples (rs =0.287, P = 0.085; Figure 3O). Such a relationship was not observed for miR-34b and miR-210 (Figure 3L, N).
miR methylation correlated to clinicopathological characteristics of GC patients
To study the possibility of using miR methylation as a prognosis predictor of GCs, we determined the prevalence of methylation of 7 miR genes and analyzed the relationship between the clinicopathological features of GC and the corresponding methylation levels of these miR CpG islands studied above for all 112 GC patients (Table 2 and Additional file 1: Table S6). Higher miR-9-1 methylation was consistently observed in well/moderately differentiated, non-metastatic/less-invasive GCs, but this was not statistically significant. The proportion of demethylated miR-200b was higher in lymph metastatic GCs than in non-metastatic GCs (median, 45% versus 53%; Mann-Whiney U-test, P = 0.021). In contrast, the positive rate of miR-137 methylation positively correlated with the depth of invasion, pTNM, lymph metastasis, and vessel embolus of GCs (trend-test for depth of invasion, P = 0.012; Pearson’s Chi-square test for pTNM/ lymph metastasis/ vascular embolus, P = 0.024/ 0.028/ 0.031). Similarly, higher miR-210 methylation was observed in GCs with distant metastasis than in GCs without distant metastasis (P = 0.048). In addition, the positive rate of miR-9-1 methylation and the proportion of methylated miR-137 in elderly GC patients were significantly higher than in younger patients (Pearson’s Chi-square test, miR-9-1, P = 0.005; Student’s t-test, miR-137, P = 0.009).
Table 2.
Comparison of methylation of 7 miR CpG islands in 112 GC samples from patients with various clinicopathological characteristics
| Clinicopathological features |
miR-9-1 methylation |
miR-9-3 methylation |
miR-137 methylation |
miR-34b methylation |
miR-200b methylation |
miR-375 methylation |
miR-210 methylation |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive rate (%) | Proportion, Median * [25-75%] | Positive rate (%) | Proportion, Median [25-75%] | Positive rate (%) | Proportion, Median [25-75%] | Positive rate (%) | Proportion, Median [25-75%] | Positive rate (%) | Proportion, Median [25-75%] | Positive rate (%) | Proportion, Median [25-75%] | Positive rate (%) | Proportion, Median [25-75%] | ||
| Age |
≤60 (n = 49) |
46.9a |
21 [14–38] |
68.8 |
35 [31–42] |
95.9 |
34 [16–48]b |
62.5 |
5 [2–16] |
92.9 |
49 [43–55] |
54.3 |
3 [2–14] |
65.1 |
9 [5–27] |
| |
>60 (n = 63) |
73.0 |
33 [18–44] |
61.0 |
36 [30–44] |
93.3 |
47 [29–61] |
54.2 |
10 [3–23] |
87.7 |
48 [40–56] |
38.3 |
3 [2–14] |
72.9 |
10 [5–17] |
| Sex |
Male (n = 80) |
61.2 |
31 [14–45] |
62.7 |
35 [30–42] |
96.1 |
40 [24–54] |
53.8 |
7 [3–18] |
88.4 |
48 [40–56] |
44.6 |
3 [2–7] |
69.9 |
9 [5–17] |
| |
Female (n = 32) |
62.5 |
27 [17–36] |
68.8 |
41 [30–43] |
90.6 |
43 [18–54] |
69.0 |
8 [2–26] |
93.3 |
51 [46–55] |
46.9 |
8 [2–19] |
69.0 |
12 [5–22] |
| Preoperative chemotherapy |
No (n = 100) |
63.0 |
32 [16–44] |
63.2 |
35 [30–43] |
93.8 |
39 [23–56] |
58.9 |
7 [3–18] |
91.0 |
49 [41–56] |
44.7 |
4 [2–16] |
69.6 |
9 [5–19] |
| Yes (n = 9) |
55.6 |
21 [12–31] |
66.7 |
37 [31–54] |
100 |
49 [30–53] |
44.4 |
5 [2–16] |
71.4 |
49 [42–76] |
44.4 |
2 [2–4] |
57.1 |
13 [10–24] |
