Abstract
The classic 18.5 kDa isoform of myelin basic protein (MBP) is central to maintaining the structural homeostasis of the myelin sheath of the central nervous system. It is an intrinsically disordered, promiscuous, multifunctional, peripheral membrane protein, whose conformation adapts to its particular environment. Its study requires the selective and complementary application of diverse approaches, of which solution and solid-state NMR spectroscopy are the most powerful to elucidate site-specific features. We review here several recent solution and solid-state NMR spectroscopic studies of 18.5 kDa MBP, and the induced partial disorder-to-order transitions that it has been demonstrated to undergo when complexed with calmodulin, actin, and phospholipid membranes.
Keywords: myelin basic protein, intrinsically disordered protein, membrane protein, calmodulin, actin, lipid bilayer, NMR spectroscopy, induced folding
Introduction
Multiple transcription start sites and differential exon splicing of the genes of the oligodendrocyte lineage (Golli) produce several developmentally regulated size isomers of the myelin basic protein (MBP) family (Pribyl et al. 1993; Givogri et al. 2000; Campagnoni and Campagnoni 2008). The 18.5 kDa isoform of classic myelin basic protein (herein referred to as MBP, with murine sequence numbering) is the predominant isoform found in mature central nervous system myelin, and serves as our familial prototype. Its primary function in vivo (it is primarily peripheral membrane associated) is to facilitate and maintain the compaction of the mature myelin sheath of the central nervous system (CNS) (Boggs 2006). In addition, MBP may link the cytoskeleton and the oligodendrocyte membrane, and potentially both to SH3 domain containing proteins (Boggs 2008; Polverini et al. 2008; Harauz and Libich 2009; Homchaudhuri et al. 2009), associations that may be modulated by calmodulin (CaM) (Libich et al. 2003b; Polverini et al. 2004). Other proposed functional roles include signal transduction during myelin development (DeBruin and Harauz 2007) or repair (Moscarello et al. 1997), and participation in molecular recognition and networking events (Boggs 2006; Homchaudhuri et al. 2009). Extensive post-translational modifications create a dynamic molecular barcode to regulate the protein’s targeting, and its association with a diversity of other proteins and ligands (Harauz et al. 2004; Boggs 2006; Harauz et al. 2009; Harauz and Libich 2009). Aberrant post-translational modifications of MBP are linked to the pathophysiology of the demyelinating disease multiple sclerosis (Moscarello et al. 2007; Musse and Harauz 2007; Musse et al. 2008). It is this vibrant, multifunctional context in which structural studies of MBP need to be considered. Here, we review some recent results of our complementary solution and solid-state NMR spectroscopic investigations of 18.5 kDa MBP in specific contexts, namely in association with calmodulin, actin, and lipids.
Intrinsic disorder and MBP
All members of the MBP family can be classified as intrinsically disordered proteins (IDPs) (Harauz et al. 2004; Dunker et al. 2008). Strictly speaking, however, MBP contains alternating regions of predicted disorder and order (Libich and Harauz 2008b). Structurally, IDPs lack a stable hydrophobic core and exist as an ensemble of interchangeable conformations that exhibit dynamic φ and ψ angles (Fink 2005; Dyson and Wright 2006). Although often referred to as “random coil”, IDPs do not behave as true random coils, since they have an inherent propensity to form localized, ordered, secondary structure elements (Shortle 2002). These regions in IDPs are often transient in nature, and are highly dependent on environment, such as the type of solvent or the presence of binding partners (e.g., other proteins or small molecules) and (or) interacting surfaces (e.g., phospholipid membranes) (Liu et al. 2006; Tompa et al. 2009).
Disordered proteins have multiple degrees of freedom in conformational searches, and can regulate intermolecular distance upon association (Dyson and Wright 2005). Molecular recognition is a concept whereby an IDP folds (or becomes more ordered) upon association with its target, which effectively decreases its conformational entropy (Vacic et al. 2007). This disorder-to-order transition serves to uncouple binding strength from specificity, allowing for highly specific, yet readily reversible interactions (Mészáros et al. 2007; Mittag et al. 2009; Wright and Dyson 2009). Thus, IDPs are able to adopt multiple conformations in response to different, structurally diverse binding partners or external stimuli. This property is referred to variously as promiscuity, moonlighting, or one-to-many signalling (Dunker et al. 2001; Tompa 2005; Hazy and Tompa 2009), and is central to an IDP’s function as a signalling or control hub protein (Dunker et al. 2008). These characteristics of IDPs are all exemplified in MBP, which may be categorized partly as a structural linker, and partly as an induced-fit protein target (Harauz and Libich 2009).
Nuclear magnetic resonance (NMR) spectroscopy of IDPs
Many different computational (e.g., sequence analysis, disorder predictors), biochemical (e.g., enzymatic digestion), hydrodynamic (e.g., SEC (size-exclusion chromatography), SAXS (small angle X-ray scattering), DLS (dynamic light scattering, sedimentation analysis)), and spectroscopic (e.g., CD (circular dichroism), FTIR (Fourier transform infrared), fluorescence, NMR (nuclear magnetic resonance)) approaches have been used to characterize the inherent level of residual structure of IDPs (Receveur-Bréchot et al. 2006; Radivojac et al. 2007). In particular, NMR spectroscopy is the technique best able to provide site-specific, atomic-level details on the conformation and dynamics of such proteins, alone and in complex with other biomolecules (Eliezer 2009; Wright and Dyson 2009).
The basis of NMR spectroscopy is that nuclei with a nonzero magnetic moment can interact with each other and their surroundings, yielding measurable correlations. In biomolecular NMR spectroscopy, the active nuclei of interest are 1H, 31P, 15N, and 13C (Kanelis et al. 2001). The isotropic chemical shift (commonly referred to as chemical shift) is the primary NMR observable, and reports on the specific environment of individual atoms in a molecule. Furthermore, the lifetime of the excited state can be orders of magnitude longer in NMR than in other spectroscopic techniques, which allows for the assessment of dynamics over a wide range of time-scales. As well, resonance frequency information may be transferred between nuclei, a technique that forms the cornerstone of heteronuclear NMR experiments (Rule and Hitchens 2006).
The ability of NMR spectroscopy to gather detailed, high-resolution information in a dynamic context is particularly useful for the study of IDPs (Daughdrill et al. 2005; Eliezer 2009). Sequence-specific assignments of the observable resonances of isotopically labelled proteins are accomplished through the use of a series of multidimensional, heteronuclear correlation experiments (Kanelis et al. 2001). In contrast to well-folded proteins, conformational averaging of IDPs results in poor chemical shift dispersion, which may be partially alleviated by the superior chemical shift dispersion of 13C′ resonances (Yao et al. 1997). Nonetheless, overlapping peaks may not be unambiguously assigned, but even with partial resonance assignments, valuable structural information may be surmised (Dyson and Wright 2004).
The most accessible data for accessing secondary structure in proteins are chemical shifts, primarily of 1Hα, 13Cα, and 13Cβ, although chemical shift index (CSI) analysis has been extended to all backbone nuclei (Wishart and Nip 1998; Schwarzinger et al. 2000). The 13Cα and 13Cβ chemical shifts are determined primarily by the φ and ψ dihedral angles of the peptide bond (de Dios et al. 1993). Coupled with the extensive tabulation of random coil values for amino acids (Schwarzinger et al. 2001), sequence-corrected CSI plots offer a picture of the residual secondary structure of an IDP. Moreover, the measurement of backbone dynamics provides important insights into internal motions that identify regions of compaction and (or) secondary structure in IDPs (Eliezer 2009). The relaxation parameters (R1, R2, and the heteronuclear NOE (nuclear Overhauser effect); Jeener et al. 1979) for the backbone amides of IDPs can be measured and expressed within the framework of the model-free formalism, albeit with some caveats (Barbar 1999), and offer insights into the conformational dynamics on fast (ps–ns) and slow (μs–ms) time scales (Palmer 2004).
