Abstract
The prominent host muramidase lysozyme cleaves bacterial peptidoglycan (PG), and the enzyme is abundant in mucosal secretions. The lytic enzyme susceptibility of Gram-negative bacteria and mechanisms they use to thwart lytic enzyme activity are poorly studied. We previously characterized a Helicobacter pylori PG modification enzyme, an N-deacetylase (PgdA) involved in lysozyme resistance. In this study, another PG modification enzyme, a putative PG O-acetyltransferase (PatA), was identified. Mass spectral analysis of the purified PG demonstrated that a patA strain contained a greatly reduced amount of acetylated muropeptides, indicating a role for PatA in H. pylori PG O-acetylation. The PG modification mutant strains (pgdA, patA, or pgdA patA) were more susceptible to lysozyme killing than the parent, but this assay required high lysozyme levels (up to 50 mg/ml). However, addition of host lactoferrin conferred lysozyme sensitivity to H. pylori, at physiologically relevant concentrations of both host components (3 mg/ml lactoferrin plus 0.3 mg/ml lysozyme). The pgdA patA double mutant strain was far more susceptible to lysozyme/lactoferrin killing than the parent. Peptidoglycan purified from a pgdA patA mutant was five times more sensitive to lysozyme than PG from the parent strain, while PG from both single mutants displayed intermediate sensitivity. Both sensitivity assays for whole cells and for purified PGs indicated that the modifications mediated by PgdA and PatA have a synergistic effect, conferring lysozyme tolerance. In a mouse infection model, significant colonization deficiency was observed for the double mutant at 3 weeks postinoculation. The results show that PG modifications affect the survival of a Gram-negative pathogen.
Importance Pathogenic bacteria evade host antibacterial enzymes by a variety of mechanisms, which include resisting lytic enzymes abundant in the host. Enzymatic modifications to peptidoglycan (PG, the site of action of lysozyme) are a known mechanism used by Gram-positive bacteria to protect against host lysozyme attack. However, Gram-negative bacteria contain a thin layer of PG and a recalcitrant outer membrane permeability barrier to resist lysis, so molecular modifications to cell wall structure in order to combat lysis remain largely unstudied. Here we show that two Helicobacter pylori PG modification enzymes (PgdA and PatA) confer a clear protective advantage to a Gram-negative bacterium. They protect the bacterium from lytic enzyme degradation, albeit via different PG modification activities. Many pathogens are Gram negative, so some would be expected to have a similar cell wall-modifying strategy. Understanding such strategies may be useful for combating pathogen growth.
Importance
Pathogenic bacteria evade host antibacterial enzymes by a variety of mechanisms, which include resisting lytic enzymes abundant in the host. Enzymatic modifications to peptidoglycan (PG, the site of action of lysozyme) are a known mechanism used by Gram-positive bacteria to protect against host lysozyme attack. However, Gram-negative bacteria contain a thin layer of PG and a recalcitrant outer membrane permeability barrier to resist lysis, so molecular modifications to cell wall structure in order to combat lysis remain largely unstudied. Here we show that two Helicobacter pylori PG modification enzymes (PgdA and PatA) confer a clear protective advantage to a Gram-negative bacterium. They protect the bacterium from lytic enzyme degradation, albeit via different PG modification activities. Many pathogens are Gram negative, so some would be expected to have a similar cell wall-modifying strategy. Understanding such strategies may be useful for combating pathogen growth.
Introduction
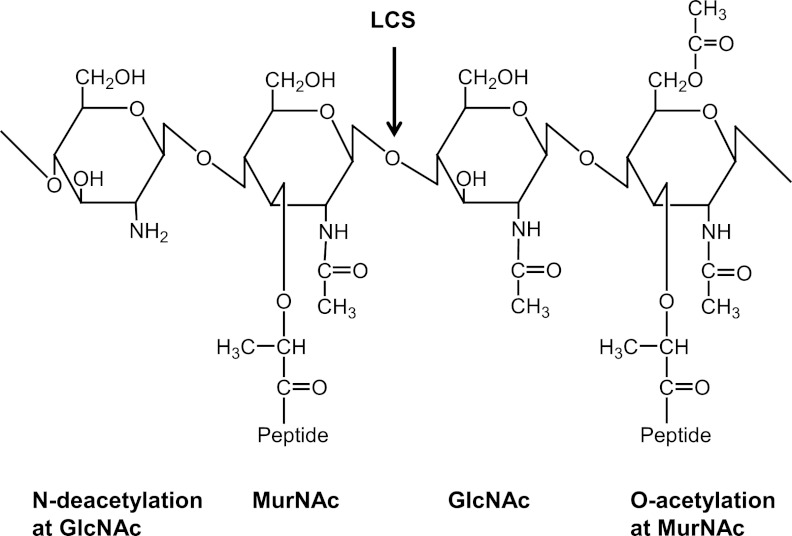
Peptidoglycan (PG) is one of the protective barriers of the bacterial cell wall. PG consists of glycan strands of alternating beta-1,4-linked N-acetylglucosamine (GlucNAc) and N-acetylmuramic acid (MurNAc), which are cross-linked by short peptide chains (1) (Fig. 1). Given the inherent importance of PG in bacteria, it is not surprising that lysozyme, one of the important components of the innate immunity of eukaryotic hosts, targets this polymer (2). Lysozyme is a muramidase that cleaves bacterial PG between the glycosidic beta-1,4-linked MurNAc and GlcNAc. Lysozyme is produced by the epithelia and is also a major component of the granules of neutrophils and macrophages, often being recruited when the mucosa is acutely inflamed (3, 4). The lytic enzyme is induced in numerous types of cells, e.g., monocytes and macrophages (5) and colonic epithelial cells (6). To establish infection, pathogenic bacteria have developed strategies to counteract the hydrolytic activity of lysozyme.
FIG 1 .
Structure of the GlcNAcβ 1,4-linked MurNAc repeating disaccharide of bacterial peptidoglycan, also showing modifications that confer lysozyme resistance (de-N-acetylation at GlcNAc and O-acetylation at MurNAc). LCS, lysozyme cleavage site.
Some bacterial pathogens modify their PG to resist host lysozyme (7). Two main types of PG modifications conferring lysozyme resistance have been identified: O-acetylation occurring at the C-6 hydroxyl moiety of the MurNAc residues (8, 9) and N-deacetylation of the GlcNAc residues (10–13) (Fig. 1). These have been studied in Gram-positive bacteria, and the genes responsible for these activities are oatA (for O-acetyltransferase) and pgdA (for PG N-deacetylase), respectively (7). PG modifications in Gram-negative bacteria and their involvement in pathogenesis have been rarely investigated.