|
| Location |
Cardiac (n = 34) |
64.7 |
37 [21–47] |
65.6 |
43 [35–51]b |
90.9 |
49 [28–61]b |
57.6 |
8 [3–23] |
75.0d |
43 [38–52] |
31.2 |
3 [1–8] |
64.5 |
9 [4–17] |
| |
Non-cardiac (n = 78) |
60.3 |
28 [14–42] |
64.0 |
34 [30–42] |
96.1 |
38 [20–52] |
58.1 |
6 [3–18] |
97.0 |
50 [42–56] |
51.4 |
3 [2–16] |
71.8 |
10 [5–21] |
| Differentiation |
Well/Mod. (n = 29) |
72.4a |
27 [9–44] |
69.0 |
37 [30–50] |
86.2 |
45 [31–53] |
44.4 |
9 [3–16] |
84.6 |
53 [42–58] |
40.7 |
3 [2–4] |
73.1 |
9 [4–17] |
| |
Poor (n = 78) |
56.4 |
31 [19–42] |
63.0 |
35 [30–42] |
97.3 |
38 [22–55] |
64.0 |
6 [3–19] |
91.3 |
48 [40–54] |
45.9 |
3 [2–14] |
66.2 |
10 [5–21] |
| Vascular embolus |
No (n = 60) |
65.0 |
26 [14–45] |
70.2 |
35 [31–43] |
89.7d |
45 [26–59] |
55.0 |
10 [3–19]c |
92.5 |
50 [42–57] |
51.7 |
3 [2–13] |
70.2 |
9 [5–20] |
| Yes (n = 49) |
59.2 |
31 [19–39] |
55.3 |
37 [30–43] |
100 |
39 [22–50] |
65.9 |
3 [2–11] |
86.0 |
48 [40–53] |
40.0 |
5 [2–20] |
66.7 |
11 [5–20] |
|
| pTNM stage |
I-II (n = 40) |
70.0 |
30 [14–47] |
67.6 |
37 [33–45] |
89.7d |
46 [35–59] |
59.0 |
10 [3–28]c |
91.9 |
49 [41–56] |
51.3 |
3 [2–8] |
64.1 |
9 [5–19] |
| |
III-IV (n = 59) |
59.3 |
30 [19–40] |
60.3 |
39 [32–43] |
100 |
40 [22–53] |
58.2 |
4 [2–17] |
87.8 |
49 [41–54] |
41.8 |
5 [2–20] |
70.0 |
12 [5–28] |
| Depth of invasion |
T1-2 (n = 25) |
68.0 |
34 [11–52] |
72.7 |
36 [31–44] |
84.0e |
48 [36–59] |
73.9 |
10 [3–21] |
86.4 |
52 [44–56] |
47.8 |
2 [2–4] |
70.8 |
9 [3–19] |
| T3 (n = 53) |
62.3 |
28 [17–41] |
58.5 |
35 [30–43] |
96.1 |
43 [28–53] |
49.0 |
7 [2–24] |
91.3 |
46 [36–56] |
48.0 |
5 [2–18] |
68.8 |
10 [5–18] |
|
| T4 (n = 25) |
60.0 |
30 [16–38] |
62.5 |
42 [33–44] |
100 |
31 [15–53] |
66.7 |
5 [2–17] |
90.9 |
49 [41–55] |
36.0 |
8 [3–20] |
66.7 |
14 [5–32] |
|
| Lymph node metastasis |
N0 (n = 57) |
68.4 |
26 [13–47] |
67.3 |
35 [32–43] |
89.1d |
45 [22–58] |
61.8 |
10 [3–20] |
90.6 |
53 [43–57]c |
46.4 |
2 [2–10] |
67.3 |
9 [5–19] |
| N1-3 (n = 51) |
56.9 |
31 [21–41] |
58.3 |
39 [30–44] |
100 |
39 [26–52] |
58.3 |
4 [2–16] |
88.1 |
45 [37–52] |
47.8 |
5 [2–20] |
69.8 |
11 [5–23] |
|
| Distant metastasis | M0 (n = 77) |
67.5a |
30 [14–44] |
64.4 |
37 [32–43] |
92.0 |
42 [24–56] |
60.0 |
8 [3–19] |
91.5 |
49 [41–56] |
46.7 |
3 [2–13] |
68.1 |
9 [5–17]c |
| M1 (n = 31) | 51.6 | 30 [21–40] | 60.0 | 36 [30–43] | 100 | 43 [24–53] | 60.7 | 6 [2–17] | 83.3 | 49 [40–53] | 48.1 | 6 [3–20] | 69.2 | 15 [9–28] | |
* %; [25-75%], 25%-75% percentiles; a, Pearson’s Chi-square test, miR-9-1 for age, differentiation, distant metastasis: χ2 = 7.924, 2.271, 2.402, and P = 0.005, 0.132, 0.121, respectively; b, Student t-test: miR-9-3 for location [42 ± 2 (Mean ± SD) vs 34 ± 1], t = 3.303, P = 0.002; miR-137 for age [34 ± 3 vs 44 ± 3], t = −2.651, P = 0.009; for location [46 ± 3 vs 37 ± 2], t = 2.103, P =0.038; c, Mann–Whitney U-test: miR-34b for pTNM stage/ vascular embolus, U = 239.000/320.000, P = 0.028/0.025, miR-200b for lymph node metastasis, U = 627.000, P = 0.021; miR-210 for distant metastasis, U = 301.000, P = 0.048; d, Fisher’s exact test: miR-137 for vascular embolus/pTNM/lymph metastasis, P = 0.031/0.024/0.028; miR-200b for location, P = 0.002; e,Trend test: miR-137 for depth of invasion, χ2 = 6.330, P = 0.012.