The conformational flexibility inherent to IDPs makes quantitative interpretation of classically derived NMR structural restraints such as coupling constants or medium- and long-range NOEs, ambiguous in many instances (Crowhurst et al. 2002). In some cases, residual dipolar couplings (RDCs), have been used successfully to interpret the backbone orientation and topology of IDPs, although caution should be used in choosing the aligning medium for such studies (Shortle and Ackerman 2001; Bax 2003; Marsh et al. 2008). A notable and versatile technique for assessing the conformation of IDPs is the use of a sequence-specific paramagnetic probe such as the nitroxide spin label PROXYL (1-oxyl-2,2,5,5-tetramethyl-3-pyrrolidinyl) (Gillespie and Shortle 1997). By comparing the intensities or line widths between labelled and unlabelled samples, close contacts of the polypeptide chain may be identified (Eliezer 2009).
Compare and contrast: solution and solid-state NMR spectroscopy
Experiments for solution-based NMR spectroscopy are much more developed and accessible than for solid-state NMR (ssNMR) spectroscopy, and have hitherto been more widely employed for protein structural studies (Kanelis et al. 2001). The fundamental difference between solution and solid-state samples is the time scale and extent of molecular motions of the proteins. In solution, molecules are able to tumble isotropically on a time scale that eliminates anisotropic line-broadening mechanisms such as chemical shift anisotropy and dipolar couplings. Thus, sharp and well-resolved lines are observed for all nuclei (1H, 13C, and 15N). As the size of the molecule or complex increases, the overall tumbling rate decreases, and the effects of these relaxation mechanisms are reintroduced into the spectrum. Consequently, there is a fundamental size limit for solution NMR spectroscopy of approximately 30 kDa. Specialized pulse sequences (e.g., TROSY (transverse relaxation optimized spectroscopy)), combined with selective isotopic labelling schemes have extended the effective size of proteins that can be studied by solution NMR spectroscopy (Tugarinov et al. 2004), but for many systems this size barrier remains.
In contrast to solution samples, the molecular motions in solid-state assemblies are either absent or slow, and consequently magic angle spinning (MAS) is required to obtain high-resolution spectra of 13C and 15N nuclei (Andrew et al. 1958). The lack of proton detection in ssNMR spectroscopy results in a serious loss of sensitivity, and must be overcome before the technique can be widely applied to high-resolution structural studies of proteins (Agarwal et al. 2006; Chevelkov et al. 2006). Although much effort can be invested in mimicking a protein’s in vivo environment in solution samples, many proteins function as parts of larger complexes or are embedded in the lipid bilayer—situations that are difficult or impossible to replicate in solution. The real advantage of ssNMR over solution NMR spectroscopy is that proteins can be reconstituted in environments that are more representative of their native states, which is particularly important for IDPs that are so structurally sensitive to their immediate surroundings (Libich and Harauz 2008a). Nonetheless, the powerful repertoire of solution NMR spectroscopic experiments currently available means that the two approaches complement each other (e.g., Traaseth et al. (2009)).
In this mini-review, we present three structural investigations of the 18.5 kDa MBP using both solution and solid-state NMR spectroscopy (Zhong et al. 2007; Libich and Harauz 2008b; Ahmed et al. 2009). There are distinct advantages for combining different NMR methods to conformational analyses of this protein (Harauz and Ladizhansky 2008). Although some MBP isoforms translocate between the cytoplasm and nucleus, the primary in vivo environment of the 18.5 kDa isoform is peripherally associated with the cytoplasmic leaflet of the oligodendrocyte membrane. Thus, it is impossible to prepare a sample of lipid-associated MBP that would be amenable to solution NMR spectroscopy (Libich and Harauz 2008a). Similarly, MBP binds and bundles actin to form large, filamentous assemblies. On the other hand, the binding site of CaM on MBP is best elucidated using solution NMR methods, since the sequence-specific high-resolution information needed is not easily obtained with ssNMR. Furthermore, interpretation of some of the ssNMR data can be aided by having available the solution-based resonance assignments of MBP (Libich et al. 2004; Libich et al. 2007).
Ca2+-CaM binds primarily to a C-terminal target on MBP
Myelin basic protein interacts with CaM in a specific, calcium-dependent manner, with a 1:1 stoichiometry, and with a dissociation constant of 144 ± 76 nmol·L−1 (Libich et al. 2003b). Initial studies suggested that CaM bound to a predicted target site at the C-terminus of MBP, until a deletion mutant lacking this region also demonstrated CaM binding (Libich and Harauz 2002; Libich et al. 2003a). A reduced charge variant of MBP was also shown to bind CaM in a manner suggestive of a second binding site (Libich et al. 2003b). Furthermore, site-directed spin-labelling (SDSL) and electron paramagnetic resonance (EPR) spectroscopic experiments indicated that residues along the entire length of MBP were immobilized upon CaM association (Polverini et al. 2004). Taken together, these diverse data suggested that there could be multiple binding sites for CaM on MBP.
To definitively identify the CaM-binding target, solution NMR studies of the MBP:CaM complex were undertaken (Libich and Harauz 2008b). It is common in such interaction studies to titrate an unlabelled version of one binding partner into a solution of a 15N-labelled version of the other partner. In this way, the chemical shift perturbations of the backbone amides may be tracked using a standard 1H–15N HSQC (heteronuclear single quantum coherence) experiment, and the binding site thereby identified. However, we found that there were severe problems with this routine approach in our study of MBP:CaM binding. First, the conformational heterogeneity of MBP made it extremely difficult to add CaM while limiting precipitation, even though the heterocomplex was relatively stable at lower protein concentrations (Libich et al. 2003b; Polverini et al. 2004). Secondly, the relatively weak dissociation constant suggested that there was an equilibrium condition where multiple states of MBP (and CaM) could be present, thus further complicating the observed spectra. For these reasons, the more time-consuming approach of resonance assignment of the bound state of MBP was employed, the advantage being increased (and reproducible) sample stability and the ability to control the equilibrium state of the system (Libich and Harauz 2008b).
Spectra of 15N-CaM bound to unlabelled MBP showed chemical shift perturbations involving residues primarily located in the EF-hand motifs and central linker region of CaM. This result is consistent with what has been previously observed for CaM binding to various other peptide targets, and did not indicate unfolding or precipitation (Hayashi et al. 2002). Conversely, there were major chemical shift perturbations of 15N-MBP in the presence of unlabelled CaM, with approximately 40 cross-peaks shifted upfield in the 1H dimension and downfield in the 15N dimension relative to 15N-MBP alone (Fig. 1A). The three-dimensional experiments (CBCA(CO)NH, CBCANH, and HCC(CO)NH) were analyzed using a conventional backbone assignment strategy (Kay 1995) that identified over 94% of the backbone resonances expected from MBP. The majority of the cross-peaks that displayed large chemical shift differences were identified as hydrophobic residues (Phe, Tyr, and Leu) dispersed throughout the peptide chain. This observation was consistent with the results of the SDSL/EPR investigations of the MBP:CaM studies, which showed the greatest immobilization of the spin label when it was placed near hydrophobic clusters on MBP (Polverini et al. 2004). This effect could be indicative of either specific interactions, and (or) the sequestering of hydrophobic residues from the bulk solvent. Indeed, the unbound portion of MBP would be expected to retain considerable flexibility, and thus multiple hydrophobic interactions could be expected (Mészáros et al. 2007).
