Abstract
The estrogen receptor (ER)β1 is successively lost during cancer progression, whereas its splice variant, ERβ2, is expressed in advanced prostate cancer. The latter form of cancer often metastasizes to bone, and we wanted to investigate whether the loss of ERβ1 and/or the expression of ERβ2 affect such signaling pathways in prostate cancer. Using PC3 and 22Rv1 prostate cancer cell lines that stably express ERβ1 or ERβ2, we found that the ERβ variants differentially regulate genes known to affect tumor behavior. We found that ERβ1 repressed the expression of the bone metastasis regulator Runx2 in PC3 cells. By contrast, RUNX2 expression was up-regulated at the mRNA level by ERβ2 in PC3 cells, whereas Slug was up-regulated by ERβ2 in both PC3 and 22Rv1 cells. In addition, the expression of Twist1, a factor whose expression strongly correlates with high Gleason grade prostate carcinoma, was increased by ERβ2. In agreement with the increased Twist1 expression, we found increased expression of Dickkopf homolog 1; Dickkopf homolog 1 is a factor that has been shown to increase the RANK ligand/osteoprotegerin ratio and enhance osteoclastogenesis, indicating that the expression of ERβ2 can cause osteolytic cancer. Furthermore, we found that only ERβ1 inhibited proliferation, whereas ERβ2 increased proliferation. The expression of the proliferation markers Cyclin E, c-Myc, and p45Skp2 was differentially affected by ERβ1 and ERβ2 expression. In addition, nuclear β-catenin protein and its mRNA levels were reduced by ERβ1 expression. In conclusion, we found that ERβ1 inhibited proliferation and factors known to be involved in bone metastasis, whereas ERβ2 increased proliferation and up-regulated factors involved in bone metastasis. Thus, in prostate cancer cells, ERβ2 has oncogenic abilities that are in strong contrast to the tumor-suppressing effects of ERβ1.
Prostate cancer is the most frequently diagnosed cancer in men in the Western world and the second leading cause of cancer-related death in men. Unlike many other cancer types, prostate cancer is a slow-progressing disease and usually takes many years to manifest. In the early stages of prostate cancer, androgen ablation is the frontline adjuvant treatment. In the advanced stage, prostate cancer becomes androgen independent and exhibits an increased propensity to metastasize to bone, resulting in debilitating skeletal complications (1). Gene polymorphisms in the estrogen receptor (ER)β1 locus have been shown to be associated with prostate cancer risk (2). The ERβ1 knockout mouse exhibits increased hyperplasia in the prostate, indicating the importance of ERβ1 for maintaining a normal prostate (3). ERβ1 has also been shown to act as a tumor suppressor in the prostate (4), and its expression declines during the progression of cancer (5, 6). Furthermore, the loss of ERβ1 is sufficient to promote the epithelial-to-mesenchymal transition (4), indicating that ERβ1 is antimetastatic. ERβ has several splice variants; ERβ1 is the main variant, also referred to as ERβ wild type, and ERβ2 and ERβ5 are the most studied splice variants (7, 8). The ERβ2 splice variant is exclusive for primates, and this variant has been shown to be related to poor prognosis and to promote cell invasion by the prostate cancer cell line PC3 (8). ERβ2 differs from ERβ1 at the C terminus, where the ligand-binding domain has been truncated and partially replaced by a new exon specific for ERβ2 (9). Although this compromises the ligand-binding domain, the functional DNA-binding domain and the intact N-terminal domain suggest that ERβ2 can participate in gene regulation.
The prostate cancer cell line PC3 is often used as a model to study bone metastasis and was originally isolated from a bone-metastatic prostate cancer (10). This cell line expresses high levels of Runt-related transcription factor (RUNX2), an osseous master transcription factor that is important during bone metastasis of prostate cancer (11, 12). The basic helix-loop-helix transcription factor, Twist1, is highly increased in malignant prostate cancer, and its expression correlates with a higher Gleason grade (13). Twist1 also promotes prostate cancer metastasis to bone by promoting osteoclast differentiation, partly by regulating the expression of Dickkopf homolog 1 (DKK1) (14). DKK1 is a soluble inhibitor of Wnt signaling, and its expression decreases bone formation by inhibition of osteoprotegerin secretion from osteoblasts present in the bone leading to stimulation of osteoclasts (15). We set out to further dissect the mechanisms underlying the effects of ERβ1 and ERβ2 on proliferation and metastatic ability using PC3 cells as a model system for androgen receptor (AR) negative prostate cancer cells and the 22Rv1 cell line as an AR positive prostate cancer cell line, which in addition is a model for castration-resistant prostate cancer (16). Furthermore, in this cell line, AR is present in two forms, where one is a full-length and another is a truncated splice variant, explaining its androgen independent growth (16).
Materials and Methods
Cell culture and generation of stable ERβ-expressing cells
The PC3 and 22Rv1 cell lines was obtained from the American Type Culture Collection (Manassas, VA) and maintained in RPMI 1640 (Invitrogen, Inc., Carlsbad, CA) medium supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO), 2 mm l-glutamine, and 25 mm HEPES buffer (Invitrogen, Inc.). For ligand treatment, the medium was changed to phenol red-free RPMI 1640 supplemented with 10% dextran-coated charcoal-treated FBS (Sigma). All experiments used the cells below passage 30.
Construction of an inducible system for ERβ1 and ERβ2
Prostate cancer cell lines contain minute amounts of endogenous ERβ1 or ERβ2. Therefore, a transposon-based system was used to stably transfect PC3 prostate cancer cells with the ERβ constructs (PC3-ERβ1 and PC3-ERβ2) or with empty vector (PC3-control) as a control. The transposon-based system consisted of two plasmids: 1) pIR-tetracycline response element (TRE), which contains a pair of transposon elements (inverted repeat, IR) to allow the efficient integration of the plasmid and the gene (ERβ1 or ERβ2) into the genomic DNA, and 2) piggybac, which expresses the transposase enzyme required for the integration of the transposon element. The pIR-TRE plasmid also contained a bidirectional promoter expressing the green fluorescence protein gene in one direction and the tetracycline-regulated ERβ (Tet-off) gene in the other direction. The transactivator for Tet-off system regulation via the TRE was from a third plasmid, pTAN. The ERβ genes were cloned into the PvuII/NheI (Promega, Madison, WI) site of pIR-TRE. All of the plasmids were transfected using Lipofectamine 2000 (Invitrogen, Inc.) in Opti-MEM medium (Invitrogen, Inc.) according to the manufacturer's protocol. Complete RPMI 1640 medium with 10% FBS medium was added after 6 h of incubation with the transfection reagents.
Cell sorting
The cells were initially grown in normal medium containing 250 ng/ml doxycycline and 0.5 μg/ml puromycin to select stable integration. Before cell sorting, the doxycycline and puromycin were removed from the cells for 24 h to allow ERβ expression. For cell sorting, the cells were trypsinized and incubated for 5 min in a 37 C incubator. The trypsin was neutralized by adding complete RPMI 1640 medium followed by centrifugation of the cells at 1500 × g for 5 min. The cell pellets were then resuspended in 1% FBS in PBS with penicillin-streptomycin and sorted for green fluorescence protein expression using FACS (BD FACSAria II; BD Biosciences, San Jose, CA) under aseptic conditions. The cells were collected in RPMI 1640 medium with penicillin-streptomycin. The sorted cells were further centrifuged at 1500 × g for 5 min and plated in flasks with complete RPMI 1640 medium containing 0.5 μg/ml puromycin and 250 ng/ml doxycycline to expand the cell number.
Transient transfections
PC3 or 22Rv1 cells at 60–70% confluency were transfected in 12-well plates. Two hours before the transfection, the cells were fed with serum-free Opti-MEM medium. The transfection was performed using Lipofectamine 2000 (Invitrogen, Inc.) according to the manufacturer's protocol. Six hours after the transfection, the cells were fed with complete RPMI 1640 medium containing 10% FBS or 10% dextran-coated charcoal serum according to the assay requirements. The cells were harvested after 24 h for analysis.
Cell proliferation assay
To investigate the ERβ1/ERβ2-mediated effects on cellular proliferation, PC3 and 22Rv1 cells stably expressing ERβ1, ERβ2, or the control vector were plated in 12-well cell culture plates in RPMI 1640 either with 0 or 10% FBS. The plating efficiency was similar among the different plates. After 48 h, the cells were counted both manually with a Neubauer chamber and also with a Countess Automated Cell Counter (Invitrogen, Inc.). In addition, cell proliferation was quantified by an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay on a 96-well plate, as described in the manufacturer's protocol (Promega). The experiments were performed in triplicate and were repeated three times.
Protein extract preparation
Nuclear extracts from PC3 cells were prepared as described previously (17). To prepare whole-cell extracts, the cells were washed twice with PBS, lysed in 10 times packed cell volume of lysis buffer [0.1% Nonidet P-40 (NP-40), 250 mm KCl, 5 mm HEPES (pH 7.9), 10% (vol/vol) glycerol, 4 mm NaF, 4 mm sodium orthovanadate, 0.2 mm EDTA, 0.2 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor cocktail, and PhosStop (Roche, Indianapolis, IN)] for 30 min on ice and then centrifuged at 14,000 × g for 10 min.