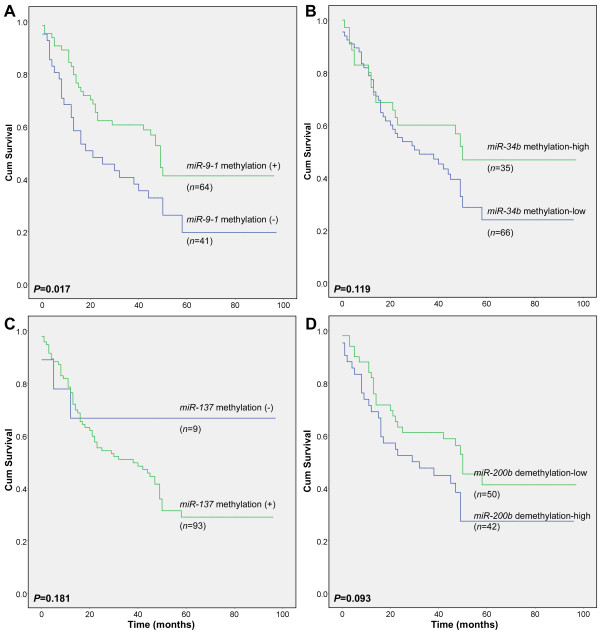
Kaplan-Meier analysis showed that the overall survival of GC patients with miR-9-1 methylation was significantly longer than in those without methylation (P = 0.017; Figure 4A). Similarly, the overall survival of GC patients with high miR-34b methylation (the proportion of methylated miR-34b > 4%) was likely longer than those with low miR-34b methylation (P = 0.119; Figure 4B). In contrast, the overall survival of GC patients with miR-137 methylation was shorter than for those without methylation, but was not statistically significant (P = 0.181; Figure 4C). Interestingly, the overall survival of GC patients with high levels of demethylated miR-200b (proportion of methylated miR-200b <45%) was shorter than for low demethylation patients (P = 0.093; Figure 4D). In addition to miR-9-1 methylation, vessel embolus, pTNM stages, invasion, node and distant metastasis are also significant survival predictors in univariate analysis (Additional file 1: Table S7). After adjustment for age, sex, differentiation, location, preoperative chemotherapy, pTNM stage, and vascular embolus, miR-9-1 methylation was still an adequate prognostic indicator in multivariate analysis (adjusted P = 0.014; hazard ratios = 0.475 [95% CI, 0.262–0.860]).
Figure 4.
Overall survival curves of GC patients with different methylation status of 4 miR CpG islands in Kaplan-Meier analysis. (A) miR-9-1 methylation used as the classifier; (B) miR-34b methylation used as the classifier; (C) miR-137 methylation used as the classifier; (D) miR-200b demethylation used as the classifier.
Discussion
The expression patterns exhibited by the miR genes reflect cell lineages and differentiation status of tumor tissue [34]. Increasing attention is currently being focused on miRNAs due to their contribution to maintaining the pluripotency of cancer stem/initiating cells and their application potential as molecular therapy targets and biomarkers. However, the regulatory mechanisms of miR expression in cells and alterations of miRNA levels in tumors are far from clear. In the present study, we characterized the methylation status of 9 representative miR CpG islands in human gastric tissues with various pathological changes and found that abnormal methylation of 5 miR CpG islands (miR-9-1, miR-9-3, miR-34b/c, miR-137, and miR-210) and demethylation of miR-200b CpG island correlated with the development of GC. Furthermore, methylation of the miR-203 and miR-375 CpG islands might be one kind of host adaptations to gastric carcinogenesis. Most importantly, we found that methylation of all these CpG islands was inversely correlated with the expression of these miR genes in a panel of cultured cell lines. To the best of our knowledge, this is the first comprehensive study to illustrate the inverse relationship between the methylation of miR CpG islands and their expression.
Transcription of protein-coding genes is mainly regulated by transcription factors and the accessibility of their binding sites in the promoter regions. It is well known that methylation of CpG islands around the TSS can block transcription factor binding by decreasing the accessibility of these sites, thus inactivate gene expression epigenetically. Bioinformatic analysis shows that 129 of 721 (18%) of the human miR genes in the miRBase (Release 14.0) are CpG island-related (Additional file 1: Table S1). Of the CpG related miR genes, 50 locate within CpG islands and 9 closely flank CpG islands with a <600-bp CpG-sporadic interval; an additional 70 miR genes are near CpG islands with a 0.6 ~ 10-kb CpG-sporadic interval. It has been suggested that methylation of some miR CpG islands might be inversely correlated to their expression [12-17]. However, a solid relationship between miR methylation and expression has not been thoroughly established as only weak supporting evidence has been provided in many of the previous studies, as we have summarized for 9 tested miR genes/clusters (extragenic miR-9-3, miR-137, miR-200b/200a/429, miR-203, miR-375; intragenic miR-9-1, miR-34b/c, miR-193b/365-1, and miR-210) in this present study (Additional file 1: Table S2) [19-27]. One frequently used evidence is that methylated miR genes could be reactivated by inhibitors of DNA methyltransferase (5-aza-cytosine or 5-aza-deoxycytosine) because these cytosine analogues, which cannot be methylated in DNA, inhibit maintenance methylation through direct replacement of methylation targets (cytosine residues). However, this method is not adequate as methyltransferases inhibition that may indirectly reactivate transcription of both CpG island-free genes and methylated genes through DNA damage and repair pathways [35].
To investigate systemically if methylation of miR CpG islands represses transcription of miR genes, various cell lines and tissue samples with different methylation status of representative miR CpG islands must be used. Thus, instead of randomly selecting candidates from the 129 miR CpG islands, we first screened a set of CpG islands of miR genes that may be abnormally methylated during gastric carcinogenesis. Initially the methylation status of 9 miR CpG islands in human gastric tissues with various pathological changes was analyzed with DHPLC. As is consistent with others’ reports [14,15,19,25,27,36-40], miR-9-1, miR-9-3, miR-34b, miR-137, and miR-375 methylation was observed in gastric carcinogenesis in the present study. We also observed methylation changes in miR-200b, miR-193b, miR-203, and miR-210 CpG islands in the development of GCs that has not been previously reported. Interestingly, we found that the proportion of demethylated miR-200b gradually increased significantly in gastric tissues along with the severity of these changes. This suggests that miR-200b demethylation (or hypomethylation) may be involved in gastric carcinogenesis. In addition, miR-203 and miR-375 methylation increased gradually in gastritis and SM samples, but decreased in GC samples. This implies that miR-203 and miR-375 methylation is not a GC-specific event, but rather a host adaptation to gastric carcinogenesis.