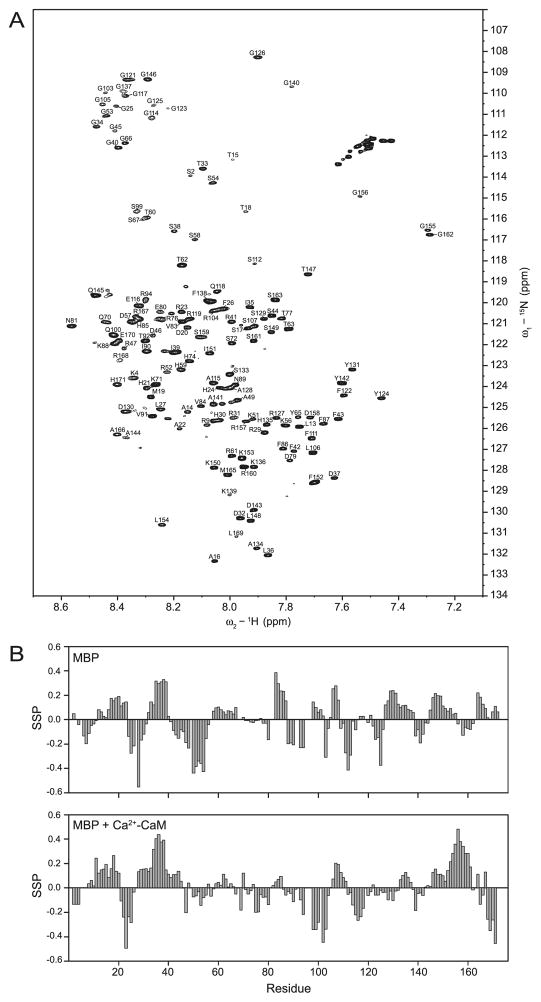
Fig. 1.
Solution NMR spectroscopy of 18.5 kDa recombinant murine MBP (176 residues including an LEH6 tag) in association with Ca2+-CaM (Libich and Harauz 2008b). (A) The 1H-15N HSQC spectrum of 15N-labelled MBP in the presence of Ca2+-CaM. Of the 164 expected backbone peaks, 145 were assigned. (B) Secondary structure propensity (SSP) of MBP alone (top panel) and bound with CaM (bottom panel). The SSP score is interpreted as the percentage of conformers in the disordered ensemble that exhibit ordered secondary structure at a given residue (Marsh et al. 2006). The redistribution of propensities of structural conformers in the C-terminus of MBP indicates increased stabilization resulting from the binding of Ca2+-CaM. (Adapted from Libich and Harauz (2008b) with permission.)
The CaM-binding site on MBP was identified using a global analysis of HN, N, Cα, and Cβ chemical shifts of MBP in the bound and unbound states using the SSP (secondary structure propensity) algorithm (Marsh et al. 2006) (Fig. 1B). Although large perturbations for HN–N shifts were observed distributed throughout the peptide chain (as discussed above), there was a clustering of perturbed shifts in the C-terminal region (Pro120–Arg160). Furthermore, 4 residues (Gly155, Gly156, Gly162, and Thr147) displayed abnormally large chemical shifts upon CaM binding (Fig. 1A).
The amphipathic α-helix represents a well-studied canonical target for CaM binding; however, the recognized targets and modes of interaction are quite diverse (Hoeflich and Ikura 2002). Indeed, there is no specific requirement for α-helical formation in MBP upon CaM binding, and this particular association may represent a different kind of disorder-to-order transition. Although no fully formed secondary structure was identified by CSI analysis in unbound MBP, there were several regions of increased stabilization that coincided with relaxation measurements (Libich and Harauz 2008b). Conversely, when bound to CaM, the stabilized regions observed within MBP were redistributed, suggesting that the source of stabilization was CaM. Particularly, the C-terminal region (containing the aforementioned residues) showed an increased propensity for α-helix formation (Fig. 1B). Collectively, these observations indicated that the primary interaction site for CaM was in the C-terminal region of MBP. Future investigations using editing- or filtering-type pulse sequences would serve to define the intra-molecular interactions more precisely (Lee et al. 1994), whereas isotope-editing experiments would provide information on intermolecular associations (Folmer et al. 1995).
Association with actin stabilizes ordered secondary structure in MBP
Myelin basic protein is able to polymerize G-actin to F-actin (filaments) in vitro and subsequently assemble F-actin into bundles, a function that is modulated by post-translational modifications such as deimination and phosphorylation, and can be reversed by the calcium-dependent binding of CaM to MBP, as recently reviewed in Boggs (2008). The large filamentous complexes formed by F-actin associated with MBP are not amenable to solution NMR techniques, and we have employed ssNMR spectroscopy (augmented by Fourier transform infrared spectroscopy) to study the dynamics and formation of ordered secondary structure in MBP in this assembly (Ahmed et al. 2009).
Cross-polarization (CP; Pines et al. 1973) MAS spectra revealed a temperature-dependent differential mobility of MBP in the complex. Spectra recorded at lower temperatures (250 K) displayed broad peaks that were more intense than sharper peaks in the spectra recorded at higher temperatures (Fig. 2A). Both the intensity and resolution of the resonances were indicative of the molecular motions resulting from the conformational averaging of MBP in association with F-actin. In contrast to the CP-MAS spectra, the intensity and resolution of peaks arising from protein resonances in insensitive nuclear enhancement by polarization transfer (INEPT; Morris and Freeman 1979) MAS spectra increased with increasing temperature (Fig. 2A). These experiments established that even in complex with actin, MBP remained conformationally heterogeneous, consistent with observations of MBP in complex both with CaM and with phospholipid bilayers (Zhong et al. 2007; Libich and Harauz 2008b).
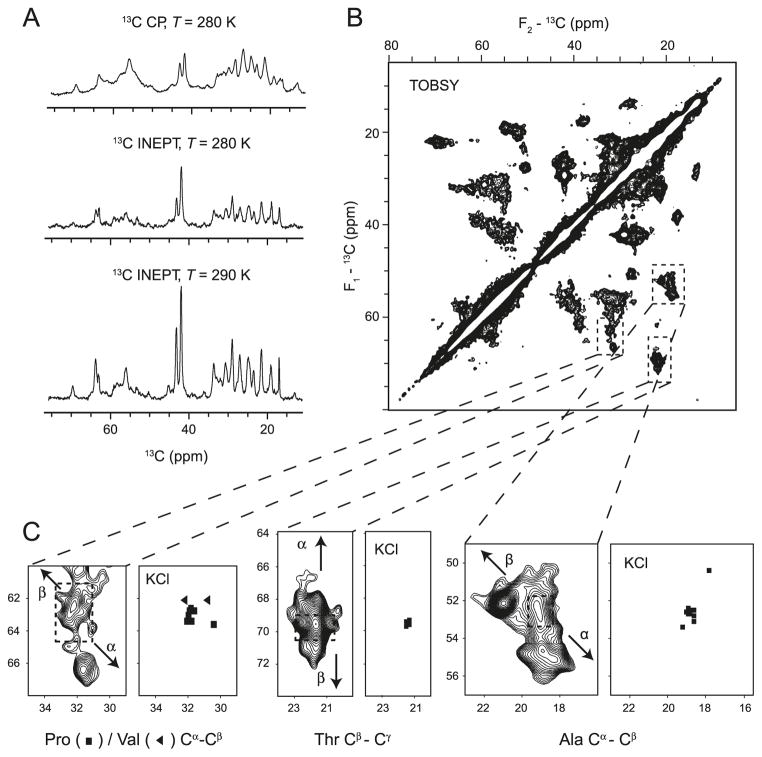
Fig. 2.
