ABSTRACT
The rodent Peromyscus leucopus is a major natural reservoir for the Lyme disease agent Borrelia burgdorferi and a host for its vector Ixodes scapularis. At various locations in northeastern United States 10 to 15 B. burgdorferi strains coexist at different prevalences in tick populations. We asked whether representative strains of high or low prevalence differed in their infections of P. leucopus. After 5 weeks of experimental infection of groups with each of 6 isolates, distributions and burdens of bacteria in tissues were measured by quantitative PCR, and antibodies to B. burgdorferi were evaluated by immunoblotting and protein microarray. All groups of animals were infected in their joints, ears, tails, and hearts, but overall spirochete burdens were lower in animals infected with low-prevalence strains. Animals were similar regardless of the infecting isolate in their levels of antibodies to whole cells, FlaB, BmpA, and DbpB proteins, and the conserved N-terminal region of the serotype-defining OspC proteins. But there were strain-specific antibody responses to full-length OspC and to plasmid-encoded VlsE, BBK07, and BBK12 proteins. Sequencing of additional VlsE genes revealed substantial diversity within some pairs of strains but near-identical sequences within other pairs, which otherwise differed in their ospC alleles. The presence or absence of full-length bbk07 and bbk12 genes accounted for the differences in antibody responses. We propose that for B. burgdorferi, there is selection in reservoir species for (i) sequence diversity, as for OspC and VlsE, and (ii) the presence or absence of polymorphisms, as for BBK07 and BBK12.
IMPORTANCE
Humans are dead-end hosts for Borrelia agents of Lyme disease (LD), and, thus, irrelevant for the pathogens’ maintenance. Many reports of human cases and laboratory mouse infections exist, but less is known about infection and immunity in natural reservoirs, such as the rodent Peromyscus leucopus. We observed that high- and low-prevalence strains of Borrelia burgdorferi were capable of infecting P. leucopus but elicited different patterns of antibody responses. Antibody reactivities to the VlsE protein were as type-specific as previously characterized reactivities to serotype-defining OspC proteins. In addition, the low-prevalence strains lacked full-length genes for two proteins that (i) are encoded by a virulence-associated plasmid in some high-prevalence strains and (ii) LD patients and field-captured rodents commonly have antibodies to. Immune selection against these genes may have led to null phenotype lineages that can infect otherwise immune hosts but at the cost of reduced fitness and lower prevalence.
Introduction
Lyme disease (LD) is a common infection in parts of the North America and Eurasia continents. But humans, aside from our collective effects on forestation, climate, and biodiversity, are irrelevant for maintaining the etiologic agents in nature. Unlike the plague agent Yersinia pestis, which can be transmitted between humans as well as from animals, Borrelia burgdorferi is obligately vector-borne and depends entirely on other vertebrates for its reservoirs. If humans disappeared from North America, B. burgdorferi would persist on this continent in a variety of mammals and birds, as it has for thousands of years (1).
If the research goal is understanding disease in humans, focusing on pathogenesis is rewarding. If the objective is a vaccine, results from the laboratory mouse, Mus musculus, are sufficiently predictive to justify clinical trials (2). But for questions on the ecology of this pathogen, predictions about its geographic spread, and field-based prevention strategies, insights gained from studies of human cases or laboratory rodents may be less informative than studies of the actual reservoirs of B. burgdorferi.
In regions of North America where LD has its highest incidences, Peromyscus leucopus is a common reservoir for B. burgdorferi and a preferred host for Ixodes scapularis, its tick vector (3). In these areas the majority of P. leucopus become infected with B. burgdorferi (4, 5). Its common name of “white-footed mouse” belies its phylogenetic distance from the house mouse, Mus musculus. Peromyscus belongs to the taxon Cricetidae, together with voles and hamsters, and not Muridae, the family for laboratory mice and rats (6). While B. burgdorferi-infected M. musculus mice exhibit arthritis and carditis (7), such lesions were not observed in P. leucopus (8, 9), Peromyscus maniculatus (10), or the related species Mesocricetus auratus, the Syrian hamster (11). P. leucopus and Syrian hamsters may be persistently infected with B. burgdorferi (4, 12–14) but, unlike M. musculus (7), without discernible evidence of disease or disability (9, 10, 15).
Differences in disease expression in the host may also be attributable to traits of the infecting microbe. The concept of strain differences in infectiousness or virulence was suggested by epidemiological studies of human cases whose B. burgdorferi isolates were genotyped (16–18). Similar differences in propensities to disseminate in M. musculus were reported for limited strain sets (19, 20). But in another study of different strains, bacterial densities in the blood were not predictive of tissue burdens or pathology in the laboratory mouse (21). Derdáková et al. and Hanincová et al. infected P. leucopus with two different strains and reported differences in durations of transmissibility to naive mice (22, 23), but tissue distributions of the bacteria and the antigen-specific immune responses were not assessed.