Western blotting and antibodies
Fifty micrograms of protein were loaded on an SDS-PAGE 4–20% Bis-Tris gradient gel or a 10% Bis-Tris gel with Tris running buffer (18) and transferred to a nitrocellulose membrane after electrophoretic separation. The membranes were blocked with 5% nonfat powdered milk in 0.1% Tris-buffered saline with Tween 20 buffer. The blots were then probed with various primary antibodies, such as ERβ (clone 14C8; Abcam, Cambridge, MA), Twist1 (sc-15393), Slug (sc-166476), Cyclin E (sc-247), β-catenin (sc-7963), p45Skp2 (sc-7164), p21WAF1/CIP1 (sc-817), p27Kip1 (sc-1641), and DKK1 (sc-25516) (all from Santa Cruz Biotechnology, Inc., Santa Cruz, CA), the p-retinoblastoma (Rb)-antibody sampler kit (Cell Signaling, Danvers, MA), phospho-p27Kip1 (R&D Systems, Minneapolis, MN), and Runx2/Cbfal (MBL D130-3) and Cyclin D1 (RB-212-P1; LabVision, Thermo Fisher Scientific, Inc., Fremont, CA). The primary antibodies were used at dilutions of 1:200–1000, and the secondary antibody was used at a dilution of 1:10,000.
RNA extraction and quantitative real-time PCR (qPCR)
RNA extraction was performed with TRIzol reagent (Invitrogen, Inc.) and chloroform extraction according to the manufacturer's protocol. cDNA was synthesized from 1 μg of the total RNA with the Superscript First-Strand System according to the standard protocol (Invitrogen, Inc.). Real-time PCR was performed with the SYBR Green I dye master mix (Applied Biosystems, Foster City, CA). The following primers were used: 18s rRNA forward (F), 5′-CCT GCG GCT TAA TTT GAC TCA-3′ and reverse (R), 5′-AGC TAT CAA TCT GTC AAT CCT GTC C-3; glyceraldehyde-3-phosphate dehydrogenase F, 5′-TGA CAA CTT TGG TAT YCG TGG AAG G-3′ and R, 5′-AGG CAG GGA TGA TGT TCT GGA GAG-3′ (as reference genes); ERβ1 F, 5′-ACT TGC TGA ACG CCG TGA CC-3′ and R, 5′-CAG ATG TTC CAT GCC CTT GTT-3′; ERβ2 F, 5-ACT TGC TGA ACG CCG TGA CC-3 and R, 5′-CCA TCG TTG CTT CAG GCA A-3′; Slug F, 5′-AGC AGT TGC ACT GTG ATG CC-3′ and R, 5′-ACA CAG CAG CCA GAT TCC TC-3′; β-catenin F, 5′-TTA AAC TCC TGC ACC CAC CAT-3′ and R, 5′-AGG GCA AGG TTT CGA ATC AA-3′; Twist1 F, 5′-GGA GTC CGC AGT CTT ACG AG-3′ and R, 5′-TCT GGA GGA CCT GGT AGA GG-3′; c-MYC F, 5′-CTG GTG CTC CAT GAG GAG AC-3′ and R, 5′-CTT TTC CAC AGA AAC AAC ATC-3′; Cyclin D1 F, 5′-CCG TCC ATG CGG AAG ATC-3′ and R, 5′-ATG GCC AGC GGG AAG AC-3′; RUNX2 F, 5′-AAC TCA AGT CCC CCG CCT CCC-3′ and R, 5′-GCC ACG GGC AGG GTC TTG TT-3′; and DKK1 F, 5′-AAC GCT GCA TGC GTC ACG CTA-3′ and R, 5′-TCC TGA GGC ACA GTC TGA TGA CCG-3′. All the primers were ordered from Integrated DNA Technologies, Inc. (Coralville, IA). The qPCR reactions were performed with a 7500 Fast Real-Time PCR System (Applied Biosystems) using optimized conditions for the SYBR Green I dye system: 50 C for 2 min, 95 C for 10 min, followed by 40–50 cycles at 95 C for 15 sec and 60 C for 50 sec. The optimum concentration of primers was determined in preliminary experiments, and the amplification specificity was confirmed by dissociation curve analysis.
Immunocytochemistry
Immunostaining was performed as described previously (19). In brief, the cells were grown on autoclaved coverslips. Twenty-four hours after treatment, the cells were washed twice with PBS. The cells were treated with 4% formalin in PBS followed by 0.25% Triton X-100. The cells were then blocked with 3% BSA. The coverslips containing the cells were placed in a humidified chamber, and primary antibodies were added at a 1:50–200 dilution. The cells were incubated at 37 C for 1 h. After incubation, the cells were washed three times with PBS. Fluorescence-labeled (Texas red or Cy3 dye) secondary antibody was added at a 1:1000 dilution and incubated at 37 C for 1 h. The cells were further washed five times with PBS and nuclear stained with 4′,6-diamidino-2-phenylindole (DAPI) for 1 min. The coverslips were then mounted on a slide with fluorescence mounting solution (Dako, Carpinteria, CA). Fluorescent images were captured using an Olympus BX51 microscope fitted with XM10 and BP25 cameras (Olympus, Tokyo, Japan). Primary antibodies used for immunocytochemistry are custom-made ERβ1 (503) and ERβ2 (3257).
Immunohistochemistry
To examine the number of proliferating cells in ERβ1/β2-expressing tumor xenografts, immunohistochemistry with a Ki67 antibody was performed. Antigen retrieval was done in PT module (ThermoScientific) in 0.01 mol/liter citric acid (pH 6.0) for 10 min. Endogenous peroxidase activity was blocked by incubating sections shaking in 3% hydrogen peroxide in 50% methanol for 30 min and then blocked in 3% BSA and 0.1% NP-40 for 10 min at room temperature. Primary antibody Ki67 (Abcam) was diluted 1:200 in 3% BSA and 0.1% NP-40 and incubated overnight at 4 C. Sections were then washed consecutively in 0.1% NP-40 for 30 min, and with PBS, followed by incubation with a rabbit-on-rodent horseradish peroxidase polymer (Biocare Medical, Concord, CA) for 30 min at room temperature. After this, sections were washed consecutively in 0.1% NP-40 for 30 min, and with PBS for 10 min. The slides are then stained in 3,3′-diaminobenzidine and counterstained with Mayer hematoxylin (Sigma) before dehydration through ethanol, and mounted in Pertex (Histolab, Göteborg, Sweden). The number of Ki67-positive cells was counted in three independent fields per slide with ×20 objective. The average number of positive cells and sd were calculated for each group.
Immunoprecipitation
Equal concentrations of protein lysates were incubated with 1 μg of rabbit ERβ LBD (ligand binding domain) antibody (produced in the laboratory) overnight, and the immune complexes were adsorbed onto protein A-sepharose beads for 4 h. The beads were pelleted at 5000 × g for 30 sec on a bench-top centrifuge at 4 C. The supernatant was removed and the pellet washed three times with ice-cold Tris-buffered saline buffer [50 mm Tris HCL and 150 mm NaCl (pH 7.4)]. The immune complexes were then boiled with 20 μl of 2× sample buffer [125 mm Tris HCl (pH 6.8), with 4% SDA, 20% (vol/vol) glycerol, and 0.004% bromophenol blue] and subjected to SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis). The proteins were then transferred to a nitrocellulose membrane for Western blot analyses. The blot was then probed with ERβ antibody (clone 14C8; Abcam). The primary antibodies were used at a dilution of 1:1000, and the secondary antibody was used at a dilution of 1:10 000.
Bromodeoxyuridine (BrdU) incorporation assay
The BrdU incorporation assay was performed as described previously (20). Briefly, the cells were grown for 24 h on Nunc Lab-Tek chamber slides (Thermo Scientific, Rockford, IL) without doxycycline. The cells were pulse labeled with 10 μm BrdU (Sigma) for 5 h and were then fixed with 4% paraformaldehyde at room temperature. The DNA was denatured with 2 m HCl in PBS containing 0.5% Triton X-100 at 37 C for 30 min. The cells were then blocked with 3% BSA in PBS and incubated with a Texas red-conjugated primary BrdU antibody (K404-60; BioVision, Mountain View, CA) in 3% BSA at 37 C for 1 h. The cells were counterstained with DAPI (Invitrogen, Inc.). Images were captured with Olympus BX51 microscope fitted with XM10 and BP25 cameras.