Solid-state NMR spectroscopy of 18.5 kDa recombinant murine MBP in association with actin filaments and bundles (Ahmed et al. 2009). (A) The 13C CP-MAS spectra and 13C INEPT-MAS spectra of 13C,15N-labelled MBP in complex with actin shown as a function of temperature. (B) Aliphatic region of TOBSY (total through-bond correlation spectroscopy) 13C–13C correlation spectrum of MBP in complex with actin collected at 800 MHz. (C) Expansions of the Ala Cα–Cβ region, the Thr Cβ–Cγ region, and the Pro/Val Cα–Cβ region. The dashed squares indicate random coil regions, and arrows indicate the trends of the change in chemical shift as a function of type of secondary structure (α helix or β strand). The chemical shifts observed in the TOBSY spectrum are compared with the predominantly random coil chemical shifts observed for isolated MBP dissolved in 100 mmol·L−1 KCl (Libich et al. 2007). (Adapted from Ahmed et al. (2009) with permission.)
To further assess the mobility, conformational heterogeneity, and secondary structure formation of MBP bound to actin, a series of two-dimensional 13C–13C correlation spectra were recorded using both through-bond (TOBSY (through-bond correlation spectroscopy); Hardy et al. 2001) and through-space (DARR (dipolar-assisted rotational resonance; Takegoshi et al. 2001) or SPC53 (Hohwy et al. 2002)) interactions as mechanisms to establish molecular correlations. Through-space spectra acquired using either DARR mixing (50 ms) or the shorter mixing time of SPC53 (1.26 ms) supported the conclusions derived from the one-dimensional CP-MAS and INEPT-MAS spectra. At lower temperatures, most of the backbone–backbone, backbone – side chain, and side chain – side chain correlations were observable, albeit with dramatically decreased resolution compared with spectra recorded at higher temperatures. Because of the length of the DARR mixing scheme, it was likely that multiple-bond and inter-residue correlations were observed in addition to single-bond correlations. Using the shorter SPC53 mixing scheme, only one-bond correlations were observed, but these cross-peaks were very broad, reflecting the conformational heterogeneity of MBP in complex with actin. Thus, the increased mobility of MBP at elevated temperatures caused the averaging of carbon–carbon dipolar interactions, in turn decreasing the polarization transfer efficiency, and resulted in less intense but better-resolved spectra than what was observed at lower temperatures.
Through-bond correlations using a TOBSY pulse sequence with an appropriately chosen mixing time (8.4 ms) resulted in spectra displaying mainly one-bond correlations. Although the resolution of these spectra was greater than in the through-space experiments, the transfer efficiency was markedly different among the particular correlations. For example, very weak Cα–Cβ correlations and pronounced Cβ–Cγ correlations were observed for threonine residues (Fig. 2B). Nonetheless, we were able to establish amino acid specific correlations, assigning particular cross-peaks to specific residue types, and thus draw conclusions about the stabilization of both α-helical and β-strand secondary structure elements occurring in MBP upon association with actin.
Even without sequence-specific residue assignments, the information content of the TOBSY spectra was sufficient to extract distinct chemical shift ranges for specific atoms of particular amino acid types. Particularly well-isolated cross-peaks from the Ala Cα–Cβ, Thr Cβ–Cγ, and Pro/Val Cα–Cβ regions were identified (Fig. 2C). The chemical shift ranges observed for these cross-peaks were considerably more disperse than random coil values, and indicated the formation of both α-helical and β-strand type secondary structure elements. For example, the ranges of TOBSY-observed Cα and Cβ chemical shifts for Ala residues were 50.6–56.0 ppm and 17.8–22.0 ppm, respectively, which were considerably more dispersed than the random coil shifts (52.7 ± 0.7 ppm for Cα, and 19.0 ± 0.7 ppm for Cβ). Likewise, similar dispersion was observed for the Thr and Pro/Val regions, and the presence of both upfield and downfield chemical shifts was indicative of both α-helical and β-strand secondary structure (Wishart and Nip 1998).
Solution NMR assignments of MBP dissolved in 100 mmol·L−1 KCl had reported chemical shifts that were predominantly random coil in nature (Libich and Harauz 2008a). In particular, the KCl solution chemical shifts of the 11 alanines’ Cα or 10 threonines’ Cβ nuclei were predominantly near random coil values, in stark contrast to the 5–6 ppm dispersion observed for these resonances in the TOBSY spectra of MBP:actin (Fig. 2C). This observation unambiguously demonstrated the presence of ordered secondary structure in actin-bound MBP. Extending this comparison, the probable locations of the secondary structure elements formed in actin-bound MBP could be determined. Of the 11 alanines, 5 are located in the N-terminal (residues Ala1–Ala49) and 6 are in the C-terminal segment (Ala115–Arg168) of MBP. Similarly, 7 threonines are located in the N-terminal (Thr15–Thr77) and 1 in the C-terminal (Thr147) segment, whereas the remaining 2 (Thr92 and Thr95) are centrally located in the protein. Thus, the high population of these resonances in both terminal segments of MBP suggests that stabilized secondary structure lies therein. A precise and unequivocal delineation of the actin-binding regions of MBP awaits residue-specific assignments.
MBP remains partially solvent-exposed when bound to lipid membranes
The primary physiological function of MBP is as a membrane linker, binding to and stabilizing the lipid bilayers that form the myelin sheath (Harauz et al. 2004; Boggs 2006). Numerous studies have shown an increase in stabilized secondary structure of MBP in a lipid environment (reviewed in Smith (1992) and Harauz et al. (2004)). Thus, there is a need to investigate the structural properties of MBP bound to lipids not only to mimic the primary in vivo environment of the protein, but to also provide context to the various in vitro biophysical studies conducted on MBP in the absence of lipids. Although some solution NMR studies have been successful with short peptides derived from MBP bound to detergent micelles (e.g., Farès et al. (2006)), the aggregation propensity of full-length MBP for lipid vesicles makes an MBP:lipid sample suitable only for ssNMR spectroscopic investigations.
Peripheral membrane-associated MBP represents a very different system from that of the microcrystalline (Igumenova et al. 2004), amyloid fibril (Siemer et al. 2006), or transmembrane protein (Frericks et al. 2006; Li et al. 2008; Shi et al. 2009) samples for which ssNMR spectroscopic residue-specific assignments have been achieved. Due to the fast molecular motions inherent in MBP, which averaged the dipolar interactions, traditional ssNMR techniques were not applicable and necessitated the development of a complementary suite of NMR experiments to obtain sequence-specific assignments (Zhong et al. 2007). These experiments combined the solution INEPT methodology with solid-state MAS and high-power decoupling to study the mobile portions of MBP. The assignment process was facilitated by a set of complementary three-dimensional constant-time experiments. The NCOCX experiment correlated the amide nitrogen chemical shift with the preceding residue’s C′ and CX chemical shifts (i.e., N[i]-C′ [i-1]-CX[i-1]), whereas the NCACX experiment provided both inter- and intra- residue correlations (i.e., N[i]-Cα[i-1]-CX[i-1] and N[i]-Cα[i]-CX[i]). Additionally, C′ [i-1]-N[i]-Cα[i]/CX[i] and Cα[i]-N[i]-C′ [i-1]/CX[i-1] correlations were provided by the CONCACX and CAN(CO)CX experiments, respectively (Fig. 3A).