One basis for isolate selection for studies of strain differences in a natural reservoir is their relative prevalences, which can be estimated by counting genotypes of B. burgdorferi among nymphal I. scapularis ticks (1, 24, 25). Genotypes can be defined by sequences of 16S-23S rRNA intergenic spacers (IGS), the genes for the outer membrane protein OspC, and multilocus sequence typing (MLST). For a given geographic area, these different schemes are congruent (1, 25–27). Most genotype surveys have found a log-normal distribution of strains, which has suggested the action of balancing selection (28).
In previous studies of P. leucopus captured in the field, sera from infected animals had antibody reactivity profiles that were similar to those of LD patients (4, 29). This included a high frequency of reactivity to the OspC, VlsE, BBK07, and BBK12 proteins of a single strain, B31. The OspC proteins are highly polymorphic and largely strain-specific (24), but contain epitopes shared among the protein family (30). Magnarelli et al. also reported a high frequency of antibody to the VlsE protein among captured P. leucopus in Connecticut (31). The VlsE protein is associated with antigenic variation in B31 and other strains (32), but it or a component peptide is also a basis for immunodiagnostic assays with high sensitivity and specificity for LD (33). The BBK07 and BBK12 proteins are encoded by a virulence-associated linear plasmid, lp36, of strain B31 (34) and are commonly bound by antibodies from patients with LD (29, 35).
The present study of P. leucopus was carried out in a laboratory setting with quantitative PCR, immunoblotting, and protein microarray analysis. We found that six B. burgdorferi isolates, which represented strains with different prevalences in the northeastern United States, were infectious for P. leucopus and elicited similar antibody responses to conserved antigens. However, different isolates elicited different antibody reactivities to other antigens, notably the plasmid-encoded OspC, VlsE, BBK07, and BBK12 proteins. These strain-specific responses led to a deeper study of the heterogeneity of the genetic loci for these antigens.
RESULTS
Strains for infections.
Table 1 summarizes nymphal prevalences and selected genetic characteristics of the isolates: 5 clonal populations (Sh-2-82, TBO2, HB19, 27577, and WQR) and one mixed-population isolate, IDS, which was discovered to be a mixture comprising strains B31 and TBO2 after experiments began (see Text S1 in the supplemental material). The strains represented in the study varied in prevalence from 4% to 19% among infected I. scapularis nymphs in a large survey of ticks in the northeastern United States (24, 27). For this study, Sh-2-82 (here abbreviated SH2), B31, and TBO2 were classified as high-prevalence strains, and HB19, 27577, and WQR as low-prevalence strains. We carried out next-generation sequencing of the genomes of SH2, TBO2, HB19, 27577, and WQR, as well as resequencing of the genome of strain B31. We had previously noted that the ospC and IGS types of strains SH2 and HB19 were identical to corresponding loci in strains 297 and WI91-23, respectively (25, 27). Additional sequence data revealed that strains SH2 and 297 had the same MLST type, but the MLST type of strain HB19, which was from the northeastern U. S, differed from that of WI91-23, which was from the north-central region. Two other strains, 27577, which has the type F ospC allele, and WQR, which has the type T allele, were characterized for this study (see Table S2 in the supplemental material). Strains BL206 and B348 have the same OspC type, IGS sequence, and MLST type as strains B31 and N40, respectively (http://borrelia.mlst.net), and for this study were considered to belong to the same lineages.
TABLE 1 .
Borrelia burgdorferi strains, origins, prevalences, and genotypes
| Strain or isolate | State of origin | Prevalence (%)a | OspC typeb | IGS typeb,c | MLST typed | Presence or absence ofe: |
|
|---|---|---|---|---|---|---|---|
| bbe22 | bbf01 | ||||||
| SH2 | NY | 19.4 | K | 2 | 3 | + | + |
| B31/BL206 | NY | 14.4 | A | 1 | 1 | + | + |
| TBO2 | NY | 9.3 | N | 4 | 36 | + | − |
| HB19 | CT | 4.8 | I | 7 | 16 | + | + |
| 27577 | NY | 4.5 | F | 17 | 8 | + | + |
| WQR | RI | 4.3 | T | 28 | 37 | + | + |
| N40/B348 | NY | 4.0 | E | 9 | 19 | + | + |
| IDS | NY | 14.4, 9.3 | A, N | 1, 4 | 1, 36 | + | + |
Prevalence in the northeastern United States (reference 24).
Reference 27.
IGS, 16S-23S intergenic spacer.
MLST, multilocus sequence type, according to reference database at http://borrelia.mlst.net.
Determined by mapping of sequence reads as described in the text (for results, see Table S2 in the supplemental material).
Experimental infections.