Expression of some intragenic miR genes is coordinately regulated by the transcriptional mechanism of their host genes. However, a certain proportion of intragenic miR genes has their own TSS and transcribe in a host gene-independent pattern [9,41-43]. Unfortunately, the TSS sites have not been characterized for most extragenic/intergenic miR genes. It is reported that expression levels of some miR genes (including intragenic miR-152 and miR-34a/b/c and extragenic miR-203, miR-124-1/124-2, miR-129-2, and miR-181c) inversely correlate with methylation of their corresponding CpG islands [11,13,14,16,27,38,44]. In the present study, we found that methylation or demethylation of all 7 tested miR CpG islands (GC-related miR-9-1, miR-34b, miR-9-3, miR-137, miR-210, miR-200b and host-related miR-375) was consistently, inversely correlated to a statistically significant level with their corresponding miRNA levels in a number of human cell lines in vitro. Such an inverse relationships could also be observed for the miR-9-1, miR-9-3, miR-137, and miR-200b CpG islands in gastric tissue samples in vivo. Although expression of intragenic miR-9-1 is host gene-independent [9], we cannot exclude the possibility that the expression of miR-34b and miR-210 genes may also be coordinately controlled by regulatory mechanisms of their host genes. An unbiased correlation study using a customized, oligo microarray to detect methylation of all miR CpG islands and expression of intragenic miR genes and their host genes in various cell lineages at different differentiation stages may be useful to clarify which miR genes may be expressed in a host-gene dependent way. Taken together, these data strongly suggest that the methylation status of miR CpG islands could play a crucial role in the regulation of the expression of the related miR genes.
Generally, transcriptional inactivation of a miR gene by methylation in a few cells should not lead to a visible decrease in the miRNA level in a tissue sample because the majority of cells will still exhibit normal miRNA expression. This help explain why a significant inverse methylation-expression relationship was not observed for miRNA-34b, miRNA-210, or miRNA-375 as the average proportion of methylated miR-34b, miR-210, or miR-375 in SM and GC samples was relatively low (2% ~ 15%). Hypermethylation of genes in some cell populations and the concomitant over-expression of these genes in other cell populations are often reported in the same tissue samples with chronic inflammation [18]. Therefore, both the prevalence of miR methylation and the total mature miRNA levels may be useful predictors for miR inactivation and expression in tissue samples, especially in the case of low proportion of methylated miR. Although other factors certainly affect the miRNA expression levels in cells, the inverse relationship consistently observed in this present study between miR methylation and mature miRNA expression level in a number of monoclonal cell lines suggests that the cellular heterozygosity may account for the inconsistent methylation-expression relationship of some miR in tissue samples.
The miR-9-1, miR-9-2, and miR-9-3 genes all encode the same mature miRNA-9 that affects cell migration and proliferation in a tumor type-specific pattern through the NF-κB1SnailE-cadherin[45,46]. Abnormal miR-9-1 and miR-9-3 methylation is frequently reported in many cancers including GCs [15,19,40]. In the present study, we found that the positive rate of miR-9-1 and miR-9-3 methylation for all 112 GCs was significantly higher than that in 50 non-malignant tissues collected from 37 gastritis patients and 13 healthy controls. In all the tested samples, the prevalence of miR-9-3 methylation is significantly higher than that of miR-9-1. It has been reported that miR-9-3 methylation correlates with poor clinical outcomes for GC patients [40]. However, our study failed to make the same correlation. In contrast, a significant higher of the positive rate and proportion of methylated miR-9-1, but not methylated miR-9-3, was observed in GCs than SMs. This data indicates that miR-9-1 methylation may be a late cancer-specific event, while miR-9-3 methylation may be an early field effect during gastric carcinogenesis. Using miR-9-1 methylation as a biomarker for GC detection, we found a sensitivity and specificity 62% (69 of 112) and 96% (46 of 48) could be achieved, respectively. Because the positive rate of miR-9-1 methylation in GCs or SMs from stage I ~ II GC patients was similar to that of stage III ~ IV GC patients (GCs: 70% versus 59%; SMs: 40% versus 25%), it would be clinically beneficial to determine if miR-9-1 methylation in gastric juice or peripheral blood plasma could be used as a biomarker for prediction of malignant transformation of precancerous lesions of the stomach and early diagnosis of GCs. Although miR-9-1 methylation does not significantly correlate with differentiation, local invasion, metastasis, and pTNM stages in the screening cohort, it does significantly correlate with a longer overall survival of GC patients in univariate and multivariate analysis. Combination with the methylation status of other miR genes failed to strengthen the prediction power of miR-9-1 methylation on the survival of these patients. Therefore, miR-9-1 methylation might be an independent predictor of survival.