Helicobacter pylori, a Gram-negative bacterium, is an important human gastric pathogen that chronically infects 50% of the world’s population (14). During the process of colonizing the host, H. pylori induces a strong inflammatory response. However, H. pylori survives the host immune response to persistently colonize the gastric mucosa, often, it is believed, for the life span of the host (15). The mechanisms by which H. pylori evades host immune responses are poorly understood. Recently, we identified and characterized an H. pylori protein (HP310) whose expression was significantly induced under oxidative stress conditions (16). HP310 turned out to be an enzyme catalyzing PG modification: PG N-deacetylase (PgdA). With a limited sequence homology to PgdAs of Gram-positive bacteria, H. pylori PgdA is a representative of a new subfamily of bacterial PG deacetylases. We have shown that the contact of H. pylori with host immune cells (macrophages) also induces overexpression of PgdA and that H. pylori PG N-deacetylation is an important mechanism for mitigating host immune responses, contributing to pathogen persistence in the host (17). A significant phenotype of H. pylori pgdA mutants in vitro is a decrease in lysozyme tolerance compared to its parent strain, if high levels of lysozyme are used in the assay.
Neither a homolog nor a paralog of OatA of Gram-positive bacteria have been found in Gram-negative bacteria (1). Instead, a putative O-acetyltransferase gene (pacA or patA) in Neisseria was suggested to be involved in lysozyme resistance (18, 19). Purified PG from a Neisseria gonorrhoeae pacA mutant strain was more sensitive to lysozyme than the wild-type PG, but whole cells of the pacA mutant and the wild type were equally lysozyme resistant (18). In this study, we characterized a PatA homolog (HP0855) of H. pylori, and we show that PgdA and PatA have overlapping roles, together conferring lysozyme resistance.
Gram-negative bacteria are not generally susceptible to lysozyme because their outer membrane prevents access of the secreted enzyme to the PG layer. However, this barrier can be overcome in some cells via host accessory proteins, such as lactoferrin, that permeabilize the bacterial outer membrane (20). In the present study, we demonstrate that H. pylori cells become sensitive to lysozyme at physiologically relevant lytic enzyme concentrations when a membrane permeabilizer lactoferrin is present. From the results obtained with a mouse infection model, we conclude that PgdA- and PatA-mediated PG modifications contribute to H. pylori’s survival in the host.
RESULTS
Construction and primary characterization of a patA mutant of H. pylori.
The gene HP0855 in the published genome sequence (strain 26695) was annotated as a homolog of AlgI (21), which has 28% amino acid identity to AlgI in Pseudomonas aeruginosa. P. aeruginosa AlgI is involved in O-acetylation of the exopolysaccharide alginate. Given that N. gonorrhoeae and H. pylori do not produce alginate, Weadge et al. (19) proposed that HP0855, a hypothetical membrane protein, functions to O-acetylate peptidoglycan and therefore named the gene patA, for peptidoglycan O-acetyltransferase A. However, the recent study on PG O-acetylation in N. gonorrhoeae revealed that the gene downstream of patA encodes a protein (named PatB) (Fig. 2) which localizes in the periplasm and has a PG O-acetyltransferase activity (22). The transmembrane protein PatA in N. gonorrhoeae remains uncharacterized, but was proposed to function in translocation of acetyl groups across the cytoplasmic membrane to the periplasm to be used by PatB (22, 23). Downstream of the patA gene in the H. pylori genome there exists a hypothetical gene (HP0856) (Fig. 2). H. pylori PatA has 30% amino acid identity to N. gonorrhoeae PatA, but HP0856 has no significant homology to N. gonorrhoeae PatB. However, HP0855 and HP0856 appear to always coexist in certain H. pylori strains (e.g., strains 26695, X47, B38, and v225d). In those strains they exist contiguously and at the same place in the genome. Interestingly, many strains of H. pylori (such as J99 and B128) do not contain these two genes (Fig. 2). The evolutionary origin of these two genes is unknown, and no evidence of horizontal gene transfer can be found from the genetic organization.
FIG 2 .
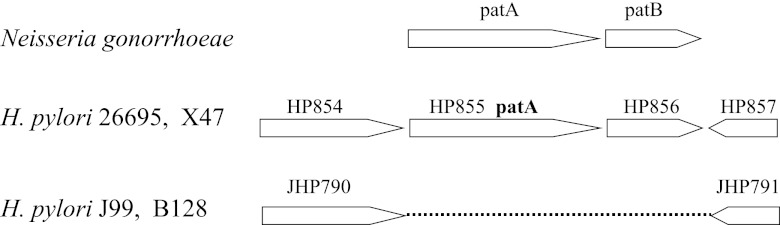
Organizations of the O-acetylpeptidoglycan (OAP) gene cluster in N. gonorrhoeae and H. pylori. HP854 and JHP790 are annotated as guaC, coding for GMP reductase; HP857 and JHP791 are annotated as gmhA, coding for phosphoheptose isomerase.
To study the physiological roles of H. pylori PatA, we constructed patA::cat mutants. The mutants were obtained in strains 26695 and X47; the growth of the patA mutants under normal growth conditions was similar to that of the wild type strain (data not shown). As an initial characterization, we examined the lysozyme sensitivity of strain 26695 patA::cat, compared to its WT parent strain (Table 1). H. pylori (WT or patA mutant) cells were grown to late log phase. Approximately 108 cells (in a volume of 1 ml) were treated with 50 mg/ml lysozyme, and the numbers of surviving cells were determined after 0, 2, 4, or 6 h. During the lysozyme treatment, the number of viable patA mutant cells decreased more rapidly than the number of wild-type (WT) cells. Six hours after the treatment, about 105 viable cells of the wild-type strain survived, while only 500 cells of the patA mutant remained viable. This indicated that the patA mutant is more sensitive to lysozyme than the wild type. Although this high concentration of lysozyme (50 mg/ml) used in vitro is not physiologically relevant, PatA seems to play a significant role in protecting H. pylori PG from lysozyme digestion. As X47 is a well-adapted strain for mouse colonization (24) and it is frequently used in our lab for mouse colonization studies (17, 25), the following assays reported herein were done with X47 and its isogenic patA mutant.
TABLE 1 .