P. leucopus animals were inoculated i.p. with 50 to 100 B. burgdorferi cells of each of 6 isolates; inoculum sizes were close to estimates of numbers of spirochetes transmitted by ticks (36). All 6 populations had been passed through immunodeficient mice before the P. leucopus infections. Each had the pncA gene of the lp25 plasmid and, with the exception of strain TBO2, the bbf01 gene of lp28-1 of strain B31 (Table 1; also, see Table S2 in the supplemental material) (37, 38).
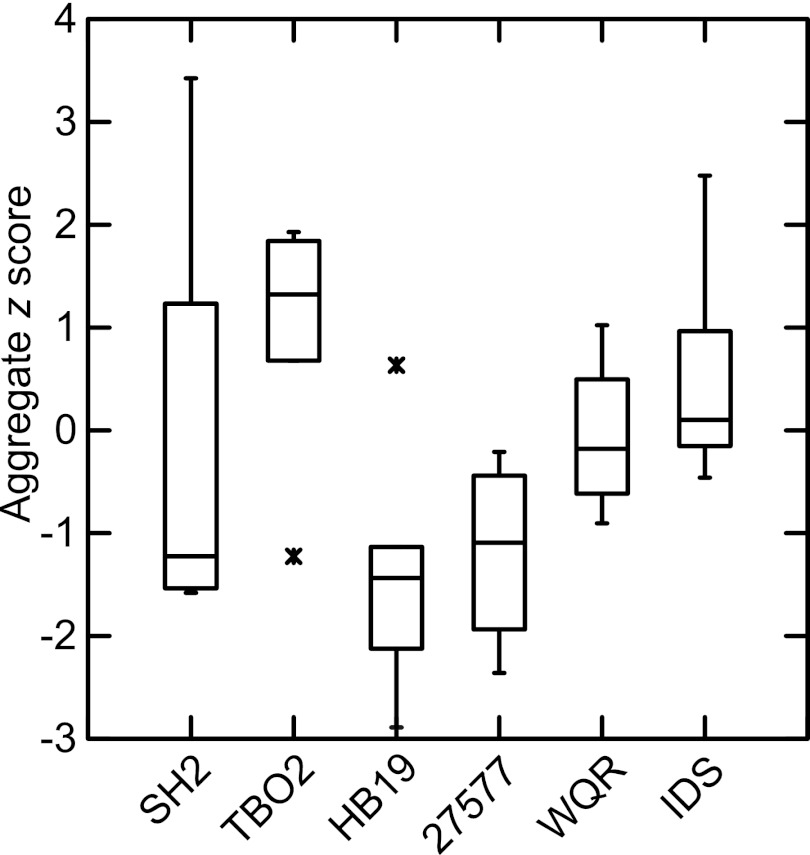
At the end of 5 weeks of infection, tissue and blood were collected from animals, and B. burgdorferi cells were quantitated by qPCR. All 30 animals became infected by the criterion of ≥2 tissues out of 4 being positive by qPCR (see Table S3 in the supplemental material). Table 2 summarizes the burdens of B. burgdorferi in the ankle joints, distal tails, ear punch biopsy specimens, and heart tissues in the 28 (93%) of 30 animals that were also seropositive by immunoblot at 5 weeks (Fig. 1). Clear distinctions between strains with regard to their burdens in the animals were not discernible on a tissue-by-tissue basis. But intrastrain as well as interstrain variances between these outbred animals were high, with coefficients of variation ranging between 1 and 2 across all strains by tissue. By normalizing the burdens in tissues by z scores (Table S3 in the supplemental material), commensurate burdens in the four tissues could be determined for each mouse (Fig. 2). Aggregate z scores, which were the sum of z scores for each tissue, for the 15 mice infected with high-prevalence strains were higher overall than those for 13 seropositive mice infected with low-prevalence strains (odds ratios [OR; 95% confidence intervals are in brackets], 2.15 [1.18 to 4.67]; R2 = 0.23; Kruskal-Wallis rank test, P = 0.018; ANOVA, P = 0.009).
TABLE 2.
Burdens of spirochetes of different strains of B. burgdorferi in tissues of seropositive P. leucopus mice
| Strain or isolate | Mean no. of 16S rRNA gene copies/µg total DNAa |
|||
|---|---|---|---|---|
| Ankle | Tail | Ear | Heart | |
| SH2 | 346 (175–685) [5/5] | 1,248 (64–24,419) [5/5] | 370 (172–798) [5/5] | 3 (2–6) [2/5] |
| TBO2 | 1,467 (575–3,738) [5/5] | 196 (80–481) [5/5] | 249 (75–832) [3/5] | 27 (13–52) [5/5] |
| HB19 | 802 (350–1,836) [5/5] | 47 (23–96) [5/5] | 121 (23–636) [5/5] | 19 (9–40) [2/5] |
| 27577 | 1231 (540–2,804) [4/4] | 551 (92–3,283) [4/4] | 181 (61–533) [4/4] | 4 (2–8) [4/4] |
| WQR | 3,722 (2,037–6,800) [4/4] | 86 (167–474) [3/4] | 69 (17–280) [4/4] | 3 (1–7) [3/4] |
| IDS | 1,082 (535–2190) [5/5] | 2,687 (1,371–5,266) [5/5] | 193 (42–889) [5/5] | 7 (3–17) [5/5] |
Values in parentheses are antilogarithms of 95% confidence intervals for log-transformed values; values in brackets are numbers of mice with >10 gene copies in tissue/number of mice of that strain examined.