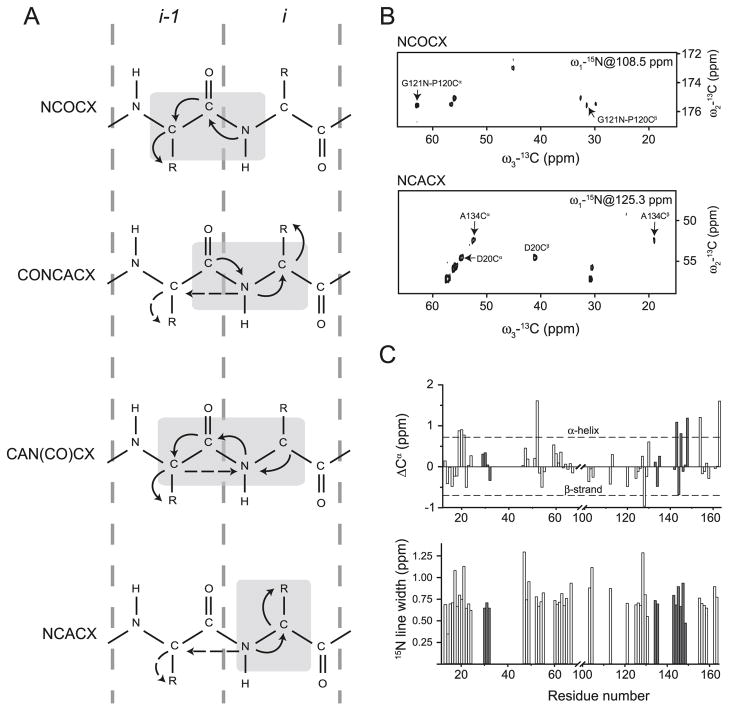
Fig. 3.
Solid-state NMR spectroscopy of 18.5 kDa recombinant murine MBP in association with DMPC:DMPG membranes (Zhong et al. 2007). (A) Schematic representation of the triple-resonance experiments used to obtain sequential assignments of lipid bilayer-associated 13C,15N-MBP. Light-grey boxes enclose correlated residues with one-bond and two-bond (N[i]-Cα[i-1]) polarization transfers indicated by solid and dashed arrows, respectively. (B) Two-dimensional 13C-13C planes extracted from the three-dimensional NCOCX and NCACX experiments. Sequence-specific assignments are indicated by the solid arrows. (C) Chemical shift index analysis of assigned Cα resonances and 15N line-width analysis for assigned residues. Tentatively-assigned residues in both panels are shown in grey. (Adapted from Zhong et al. (2007) with permission.)
To maintain the long-term stability of the sample and reduce the complexity of the MBP:lipid system, the synthetic short-chain lipids DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) and DMPG (1,2-dimyristoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]) were chosen. The one-dimensional experiments – CP-MAS, or INEPT – exploited different polarization transfer mechanisms, and thus could report on regions of MBP with differing dynamic properties (e.g., immobile vs. mobile). At low temperatures, the CP-MAS transfer efficiency increased, whereas the INEPT efficiency decreased similar to what was observed for MBP:actin complexes (Ahmed et al. 2009). This observation was indicative of the mobile portions of MBP becoming more restrained, and thus confirmed previous observations that when associated with lipids, portions of MBP remain highly conformationally flexible (Bates et al. 2003; Musse et al. 2006).
Using the aforementioned three-dimensional NCOCX/NCACX experiments, three-bond correlations could be established that allowed for the classification of spin systems by residue type (Fig. 3B). Specifically, 80 unique spin systems could be identified, and many more systems recognised, though they remain ambiguous due to chemical-shift degeneracy. The residues of MBP observed in these experiments lay within the solvent-exposed regions, and were thus likely to be flexible. This motion resulted in limited dispersion and significant degeneracy, particularly of the Cα chemical shifts. These problems (which are inherent to disordered proteins) were compounded by the fact that the NCOCX/NCACX experiments only correlated two backbone carbons from one particular residue, making residue-type assignment ambiguous for many amino acids. To overcome these difficulties, the three-bond correlations provided by the CON-CACX/CAN(CO)CX experiments were able to remove the ambiguity from some of the identified spin systems, and allowed for the sequential linking of spin systems by the establishment of the Cα[i]-Cα[i-1] connectivity (Fig. 3A). Using this assignment strategy, 62 of the 80 identified spin systems could be linked in non-contiguous fragments, of which 49 were assigned to particular segments within MBP, whereas the remainder were ambiguous due to sequence degeneracy (Zhong et al. 2007).
Chemical shift index analysis of the identified residues was employed to assess the relative proportion of ordered secondary structure (Fig. 3C). Consistent with the high degree of disorder of MBP, the majority of residues displayed random-coil like values with a few exceptions. Specifically the His21-Arg23 stretch, along with Asp143, Gln145, Leu148, and Leu154 had slight α-helical propensity based on their Cα chemical shifts, consistent with what was previously observed with the protein dissolved in 30% TFE-d2 (perdeuterated trifluoroethanol) (Libich et al. 2004). In an attempt to gather more sequential assignments, a set of NOE spectra was collected by incorporating NOE-mixing into an HHCC experiment. Although no inter-residue correlations were observed, cross-peaks were observed between water and nearly all identified residues, indicating that these fragments of MBP were solvent-exposed, and thus located either outside the lipid bilayer or in its hydrophilic portion. Further evidence for the mobility of the observable portions of MBP was derived from analysis of the 15N line-widths (Fig. 3C), which were in the 0.5–0.8 ppm range for the majority of residues, much larger than the expected 0.15–0.2 ppm range. Ruling out instrumental imperfections and susceptibility effects, the most likely source of the observed line broadening was conformational heterogeneity (Zhong et al. 2007).
The mobile segments observed were in almost contiguous runs (segments Thr15–Arg23, Asp46–Ser67, Pro120–Leu148, and Leu154–Ser163), and correlated well with the disordered segments that were independently observed by solution NMR spectroscopy of MBP dissolved in 30% TFE-d2 (Libich and Harauz 2008b) (Fig. 4). Furthermore, the unobservable resonances were a direct consequence of their reduced mobility of portions of the protein, likely due to interactions with the lipid bilayer. Regions of MBP that contained hydrophobic pairs (Phe42–Phe43, Phe86–Phe87), as well as the entire Leu68–Lys102 stretch, had previously been shown by EPR spectroscopy to be embedded in the hydrophobic portion of the bilayer where they would be significantly immobilized (Bates et al. 2003), and were not observed in these experiments. In summary, the portions of MBP characterized by these ssNMR investigations exist either exposed to solvent or in the hydrophilic portion of the bilayer, and are thus available for other functions such as ligand binding. Future ssNMR spectroscopic studies of membrane-reconstituted MBP would benefit from a sensitivity improvement by full 2H13C15N-labelling of the protein (Ohki and Kainosho 2008), in conjunction with paramagnetic relaxation enhancement studies to define long-range interactions (Nadaud et al. 2009).
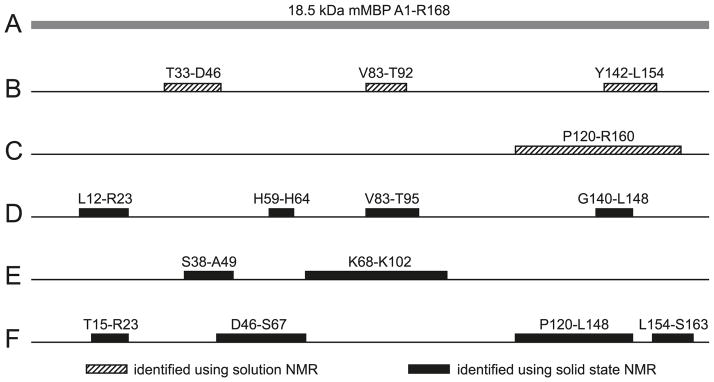
Fig. 4.