Lysozyme sensitivity
| Time (h) | Cell survivala |
|||
|---|---|---|---|---|
| 26695 (WT) |
26695 (patA::cat) |
|||
| CFU/ml | % survival | CFU/ml | % survival | |
| 0 | 3.8 × 108 | 100 | 7.5 × 107 | 100 |
| 2 | 1.2 × 108 | 32 | 6.8 × 106 | 9 |
| 4 | 2.8 × 107 | 7 | 5.0 × 105 | 0.7 |
| 6 | 2.2 × 105 | 0.06 | 5.5 × 102 | 0.0007 |
H. pylori 26695 WT or patA:cat mutant cells were treated with 50 mg/ml lysozyme for 0, 2, 4 or 6 h, and cell survival was determined by plate counting.
PatA functions in O-acetylation of H. pylori PG.
To test the hypothesis that PatA is involved in PG modification (specifically, O-acetylation), we sought to determine the extent of O-acetylation of pure PG isolated from the wild type and the patA mutant. Previously we established protocols for isolation of PG from H. pylori such that the N-deacetylation state could be assessed (16); however, those methods, which include acid treatment to remove cell wall polymers, were not useful for detection of O-acetyl groups in PG. This is because the ester bond of O-linked acetate is significantly weaker than the amide bond of N-linked acetate, so acetate is lost from O-acetylated MurNAc under acidic conditions (1). Therefore, we used a modified method to purify PG at neutral pH designed to preserve O-acetyl groups, as described by Bera et al. (8) (see Materials and Methods).
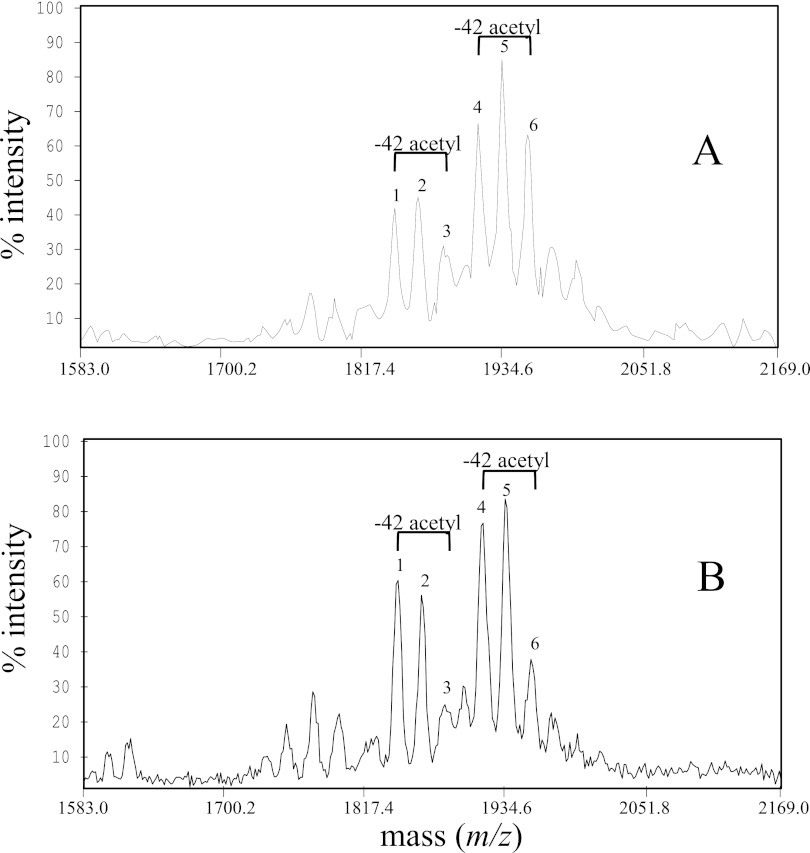
We analyzed the MALDI (matrix-assisted laser desorption ionization) mass spectra for muropeptide composition of PGs purified from H. pylori WT or the patA mutant. The mass spectral analysis for both WT and patA mutant PG revealed two main families of PG digestion products, the monomeric (disaccharide-peptide) and dimeric (disaccharide-peptide-peptide-disaccharide) muropeptides, detected as both the sodiated molecular ions (positive ion mode) and as their [M-H]−1 ions (negative ion mode) (Fig. 3). Expansion of the dimeric region revealed significant differences between the wild type and the mutant strains in the ratios and masses of peaks (Fig. 3). Many of the major molecular ions identified fit closely with ions previously reported for muropeptide components in H. pylori PG (26) and thus are likely have the same molecular formula (Table 2). We identified two major muropeptide (MP) pairs: the m/z 1847/1889 (no. 1/no. 3) pair and the m/z 1918/1960 (no. 4/no. 6) pair; each represents a difference of 42 mass units (one acetyl group). For instance, MP 6 (m/z 1960) is the acetylated form of MP 4 (m/z 1918). While the wild-type PG contained the two MP components at similar intensities, the patA mutant PG had an elevated MP 4 (m/z 1918) and a greatly reduced MP 6 (m/z 1960). As calculated from the relative ion intensities, the ratio of the nonacetylated MP (no. 4) to its corresponding acetylated MP (no. 6) is 1.00/1.19 in the WT and 1.00/0.62 in the patA mutant. This suggested that the patA mutant PG had a much smaller percentage of acetylated muropeptides than the wild-type PG. A similar difference between WT and patA mutant PG was also observed for the m/z 1847/1889 ion pair (MPs 1 and 3). The ratio of the nonacetylated MP (no. 1) to its corresponding acetylated MP (no. 3) is 1.00/1.50 in the WT and 1.00/0.79 in the patA mutant, again indicating a relative decrease in the abundance of acetylated MP in the mutant. Thus, the comparative MALDI-TOF analysis of PGs from the wild type and patA mutant indicated that PatA indeed has a role in PG O-acetylation.
FIG 3 .
Comparison of MALDI-TOF mass spectra of the dimeric muropeptide products of PGs derived from the H. pylori wild-type strain (A) and the patA::cat mutant strain (B). All spectra are displayed in the negative ion mode, and ions are of the form (M-H)−1. Some pairs of major muropeptide peaks having a difference of 42 Da in mass, the incremental mass of acetate, are labeled. Note that the ions corresponding to acetylated muropeptides are significantly reduced in the patA mutant PG compared to wild type (Table 2). The more abundant ions in the patA mutant represent muropeptides which lack an O-acetyl group (m/z 1847 and 1918). The two ion pairs (m/z 1960/1918 and 1889/1847) differ from each other by 71 mass units, the incremental mass of alanine.
TABLE 2 .