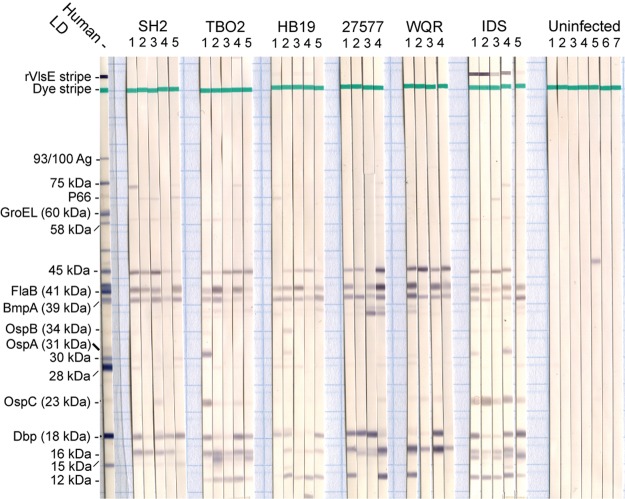
FIG 1.
Immunoblot analysis of whole-cell lysates of Borrelia burgdorferi strain B31 with sera from P. leucopus mice that were uninfected or infected with isolate IDS or strain SH2, TBO2, HB19, 27577, or WQR. A stripe of recombinant strain B31 VlsE was present on the nitrocellulose strips. Antibody binding was detected with alkaline phosphatase-labeled secondary antibody to P. leucopus IgG. The migrations of selected antigens on a reference strip, supplied by the manufacturer, with pooled sera from patients with Lyme disease (LD), is shown on the left. The numbers at the top of the aligned strips indicate the individual animals in each group of infected animals or controls.
FIG 2.
Adjusted overall tissue burdens of B. burgdorferi by isolate or strain in skin, tail, joint, and heart of P. leucopus mice after 5 weeks of infection. The box-whisker plots show aggregate z scores for all 4 tissues; their calculations are described in the text. Spirochete burdens in genome copies were estimated by quantitative PCR. Each box indicates the first and third quartiles, and the line inside the box is the median. The 1.5× interquartile range is indicated by the vertical line (whiskers) bisecting the box, and values outside this range are indicated by asterisks.
The two seronegative animals excluded from the preceding analysis were infected with the low-prevalence strains 27577 and WQR. All four tissues of these mice had markedly higher burdens than those of the seropositive animals infected with the same strain and of all but 3 animals infected with other strains (see Table S3 in the supplemental material). In the group infected with 27577, the seronegative animal had ~2,300 gene copies in the heart sample, while the mean number of gene copies for the four seropositive animals was 4.
Immunoblot analysis.
Sera collected at 5 weeks were used in a whole-cell B. burgdorferi immunoblot. In the immunoblot assay, which is shown in Fig. 1, the antigens most commonly bound by the 28 sera were the BmpA at 39 kDa (82%), FlaB at 41 kDa (75%), an unidentified 45-kDa protein (71%), either DbpA or DbpB at 18 kDa (64%), and an unidentified 16-kDa protein (64%). The binding to two other major antigens, OspC and VlsE, was more strain specific than anticipated from previous reports on field-collected P. leucopus (4, 29, 31). All 5 mice infected with the IDS isolate, which includes B31, had detectable antibody to the type A OspC of strain B31 cells this assay was based on, but only one other serum, a sample from an animal infected with strain TBO2, had discernible binding to this OspC protein. While all mice infected with IDS had detectable antibodies to recombinant VlsE of strain B31 on the blot, none of the sera from animals infected with other strains demonstrated this reactivity.
Protein microarray analysis.
For the protein microarray, 26 seropositive and 12 control P. leucopus sera were studied. In addition, we examined the binding reactions of sera from the following sources: (i) 3 P. leucopus mice infected with the relapsing fever agent Borrelia hermsii, (ii) groups of 3 M. musculus mice infected with SH2 or IDS, and (iii) 5 uninfected M. musculus mice. The binding values for these specimens against a whole-cell lysate of strain B31, recombinant antigens FlaB, BmpA, DbpB, BBK07, BBK12, OspA, and VlsE of strain B31, and B. hermsii’s Vlp17 are given in Table S4 in the supplemental material.