Microarray and bioinformatic analyses
A two-color comparative microarray technique was used that covers the fully known transcriptome of 39,600 transcripts and variants by using Operon's long-oligonucleotide spotted array. In addition, the array was complemented with 133 oligonucleotides specifically synthesized for a detailed and robust analysis of nuclear receptors, splice variants, and coregulators. Each comparison was replicated and dye swapped. In addition, we performed a duplicate dye-swap comparison and qPCR controls of the control for comparison, and to identify instances of differential expression caused by the system itself. The microarray analysis was performed essentially as described by Richter et al. (22). For each cDNA synthesis, 20 μg of total RNA were used. The samples to be cohybridized on a slide were pooled, and direct comparisons were performed using replicated samples, cDNA synthesis, and hybridizations with the Cy5/Cy3 dye assignments reversed. Five hybridizations (each comparison replicated) were performed. The human OpArray arrays were purchased from Microarrays, Inc. (Huntsville, AL). Bioinformatic analyses were performed using Pathway Studio software (Ariadne Genomics, Rockville, MD). Enrichment analysis of the gene ontology (GO) of the biological processes was performed with Fisher's exact test using the software's gene sets. The pathway pictures were also designed in Pathway Studio. The microarray data is available in NCBI's Gene Expression Omnibus through accession number GSE35090.
Experimental animals and xenograft model
PC3 prostate cancer cells expressing ERβ1 or ERβ2 were implanted into immunodeficient mice (ν/ν) to investigate tumor growth. PC3-ERβ1/ERβ2/control cells (5 × 105) were diluted in 50-μl RPMI 1640 medium + 50 μl of Matrigel (BD Biosciences) and injected sc into the flank region of 5- to 6-wk-old pathogen-free nude mice (ν/ν) (Taconic, Hudson, NY) on d 0. After 40 d, the mice were killed, and the tumor weight and size were measured. All tumors were fixed in 4% paraformaldehyde and stored in 70% ethanol. After this, tissues were paraffin embedded and subsequently sliced into 4-μm sections according to standard protocol.
Statistics
The values are expressed as the mean with 95% confidence intervals. An unpaired two-tailed t test was used to compare the differences between two groups. The significance is presented as *, P < 0.05; **, P < 0.005; and ***, P < 0.001, and nonsignificant differences are presented as ns.
Results
The expression of ERs in the prostate cancer cell line PC3
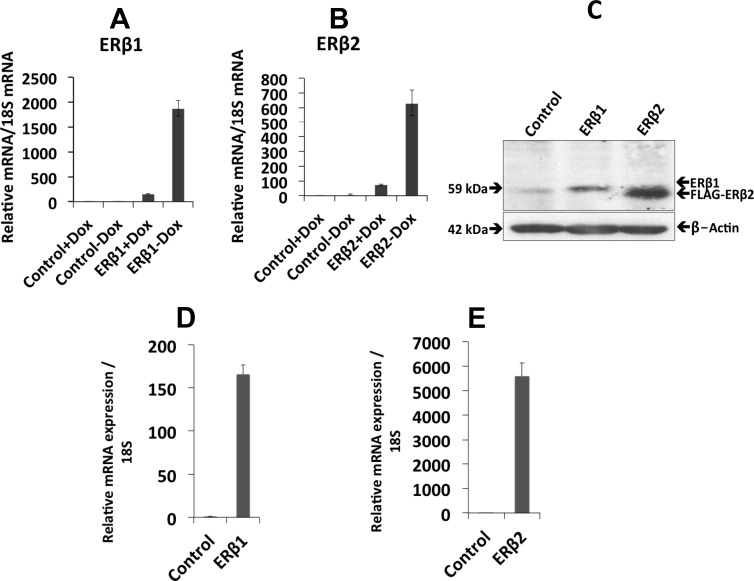
Prostate cancer cells lose the expression of ERβ1 during disease progression (21), and cells isolated from prostate cancer only express minute amounts of ERβ1. ERβ2, however, is expressed in advanced prostate cancer (8). To study how ERβ1 and its splice variant ERβ2 regulate genes involved in prostate cancer progression and metastasis to bone, we developed a doxycycline-regulated expression system for these proteins in PC3 cells. After the removal of doxycycline, we observed a robust induction of ERβ1 and ERβ2 at the mRNA level by qPCR in PC3 cells (Fig. 1, A and B), in 22Rv1 cells (Fig. 1, D and E), and at the protein level in PC3 cells by Western blotting (Fig. 1C). Immunocytochemistry of ERβ1- and ERβ2-expressing cells showed nuclear localization of the proteins (Supplemental Fig. 1, C and D, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). A new custom-made ERβ2 antibody was verified using the 22Rv1 cells stably expressing ERβ2 by Western blotting and immunocytochemistry (Supplemental Fig. 1, A and B). In addition, the expression of ERβ1 and ERβ2 in the stably transfected PC3 cells was verified by immunoprecipitation followed by Western blotting using ERβ (14c8) antibody (Supplemental Fig. 2).
Fig. 1.
Stable expression of ERβ1 and ERβ2. A and B, qPCR analysis of ERβ1 and ERβ2 mRNA in stably transfected PC3 cells. Control +Dox (Doxycycline) (bar 1), control −Dox (bar 2), ERβ1/β2 +Dox (bar 3), and ERβ1/β2 −Dox (bar 4). C, The protein expression of ERβ1 and ERβ2 was verified by Western blotting using the ERβ antibody 14C8 (Abcam). Lane 1 represents the control cells, lane 2 ERβ1-expressing cells, and lane 3 ERβ2-expressing cells. The FLAG-ERβ2 band is slightly lower than the ERβ1 band. β-Actin was used as a loading control. D and E, qPCR analysis of ERβ1 and ERβ2 mRNA in stably transfected 22Rv1 cells. All the experiments were performed n = 3.
Genome-wide gene regulation mediated by ERβ1 and ERβ2
For an extensive comparison of ERβ1 and ERβ2 functions, we performed microarray analyses of PC3 cells. For ERβ1, we compared the transcriptome changes in one mixed-clone cell population and one single-clone cell population, respectively, with control cells expressing empty vector. We observed statistically significant up-regulations of 115 genes and down-regulations of 196 genes. The top-ranked 15 most significantly regulated genes by ERβ1 are represented in Table 2. Strikingly, various proliferation-associated genes were highly overrepresented (e.g. positive regulation of cell proliferation and nucleosome assembly, P = 1.05 × 10−3 and P = 3.21 × 10−30, respectively). In the gene group “positive regulation of cell proliferation” that was down-regulated by ERβ1 (Table 1), we find cell-cycle genes, such as p45Skp2 (SKP2), Cyclin A2 (CCNA2), CDK14, CDCA7, CDCA4, and CKS1B (Supplemental Table 1). A marked decrease in growth-promoting factors was further strongly indicated by the down-regulation of numerous histone, ribosome, and chaperone family members or subunits. In addition, apoptosis-related genes were significantly overrepresented among the up-regulated genes (P = 2.60 × 10−5) (Table 1), suggesting that these functions are regulated by ERβ1. In addition, HIPK2, which encodes a p53-stabilizing kinase (23), was up-regulated. The chromatin-binding factor HMGB1 was down-regulated; HMGB1 is proposed to interact with various transcription factors, such as p53, Rb, and ERα, and its down-regulation contributes to apoptosis in LNCaP prostate cancer cells (24). Cursory microarray of ERβ1 with 17β-estradiol did not reveal any ligand-dependent gene regulation (microarray data not shown). Representative qPCR of RUNX2, HOXB13, PLAU, and LRH-1 (Supplemental Fig. 3, A–D) supports that overexpressed ERβ1 has no or little ligand-dependent gene-regulatory function in prostate cancer cells. This confirms our previous findings in colon cancer cells (25).
Table 2.
Top-ranked 15 up-regulated and down-regulated genes by ERβ1 according to the adjusted P value
| Gene symbol | Log FC | P value |
|---|---|---|
| Up-regulated | ||
| RAB5B | 0.84 | 6.65E-03 |
| STAT6 | 1.34 | 9.71E-03 |
| HIPK2 | 1.24 | 1.29E-02 |
| LTBP1 | 1.08 | 1.68E-02 |
| PIK3R3 | 1.23 | 1.75E-02 |
| KMO, OPN3 | 1.10 | 1.77E-02 |
| ARCN1 | 0.95 | 2.40E-02 |
| C10orf116 | 1.45 | 2.40E-02 |
| PNRC2 | 0.70 | 2.56E-02 |
| RTN3 | 0.86 | 2.56E-02 |
| CHPF2 | 0.59 | 2.60E-02 |
| KIAA0913 | 0.98 | 2.60E-02 |
| COX6A1 | 0.58 | 2.64E-02 |
| MAST2 | 0.64 | 2.64E-02 |
| GATS, GATSL2, GTF2IP1, LOC100093631 | 0.66 | 2.64E-02 |
| Down-regulated | ||
| HIST1H3D, HIST1H2AD | −1.33 | 6.65E-03 |
| GCSH, LOC100329108 | −1.25 | 6.65E-03 |
| HIST1H3H | −1.09 | 6.65E-03 |
| HIST2H2BF | −1.01 | 6.65E-03 |
| ATP13A3 | −0.84 | 6.65E-03 |
| GMPS | −1.01 | 9.71E-03 |
| HIST1H2AE | −1.26 | 1.29E-02 |
| HIST1H4H | −1.05 | 1.29E-02 |
| HIST1H3G | −1.34 | 1.30E-02 |
| HIST1H2AH | −1.31 | 1.68E-02 |
| RPS15A | −0.90 | 1.68E-02 |
| AKT3 | −0.90 | 1.68E-02 |
| FBN2 | −1.29 | 1.77E-02 |
| DNAJC19 | −1.00 | 1.77E-02 |
| HIST1H4E | −1.03 | 2.33E-02 |
Multiple entries on the same line indicate probe mapping to multiple locations in the genome; the first entry represents the first hit in the BLAST query. LogFC, Logarithmic fold change.