miR-137 has been reported to target the CtBP1, a co-repressor of various tumor suppressor genes [47]. Many studies have reported miR-137 methylation in head and neck and colorectal carcinomas [22,23,48,49]. We found that miR-137 methylation was also very common among gastritis lesion, SM, and GC samples (63% ~ 96%), and the positive rate of miR-137 methylation increased gradually in gastric tissues along with the severity of pathological changes. Therefore, miR-137 methylation may be another early epigenetic event that occurs during gastric carcinogenesis. Because the proportion of methylated miR-137 showed a positive correlation with the invasiveness of GCs, miR-137 methylation may also affect the progression of the tumor. When compared to the GC patients without miR-137 methylation, GC patients with miR-137 methylation were more likely to have a poor overall survival; however, this effect was not statistically significant. This is consistent previous study that has reported that miR-137 methylation (by MSP) correlated to the overall survival of 67 patients with head and neck carcinomas [49].
miR-34b and miR-34c constitute a miR gene cluster. Previous studies have suggested that mature miRNA-34b and miRNA-34c are targets of P53 [50]. As a tumor suppressor gene, miR-34b/c methylation has also frequently been reported in many carcinomas including GCs [14,51,52]. miR-34b methylation has been reported to relate to the invasiveness of non-small lung carcinoma, colorectal carcinoma, GC, and melanoma [53-55]. In the present study, we did not find a significant correlation between miR-34b methylation and GC invasion; however, a higher proportion of methylated miR-34b was observed in stage I ~ II GCs than in stage III ~ IV GCs (P = 0.025). The overall survival of GC patients with methylated miR-34b (the proportion > 4%) was likely to be longer than in those patients without the methylation. In addition, we found that the prevalence of miR-34b methylation between SM and GC samples was similar; this is consistent with a recent report that miR-34b methylation might be an early field-effect in the development of GCs [55].
The miR-200b gene is located in the miR-200b/miR-200a/miR-429 cluster. Its function is related to the processes of epithelium-mesenchymal transition (EMT) by targeting ZEB1 and ZEB2 and results in E-cadherin silencing [56-58]. There is a CpG island upstream of this cluster which contains a 4-kb CpG-sporadic interval. It has been reported that miR-200a/b is hypomethylated and over-expressed in pancreatic cancers and that circulating miRNA-200 may be used as a cancer biomarker. The positive relationship between miR-200b demethylation and over-expression has also been previously found [59]. In the present study, we observed that full demethylation of the miR-200b CpG island was a common field event in both GC and SM samples. Because the proportion of demethylated miR-200b in gastritis/normal biopsies was significantly lower than that in SM and GC samples, miR-200b demethylation might be another potential GC biomarker.
miR-210 is a hypoxia inducible gene which may inhibit cancer cell survival and proliferation through targeting FGFRL1 [60]. It has been reported that miR-210 may directly bind to vacuole membrane protein 1 (VMP1) and promote cancer metastasis [61]. miR-210 methylation has been previously reported to be presented in glioma [62]. We found that miR-210 methylation not only correlated with the severity of gastric pathogenesis, but also correlated with H. pylori infection. According to the possible cause effect relationship between H. pylori infection and the development of GCs, miR-210 inactivation by DNA methylation may play a role in gastric carcinogenesis.
miR-375 promotes cancer cell proliferation through RASD1-ERα pathway [63]. methylation has been previously reported in melonoma [64], breast cancer [63], gastric carcinoma [39] and hepatocellular carcinoma [12]. However, our present study found that for the 106 patients studied, there was a significant higher positive rate and proportion of methylated miR-375 present in SMs compared to GCs. Because significant correlation between miR-375 methylation and H. pylori infection was not observed among the tested gastric samples, we suggest that miR-375 methylation might be a unique host adaptation to the development of GCs.
Previous studies have shown that miR-203 methylation drives H. pylori-associated gastric lymphomagenesis [65]. miR-203 and miR-193b methylation has also been reported in hepatomas and prostate cancer, respectively [12,37]. We observed that the positive rate of miR-203 and miR-193b methylation increased in normal, gastritis, and SM tissues, but decreased in GCs as did miR-375 methylation pattern. However, correlation between H. pylori infection and miR-203 and miR-193b methylation was not found in any of the gastric tissue samples that were studied. Therefore, we hypothesize that miR-203 and miR-193b methylation may also be a host adaptation to the development of gastritis or GCs.