In general, the reactivities with the whole-cell lysate, FlaB, BmpA, and DbpB were similar to what we observed with the immunoblots. That is, most of the infected P. leucopus and M. musculus mice had antibodies against these antigens, regardless of the infecting strain. There was no difference between animals infected with low- and high-prevalence strains with respect to antibody binding to the lysate, FlaB, BmpA, or DbpB.
We also examined by microarray the antibody responses to the diverse set of OspC proteins represented by the infecting strains, as confirmed by mapping sequence reads to references (see Table S2 in the supplemental material): A (B31), K (SH2), N (TBO2), I (HB19), F (27577), and T (WQR). The array also included type E OspC, which is produced by strain B348. To assess the cross-reactivity attributable to the conserved N-terminal end of the processed protein and its first helix, we synthesized a peptide that included 54 amino acids of type A OspC (“first helix”). The corresponding regions of OspC types K, N, I, F, T, and E were 94 to 98% identical over this length to the reference. The results of the array analysis are given in Table S5 in the supplemental material.
Sera from all infected P. leucopus and M. musculus had antibody binding to the first helix that was significantly higher than that from uninfected controls. Antigenic cross-reactivity between the longer recombinant OspC proteins was also evident in the binding of antibodies to heterotypic proteins for both species. For the P. leucopus samples, the pairwise R2 values averaged 0.69 (0.57 to 0.81) for the first helix versus OspC A, E, F, I, K, N, and T and 0.59 (0.50 to 0.68) for these OspC types against each other. But, as we observed with the immunoblot analysis, there was also evidence of type-specific antibody responses among the infected animals of both species (see Table S5 in the supplemental material). For all groups of animals except the P. leucopus mice infected with WQR, there was more antibody binding to the homotypic OspC than to the 6 heterotypic ones, by an overall ratio of 4.3 (2.0 to 5.6), including the WQR results (see Table S5 in the supplemental material).
Antibodies to VlsE.
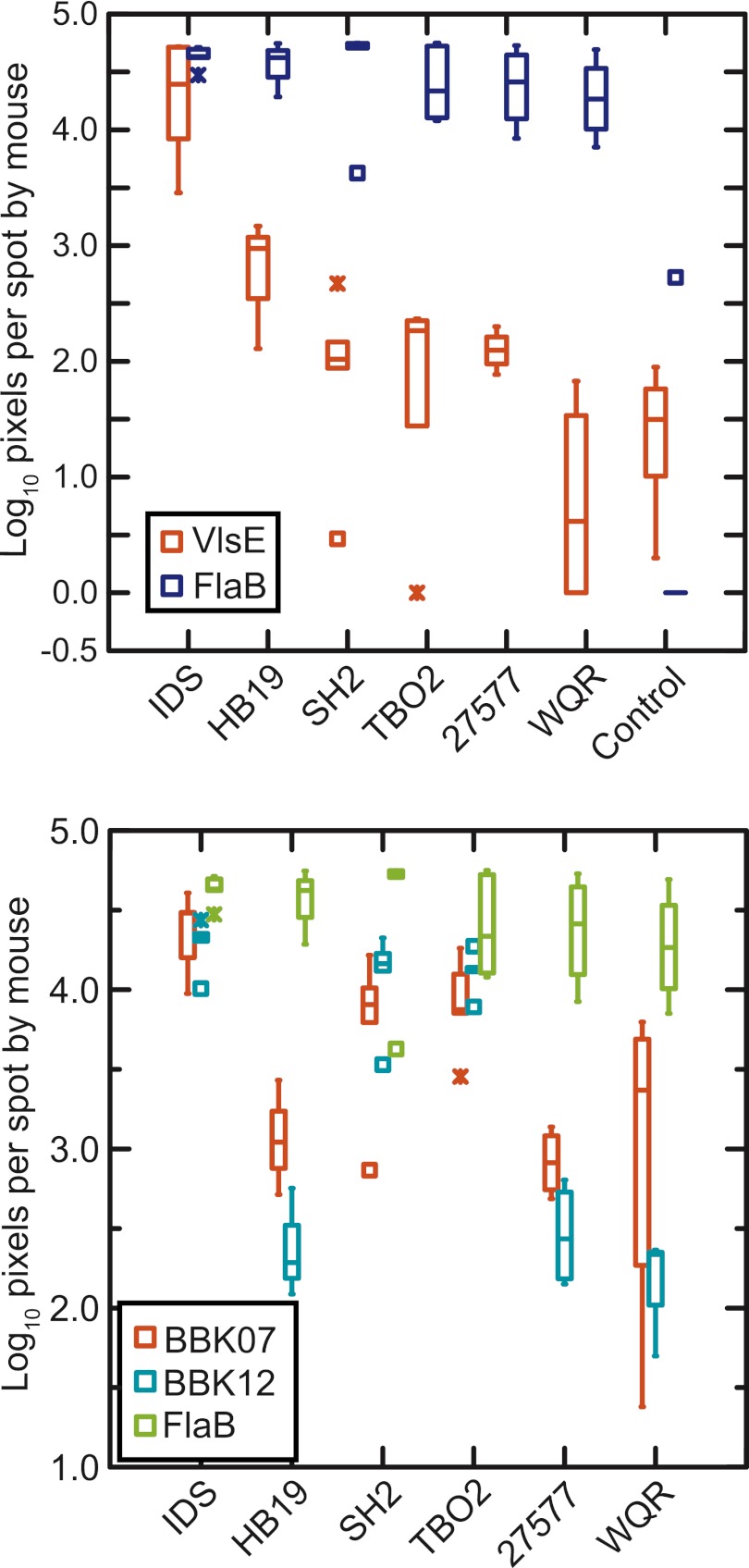
To identify and quantitate antibodies to specific antigens, we used a microarray with selected recombinant proteins. As with the immunoblotting, the microarray results were evidence of strain-specific responses of P. leucopus to the VlsE protein (see Table S4 in the supplemental material). Figure 3 summarizes these data and for comparison also shows the binding values for antibodies to FlaB. The binding of antibodies of strain WQR-infected animals was as negligible as that of control sera. The binding of antibodies of animals infected with 27577, TBO2, SH2, and HB19 was marginally greater than that of controls, but the values were still significantly lower than the binding shown by sera from IDS-infected animals.