Table 1.
Top 15 overrepresented GO biological processes among the up-regulated and down-regulated genes in the ERβ1 vs. control PC3 cells in the microarray comparison
| GO biological process | Accession | P value |
|---|---|---|
| Up-regulated | ||
| Regulation of transcription | 0006351 | 1.42E-07 |
| Transcription | 0006355 | 5.07E-07 |
| Response to testosterone stimulus | 0033574 | 4.82E-06 |
| Apoptosis | 0006915 | 2.60E-05 |
| Positive regulation of transcription from the RNA pol II promoter | 0045944 | 4.47E-05 |
| Positive regulation of apoptosis | 0043065 | 2.85E-04 |
| Artery morphogenesis | 0048844 | 8.80E-04 |
| Positive regulation of DNA binding | 0043388 | 1.91E-03 |
| Response to drug | 0042493 | 2.00E-03 |
| Nervous system development | 0007399 | 2.81E-03 |
| Endothelial cell fate specification | 0060847 | 2.95E-03 |
| Vitellogenesis | 0007296 | 2.95E-03 |
| Positive regulation of erythrocyte aggregation | 0034120 | 2.95E-03 |
| Actin polymerization-dependent cell motility | 0070358 | 2.95E-03 |
| Calcitonin catabolic process | 0010816 | 2.95E-03 |
| Down-regulated | ||
| Nucleosome assembly | 0006334 | 3.21E-30 |
| Translational elongation | 0006414 | 1.03E-04 |
| Nucleosome positioning | 0016584 | 3.52E-04 |
| Protein folding | 0006457 | 5.18E-04 |
| Organ regeneration | 0031100 | 7.14E-04 |
| Positive regulation of cell proliferation | 0008284 | 1.05E-03 |
| Protein polyubiquitination | 0000209 | 1.90E-03 |
| DNA unwinding involved in replication | 0000209 | 2.73E-03 |
| rRNA processing | 0006364 | 3.00E-03 |
| Translational initiation | 0006413 | 3.05E-03 |
| Positive regulation of caspase activity | 0043280 | 3.08E-03 |
| Translation | 0006412 | 3.17E-03 |
| Fatty acid transport | 0015908 | 3.85E-03 |
| Cell division | 0051301 | 4.58E-03 |
| Hepatic duct development | 0061011 | 4.89E-03 |
Pathway Studio enrichment analysis of GO groups with genes identified as differentially expressed as described in Materials and Methods. The P values were calculated with Fisher's exact test.
For ERβ2, we separately compared one mixed-clone cell population and one single-clone cell population with control cells. Although they shared regulatory events, we chose to focus on the single-clone population, because it expressed higher levels of ERβ2 (data not shown), thus exhibiting a more pronounced effect. The phenotypes of both populations were similar, with a dose-dependent-like response to the detected ERβ2 levels. Intriguingly, among the genes that were differentially expressed as a result of ERβ2 expression, there was an enrichment of genes associated with migratory or invasive behavior. The overrepresented biological processes included cell adhesion, angiogenesis, and the positive regulation of cell migration (P = 2.85 × 10−12, P = 1.81 × 10−8, and P = and 9.73 × 10−8, respectively) (Supplemental Table 2). Notably, THBS1, which is decreased in angiogenic tumor cells (26), was down-regulated as were 31 genes associated with cell adhesion. Furthermore, we observed the up-regulation of MGAT5, which is known to enhance the invasion ability of prostate cancer cells via the activation of matriptase (27). HOXB13 was also up-regulated by ERβ2 and has previously been shown to be estrogen responsive via ERα in breast cancer cells (28). Furthermore, HOXB13 regulates the response to androgen-regulated migration in prostate cells (29). For ERβ2, we also observed the regulation of proliferation- and apoptosis-linked genes, with an overrepresentation of negative and positive regulation of cell proliferation and negative regulation of apoptosis (P = 1.36 × 10−5, P = 4.87 × 10−5, and P = and 3.24 × 10−8, respectively). In particular, the cell-cycle regulator CDKN1A was down-regulated by ERβ2. We observed only a few similarities or opposing actions between the two ERβ splice variants in terms of regulating specific genes. For example, we noted that PDCD10, which encodes a kinase involved in ERK signaling, was up-regulated in ERβ2-expressing cells but was down-regulated in ERβ1-expressing cells. PDCD10 is implicated in apoptosis, and knockdown of its endogenous expression inhibits PC3 cell growth (30).
The effects of ERβ1 and ERβ2 expression on cellular proliferation
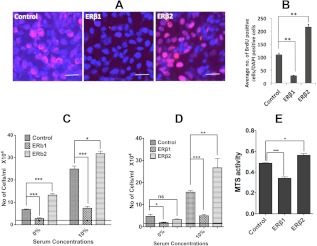
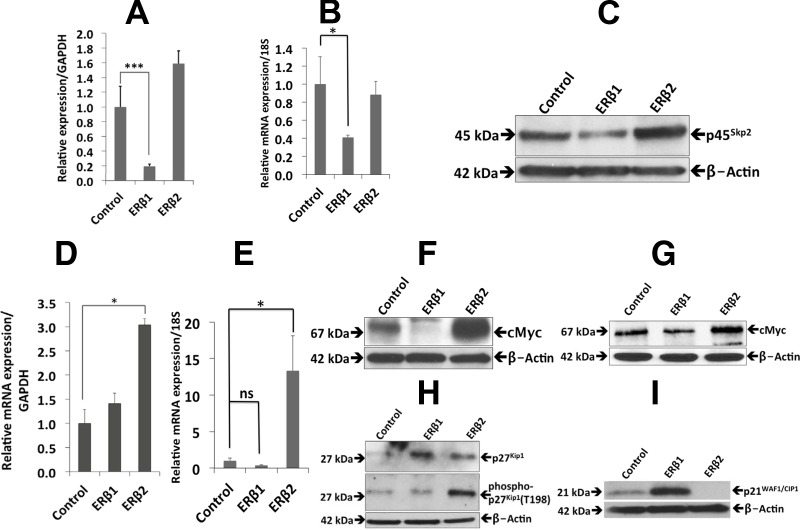
Because the transcriptome analysis indicated that proliferation was affected by the expression of ERβ1, we examined this using BrdU immunostaining. We found a decrease in BrdU incorporation in cells expressing ERβ1, whereas there was a substantial increase in BrdU incorporation in cells expressing ERβ2 compared with the control PC3 cells (Fig. 2, A and B). We further examined cell proliferation by MTS assays, which also demonstrated an antiproliferative effect of ERβ1 and a proproliferative effect by ERβ2, compared with the control PC3 cells (Fig. 2E). In addition, the effect on cell proliferation in 0 and 10% serum was assessed by cell counting. We note that although ERβ2 increased cell proliferation under both conditions, the serum-free condition revealed a substantial growth-promoting effect. The increased proliferation in media without serum indicates that ERβ2 mediates ability for cells to survive in the absence of external growth factor stimulation (Fig. 2C). Using 22Rv1 cells, we could observe a similar change of proliferation as in PC3 cells (Fig. 2D). Changes in cell proliferation genes, as detected by microarray, qPCR, and Western blotting, and their relative relationship according to the literature, are summarized in figure 9 below. Analyses of p45Skp2 and c-Myc at the mRNA and protein levels supported the inhibition of proliferation by ERβ1, although c-Myc was down-regulated only at the protein level. A similar observation was made in the 22Rv1 cell line (Fig. 3, A–F). Previously, we observed that the expression of ERβ1 strongly down-regulates the expression of the p45Skp2 and c-Myc in both breast and colon cancer cell lines (25, 31), which is in accordance with the present study. By contrast, we noted that ERβ2 increased p45Skp2 mRNA and protein expression in the PC3 cells, whereas the level remained unchanged in the 22Rv1 cells (Fig. 3, A and B). Intriguingly, although ERβ1 decreased the level of c-Myc protein without significantly changing its mRNA levels, ERβ2 increased both c-Myc mRNA and protein (Fig. 3, D–G). In addition, and in agreement with previous studies, ERβ1 increased expression of both p21WAF1/CIP1 and p27KIP1 at the protein level (Fig. 3, H and I). Unexpectedly, ERβ2 also increased p27KIP1, which is incompatible with the increased p45Skp2 expression. However, a closer examination revealed an increased phosphorylation of p27KIP1 on amino acid residue T198, which has previously been shown to protect from p45SKP2-mediated degradation (Fig. 3H) (32).