Summary of solution and solid-state NMR spectroscopic investigations of 18.5 kDa MBP. (A) The highly charged (+19 at neutral pH) recombinant murine version of the unmodified C1 component (rmC1) has been used as the prototype for the family. The C-terminal LEH6 tag is not shown here. (B) Solution NMR spectroscopic studies of MBP in 30% TFE-d2 defined 3 specific regions with strong α-helical propensity: Thr33–Asp46, Val83–Thr92, and Tyr142–Leu154 (Libich and Harauz 2008b; Harauz and Libich 2009). (C) Solution NMR investigations demonstrated that the C-terminal segment was the primary binding site for calmodulin in solution, as indicated by contiguous chemical shift perturbations (Pro120–Arg160) (Libich and Harauz 2008b). (D) Solid-state NMR studies of MBP-actin complexes indicated some induced disorder-to-order transitions in both the N- and C-terminal regions, as well as in the central immunodominant epitope (Ahmed et al. 2009). (E and F) Solid-state NMR spectroscopic investigations of MBP reconstituted with artificial membranes (Zhong et al. 2007) showed that part of the protein was immobilized by the interaction with the membrane (E), but a large portion of it was mobile (F).
Summary: structural polymorphism of MBP
We have presented a synopsis of several recent NMR spectroscopic investigations of the interactions of MBP with Ca2+-CaM, actin, and lipids—diverse yet physiologically relevant contexts. In all of these assemblies, MBP displays molecular recognition fragments (Mohan et al. 2006), and a degree of induced folding or coupled folding and binding (Hazy and Tompa 2009; Mittag et al. 2009; Wright and Dyson 2009), as well as “fuzziness”, a conceptual framework proposed by Tompa and Fuxreiter (2008). The results presented here are summarized in Fig. 4, with specific reference to the 18.5 kDa murine MBP isoform (Fig. 4A). Solution NMR spectroscopic studies of the full-length protein in the membrane-mimetic solvent (30% TFE-d2) defined 3 specific regions of MBP with strong α-helical propensity: Thr33–Asp46, Val83–Thr92, and Tyr142–Leu154 (Libich and Harauz 2008b; Harauz and Libich 2009) (Fig. 4B). These regions also have strong hydrophobic moments (Harauz et al. 2009). The central (Val83–Thr92) segment is a primary immunodominant epitope in multiple sclerosis, and has been shown by electron paramagnetic resonance (EPR) spectroscopy to form an amphipathic, membrane surface associated α helix in situ (Bates et al. 2003; Musse et al. 2006). Moreover, solution NMR investigations demonstrated that the C-terminal segment of MBP was, indeed, the primary binding site for calmodulin in solution, as indicated by contiguous chemical shift perturbations (Pro120–Arg160) (Libich and Harauz 2008b) (Fig. 4C).
Solid-state NMR studies of MBP–actin complexes indicated some induced disorder-to-order transitions in both the N- and C-terminal regions, as well as in the central immunodominant epitope (Ahmed et al. 2009). Shown in Fig. 4D are the regions of MBP most likely to form secondary structure upon actin association, but a more precise delineation awaits residue assignments. Finally, solid-state NMR spectroscopic investigations of MBP reconstituted with artificial membranes (Zhong et al. 2007) showed that part of the protein was immobilized by the interaction with the membrane, but a large portion of it was mobile (Figs. 4E and 4F). The two immobilized segments depicted in Fig. 4E corresponded to regions with large hydrophobic moments that were predicted to be α-helical in organic solvent (30% TFE-d2) by solution NMR spectroscopy (see Fig. 4B), viz., fragments Ser38–Ala49 versus Thr33–Asp46, and Pro82–Pro93 versus Leu68–Lys102, respectively. These same regions contain the Phe42–Phe43 and Phe86–Phe87 pairs that would be expected to penetrate into the hydrophobic interior of the lipid bilayer and thus effect membrane anchoring. As shown in Fig. 4F, the highly mobile fragments of membrane-reconstituted MBP were found to be in almost contiguous runs: Thr15–Arg23, Asp46–Ser67, Pro120–Leu148, and Leu154–Ser163. These mobile segments were mostly disordered and exposed to solvent, being likely located outside the lipid bi-layer, or in its hydrophilic portion. It would be expected that these mobile portions of MBP would thus be available to bind, for example, cytoskeletal and SH3-domain-containing proteins to the membrane. The extents of the first two of these regions were in good agreement with membrane-penetration results from EPR spectroscopy (Bates et al. 2003), and correlated well with disordered regions observed by solution NMR spectroscopy (Libich and Harauz 2008b).
It is unclear why the segments Pro120–Leu148 and Leu154–Ser163 were mobile in this semi-solid system (Fig. 4F)— they overlap the C-terminal segment (Tyr142–Leu154), which was shown to have a strong α-helical propensity in 30% TFE-d2 (Fig. 4B) (Libich and Harauz 2008b). Moreover, a C-terminal residue Ser159 Cys spin-labelled mutant examined by EPR was found to be buried in the bilayer (Bates et al. 2003). The differences between these various studies may be due to the different environments and lipid compositions, which were dictated in large part by experimental requirements. On the other hand, a relatively weaker membrane association of the C-terminal region of MBP may make it accessible as a Ca2+–calmodulin target (Libich et al. 2003b; Polverini et al. 2004; Libich and Harauz 2008b). In general, then, in association with lipid bilayers, long stretches of MBP remain exposed to the bulk solvent (Fig. 4) to serve as binding sites for other proteins (e.g., cytoskeleton, CaM, SH3-domain) and modifying enzymes such as kinases, thus making MBP a dynamic linker of the cytoplasm to the membrane. Further investigations using complementary NMR methodologies, such as those described here, will serve to better define the adaptability and conformational polymorphism of MBP in myelin, and to further our understanding of how a disruption of the equilibrium of associations can result in myelin instability and confound inherent attempts at repair in multiple sclerosis.
Acknowledgments
These investigations were supported by the Canadian Institutes of Health Research (CIHR, Operating Grant MOP 74468 to G.H. and V.L.), the Canada Foundation for Innovation, and the Ontario Innovation Trust. V.L. is a Canada Research Chair in Biophysics at the University of Guelph, and is a recipient of an Early Researcher Award from the Ontario Ministry of Research and Innovation. D.S.L. was supported in part by an Ontario Graduate Scholarship. M.A. was a recipient of a Doctoral Studentship from the Ministry of Higher Education and Scientific Research of Egypt. We are grateful to Ms. Valerie Robertson (University of Guelph NMR Centre) and Dr. Martine Monette (Bruker BioSpin Canada) for their support and encouragement.
Footnotes
This paper is one of a selection of papers published in this special issue entitled “Canadian Society of Biochemistry, Molecular & Cellular Biology 52nd Annual Meeting — Protein Folding: Principles and Diseases” and has undergone the Journal’s usual peer review process.
Contributor Information
David S. Libich, Centre for Structural Biology, Institute of Fundamental Sciences, Department of Physics, Massey University, Palmerston North, New Zealand; Department of Molecular and Cellular Biology, and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON, N1G 2W1, Canada.
Mumdooh A.M. Ahmed, Department of Physics, and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON, N1G 2W1, Canada
Ligang Zhong, Department of Physics, and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON, N1G 2W1, Canada.
Vladimir V. Bamm, Department of Molecular and Cellular Biology, and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON, N1G 2W1, Canada
Vladimir Ladizhansky, Department of Physics, and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON, N1G 2W1, Canada.
George Harauz, Department of Molecular and Cellular Biology, and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON, N1G 2W1, Canada.