FIG 3.
Binding of antibodies in P. leucopus sera to selected proteins of B. burgdorferi strain B31 on a protein microarray. (Top) VlsE (red) and FlaB (blue) proteins; (bottom) BBK07 (red), BBK12 (blue), and FlaB (green) proteins. The box-whisker plots (Fig. 2) show log-transformed intensity values in pixels for the binding of antibodies from uninfected control animals (top) and infected animals (top and bottom).
While we cannot exclude a confounding effect of the mixed infection, we presume that the binding of antibodies from IDS-infected animals to VlsE of strain B31 is largely attributable to the B31 component of that mixture. Evidence for this was the lack of antibody binding to VlsE shown by animals infected with TBO2 alone. The strain specificity shown by P. leucopus for VlsE protein presented in the array format contrasts with the evidence of cross-reactivity of anti-VlsE antibodies in the case of M. musculus infected with strain SH2 (see Table S4 in the supplemental material).
P. leucopus did not have detectable cross-reactive antibodies to B. hermsii’s Vlp17 protein, which is homologous to VlsE (39) (see Table S4 in the supplemental material). A reciprocal pattern was observed with B. hermsii infections: little or no antibody to FlaB, BmpA, DbpB, and VlsE of B. burgdorferi but large amounts of binding to the Vlp17 protein.
Antibodies to BBK7 and BBK12.
Another set of antigens of interest were the paralogous BBK07 and BBK12 proteins, because of their documented immunogenicity in natural infections of P. leucopus and humans (29), as well as the association of their encoding plasmid with infectiousness (40). As Fig. 3 and as Table S4 in the supplemental material show, there were marked differences between strains in whether antibodies to these proteins were elicited during infection of P. leucopus. For comparison purposes antibodies to FlaB again are shown; there was little or no detectable antibodies to these proteins in control P. leucopus or M. musculus sera (Table S4 in the supplemental material). Mice infected with IDS or one of the other two high-prevalence strains had antibodies that bound to the BBK07 and BBK12 proteins of strain B31. There was little or no detectable binding to either protein by antibodies in sera from mice infected with the low-prevalence strains HB19, 27577, and WQR.
Sera of P. leucopus mice infected by tick bites.