Fig. 2.
The effects of ERβ1 and ERβ2 on cell proliferation. A, The BrdU assay of the cells expressing ERβ1 showed decreased uptake of BrdU, whereas the cells expressing ERβ2 showed an increased incorporation of BrdU compared with the control. B, Quantification of BrdU incorporation as calculated by counting the number of BrdU-positive cells in three fields per DAPI-positive cells. The ERβ1-expressing cells showed lower BrdU incorporation compared with control cells. Data are mean values ± se, calculated using Student's t test, P < 0.003. ERβ2-expressing cells showed larger BrdU incorporation compared with control cells. Data are mean values ± se, calculated using Student's t test, P < 0.008. C, 25,000 PC3 cells were seeded in a 12-well plate with 0 or 10% FBS. After 48 h, the cells were counted with a Neubauer chamber. ERβ1 significantly decreased cell proliferation (0% FBS, P < 0.0004 and 10% FBS, P < 0.0003), whereas ERβ2 significantly increased cell proliferation both at 0 and 10% FBS, respectively (0% FBS, P < 0.0007 and 10% FBS, P < 0.017). D, 25,000 22Rv1 cells were seeded in a 12-well plate with 0 or 10% FBS. After 48 h, the cells were counted with a Neubauer chamber. ERβ1 significantly decreased cell proliferation (0% FBS, P < 0.037 and 10% FBS, P < 0.0003), whereas ERβ2 significantly increased cell proliferation at 10% FBS (P < 0.0053) and no significant change at 0% FBS. E, 10,000 cells/well were seeded in 96-well cell culture plates. After 48 h, an MTS assay kit was used to determine the rate of cell proliferation. ERβ1 decreases cellular proliferation (P < 0.0001), whereas ERβ2 enhances proliferation (P < 0.05) compared with the control. Data are mean values ± se, calculated using Student's t test. Data are mean values ± se, calculated using Student's t test. All the experiments were performed n = 3. ns, Nonsignificant.
Fig. 3.
The effects of ERβ1 and ERβ2 on the expression of proproliferation genes. A, The qPCR analysis of p45Skp2 in PC3 cells showed less expression in the ERβ1-expressing cells than in the control cells (P < 0.0006). Data are mean values ± se, calculated using Student's t test. B, The qPCR analysis of p45Skp2 in 22Rv1 cells showed less expression in the ERβ1-expressing cells than in the control cells (P < 0.022). Data are mean values ± se, calculated using Student's t test. C, Western blot analysis showed a decrease in p45Skp2 in ERβ1-expressing PC3 cells, whereas an increase was shown in cells expressing ERβ2. D, The qPCR analysis of c-Myc in PC3 cells showed increased expression in the ERβ2-expressing cells (P < 0.01) but no significant difference for ERβ1 cells compared with the control. Data are mean values ± se, calculated using Student's t test. E, The qPCR analysis of c-Myc in 22Rv1 cells showed increased expression in the ERβ2-expressing cells (P < 0.08) but no significant difference for ERβ1 cells compared with the control. Data are mean values ± se, calculated using Student's t test. F, Western blot analysis showed an increase in c-Myc in PC3 cells expressing ERβ2 and a sharp decrease in cells expressing ERβ1 compared with the control. G, Western blot analysis showed an increase in c-Myc in 22Rv1 cells expressing ERβ2 and a decrease in cells expressing ERβ1 compared with the control. H, Western blot analysis of p27Kip1 in PC3 cells showed an increase in expression, both in ERβ1- and ERβ2-expressing cells as demonstrated in the top panel. The middle panel shows phosphorylation of p27Kip1 at T198. There is a significant increase in the phosphorylation of p27Kip1 (T198) only in cells expressing ERβ2. β-Actin was used as a loading control. I, Western blotting of p21WAF1/CIP1 expression in PC3 cells showed an increase in p21WAF1/CIP1 in the cells expressing ERβ1 and a significant decrease in the cells expressing ERβ2 compared with the control. All the experiments were performed n = 3. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; ns, nonsignificant.
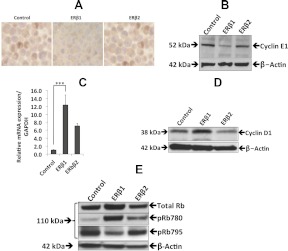
In agreement with the observed regulation of p45Skp2 and c-Myc, we observed a slight increase in expression of total Rb protein in ERβ1-expressing cells. In addition, the phosphorylation of Rb at Ser795 decreased when ERβ1 was expressed compared with the control, as was observed previously in SW480 colon cancer cells stably expressing ERβ1 (25). By contrast, the ERβ2-expressing cells did not exhibit any change in the phosphorylation of Rb at Ser795 (Fig. 4E). The phosphorylation of Rb at Ser795 by the Cdk2-Cyclin E complex is required for the cell cycle to progress from late G1 to S phase. In agreement with the decreased Rb phosphorylation at Ser795, the Cyclin E expression was decreased in ERβ1-expressing cells, whereas no change was seen in ERβ2-expressing cells (Fig. 4, A and B). Finally, we observed increased phosphorylation of Rb at Ser780 in ERβ1-expressing cells (Fig. 4E). The increased phosphorylation at Ser780 by the Cdk4/6-Cyclin D1 is in agreement with the observed increased expression of Cyclin D1 in the cells expressing ERβ1 (Fig. 4, C and D). In summary, the genome-wide analysis indicated a significant effect by ERβ1 on proliferation. Focused studies of cell-cycle regulatory proteins at the mRNA, protein, and phosphorylation levels dissected the effect of ERβ1 and ERβ2 on these parameters and defined the opposing roles of the two ER subtypes on prostate cancer cell proliferation.
Fig. 4.
The effects of ERβ1 and ERβ2 on the cell cycle. A, Immunocytochemical staining of Cyclin E in ERβ1- and ERβ2-expressing PC3 cell pellets. The left panel shows control cells, the middle panel ERβ1 cells, and the right panel ERβ2 cells. The expression of Cyclin E decreased in the cells expressing ERβ1 but remained unchanged in ERβ2-expressing cells. B, Western blot analysis showed a decrease in Cyclin E in PC3 cells expressing ERβ1 compared with control and ERβ2-expressing cells. C, The qPCR analysis of ERβ1 and ERβ2 significantly increased Cyclin D1 expression in PC3 cells (P < 0.0001). Data are mean values ± se, calculated using Student's t test. D, Western blot analysis showed an increase in Cyclin D1 expression in ERβ1 cells compared with control cells and no change in ERβ2 cells. E, Western blot analysis of Rb proteins. The top panel shows the total Rb protein. The second panel shows phosphorylation at Ser780, and the third panel shows phosphorylation at Ser795. The fourth panel is the β-actin loading control. The total Rb and pRb780 increased, whereas pRb795 decreased by ERβ1. In the ERβ2-expressing cells, total Rb and pRb795 were unchanged, and there was a small increase in pRb780. All the experiments were performed n = 3. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
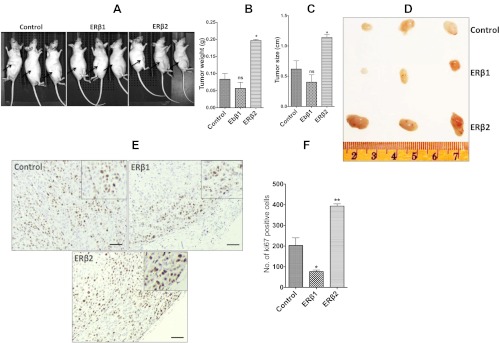
To obtain a more complete view of the role of ERβ1 and ERβ2 in cell proliferation, we investigated how ERβ1's and ERβ2's expression affected growth of PC3-xenografts (see figure 8 below). PC3-control, PC3-ERβ1, or PC3-ERβ2 cells were transplanted into the flank region of nude mice (nu/nu). At end point, the average weight of PC3-control xenografts was 83 mg, whereas average weight of PC3-ERβ1 xenografts was 56 mg, a reduction of tumor weight by 32% after ERβ1 expression; and the average weight of PC3-ERβ2 xenografts was 166 mg, an increase of tumor weight by 200% after ERβ2 expression. Immunohistochemistry with a Ki67-specific antibody was performed. The xenografts in the mice injected with ERβ1-expressing cells showed a significant down-regulation of the number of Ki67-positive cells, whereas a significant increase in the number of Ki67-positive cells was observed for ERβ2-expressing cells.