Conclusions
In conclusion, alteration of methylation status of 6 of 9 tested miR CpG islands was characterized in gastric carcinogenesis. miR-210 methylation correlated with H. pylori infection. miR-9-1 and miR-137 methylation may be a GC-specific event. Methylation of miR CpG islands may significantly down-regulate their transcription regularly.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YD participated in the design, conducted experiments, and wrote the manuscript; ZL contributed to the DHPLC assay development and gastric sample management; LG collected the clinical information; JZ extracted genomic DNA samples from tissues and detect H. pylori 23 S rDNA; B-DZ collected the gastric biopsy samples; JJ collected gastric carcinoma samples; DD designed and coordinated the study, analyzed data, and wrote the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Figure S1. Illustrations of 9 tested miR-hosting CpG islands. Open frame, location of pre-miRNA transcript. Open arrow, pre-miRNA transcript direction; Blue-underline, locations of the forward and reversed primer-matching regions used to detect methylation of CpG island with DHPLC and bisulfite sequencing; Purple vertical bars, CpG sites. Figure S2. DHPLC chromatogram of methylated and unmethylated miR-9-1 in various cell lines. UV-detector; the partial denaturing temperature, 55.4°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S3. DHPLC chromatogram of methylated and unmethylated miR-9-3 in various cell lines. UV-detector; the partial denaturing temperature, 58.5°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S4. DHPLC chromatogram of methylated and unmethylated miR-34b in various cell lines. UV-detector; the partial denaturing temperature, 56.8°C; U, peak for the unmethylated PCR products; M (partial or full), peak for the partially or fully methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S5. DHPLC chromatogram of methylated and unmethylated miR-210 in various cell lines. Fluorescence-detector; the partial denaturing temperature, 58.7°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S6. DHPLC chromatogram of methylated and unmethylated miR-137 in various cell lines. Fluorescence-detector; the partial denaturing temperature, 58.3°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S7. DHPLC chromatogram of methylated and unmethylated miR-375 in various cell lines. UV-detector; the partial denaturing temperature, 55.7°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S8. DHPLC chromatogram of methylated and unmethylated miR-200b in various cell lines. UV-detector; the partial denaturing temperature, 55.6°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S9. DHPLC chromatogram of methylated and unmethylated miR-193b in various cell lines Fluorescence-detector; the partial denaturing temperature, 56.5°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S10. DHPLC chromatogram of methylated and unmethylated miR-203 in various cell lines. UV-detector; the partial denaturing temperature, 57.7°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S11. Distribution of miR methylation in various gastric mucosa samples with and without H. pylori infection. Each line represents methylation status of one miR gene and each column represents one sample. The proportion of methylated allele is displayed step-wisely: negative (white square), positive and <30% (light gray square), or 30%–60% (deep gray square) or >60% (black square). Not informative sample is marked as “ND”. The case with or without H. pylori-specific 23 S rDNA was labeled with letter “p” (light green) and “n” (blue square). Figure S12. Comparison of levels of 7 mature miRNAs by quantitative RT-PCR in 20 pairs of fresh gastric carcinoma (GC) and the corresponding surgical margin (SM) tissue samples. The relative miRNA levels of the paired GC and SM samples from the same patient were linked by a line. Table S1. Classification of 129 CpG island-related human miR genes from miRBase (Release 14.0). Table S2. Summary of reported data on the inverse methylation-expression relationship for 9 representative miR CpG islands analyzed in the present study. Table S3. Primer sequences, amplicons, and PCR/DHPLC annealing/ denaturing temperatures used in the detection of the methylation status of 9 miR CpG islands. Table S4. Methylation status of 7 miR CpG islands in the paired gastric carcinoma (GC) and corresponding surgical margin (SM) samples from a total of 112 GC patients. Table S5. miRNA polyA primer for quantitative RT-PCR assays. Table S6. Comparison of the methylation status in 7 miR genes from the surgical margin (SM) samples of 112 GC patients with various clinicopathological characteristics. Table S7. Univariate analysis of the relationship between overall survival of GC patients and miR-9-1 methylation or other clinicopathological characteristics.
Contributor Information
Yantao Du, Email: duyantao@bjmu.edu.cn.
Zhaojun Liu, Email: liuzhaojun@bjmu.edu.cn.
Liankun Gu, Email: gulkun@163.com.
Jing Zhou, Email: jane72@163.com.
Bu-dong Zhu, Email: zhubd@tom.com.
Jiafu Ji, Email: jiafuj@hotmail.com.
Dajun Deng, Email: dengdajun@bjmu.edu.cn.
Acknowledgements
We thank Dr. Huidong Shi and Mr. James Wilson at Georgia Health Science University for English language editing. This work is supported by Natural Science Foundation of China (A3 Foresight Program No. 30921140311), National High Technology R&D Program 2006AA02A402, and National Basic Research Program of China (973 Program 2010CB529300 and 2011CB504201).