Analysis of muropeptides purified from H. pylori X47 and its patA mutant cells
| Muropeptidea | Molecular mass of [M-H] ions (Da)b |
Proposed structurec | Relative abundance of muropeptidesd |
|
|---|---|---|---|---|
| WT | patA | |||
| 1 | 1,847 | NAG-[(an)NAM]-Ala–Glu-mDap-Ala NAG-[NAM]-Ala–Glu-mDap-Ala |
2.6 | 1.9 |
| 2 | 1,867 | NAG-[NAM]-Ala–Glu-mDap-Ala NAG-[NAM]-Ala–Glu-mDap-Ala |
2.9 | 3.1 |
| 3 | 1,889 | NAG-[(an)NAM]-Ala–Glu-mDap-Ala NAG-[(Ac)NAM]-Ala–Glu-mDap-Ala |
3.9 | 1.5 |
| 4 | 1,918 | NAG-[(an)NAM]-Ala–Glu-mDap-Ala NAG-[NAM]-Ala–Glu-mDap-Ala-Ala |
4.2 | 4.0 |
| 5 | 1,938 | NAG-[NAM]-Ala–Glu-mDap-Ala NAG-[NAM]-Ala–Glu-mDap-Ala-Ala |
5.5 | 5.5 |
| 6 | 1,960 | NAG-[(an)NAM]-Ala–Glu-mDap-Ala NAG-[(Ac)NAM]-Ala–Glu-mDap-Ala-Ala |
5.0 | 2.5 |
Numbers correspond to the peaks labeled in Fig 3.
Molecular mass values measured for borodeuteride-reduced muropeptides.
For each proposed structure, the last Ala of the first PG unit is linked to the mDap of the second unit. Muropeptides best fitting to the molecular mass measured by MALDI-MS. Abbreviations: NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid; (an)NAM, (16)anhydro-N-acetylmuramic acid; (Ac)NAM, O-acetyl-N-acetylmuramic acid. According to the literature, the most common site for O-acetylation of PG is the 6 position of MurNAc, although we have not proven this for our system.
Relative abundances based on the measured relative ion intensities shown in Fig 3.
PgdA and PatA contribute synergistically to lysozyme resistance.
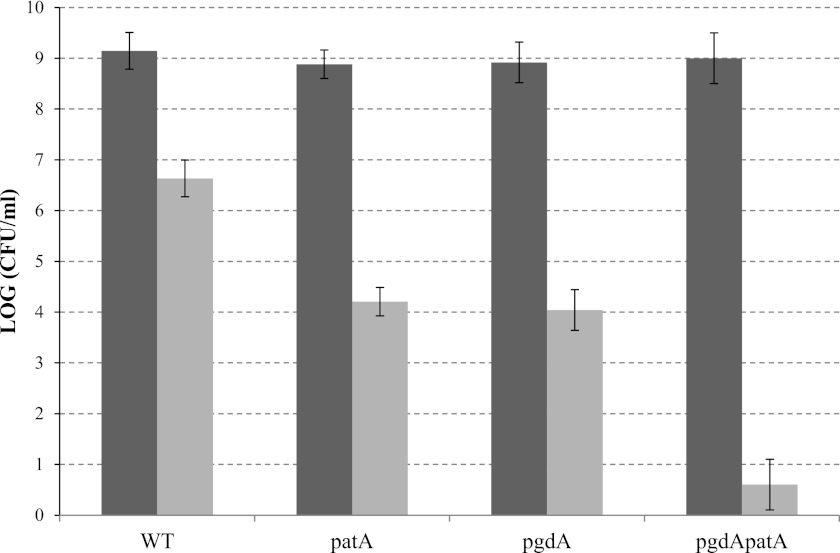
Both patA (this study) and pgdA (16) mutants showed a higher sensitivity to lysozyme than the wild type, although the concentration of lysozyme (50 mg/ml) used was much higher than levels expected to be encountered in vivo. To study the total contribution of PG modifying enzymes (PatA and PgdA) in lysozyme resistance, we constructed a H. pylori X47 pgdA patA double mutant. Lysozyme sensitivity was determined for the wild type and the patA, pgdA, and double mutants. H. pylori cells grown to late log phase (~109 cells/ml) were treated with 30 mg/ml lysozyme for 6 h, and the numbers of surviving cells were determined (Fig. 4). While more than 106 cells/ml of the wild-type strain survived, approximately 104 cells/ml of the patA or pgdA mutant strain survived, indicating that PatA and PgdA individually contribute similarly to lysozyme resistance. The same treatment (30 mg/ml lysozyme for 6 h) killed almost all of the double mutant cells (<10 cells/ml survived). This indicated that both PgdA and PatA are required for full resistance to lysozyme.
FIG 4 .
Lysozyme sensitivity of H. pylori X47 WT and the patA, pgdA and pgdA patA mutants. H. pylori cells grown to late log phase (~109 cells/ml, black bars) were treated with 30 mg/ml lysozyme for 6 h, and the numbers of surviving cells (gray bars) were determined after serial dilution and plate counting. The data are the means from three experiments, with standard deviation indicated by the error bars. According to Student’s t test, there are significant differences (P < 0.001) between the double mutant and each single mutant and between each single mutant and the wild type but not between the two single mutants.