The effects of ERβ1 and ERβ2 expression on bone metastasis genes
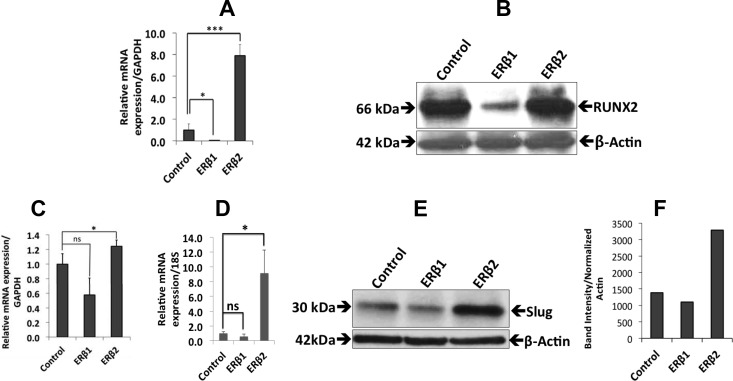
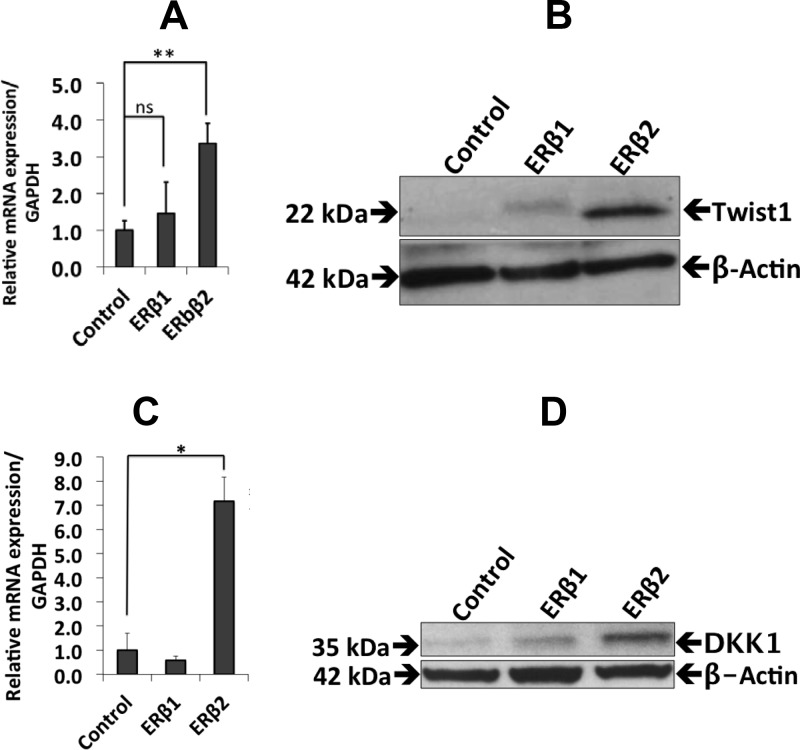
Because PC3 cells are highly metastatic to bone in a mouse model (33), and because ERβ2 has been indicated to affect cell invasion in prostate cancer cells (8), we wanted to investigate whether the expression of ERβ1 or ERβ2 in PC3 cells regulates genes involved in metastatic behavior. The osseous master transcription factor RUNX2 is expressed at high levels in PC3 cells and has been shown to increase the metastatic potential of these cells (11). The expression of ERβ1 reduced the mRNA and protein levels of RUNX2, as assessed by qPCR and Western blotting, whereas ERβ2 increased RUNX2 mRNA, but the protein level remained unchanged (Fig. 5, A and B). In addition, ERβ2 expression increased the level of Slug expression (Fig. 5, C–F). We also studied the expression of Twist1, another factor that plays an important role during prostate cancer metastasis. Twist1 was not detectable in the control cells or in the ERβ1-expressing cells but increased significantly at the protein level after ERβ2 expression. In addition, the mRNA level of Twist1 increased in ERβ2-expressing cells compared with the control cells (Fig. 6, A and B). We also found that DKK1 mRNA and protein levels were up-regulated in the ERβ2-expressing cells. DKK1 is a target gene of Twist1, and its up-regulation is consistent with the higher Twist1 protein level observed in ERβ2 cells (Fig. 6, C and D).
Fig. 5.
The effects of ERβ1 and ERβ2 on the expression of genes involved in bone metastasis. A, qPCR analysis of ERβ1 cells showed a decrease in Runx2 expression (P < 0.01), whereas the expression of Runx2 increased in ERβ2 cells (P < 0.001). Data are mean values ± se, calculated using Student's t test. B, Western blot analysis of the Runx2 protein (50 μg of nuclear extract were loaded in each lane). β-Actin was used as a loading control. Runx2 expression decreased in the ERβ1 cells, but there was no reduction in the ERβ2 cells. C, qPCR analysis of the ERβ2 cells showed an increase in Slug expression (P < 0.03), whereas there was no significant change in ERβ1 cells. D, qPCR analysis of the 22Rv1 cells expressing ERβ2 also showed an increase in Slug expression (P < 0.034), whereas there was no significant change in ERβ1 cells. Data are mean values ± se, calculated using Student's t test. E, Western blot analysis of PC3 cells showed a small increase in Slug protein in ERβ2-expressing cells but no significant change in cells expressing ERβ1. F, Quantification of the Slug protein expression by ImageJ software (National Institutes of Health, Bethesda, MD). All the experiments were performed n = 3. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; ns, nonsignificant.
Fig. 6.
The effects of ERβ1 and ERβ2 on the expression of DKK1. A, qPCR analysis showed increased Twist1 mRNA in ERβ2-expressing cells (P < 0.0033) but no significant change in ERβ1-expressing cells. Data are mean values ± se, calculated using Student's t test. B, Western blot analysis showed an increase in the Twist1 protein level in ERβ2-expressing cells. C, qPCR analysis of the cells expressing ERβ2 showed an increase in DKK1 transcripts (P < 0.09) but no significant change in ERβ1-expressing cells. D, Western blot analysis showed an increase in the DKK1 protein in ERβ2-expressing cells, whereas no significant change in DKK1 was observed in ERβ1-expressing cells. All experiments were performed n = 3. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; ns, nonsignificant.
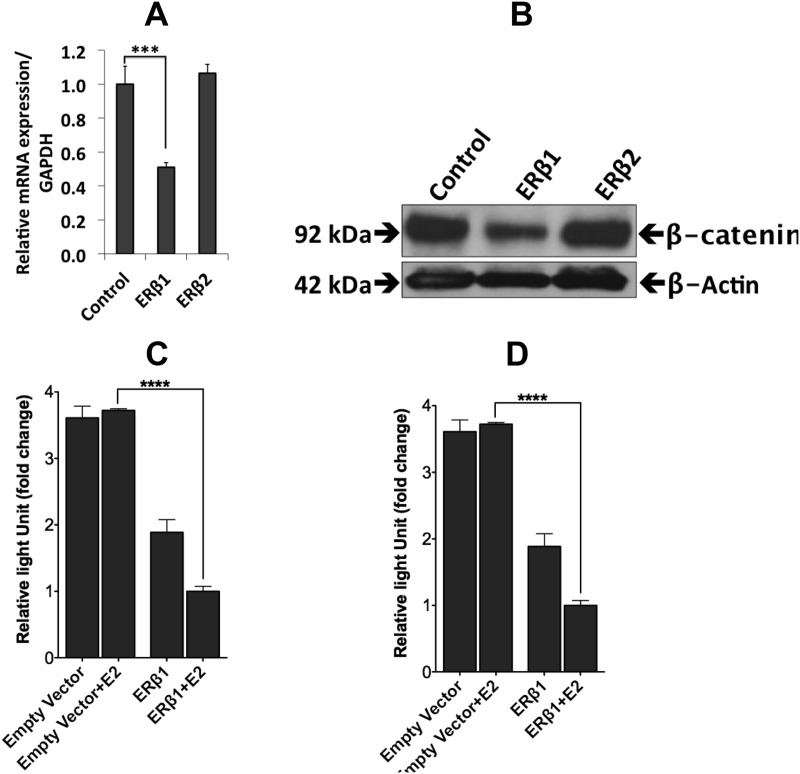
Because Runx2 is a known target gene of β-catenin signaling (34), we investigated the nuclear fraction of β-catenin. As shown in Fig. 7, A and B, β-catenin mRNA and nuclear β-catenin protein were reduced by ERβ1 expression; however, β-catenin was not affected by ERβ2 expression. Furthermore, using a TCF/LEF (T-cell factor/lymphoid enhancer factor) luciferase reporter, we found that the β-catenin activity was repressed in cells expressing ERβ1 in a 17β-estradiol-independent fashion and further repressed by addition of 17β-estradiol. In cells expressing ERβ2, however, the reporter activity was enhanced in a concentration-dependent manner (Fig. 7, C and D). Our findings by microarray, qPCR, and Western blotting, and their significance according to literature, are summarized in figure 9 below.
Fig. 7.