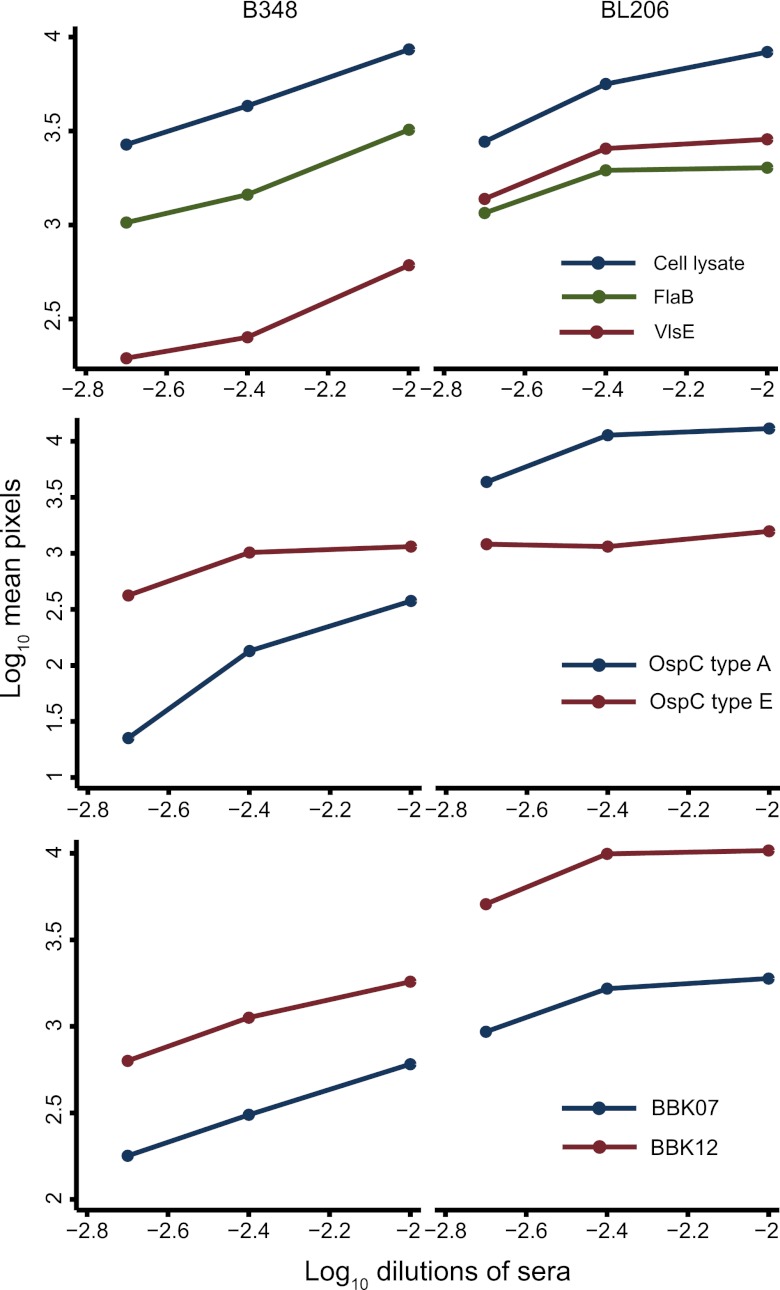
To assess whether the route of infection had an effect on some of the antibody responses, we examined sera from P. leucopus infected by tick bite with either strain BL206, which carries a type A ospC gene, or strain B348, which bears a type E ospC gene, for antibody binding to the whole-cell lysate, FlaB, VlsE, OspC type A, OspC type E, BBK07, and BBK12 on the array. The sera were diluted 1:100, 1:250, or 1:500 (Fig. 4). The sera were similar in antibody binding to the cell lysate and to FlaB but differed in binding to the other antigens. The binding of antibodies in the B348 sample to VlsE of strain B31 was severalfold lower at each serum dilution than that observed with BL206 sample. With the OspC proteins, there was evidence again of both cross-reactive and type-specific antibody responses; at each dilution, the amount of binding to the homotypic OspC was greater, up to 10-fold, than that to the heterotypic protein. With both samples there was more binding to BBK12 than to BBK07, but the amount of binding of antibodies in the B348 sample was smaller than the corresponding value for BL206 at each serum dilution.
FIG 4.
Binding of antibodies in sera from P. leucopus mice, infected by tick bites with B. burgdorferi strain B348 or BL206, to purified recombinant FlaB or VlsE (top), OspC type A or E (middle), and BBK07 or BBK12 (bottom) on arrays. The graphs are scatter plots, separated according to the infecting strain, with connecting lines of log-transformed intensity values in pixels against log-transformed serum dilutions.
VlsE genes.
In a previous study of sera from infected P. leucopus captured in the field, we observed that the majority of the animals had antibodies that bound to the VlsE protein of strain B31 (29). Here, cross-reactive antibodies to B31’s VlsE were noted in M. musculus infected with SH2, but not in P. leucopus infected with that strain or strains TBO2, HB19, 27577, or WQR (see Table S4 in the supplemental material). This finding led us to determine if these other strains had vlsE sequences.