References
- Agarwal V, Diehl A, Skrynnikov N, Reif B. High resolution 1H detected 1H,13C correlation spectra in MAS solid-state NMR using deuterated proteins with selective 1H,2H isotopic labeling of methyl groups. J Am Chem Soc. 2006;128(39):12620–12621. doi: 10.1021/ja064379m. [DOI] [PubMed] [Google Scholar]
- Ahmed MA, Bamm VV, Shi L, Steiner-Mosonyi M, Dawson JF, Brown L, et al. Induced secondary structure and polymorphism in an intrinsically disordered structural linker of the cns: solid-state NMR and FTIR spectroscopy of myelin basic protein bound to actin. Biophys J. 2009;96(1):180–191. doi: 10.1016/j.bpj.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew ER, Bradbury A, Eades RG. Nuclear magnetic resonance spectra from a crystal rotated at high speed. Nature. 1958;182(4650):1659–1659. doi: 10.1038/1821659a0. [DOI] [Google Scholar]
- Barbar E. NMR characterization of partially folded and unfolded conformational ensembles of proteins. Biopolymers. 1999;51(3):191–207. doi: 10.1002/(SICI)1097-0282(1999)51:3<191::AID-BIP3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bates IR, Boggs JM, Feix JB, Harauz G. Membrane-anchoring and charge effects in the interaction of myelin basic protein with lipid bilayers studied by site-directed spin labeling. J Biol Chem. 2003;278(31):29041–29047. doi: 10.1074/jbc. [DOI] [PubMed] [Google Scholar]
- Bates IR, Feix JB, Boggs JM, Harauz G. An immunodominant epitope of myelin basic protein is an amphipathic alpha-helix. J Biol Chem. 2003;279(7):5757–5764. doi: 10.1074/jbc.M311504200. [DOI] [PubMed] [Google Scholar]
- Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12(1):1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci. 2006;63(17):1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein interactions with actin and tubulin in vitro: Binding, assembly, and regulation. In: Boggs JM, editor. Myelin basic protein. Nova Science Publishers; New York: 2008. pp. 149–167. [Google Scholar]
- Campagnoni AT, Campagnoni CW. The properties and functions of the golli myelin basic proteins. In: Boggs JM, editor. Myelin basic protein. Nova Science Publishers; New York: 2008. pp. 1–17. [Google Scholar]
- Chevelkov V, Rehbein K, Diehl A, Reif B. Ultra-high resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew Chem Int Ed Engl. 2006;45(23):3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- Crowhurst KA, Tollinger M, Forman-Kay JD. Cooperative interactions and a non-native buried Trp in the unfolded state of an SH3 domain. J Mol Biol. 2002;322(1):163–178. doi: 10.1016/S0022-2836(02)00741-6. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of protein folding. Wiley-VCH, Verlag; Weinheim, Germany: 2005. pp. 271–353. [Google Scholar]
- de Dios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science. 1993;260(5113):1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- DeBruin LS, Harauz G. White matter rafting—membrane microdomains in myelin. Neurochem Res. 2007;32(2):213–228. doi: 10.1007/s11064-006-9137-4. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem Rev. 2004;104(8):3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. According to current textbooks, a well-defined three-dimensional structure is a prerequisite for the function of a protein. Is this correct? IUBMB Life. 2006;58(2):107–109. doi: 10.1080/15216540500484376. [DOI] [PubMed] [Google Scholar]
- Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19(1):23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farès C, Libich DS, Harauz G. Solution NMR structure of an immunodominant epitope of myelin basic protein. Conformational dependence on environment of an intrinsically unstructured protein. FEBS J. 2006;273(3):601–614. doi: 10.1111/j.1742-4658.2005.05093.x. [DOI] [PubMed] [Google Scholar]
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15(1):35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Folmer RHA, Hilbers CW, Konings RNH, Hallenga K. A 13C double-filtered NOESY with strongly reduced artefacts and improved sensitivity. J Biomol NMR. 1995;5(4):427–432. doi: 10.1007/BF00182287. [DOI] [PubMed] [Google Scholar]
- Frericks HL, Zhou DH, Yap LL, Gennis RB, Rienstra CM. Magic-angle spinning solid-state NMR of a 144 kDa membrane protein complex: E.. coli cytochrome bo3 oxidase. J Biomol NMR. 2006;36(1):55–71. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels. J Mol Biol. 1997;268(1):158–169. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- Givogri MI, Bongarzone ER, Campagnoni AT. New insights on the biology of myelin basic protein gene: the neural-immune connection. J Neurosci Res. 2000;59(2):153–159. doi: 10.1002/(SICI)1097-4547(20000115)59:2<153::AID-JNR1>3.0. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ladizhansky V. Structure and dynamics of the myelin basic protein (MBP) family by solution and solid-state NMR. In: Boggs JM, editor. Myelin basic protein. Nova Science Publishers; New York: 2008. pp. 197–232. [Google Scholar]
- Harauz G, Libich DS. The classic basic protein of myelin—conserved structural motifs and the dynamic molecular barcode involved in membrane adhesion and protein–protein interactions. Curr Protein Pept Sci. 2009;10(3):196–215. doi: 10.2174/138920309788452218. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Farès C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35(7):503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ladizhansky V, Boggs JM. Structural polymorphism and multifunctionality of myelin basic protein. Biochemistry. 2009;48(34):8094–8104. doi: 10.1021/bi901005f. [DOI] [PubMed] [Google Scholar]
- Hardy EH, Verel R, Meier BH. Fast MAS total through-bond correlation spectroscopy. J Magn Reson. 2001;148(2):459–464. doi: 10.1006/jmre.2000.2258. [DOI] [PubMed] [Google Scholar]
- Hayashi N, Matsubara M, Jinbo Y, Titani K, Izumi Y, Matsushima N. Nef of HIV-1 interacts directly with calcium-bound calmodulin. Protein Sci. 2002;11(3):529–537. doi: 10.1110/ps.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazy E, Tompa P. Limitations of induced folding in molecular recognition by intrinsically disordered proteins. Chem Phys Chem. 2009;10(9–10):1415–1419. doi: 10.1002/cphc.200900205. [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108(6):739–742. doi: 10.1016/S0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- Hohwy M, Rienstra CM, Griffin RG. Band-selective homonuclear dipolar recoupling in rotating solids. J Chem Phys. 2002;117:4973–4987. doi: 10.1063/1.1488136. [DOI] [Google Scholar]
- Homchaudhuri L, Polverini E, Gao W, Harauz G, Boggs JM. Influence of membrane surface charge and post-translational modifications to myelin basic protein on its ability to tether the fyn-SH3 domain to a membrane in vitro. Biochemistry. 2009;48(11):2385–2393. doi: 10.1021/bi8022587. [DOI] [PubMed] [Google Scholar]
- Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ. Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126(21):6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J Chem Phys. 1979;71:4546–4553. doi: 10.1063/1.438208.. [DOI] [Google Scholar]
- Kanelis V, Forman-Kay JD, Kay LE. Multidimensional NMR methods for protein structure determination. IUBMB Life. 2001;52(6):291–302. doi: 10.1080/152165401317291147. [DOI] [PubMed] [Google Scholar]
- Kay LE. Pulsed field gradient multi-dimensional NMR methods for the study of protein structure and dynamics in solution. Prog Biophys Mol Biol. 1995;63(3):277–299. doi: 10.1016/0079-6107(95)00007-0. [DOI] [PubMed] [Google Scholar]
- Lee W, Revington MJ, Arrowsmith C, Kay LE. A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett. 1994;350(1):87–90. doi: 10.1016/0014-5793(94)00740-3. [DOI] [PubMed] [Google Scholar]
- Li Y, Berthold DA, Gennis RB, Rienstra CM. Chemical shift assignment of the transmembrane helices of DsbB, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Sci. 2008;17(2):199–204. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libich DS, Harauz G. Interactions of the 18.5-kDa isoform of myelin basic protein with Ca2+-calmodulin: in vitro studies using fluorescence microscopy and spectroscopy. Biochem Cell Biol. 2002;80(4):395–406. doi: 10.1139/o02-020. [DOI] [PubMed] [Google Scholar]