The effects of ERβ1 and ERβ2 on the expression of β-catenin. A, qPCR analysis of the ERβ1 cells showed a decrease in β-catenin expression (P < 0.00001). Data are mean values ± se, calculated using Student's t test. B, Western blot analysis showed a small increase in the β-catenin protein in ERβ2-expressing cells, whereas there was a decrease in ERβ1-expressing cells. C, The luciferase (TCF/LEF) reporter assay for β-catenin showed a 17β-estradiol(E2)-independent decrease in activity for vehicle-treated ERβ1-expressing cells compared with the mock-transfected cells, which was further repressed by 17β-estradiol compared with the vehicle-treated ERβ1-transfected cells (P < 0.0001). Data are mean values ± se, calculated using Student's t test. D, The luciferase (TCF/LEF) reporter assay for β-catenin showed an ERβ2-concentration-dependent increase in activity (P < 0.001 for 200 ng) compared with the empty vector transfection. Data are mean values ± se, calculated using Student's t test. All the experiments were performed n = 3. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Discussion
The second ER, ERβ, has been known for more than 15 yr (35). An antiproliferative effect of ERβ has been found in many laboratories in cell lines and in vivo (31, 36, 37). ERβ knockout mice develop hyperplasia in the prostate (3), indicating a tumor-suppressive role of the receptor. ERβ1 expression has been shown to decrease with an increasing Gleason grade of the prostate cancer (38). Its splice variant ERβ2 is specifically expressed in advanced prostate cancer (22), but far less is known about this variant. One reason for this lack of knowledge is that ERβ2 is only expressed in humans and nonhuman primates and cannot be studied in the mouse. In this investigation, we addressed what genes are affected by ERβ1 vs. ERβ2 expression in the prostate cancer cell line PC3 to elucidate the roles these two receptors play in the context of prostate tumorigenesis.
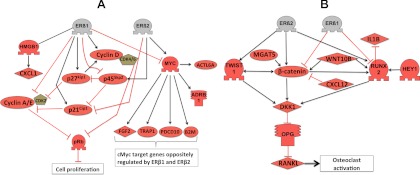
In our studies of the effects of ERβ1, the most prominent findings were the simultaneous repression of cell proliferation and bone metastasis genes. ERβ1 suppresses proliferation by down-regulating c-Myc and p45Skp2 and their proliferative actions (see figure 9 below). ERβ1 also affects multiple cell-cycle genes at either or both of the mRNA and protein levels. The down-regulation of p45Skp2 and Cyclin E/A (CCNE2/CCNA2) and the concomitant increase in the expression of p21WAF1/CIP1 (CDKN1A) and p27KIP1 (CDKN1B) mediate an overall antiproliferative effect of ERβ1 (25). Another indication of the antitumor effect of ERβ1 is the up-regulation of Rb protein in ERβ1-expressing cells. Loss of Rb expression has been correlated to castration-resistant prostate cancer (39). The observed ERβ2-induced up-regulation of both p45Skp2 and p27KIP1 is in disagreement with the inverse relationship between these two factors. However, our finding that p27KIP1 was phosphorylated on T198 indicates that the protein is protected from p45Skp2-mediated degradation (32). RSK1 is known to phosphorylate this site, and we propose that ERβ2 expression can activate RSK1 directly or via MAPK signaling.
The PC3 cell line is known for its bone-metastatic capacity in a mouse model, and the cell line was originally isolated from a bone metastasis of prostate adenocarcinoma. This led us to study how the genes involved in bone metastasis are regulated by ERβ1. Our finding that RUNX2, an important gene in bone formation and bone metastasis, is strongly down-regulated by ERβ1 indicates that ERβ1 could have a major inhibitory effect on bone metastasis in prostate cancer. Because Slug is a known target gene of Runx2 (40), we investigated the ERβ1-mediated regulation of Slug and verified that it was also repressed. Our findings corroborate a study by Leung et al. (8) describing that expression of nuclear ERβ2 in the prostate tumor correlates to a worse prognosis. Because most of the metastatic prostate cancers express AR and become castration resistant, we wanted to investigate the effect of ERβ1 and ERβ2 expression in the castration-resistant cell line 22Rv1 (16). Our finding that ERβ2 increases c-Myc expression and proliferation in this cell line indicates that ERβ2 can play a role in AR positive as well as in AR negative prostate cancers.
c-Myc is a target gene of β-catenin (41), and our observation of the increased activity of β-catenin-responsive reporter by ERβ2 (Fig. 7) is in agreement with the observation that c-Myc is up-regulated by ERβ2. We observed that the levels of c-Myc mRNA did not change upon the expression of ERβ1 but that c-Myc protein expression was strongly reduced, indicating that posttranscriptional modification of c-Myc maybe by microRNA or proteasomal degradation. The up-regulation of Twist1 mRNA and protein and the subsequent up-regulation of DKK1 in the ERβ2-expressing cells indicate that ERβ2 can stimulate osteolytic metastasis in bone. The inhibition of DKK1 by small interfering RNA in a bone metastasis model has been shown to result in a switch in the behavior of the cancer from osteolytic to osteoblastic lesion (42). Furthermore, treatment with an antibody against DKK1 was shown to reduce the tumor burden in a mouse model of prostate cancer (43). We show by using the cell lines in xenograft experiment that the effect on growth by ERβ1 and ERβ2 are extensive during tumor formation (Fig. 8). Figure 9B summarizes the metastatic factors regulated by ERβ2 in our study, describing their roles in balancing osteoblastic and osteolytic behavior, as inferred from the literature. Because of the lack of cell lines endogenously expressing ERβ2, we had to rely on cells stably transfected with ERβ2 to express the protein. Therefore, further studies in tumor cells that express ERβ2 endogenously are needed to confirm the observed regulatory networks.
Fig. 8.
Xenograft transplantation of control PC3 cells and PC3 cells expressing ERβ1 and ERβ2. Nude (nu/nu) mice transplanted with PC3 cells expressing ERβ1 (n = 3), ERβ2 (n = 3), or with empty vector control (n = 3). The cells (5 × 105) were mixed with 50% Matrigel and implanted sc into the flank region. The tumor was monitored regularly and after 40 d, the mice were killed and photographed. A, PC3-control, PC3-ERβ1, and PC3-ERβ2 cells transplanted mice, with arrows pointing at the injection site and the picture of the excised xenografts at the end point. Scale in centimeters. B, Tumor weight at the end point. C, Tumor size at the end point. D, Representative pictures of Ki67 immunohistochemically stained slides from PC3-control, PC3-ERβ1, and PC3-ERβ2 xenografts. E, Number of Ki67-positive cells in xenograft slides per visual field (×20 objective) identified by immunohistochemistry. Bar 1, PC3-control; bar 2, PC3-ERβ1; and bar 3, PC3-ERβ2 xenografts. Number of positive cells was counted in three independent areas of each xenograft slide. Columns, average number; bars, sd. PC3-control vs. PC3-ERβ1, P < 0.01 and PC3-control vs. PC3-ERβ1, P < 0.001.
Fig. 9.
The regulation of proliferation and of bone-metastasis genes by ERβ1 and ERβ2. This figure incorporates data of the genes that were found to be regulated by the expression of ERβ1 or ERβ2, together with data of their relative relationships from the proprietary software database. A, Model of the antiproliferative circuit promoted by ERβ1. The down-regulation of c-Myc, Cyclin E, and p45SKP2 and the up-regulation of Cyclin D1 result in an overall inhibition of cell growth. Also shown is the ERβ2-mediated up-regulation of c-Myc and p45SKP2, which causes ERβ2 cells to proliferate faster than the control cells. B, Pathway diagram showing how ERβ2 regulates factors that control osteoblastic and osteolytic activity by up-regulating RUNX2, Twist1, and β-catenin, resulting in increased DKK1, which controls the RANK ligand/osteoprotegerin (OPG) ratio. In addition, other genes that are either up-regulated or down-regulated by ERβ2 may have possible net effects that contribute to ERβ2-mediated effects.
In conclusion, in PC3 cells, we found that ERβ1 opposes genes involved in bone metastasis and acts as a tumor suppressor, whereas its splice variant ERβ2 acts in the opposite fashion. In addition, the ERβ2-mediated up-regulation of Twist1 and its target gene DKK1 indicates that the expression of ERβ2 in a prostate bone-metastatic tumor will cause the tumor to become osteolytic. It remains to be proven whether this is the cause for the reduced overall survival of patients who express ERβ2 in their tumors (8). This study potentially identifies ERβ2 as a possible new target for prostate cancer treatment.
Supplementary Material
Acknowledgments
We thank Dr. Shaun Zhang for providing reagents allowing us to develop the expression system and Dr. Xinping Fu for help with establishing FACS sorting of cells. We are also thankful to Dr. Armando Rivera, Dr. Lena Herbst, and Dr. Kaberi Das Panja for their help with the mice xenograft experiments.
This work was supported by The Cancer Prevention and Research Institute of Texas Grants HIRP100680 and RP110444, the Texas Emerging Technology Fund under Agreement 300-9-1958, the Robert A. Welch Foundation Grant E-0004, and the Swedish Cancer Fund.
Disclosure Summary: The authors have nothing to disclose.
NURSA Molecule Pages:
Annotations provided by Nuclear Receptor Signaling Atlas (NURSA) Bioinformatics Resource. Molecule Pages can be accessed on the NURSA website at www.nursa.org.