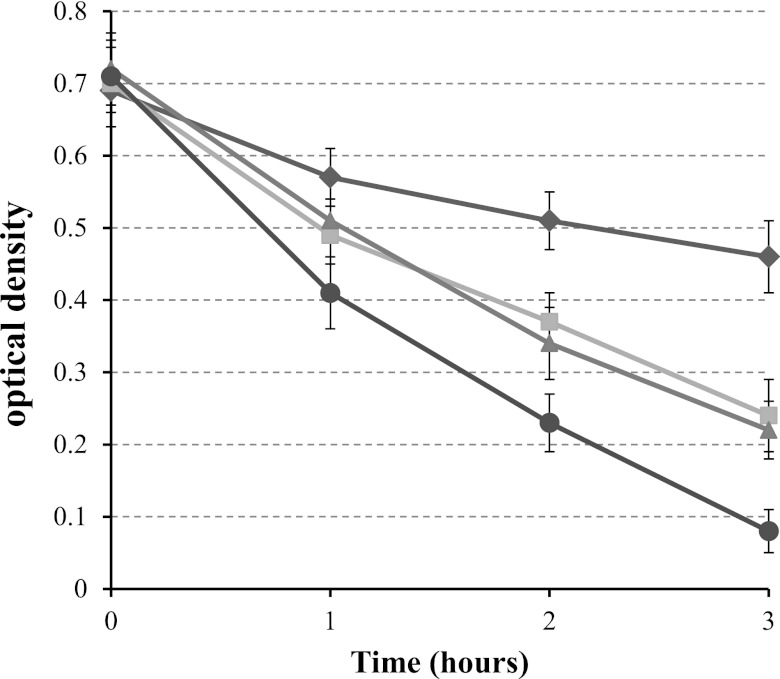
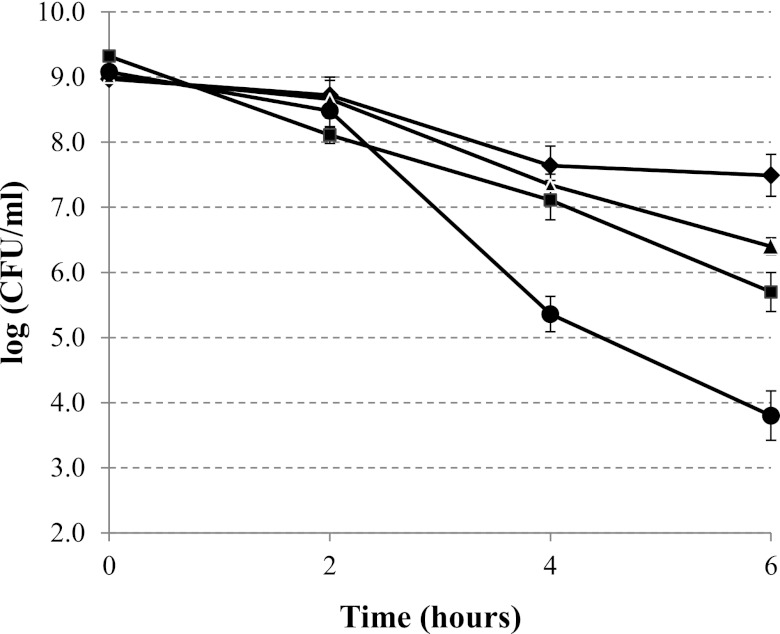
To further address the contribution of PatA and PgdA to protecting PG from lysozyme digestion, we purified PGs from different strains (WT and the patA, pgdA, and double mutants) and assessed their sensitivity to lysozyme directly. Purified PGs were treated with 0.3 mg/ml lysozyme for 1, 2, or 3 h, and the optical density at 600 nm was determined as an indication of PG degradation (Fig. 5). Upon treatment with lysozyme, a slight decrease of the optical density was observed for the WT PG. In contrast, the optical density decreased more rapidly for the pure PG from the pgdA or patA single mutant. According to the Student t test, the differences between PG susceptibilities of the WT and mutants at all three time points were significantly different (P < 0.01), while there was no significant difference between those of the pgdA and patA mutants. Strikingly, the PG purified from the double mutant was much more sensitive to lysozyme digestion than the WT PG or the two single mutant PGs. The results support our hypothesis that PgdA and PatA have overlapping roles, synergistically contributing to lysozyme resistance, albeit through different modification mechanisms.
FIG 5 .
Lysozyme sensitivity of purified PGs from H. pylori strains. Purified PGs were treated with 0.3 mg/ml lysozyme for the indicated times, and the optical density at 600 nm was determined as an indication of PG degradation. The data are the means and standard deviations from three experiments. Diamonds, WT; squares, pgdA mutant; triangles, patA mutant; circle, pgdA patA mutant.
H. pylori cells become sensitive to lysozyme in the presence of a membrane permeabilizer.
H. pylori cells in vitro are highly resistant to lysozyme killing (they survive for hours in up to 50 mg/ml), but that lysozyme level seems too high to allow us to draw conclusions on the susceptibility of H. pylori to lysozyme within the host. In vivo, however, lysozyme may gain access to the cell wall of Gram-negative bacteria through outer membrane disruptions caused by lactoferrin (20). While each protein (lactoferrin or lysozyme) alone is bacteriostatic, together they can be bactericidal for certain Gram-negative bacteria, such as Escherichia coli and Vibrio cholerae (20). We sought to determine whether this is the case in H. pylori and whether H. pylori PatA and/or PgdA contributes to whole-cell resistance at physiological levels of lysozyme (in the presence of lactoferrin). Lactoferrin levels in vivo have been measured up to 7 mg/ml (27). We chose to use a regimen of 0.3 mg/ml lysozyme plus 3 mg/ml lactoferrin in the assays to test the viability of H. pylori strains. Each component alone (0.3 mg/ml lysozyme or 3 mg/ml lactoferrin) had no significant effect on viability, but over 90% of WT cells lost viability by treatment for 3 h with the lysozyme/lactoferrin mixture (data not shown). This indicated that H. pylori cells become sensitive to a physiological level of lysozyme in the presence of lactoferrin.
The sensitivities of the strains to the lysozyme/lactoferrin mixture were compared (Fig. 6). H. pylori cell suspensions (~109 cells/ml) were treated for 2, 4, or 6 h, and the numbers of surviving cells were determined. After 2 hours of lysozyme/lactoferrin treatment, a slight and statistically insignificant difference in viability between all the strains was observed. However, at later time points, the viability of the double mutant cells decreased much faster than that of the WT and single mutants. At the 4-h time point, the viable cell number of the double mutant was more than 2 logs lower than that of the WT (P < 0.001), while the viability of the patA or pgdA single mutant cells was slightly lower than that of the WT but did not reach a statistically significant level. Still, statistically significantly (P < 0.01) greater killing of the single mutant cells compared to that of the WT was observed at the 6-h time point, indicating that PgdA or PatA separately confers resistance to the physiological level of lysozyme in the presence of lactoferrin. The surviving cell number of the double mutant cells was extremely low at the 6-h time point, supporting the notion that both PgdA and PatA are required for full resistance to lysozyme.
FIG 6 .
Lysozyme sensitivity in the presence of lactoferrin. H. pylori cell suspensions (~109 cells/ml) were treated with 0.3 mg/ml lysozyme plus 3 mg/ml lactoferrin for the indicated times, and the numbers of surviving cells were determined. The data are the means and standard deviations from three experiments. Diamonds, WT; triangles, pgdA mutant; squares, patA mutant; circles, pgdA patA mutant.
PG modifications affect H. pylori’s ability to colonize mouse stomachs.
Previously we examined the mouse colonization ability of the pgdA mutant in parent strain X47 background (17). Up to 3 weeks after inoculation, there was no difference in colonization between the wild type and the mutants (similar number of H. pylori cells were detected in mouse stomachs). This suggested no role for PgdA (alone) in protecting H. pylori from direct killing by lysozyme. At 9 weeks postinoculation, the pgdA mutant showed a significantly attenuated ability to colonize mouse stomachs, which was attributed to likely augmented host immune recognition (17). Based on the results presented above, however, PgdA and PatA together have a significant role in combating lysozyme killing. Therefore, we hypothesize that the early colonization ability, when the bacterium encounters the mucosa (milieu), could be deficient in the pgdA patA strain. In this study, mouse colonization assays (using C57BL mice) were performed with the patA and pgdA patA strains and, at the same time, with the WT and the pgdA mutant as controls. The numbers of H. pylori cells colonizing mouse stomachs were determined at 3 weeks after inoculation (Table 3).