References
- Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114(3):457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Lu Y, Ryan SL, Elliott DJ, Bignell GR, Futreal PA, Ellison DW, Bailey S, Clifford SC. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS One. 2009;4(7):e6159. doi: 10.1371/journal.pone.0006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5–6):397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54(10):1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- Corthals SL, Jongen-Lavrencic M, de Knegt Y, Peeters JK, Beverloo HB, Lokhorst HM, Sonneveld P. Micro-RNA-15a and micro-RNA-16 expression and chromosome 13 deletions in multiple myeloma. Leuk Res. 2010;34(5):677–681. doi: 10.1016/j.leukres.2009.10.026. [DOI] [PubMed] [Google Scholar]
- Dijkstra MK, van Lom K, Tielemans D, Elstrodt F, Langerak AW, van ’t Veer MB, Jongen-Lavrencic M. 17p13/TP53 deletion in B-CLL patients is associated with microRNA-34a downregulation. Leukemia. 2009;23(3):625–627. doi: 10.1038/leu.2008.264. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y. et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6(3):e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010;38(20):6895–6905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67(21):10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31(5):766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, Susumu N, Aoki D, Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011;71(20):6450–6462. doi: 10.1158/0008-5472.CAN-11-0364. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, Kao HW, Fang WL, Huang KH, Chan WC. et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129(11):2600–2610. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29(42):5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Akiyama Y, Otsubo T, Shimada S, Yuasa Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 2010;31(5):777–784. doi: 10.1093/carcin/bgq013. [DOI] [PubMed] [Google Scholar]
- Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L, Wang Z. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124(2):325–334. doi: 10.1016/j.ygyno.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Deng D, Liu Z, Du Y. Epigenetic alterations as cancer diagnostic, prognostic, and predictive biomarkers. Adv Genet. 2010;71:125–176. doi: 10.1016/B978-0-12-380864-6.00005-5. [DOI] [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, Länger F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214(1):17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Chen X, Hu H, Guan X, Xiong G, Wang Y, Wang K, Li J, Xu X, Yang K, Bai Y. CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. Int J Cancer. 2012;130(7):17–24. doi: 10.1002/ijc.26171. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70(16):6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci. 2011;56(7):2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci. 2011;56(10):2849–2856. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T. et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31(12):2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Toyooka S, Soh J, Kubo T, Yamamoto H, Maki Y, Muraoka T, Shien K, Furukawa M, Ueno T. et al. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer. 2012;76(1):32–38. doi: 10.1016/j.lungcan.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Liu Z, Shen J, Zhang L, Shen L, Li Q, Zhang B, Zhou J, Gu L, Feng G, Ma J. et al. Prevalence of A2143G mutation of H. pylori-23S rRNA in Chinese subjects with and without clarithromycin use history. BMC Microbiol. 2008;8:81. doi: 10.1186/1471-2180-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh J, Lewington S, Thompson SG, Lowe GD, Collins R, Kostis JB, Wilson AC, Folsom AR, Wu K, Benderly M. et al. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: an individual participant meta-analysis. JAMA. 2005;294(14):1799–1809. doi: 10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- Eads CA, Laird PW. Combined bisulfite restriction analysis (COBRA) Methods Mol Biol. 2002;200:71–85. doi: 10.1385/1-59259-182-5:071. [DOI] [PubMed] [Google Scholar]
- Deng DJ, Deng GR, Smith MF, Zhou J, Xin HJ, Powell SM, Lu YY. Simultaneous detection of CpG methylation and single nucleotide polymorphism by denaturing high performance liquid chromatography. Nucleic Acids Res. 2002;30(3):13E. doi: 10.1093/nar/30.3.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami AR, Dickman MJ, Matin MM, Ashby JR, Brown PE, Conroy MJ, Fowler GJ, Rose JP, Sheikh QI, Yeung AT. et al. Use of fluorescent DNA-intercalating dyes in the analysis of DNA via ion-pair reversed-phase denaturing high-performance liquid chromatography. Anal Biochem. 2002;309(2):248–252. doi: 10.1016/S0003-2697(02)00306-8. [DOI] [PubMed] [Google Scholar]
- Luo D, Zhang B, Lv L, Xiang S, Liu Y, Ji J, Deng D. Methylation of CpG islands of p16 associated with progression of primary gastric carcinomas. Lab Invest. 2006;86(6):591–598. doi: 10.1038/labinvest.3700415. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Li L, Liu X, Wang D, Tu B, Wang L, Wang H, Zhu WG. 5-Aza-2'-deoxycytidine reactivates gene expression via degradation of pRb pocket proteins. FASEB J. 2011;26(1):449–459. doi: 10.1096/fj.11-190025. [DOI] [PubMed] [Google Scholar]
- Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71(8):3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhala HE, Jalava SE, Isotalo J, Bracken H, Lehmusvaara S, Tammela TL, Oja H, Visakorpi T. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127(6):1363–1372. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- Chim CS, Wong KY, Leung CY, Chung LP, Hui PK, Chan SY, Yu L. Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J Cell Mol Med. 2011;15(12):2760–2767. doi: 10.1111/j.1582-4934.2011.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M. et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70(6):2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL. et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6(10):1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz R, Silva J, García JM, Lorenzo Y, García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F. et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- Isik M, Korswagen HC, Berezikov E. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence. 2010;1(1):5. doi: 10.1186/1758-907X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikand K, Slane SD, Shukla GC. Intrinsic expression of host genes and intronic miRNAs in prostate carcinoma cells. Cancer Cell Int. 2009;9:21. doi: 10.1186/1475-2867-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6(4):323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kumar SM, Lu H, Liu A, Yang R, Pushparajan A, Guo W, Xu X. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-κB1-Snail1 pathway in melanoma. J Pathol. 2012;226(1):61–72. doi: 10.1002/path.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Deng H, Bi F, Liu J, Bemis LT, Norris D, Wang XJ, Zhang Q. MicroRNA-137 targets carboxyl-terminal binding protein 1 in melanoma cell lines. Int J Biol Sci. 2011;7(1):133–137. doi: 10.7150/ijbs.7.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin SM, Stone RA, Bunker CH, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 2010;31(5):864–870. doi: 10.1093/carcin/bgq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin SM, Stone RA, Bunker CH, Lyons-Weiler MA, Laframboise WA, Kelly L, Seethala RR, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer. 2011;117(7):1454–1462. doi: 10.1002/cncr.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Cho WC. Epigenetic alteration of microRNAs in feces of colorectal cancer and its clinical significance. Expert Rev Mol Diagn. 2011;11(7):691–694. doi: 10.1586/erm.11.57. [DOI] [PubMed] [Google Scholar]