- Libich DS, Harauz G. Solution NMR and CD spectroscopy of an intrinsically disordered, peripheral membrane protein: evaluation of aqueous and membrane-mimetic solvent conditions for studying the conformational adaptability of the 18.5 kDa isoform of myelin basic protein (MBP) Eur Biophys J. 2008a;37(6):1015–1029. doi: 10.1007/s00249-008-0334-8. [DOI] [PubMed] [Google Scholar]
- Libich DS, Harauz G. Backbone dynamics of the 18.5 kDa isoform of myelin basic protein reveals transient alpha-helices and a calmodulin-binding site. Biophys J. 2008b;94(12):4847–4866. doi: 10.1529/biophysj.107.125823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libich DS, Hill CM, Haines JD, Harauz G. Myelin basic protein has multiple calmodulin-binding sites. Biochem Biophys Res Commun. 2003a;308(2):313–319. doi: 10.1016/S0006-291X(03)01380-9. [DOI] [PubMed] [Google Scholar]
- Libich DS, Hill CMD, Bates IR, Hallett FR, Armstrong S, Siemiarczuk A, Harauz G. Interaction of the 18.5-kD isoform of myelin basic protein with Ca2+-calmodulin: effects of deimination assessed by intrinsic Trp fluorescence spectroscopy, dynamic light scattering, and circular dichroism. Protein Sci. 2003b;12(7):1507–1521. doi: 10.1110/ps.0303603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libich DS, Robertson VJ, Monette MM, Harauz G. Backbone resonance assignments of the 18.5 kDa isoform of murine myelin basic protein (MBP) J Biomol NMR. 2004;29(4):545–546. doi: 10.1023/B:JNMR.0000034348.99658.d7. [DOI] [PubMed] [Google Scholar]
- Libich DS, Monette MM, Robertson VJ, Harauz G. NMR assignment of an intrinsically disordered protein under physiological conditions: the 18.5 kDa isoform of murine myelin basic protein. Biomol NMR Assign. 2007;1(1):61–63. doi: 10.1007/s12104-007-9016-1. [DOI] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15(12):2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Baker JM, Tollinger M, Forman-Kay JD. Calculation of residual dipolar couplings from disordered state ensembles using local alignment. J Am Chem Soc. 2008;130(25):7804–7805. doi: 10.1021/ja802220c. [DOI] [PubMed] [Google Scholar]
- Mészáros B, Tompa P, Simon I, Dosztányi Z. Molecular principles of the interactions of disordered proteins. J Mol Biol. 2007;372(2):549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2009 doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362(5):1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Morris GA, Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J Am Chem Soc. 1979;101(3):760–762. doi: 10.1021/ja00497a058. [DOI] [Google Scholar]
- Moscarello MA. Myelin basic protein, the “executive” molecule of the myelin membrane. In: Juurlink BHJ, Devon RM, Doucette JR, Nazarali AJ, Schreyer DJ, Verge VMK, editors. Cell biology and pathology of myelin: evolving biological concepts and therapeutic approaches. Plenum Press; New York: 1997. pp. 13–25. [Google Scholar]
- Moscarello MA, Mastronardi FG, Wood DD. The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem Res. 2007;32(2):251–256. doi: 10.1007/s11064-006-9144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musse AA, Harauz G. Molecular “negativity” may underlie multiple sclerosis: role of the myelin basic protein family in the pathogenesis of MS. Int Rev Neurobiol. 2007;79:149–172. doi: 10.1016/S0074-7742(07)79007-4. [DOI] [PubMed] [Google Scholar]
- Musse AA, Boggs JM, Harauz G. Deimination of membrane-bound myelin basic protein in multiple sclerosis exposes an immunodominant epitope. Proc Natl Acad Sci USA. 2006;103(12):4422–4427. doi: 10.1073/pnas.0509158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musse AA, Li Z, Ackerley CA, Bienzle D, Lei H, Poma R, et al. Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis Model Mech. 2008;1(4–5):229–240. doi: 10.1242/dmm.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadaud PS, Helmus JJ, Kall SL, Jaroniec CP. Paramagnetic ions enable tuning of nuclear relaxation rates and provide long-range structural restraints in solid-state NMR of proteins. J Am Chem Soc. 2009;131(23):8108–8120. doi: 10.1021/ja900224z. [DOI] [PubMed] [Google Scholar]
- Ohki SY, Kainosho M. Stable isotope labeling methods for protein NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2008;53(4):208–226. doi: 10.1016/j.pnmrs.2008.01.003. [DOI] [Google Scholar]
- Palmer AG., 3rd NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104(8):3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59(2):569–590. doi: 10.1063/1.1680061. [DOI] [Google Scholar]
- Polverini E, Boggs JM, Bates IR, Harauz G, Cavatorta P. Electron paramagnetic resonance spectroscopy and molecular modelling of the interaction of myelin basic protein (MBP) with calmodulin (CaM)-diversity and conformational adaptability of MBP CaM-targets. J Struct Biol. 2004;148(3):353–369. doi: 10.1016/j.jsb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Polverini E, Rangaraj G, Libich DS, Boggs JM, Harauz G. Binding of the proline-rich segment of myelin basic protein to SH3 domains: spectroscopic, microarray, and modeling studies of ligand conformation and effects of posttranslational modifications. Biochemistry. 2008;47(1):267–282. doi: 10.1021/bi701336n. [DOI] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. The human myelin basic protein gene is included within a 179-kilobase transcription unit: expression in the immune and central nervous systems. Proc Natl Acad Sci USA. 1993;90(22):10695–10699. doi: 10.1073/pnas.90.22.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Receveur-Bréchot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62(1):24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Rule GS, Hitchens TK. Fundamentals of protein NMR spectroscopy. Springer; Dordrecht, Netherlands: 2006. [Google Scholar]
- Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J Biomol NMR. 2000;18(1):43–48. doi: 10.1023/A:1008386816521. [DOI] [PubMed] [Google Scholar]
- Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123(13):2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- Shi LC, Ahmed MAM, Zhang WR, Whited G, Brown LS, Ladizhansky V. Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump—structural insights. J Mol Biol. 2009;386(4):1078–1093. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Shortle D. The expanded denatured state: an ensemble of conformations trapped in a locally encoded topological space. Adv Protein Chem. 2002;62:1–23. doi: 10.1016/S0065-3233(02)62003-0. [DOI] [PubMed] [Google Scholar]
- Shortle D, Ackerman MS. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293(5529):487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128(40):13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- Smith R. The basic protein of CNS myelin: its structure and ligand binding. J Neurochem. 1992;59(5):1589–1608. doi: 10.1111/j.1471-4159.1992.tb10989.x. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344(5–6):631–637. doi: 10.1016/S0009-2614(01)00791-6. [DOI] [Google Scholar]
- Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays. 2009;31(3):328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci USA. 2009;106(25):10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Hwang PM, Kay LE. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem. 2004;73(1):107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Nip AM. Protein chemical shift analysis: a practical guide. Biochem Cell Biol. 1998;76(2–3):153–163. doi: 10.1139/bcb-76-2-3-153. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Dyson HJ, Wright PE. Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS Lett. 1997;419(2–3):285–289. doi: 10.1016/S0014-5793(97)01474-9. [DOI] [PubMed] [Google Scholar]
- Zhong L, Bamm VV, Ahmed MA, Harauz G, Ladizhansky V. Solid-state NMR spectroscopy of 18.5 kDa myelin basic protein reconstituted with lipid vesicles: spectroscopic characterisation and spectral assignments of solvent-exposed protein fragments. Biochim Biophys Acta. 2007;1768(12):3193–3205. doi: 10.1016/j.bbamem.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]