TABLE 3 .
Mouse colonization abilities of H. pylori X47 and its isogenic mutant strainsa
| Strain genotype | No. of colonized mice/totalb |
Bacterial load (mean CFU/mg stomach ± SD)c |
|---|---|---|
| WT | 10/10 | 691 ± 103 |
| pgdA | 10/10 | 598 ± 184 |
| patA | 10/10 | 623 ± 127 |
| pgdA patA | 6/10 | 60 ± 52 |
Groups of 10 mice were inoculated with H. pylori two times (two days apart) with a dose of 1.5 × 108 viable cells administered per animal each time. Colonization of H. pylori in mouse stomachs was examined 3 weeks after the first inoculation.
Number of mouse stomachs from which H. pylori was recovered/total number of stomachs assayed.
Number of H. pylori cells colonized in mouse stomach averaged from the 10 mice with standard deviation. The detection limit of the assay is 0.5 CFU/mg stomach.
Similar to the pgdA mutant, the patA mutant showed no defect in mouse colonization ability. In contrast, a greatly decreased colonization number was observed for the double mutant at 3 weeks postinoculation. Six of 10 mice were found to be colonized by the pgdA patA double mutant strain, and the mean bacterial load for the double mutant was more than 10-fold lower than that for the WT strain. According to Wilcoxon rank test analysis, the ranges of colonization values of the double mutant strain is significantly lower than that of the wild type at the 99% confidence level (P < 0.01). These results indicate that both PgdA and PatA together contribute to H. pylori survival/colonization in the host within a period of 3 weeks.
DISCUSSION
Lysozyme, which targets the bacterial cell wall component peptidoglycan (PG), is one of the first lines of host defense against bacterial infections. In Gram-negative bacteria, PG is protected from lysozyme digestion due to an outer membrane; such cells (like H. pylori) are highly resistant to lysozyme compared to Gram-positive bacteria. Molecular mechanisms conferring lysozyme resistance and putative roles in pathogenesis have been infrequently studied in Gram-negative bacteria. Some bacteria produce lysozyme inhibitors, which include the Escherichia coli Ivy protein (28) and the Salmonella enterica serovar Enteritidis PliC (29). However, homologues of these are absent in H. pylori. Previously we characterized a H. pylori PG N-deacetylase (PgdA) that is implicated in lysozyme resistance (16). In the present study, we investigated the possibility that another PG modifier (the putative PG O-acetyltransferase PatA) also confers lysozyme resistance and that the lysozyme resistance mediated by both PgdA and PatA plays a significant role in H. pylori survival in vivo.
PG O-acetylation has been studied in very few Gram-negative bacteria (1). In the present study, we used a modified protocol for isolation of PG allowing detection of the labile O-acetyl groups in PG. O-Acetylation has so far not been detected from the muropeptide compositions of H. pylori PG by other studies focusing on the structural roles of PG (26, 30–34). Most likely, many of the strains used in those studies do not contain patAB genes. Indeed, the patAB genes are present in only 4 of 30 H. pylori strains whose whole-genome sequences are available. The comparative MALDI--time-of-flight (TOF) analysis of pure PGs from the patA mutant and the WT strain suggested a role for PatA in H. pylori PG O-acetylation, as the patA mutant contained a greatly reduced amount of the acetylated muropeptides. One may ask why the peaks representing structures with acetyl groups (m/z 1889 and 1960) are not completely eliminated in the mutant. First, the observed ion peaks may arise from more than one compound, i.e., from any muropeptide (or other molecule) having the same mass. Also, it is possible that there exist other O-acetyltransferase systems in H. pylori, providing a degree of redundancy for this important modification.
A clue to the role for H. pylori PG modifications was provided by the pgdA and patA mutant strains, which displayed lysozyme resistance in vitro. A concentration of 30 or even 50 mg/ml lysozyme was required to differentiate a lysozyme-mediated killing effect between WT and pgdA or patA strains. For comparison, lysozyme at a concentration of less than 0.1 mg/ml was used for sensitivity tests for Gram-positive bacteria (8, 10, 12, 13). However, purified PGs from the H. pylori pgdA or patA mutant strains are more sensitive to lysozyme digestion than the purified PG from the WT strain, using lysozyme at an in vivo physiological concentration (0.3 mg/ml) in the assay. Most importantly, our results showed that the pgdA patA cells were extremely sensitive to physiological levels of lysozyme plus lactoferrin. For the first time, we provide evidence that H. pylori cells can be sensitive to lysozyme at physiologically relevant host enzyme concentrations. Human immune cells produce a battery of antibacterial peptides, including lysozyme and membrane permeabilizers. Thus, PG modifications in Gram-negative bacteria may play a significant role in protecting cells from killing by host lysozyme.
Several lines of evidence indicate that PatA and PgdA have a similar contributions to the overall lysozyme resistance in H. pylori (at least in the strain used in this study), and both enzymes have an additive effect to confer resistance. Many bacterial species modify their PG by either O-acetylation or N-deacetylation, but few organisms have thus far been described as having both modifications (1). While O-acetylation is the major determinant of lysozyme resistance in some bacteria, N-deacetylation plays a dominant role in others (8, 9, 35). Our results for H. pylori suggest that both modifications may be needed for full resistance to lysozyme. However, many H. pylori strains do not have a patA gene; in those strains, PgdA may be the only determinant of lysozyme resistance. We are investigating lysozyme resistance by comparing the strains that have patA gene (e.g., X47) to the strains that have no patA gene (e.g., B128).
The role of PG modifications in bacterial pathogenesis has thus far been studied in only a few invasive, Gram-positive bacteria (7). It was unclear whether PG modifications (N-deacetylation and O-acetylation) in H. pylori would provide protection from lysozyme killing in vivo. Mucosal lysozyme may have a direct effect on attenuating colonization through its ability to lyse the H. pylori pgdA patA double mutant. Alternatively, degradation of PG could release specific PG fragments recognized by the host which induce the Nod1-mediated immune response (36, 37) and indirectly stimulate bacterial clearance. H. pylori was shown to be able to inject its PG into host epithelial cells by a bacterial type IV secretion system encoded by the cag pathogenicity island (38) and via outer membrane vesicles (39). In the mouse colonization assay used here, we used the strain X47, which lacks the major pathway (cag pathogenicity island [PAI]) for PG delivery. Thus, the survival attenuation of the pgdA patA double mutant is at least partly due to lysozyme killing. Overall, the survival ability of the double mutant was more attenuated than that of WT or single mutants. Thus, PatA and PgdA are nonredundant virulence determinants of H. pylori that contribute to persistence in the host.