- AR
- Androgen receptor
- BrdU
- bromodeoxyuridine
- DAPI
- 4′,6-diamidino-2-phenylindole
- DKK1
- Dickkopf homolog 1
- ER
- estrogen receptor
- F
- forward
- FBS
- fetal bovine serum
- GO
- gene ontology
- IR
- inverted repeat
- MTS
- (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)
- NP-40
- Nonidet P-40
- qPCR
- quantitative real-time PCR
- R
- reverse
- Rb
- retinoblastoma
- RUNX2
- Runt-related transcription factor
- TCF/LEF
- T-cell factor/lymphoid enhancer factor
- TRE
- tetracycline response element.
References
- 1. Suva LJ, Washam C, Nicholas RW, Griffin RJ. 2011. Bone metastasis: mechanisms and therapeutic opportunities. Nat Rev Endocrinol 7:208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Safarinejad MR, Safarinejad S, Shafiei N. 6 January 2012. Estrogen receptors α (rs2234693 and rs9340799), and β (rs4986938 and rs1256049) genes polymorphism in prostate cancer: evidence for association with risk and histopathological tumor characteristics in Iranian men. Mol Carcinog 10.1002/mc.21870 [DOI] [PubMed] [Google Scholar]
- 3. Weihua Z, Makela S, Andersson LC, Salmi S, Saji S, Webster JI, Jensen EV, Nilsson S, Warner M, Gustafsson JA. 2001. A role for estrogen receptor β in the regulation of growth of the ventral prostate. Proc Natl Acad Sci USA 98:6330–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, Gouvin LM, Sharma VM, Mercurio AM. 2010. ERβ impedes prostate cancer EMT by destabilizing HIF-1α and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell 17:319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan M, Rayoo M, Takano EA, Fox SB. 2011. Nuclear and cytoplasmic expressions of ERβ1 and ERβ2 are predictive of response to therapy and alters prognosis in familial breast cancers. Breast Cancer Res Treat 126:395–405 [DOI] [PubMed] [Google Scholar]
- 6. Shaaban AM, Green AR, Karthik S, Alizadeh Y, Hughes TA, Harkins L, Ellis IO, Robertson JF, Paish EC, Saunders PT, Groome NP, Speirs V. 2008. Nuclear and cytoplasmic expression of ERβ1, ERβ2, and ERβ5 identifies distinct prognostic outcome for breast cancer patients. Clin Cancer Res 14:5228–5235 [DOI] [PubMed] [Google Scholar]
- 7. Leung YK, Mak P, Hassan S, Ho SM. 2006. Estrogen receptor (ER)-β isoforms: a key to understanding ER-β signaling. Proc Natl Acad Sci USA 103:13162–13167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung YK, Lam HM, Wu S, Song D, Levin L, Cheng L, Wu CL, Ho SM. 2010. Estrogen receptor β2 and β5 are associated with poor prognosis in prostate cancer, and promote cancer cell migration and invasion. Endocr Relat Cancer 17:675–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, Muramatsu M. 1998. Molecular cloning and characterization of human estrogen receptor βcx: a potential inhibitor ofestrogen action in human. Nucleic Acids Res 26:3505–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. 1979. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol 17:16–23 [PubMed] [Google Scholar]
- 11. Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA, Frenkel B. 2010. Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer 9:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR, Altieri DC, Pratap J, Keller E, Stein GS, Lian JB. 2010. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene 29:811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, Wong YC, Wang X. 2005. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res 65:5153–5162 [DOI] [PubMed] [Google Scholar]
- 14. Yuen HF, Kwok WK, Chan KK, Chua CW, Chan YP, Chu YY, Wong YC, Wang X, Chan KW. 2008. TWIST modulates prostate cancer cell-mediated bone cell activity and is upregulated by osteogenic induction. Carcinogenesis 29:1509–1518 [DOI] [PubMed] [Google Scholar]
- 15. Pinzone JJ, Hall BM, Thudi NK, Vonau M, Qiang YW, Rosol TJ, Shaughnessy JD., Jr 2009. The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood 113:517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. 2008. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 68:5469–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dignam JD, Lebovitz RM, Roeder RG. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hartman J, Müller P, Foster JS, Wimalasena J, Gustafsson JA, Ström A. 2004. HES-1 inhibits 17β-estradiol and heregulin-β1-mediated upregulation of E2F-1. Oncogene 23:8826–8833 [DOI] [PubMed] [Google Scholar]
- 19. Helguero LA, Hedengran Faulds M, Förster C, Gustafsson JA, Haldosén LA. 2006. DAX-1 expression is regulated during mammary epithelial cell differentiation. Endocrinology 147:3249–3259 [DOI] [PubMed] [Google Scholar]
- 20. Dong JT, Chen C. 2009. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci 66:2691–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ji Q, Liu PI, Elshimali Y, Stolz A. 2005. Frequent loss of estrogen and progesterone receptors in human prostatic tumors determined by quantitative real-time PCR. Mol Cell Endocrinol 229:103–110 [DOI] [PubMed] [Google Scholar]
- 22. Richter K, Wirta V, Dahl L, Bruce S, Lundeberg J, Carlsson L, Williams C. 2006. Global gene expression analyses of hematopoietic stem cell-like cell lines with inducible Lhx2 expression. BMC Genomics 7:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puca R, Nardinocchi L, Sacchi A, Rechavi G, Givol D, D'Orazi G. 2009. HIPK2 modulates p53 activity towards pro-apoptotic transcription. Mol Cancer 8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. 2010. High-mobility group box 1 and cancer. Biochim Biophys Acta 1799:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartman J, Edvardsson K, Lindberg K, Zhao C, Williams C, Ström A, Gustafsson JA. 2009. Tumor repressive functions of estrogen receptor β in SW480 colon cancer cells. Cancer Res 69:6100–6106 [DOI] [PubMed] [Google Scholar]
- 26. Kazerounian S, Yee KO, Lawler J. 2008. Thrombospondins in cancer. Cell Mol Life Sci 65:700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsui KH, Chang PL, Feng TH, Chung LC, Sung HC, Juang HH. 2008. Evaluating the function of matriptase and N-acetylglucosaminyltransferase V in prostate cancer metastasis. Anticancer Res 28:1993–1999 [PubMed] [Google Scholar]
- 28. Rodriguez BA, Cheng AS, Yan PS, Potter D, Agosto-Perez FJ, Shapiro CL, Huang TH. 2008. Epigenetic repression of the estrogen-regulated Homeobox B13 gene in breast cancer. Carcinogenesis 29:1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, Joseph JD, McDonnell DP. 2009. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell 36:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma X, Zhao H, Shan J, Long F, Chen Y, Chen Y, Zhang Y, Han X, Ma D. 2007. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell 18:1965–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ström A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. 2004. Estrogen receptor β inhibits 17β-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci USA 101:1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larrea MD, Hong F, Wander SA, da Silva TG, Helfman D, Lannigan D, Smith JA, Slingerland JM. 2009. RSK1 drives p27Kip1 phosphorylation at T198 to promote RhoA inhibition and increase cell motility. Proc Natl Acad Sci USA 106:9268–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angelucci A, Gravina GL, Rucci N, Festuccia C, Muzi P, Vicentini C, Teti A, Bologna M. 2004. Evaluation of metastatic potential in prostate carcinoma: an in vivo model. Int J Oncol 25:1713–1720 [PubMed] [Google Scholar]
- 34. Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2005. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem 280:33132–33140 [DOI] [PubMed] [Google Scholar]
- 35. Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. 1996. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA 93:5925–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. 2004. Estrogen receptor β inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res 64:423–428 [DOI] [PubMed] [Google Scholar]
- 37. Wada-Hiraike O, Hiraike H, Okinaga H, Imamov O, Barros RP, Morani A, Omoto Y, Warner M, Gustafsson JA. 2006. Role of estrogen receptor β in uterine stroma and epithelium: insights from estrogen receptor β−/− mice. Proc Natl Acad Sci USA 103:18350–18355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang GS, Wang Y, Wang P, Chen ZD. 2007. Expression of oestrogen receptor-α and oestrogen receptor-β in prostate cancer. Chin Med J 120:1611–1615 [PubMed] [Google Scholar]
- 39. Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE. 2010. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 120:4478–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lambertini E, Lisignoli G, Torreggiani E, Manferdini C, Gabusi E, Franceschetti T, Penolazzi L, Gambari R, Facchini A, Piva R. 2009. Slug gene expression supports human osteoblast maturation. Cell Mol Life Sci 66:3641–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509–1512 [DOI] [PubMed] [Google Scholar]
- 42. Hall CL, Bafico A, Dai J, Aaronson SA, Keller ET. 2005. Prostate cancer cells promote osteoblastic bone metastases through Wnts. Cancer Res 65:7554–7560 [DOI] [PubMed] [Google Scholar]
- 43. Yaccoby S, Ling W, Zhan F, Walker R, Barlogie B, Shaughnessy JD., Jr 2007. Antibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood 109:2106–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.