- Kubo T, Toyooka S, Tsukuda K, Sakaguchi M, Fukazawa T, Soh J, Asano H, Ueno T, Muraoka T, Yamamoto H. et al. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin Cancer Res. 2011;17(15):4965–4974. doi: 10.1158/1078-0432.CCR-10-3040. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Emoto N, Hamano E, Sunohara M, Kawakami M, Kage H, Kitano K, Nakajima J, Goto A, Fukayama M. et al. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer. 2012;130(11):2580–2590. doi: 10.1002/ijc.26254. [DOI] [PubMed] [Google Scholar]
- Mazar J, Khaitan D, DeBlasio D, Zhong C, Govindarajan SS, Kopanathi S, Zhang S, Ray A, Perera RJ. Epigenetic regulation of microRNA genes and the role of miR-34b in cell invasion and motility in human melanoma. PLoS One. 2011;6(9):e24922. doi: 10.1371/journal.pone.0024922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T. et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31(12):2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D. et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15(16):5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31(16):2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, Borges M, Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70(13):5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C. et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54(6):2064–2075. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286(1):420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lages E, Guttin A, El Atifi M, Ramus C, Ipas H, Dupré I, Rolland D, Salon C, Godfraind C, de Fraipont F. et al. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One. 2011;6(5):e20600. doi: 10.1371/journal.pone.0020600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H. et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70(22):9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- Mazar J, DeBlasio D, Govindarajan SS, Zhang S, Perera RJ. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011;585(15):2467–2476. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Craig VJ, Cogliatti SB, Rehrauer H, Wündisch T, Müller A. Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res. 2011;71(10):3616–3624. doi: 10.1158/0008-5472.CAN-10-3907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Illustrations of 9 tested miR-hosting CpG islands. Open frame, location of pre-miRNA transcript. Open arrow, pre-miRNA transcript direction; Blue-underline, locations of the forward and reversed primer-matching regions used to detect methylation of CpG island with DHPLC and bisulfite sequencing; Purple vertical bars, CpG sites. Figure S2. DHPLC chromatogram of methylated and unmethylated miR-9-1 in various cell lines. UV-detector; the partial denaturing temperature, 55.4°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S3. DHPLC chromatogram of methylated and unmethylated miR-9-3 in various cell lines. UV-detector; the partial denaturing temperature, 58.5°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S4. DHPLC chromatogram of methylated and unmethylated miR-34b in various cell lines. UV-detector; the partial denaturing temperature, 56.8°C; U, peak for the unmethylated PCR products; M (partial or full), peak for the partially or fully methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S5. DHPLC chromatogram of methylated and unmethylated miR-210 in various cell lines. Fluorescence-detector; the partial denaturing temperature, 58.7°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S6. DHPLC chromatogram of methylated and unmethylated miR-137 in various cell lines. Fluorescence-detector; the partial denaturing temperature, 58.3°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S7. DHPLC chromatogram of methylated and unmethylated miR-375 in various cell lines. UV-detector; the partial denaturing temperature, 55.7°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S8. DHPLC chromatogram of methylated and unmethylated miR-200b in various cell lines. UV-detector; the partial denaturing temperature, 55.6°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S9. DHPLC chromatogram of methylated and unmethylated miR-193b in various cell lines Fluorescence-detector; the partial denaturing temperature, 56.5°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S10. DHPLC chromatogram of methylated and unmethylated miR-203 in various cell lines. UV-detector; the partial denaturing temperature, 57.7°C; U, peak for the unmethylated PCR products; M, peak for the methylated PCR products; B, peripheral blood DNA; MB, M.sssI-methylated blood DNA. Figure S11. Distribution of miR methylation in various gastric mucosa samples with and without H. pylori infection. Each line represents methylation status of one miR gene and each column represents one sample. The proportion of methylated allele is displayed step-wisely: negative (white square), positive and <30% (light gray square), or 30%–60% (deep gray square) or >60% (black square). Not informative sample is marked as “ND”. The case with or without H. pylori-specific 23 S rDNA was labeled with letter “p” (light green) and “n” (blue square). Figure S12. Comparison of levels of 7 mature miRNAs by quantitative RT-PCR in 20 pairs of fresh gastric carcinoma (GC) and the corresponding surgical margin (SM) tissue samples. The relative miRNA levels of the paired GC and SM samples from the same patient were linked by a line. Table S1. Classification of 129 CpG island-related human miR genes from miRBase (Release 14.0). Table S2. Summary of reported data on the inverse methylation-expression relationship for 9 representative miR CpG islands analyzed in the present study. Table S3. Primer sequences, amplicons, and PCR/DHPLC annealing/ denaturing temperatures used in the detection of the methylation status of 9 miR CpG islands. Table S4. Methylation status of 7 miR CpG islands in the paired gastric carcinoma (GC) and corresponding surgical margin (SM) samples from a total of 112 GC patients. Table S5. miRNA polyA primer for quantitative RT-PCR assays. Table S6. Comparison of the methylation status in 7 miR genes from the surgical margin (SM) samples of 112 GC patients with various clinicopathological characteristics. Table S7. Univariate analysis of the relationship between overall survival of GC patients and miR-9-1 methylation or other clinicopathological characteristics.