MATERIALS AND METHODS
H. pylori culture conditions.
H. pylori was cultured on brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood or 5% fetal bovine serum (called BA plates). Chloramphenicol (50 mg/ml) or kanamycin (40 mg/ml) was added to the medium for culturing mutants. Cultures of H. pylori were grown microaerobically at 37°C in an incubator under continuously controlled levels of oxygen (4% partial pressure O2, 5% CO2, and the balance was N2).
Construction of the H. pylori patA and pgdA patA mutants.
An 880-bp fragment containing the H. pylori patA gene and the upstream flanking region was amplified by PCR from genomic DNA of strain 26695 using the primer pair patAF1 (5′ TCAATCTCCTATGCAGGTGG 3′) and patAR1 (5′-GCATCATCTCGCTATGATGC-3′). The PCR product was cloned into the pGEM-T vector to generate pGEM-patA. Subsequently, a chloramphenicol acetyltransferase (cat) cassette was inserted at the unique HindIII site within the patA sequence of pGEM-patA. The disrupted patA gene was then introduced into H. pylori wild-type strains by natural transformation via allelic exchange, and chloramphenicol-resistant colonies were isolated (patA::cat). The disruption of the gene in the genome of the mutant strain was confirmed by PCR showing an increase in the expected size of the PCR product and by direct sequencing of the PCR fragment. To construct the pgdA patA double mutant, the DNA fragment of patA::cat was used to transform the pgdA::aphA mutant strain, followed by selection for resistance to both chloramphenicol and kanamycin.
Isolation and preparation of peptidoglycan.
H. pylori cells were harvested, washed with ice-cold 100 mM Tris-HCl (pH 6.8), centrifuged at 8,000 × g for 20 min at 4°C, and then resuspended in 10 ml of the same buffer. The suspension was added dropwise to 10 ml of boiling 4% SDS buffered with 100 mM Tris-HCl (pH 6.8) and boiled for 30 min (the reaction was carried out in such a way as to allow constant stirring in order to prevent spilling of the solution from the hot flask). The samples were then cooled to room temperature overnight, and SDS-insoluble material was collected by centrifugation at 65,000 rpm for 1 h. The pellet was washed 5 times with warm water and resuspended in 5 ml of 100 mM Tris-HCl (pH 7.5) and 10 mM NaCl. DNase (10 µg/ml) and 50 µg/ml of RNase (Sigma) were added and incubated at 37°C with shaking for 2 h. Proteinase K (Invitrogen) at 50 µg/ml was added to the reaction mixture and incubated at 37°C with shaking overnight. The SDS-insoluble material was reextracted by boiling in 1% SDS for 15 min and collected and washed by centrifugation 4 times with warm water. The peptidoglycan pellet was resuspended in distilled water and lyophilized.
Preparation of muropeptides.
Lyophilized peptidoglycan was suspended in 200 μl of 100 mM Tris-HCl (pH 6.8), using sonication in a bath-type sonicator for 3 h at 40°C to assist resuspension. Following sonication, the suspensions were digested with 50 μg/50 μl of muramidase (lysozyme EC 3.2.1.17, from chicken egg white; Sigma L-7001, 40,800 units/mg solid) 18 h at 37°C. The digestions were placed in a 100°C bath for 3 min, cooled, and then centrifuged (14,000 × g for 3 min) to remove insoluble material (contaminants and any undigested peptidoglycan). Supernatants, containing the digested peptidoglycan, were mixed with an equal volume of 0.5 M borate buffer (pH 8.0) containing freshly dissolved sodium borodeuteride (10 mg/ml) and treated for 1 h at room temperature to reduce the free reducing ends of the muropeptide oligosaccharide moiety. The reactions were then filtered (0.2-μm nylon spin filters) prior to high-pressure liquid chromatography (HPLC) analysis.
Analysis of lysozyme digestion products of PG by mass spectrometry.
Portions of the reaction mixtures resulting from lysozyme digestion were reduced with borodeuteride as described above and then desalted using a Superdex-Peptide HR 10/30 FPLC (fast-performance liquid chromatography) column (Amersham Biosciences) eluted with 50 mM ammonium acetate, pH 6.0, collecting 0.4-ml fractions. The eluant was monitored at 206 nm, and fractions containing peptides and muropeptides (eluting prior to the salt peak) were combined, concentrated by rotary evaporation, and analyzed by MALDI-TOF mass spectrometry. MALDI mass spectrometry was performed on a Voyager-DE TOF spectrometer (Applied Biosystems, Boston, MA) in the positive and negative modes, using a matrix of 100 mM 2,5-dihydroxybenzoic acid in 90% methanol. The instrument was operated at an accelerating voltage of 25 kV with an extraction delay time of 150 ns. Samples were desorbed with a nitrogen laser (λ= 337 nm) and the detector sensitivity was 1,000 mV full scale. Mass spectra were recorded over an m/z range of 500 to 20,000; spectra are the summation of 200 acquisitions.
Maltooligosaccharides, standard peptides, and muramyldipeptide (MDP, N-acetylmuramic acid-l-alanyl-d-isoglutamine; Sigma A-9519) were used as calibration standards.
Cell survival assay for assessing lysozyme sensitivity.
H. pylori strains were grown on BA plates to late log phase, and the cells were suspended in PBS (phosphate-buffered saline) to a concentration of ~109 cells/ml. Upon addition of lysozyme (final concentration of 30 or 50 mg/ml), the cell suspensions were incubated at 37°C under 2% O2 with occasional shaking. Samples were then removed at various time points (0, 2, 4, and 6 h), serially diluted, and spread onto BA plates. Colony counts were recorded after 4 days of incubation in a microaerobic atmosphere (2% O2) at 37°C. To examine the sensitivity to lysozyme in the presence of lactoferrin, a mixture of lysozyme (0.3 mg/ml) and lactoferrin (3 mg/ml) was used in the assay as described above.
Turbidometric assay of peptidoglycan.
The susceptibility of the isolated peptidoglycan to lysozyme was analyzed with a turbidometric assay (8, 16). The peptidoglycan of the wild-type and mutant strains was sonicated and diluted to 0.5 mg ml−1 80 mM PBS (pH 6.4). After addition of 0.3 mg/ml lysozyme, the optical density at 600 nm was monitored every hour.
Mouse colonization.
Mouse colonization assays have been well described by our lab (17, 40). The conventional procedure is briefly described as follows. H. pylori cells were harvested after 48 h of growth on BA plates (37°C, 5% oxygen) and suspended in PBS to an optical density at 600 nm (OD600) of 1.7. Headspace in the tube was sparged with argon gas to minimize oxygen exposure, and the tube was tightly sealed. Food and water were withheld from the mice for 1.5 to 2 h prior to inoculation. The bacterial suspensions were administered to C57BL/6J mice (3 × 108 H. pylori cells/mouse) twice, with the oral deliveries made 2 days apart. Three weeks after the first inoculation, the mice were sacrificed, and the stomachs were removed, weighed, and homogenized in argon-sparged PBS to avoid O2 exposure. Stomach homogenate dilutions (dilutions were made in argon-sparged buffer in sealed tubes) were plated onto BA plates supplemented with bacitracin (100 mg/ml), vancomycin (10 mg/ml), and amphotericin B (10 mg/ml), and the plates were rapidly transported into an incubator containing sustained 5% partial pressure O2. After incubation for 5 to 7 days, the fresh H. pylori colonies were enumerated, and the data are expressed as CFU recovered per gram of stomach.
ACKNOWLEDGMENTS
This work was supported by the University of Georgia Foundation and by the National Institutes of Health (1R21AI078096). This work was also supported by Department of Energy, grant DE-FG-02-93ER20097 (to the Complex Carbohydrate Research Center).
We thank Sue Maier for help with mouse experiments.
Footnotes
Citation Wang G, et al. 2012. Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. mBio 3(6):e00409-12. doi:10.1128/mBio.00409-12.
REFERENCES
- 1. Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32:287–306 [DOI] [PubMed] [Google Scholar]
- 2. Dziarski R. 2003. Recognition of bacterial peptidoglycan by the innate immune system. Cell. Mol. Life Sci. 60:1793–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cramer EM, Breton-Gorius J. 1987. Ultrastructural localization of lysozyme in human neutrophils by immunogold. J. Leukoc. Biol. 41:242–247 [DOI] [PubMed] [Google Scholar]
- 4. Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J. Biosci. 35:127–160 [DOI] [PubMed] [Google Scholar]
- 5. Keshav S, Chung P, Milon G, Gordon S. 1991. Lysozyme is an inducible marker of macrophage activation in murine tissues as demonstrated by in situ hybridization. J. Exp. Med. 174:1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fahlgren A, Hammarström S, Danielsson A, Hammarström ML. 2003. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin. Exp. Immunol. 131:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis KM, Weiser JN. 2011. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect. Immun. 79:562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bera A, Herbert S, Jakob A, Vollmer W, Götz F. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778–787 [DOI] [PubMed] [Google Scholar]
- 9. Hébert L, et al. 2007. Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75:5390–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vollmer W, Tomasz A. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496–20501 [DOI] [PubMed] [Google Scholar]
- 11. Meyrand M, et al. 2007. Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology 153(Pt 10):3275–3285 [DOI] [PubMed] [Google Scholar]
- 12. Boneca IG, et al. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 104:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Psylinakis E, et al. 2005. Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J. Biol. Chem. 280:30856–30863 [DOI] [PubMed] [Google Scholar]
- 14. Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19:449–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G, Olczak A, Forsberg LS, Maier RJ. 2009. Oxidative stress-induced peptidoglycan deacetylase in Helicobacter pylori. J. Biol. Chem. 284:6790–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang G, et al. 2010. Peptidoglycan deacetylation in Helicobacter pylori contributes to bacterial survival by mitigating host immune responses. Infect. Immun. 78:4660–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dillard JP, Hackett KT. 2005. Mutations affecting peptidoglycan acetylation in Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 73:5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weadge JT, Pfeffer JM, Clarke AJ. 2005. Identification of a new family of enzymes with potential O-acetylpeptidoglycan esterase activity in both Gram-positive and gram-negative bacteria. BMC Microbiol. 5:49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellison RT, 3rd, Giehl TJ. 1991. Killing of gram-negative bacteria by lactoferrin and lysozyme. J. Clin. Invest. 88:1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 22. Moynihan PJ, Clarke AJ. 2010. O-acetylation of peptidoglycan in gram-negative bacteria: identification and characterization of peptidoglycan O-acetyltransferase in Neisseria gonorrhoeae. J. Biol. Chem. 285:13264–13273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moynihan PJ, Clarke AJ. 2011. O-acetylated peptidoglycan: controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int. J. Biochem. Cell Biol. 43:1655–1659 [DOI] [PubMed] [Google Scholar]
- 24. Akada JK, et al. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 149 19011909. [DOI] [PubMed] [Google Scholar]
- 25. Wang G, Maier RJ. 2009. A RecB-like helicase in Helicobacter pylori is important for DNA repair and host colonization. Infect. Immun. 77:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Costa K, et al. 1999. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J. Bacteriol. 181:3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Farnaud S, Evans RW. 2003. Lactoferrin—a multifunctional protein with antimicrobial properties. Mol. Immunol. 40:395–405 [DOI] [PubMed] [Google Scholar]
- 28. Deckers D, et al. 2004. Periplasmic lysozyme inhibitor contributes to lysozyme resistance in Escherichia coli. Cell. Mol. Life Sci. 61:1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Callewaert L, et al. 2008. A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathog. 4:e1000019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaput C, et al. 2006. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2:e97 doi: 10.1371/journal.ppat.0020097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chaput C, Labigne A, Boneca IG. 2007. Characterization of Helicobacter pylori lytic transglycosylases Slt and MltD. J. Bacteriol. 189:422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sycuro LK, et al. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell 141:822833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sycuro LK, et al. 2012. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 8:e1002603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonis M, Ecobichon C, Guadagnini S, Prévost MC, Boneca IG. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol. Microbiol. 78:809–819 [DOI] [PubMed] [Google Scholar]
- 35. Davis KM, Akinbi HT, Standish AJ, Weiser JN. 2008. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 4:e1000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Girardin SE, et al. 2003. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584–1587 [DOI] [PubMed] [Google Scholar]
- 37. Watanabe T, et al. 2010. NOD1-mediated mucosal Host defense against Helicobacter pylori. Int. J. Inflamm. 2010:476482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viala J, et al. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5:1166–1174 [DOI] [PubMed] [Google Scholar]
- 39. Kaparakis M, et al. 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell. Microbiol. 12:372–385 [DOI] [PubMed] [Google Scholar]
- 40. Wang G, Hong Y, Olczak A, Maier SE, Maier RJ. 2006. Dual roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect. Immun. 74:6839–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]