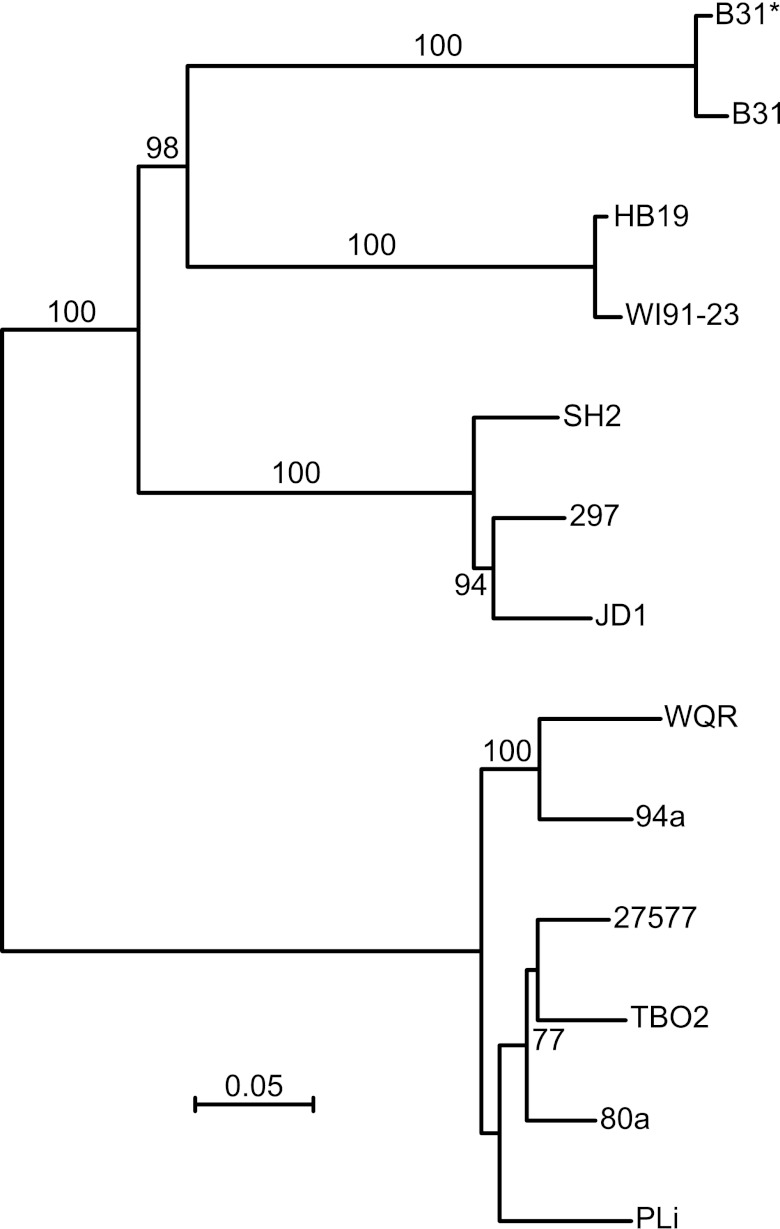
VlsE genes were identified in TBO2, HB19, SH2, 27577, and WQR by a combination of de novo assembly of the sequence reads, mapping of reads to reference vls gene sequences in the public domain, and then confirmation by sequencing of PCR products amplified from genomic DNA (see Text S1 in the supplemental material). Strain B31 was resequenced for this study to validate the sequence assembly approach. Figure 5 is a distance phylogram of a codon-based alignment of these nucleotide sequences and selected reference sequences (available at https://webfiles.uci.edu/xythoswfs/webui/_xy-8429602_1). As expected from their chromosomal loci and ospC alleles, the SH2 VlsE sequence clusters with strain 297s, and HB19’s VlsE sequence clusters with WI91-23’s protein. Both clusters are distant from the B31 sequences. More distant still are the VlsE sequences of TBO2, 27577, and WQR strains, which share similarities in VlsE sequence with strains 80a and 94a of B. burgdorferi and strain PLi of Borrelia garinii. The pairwise percent identities at ungapped sites of the deduced protein sequences ranged from 39 to 53% between the strain B31 VlsE of the proteins (see Table S5 in the supplemental material).
FIG 5.
Distance phylogram of partial vlsE nucleotide sequences deduced from assemblies of nucleotide sequences for strains SH2, HB19, WQR, TBO2, and 27577 of B. burgdorferi and reference sequences for strains B31, 297, JD1, WI91-23, 94a, and 80a of B. burgdorferi and strain PLi of B. garinii. The alignment was codon-based alignment. Nodes with support of ≥70% for 100 bootstrap iterations of the maximum-likelihood method, as carried out with the PhyML 3.0 algorithm (http://www.atgc-montpellier.fr/phyml) under the General Time Reversible model of substitution, are shown near the branches. B31* is derived from the assembly of reads from a resequencing of strain B31 by the same method as for the other strains. The scale bar indicates nucleotide distance.
We identified regions of these proteins, as well as of the Vlp17 protein, that aligned with the "C6 peptide” of immunodiagnostic applications (41) and noted that sequences of other strains differed at several positions from that of the canonical C6 peptide and corresponding sequence of strain B31 (see Fig. S2 in the supplemental material).
bbk07 and bbk12 genes.
In strain B31, the bbk07 and bbk12 genes are located on lp36, a 36-kb linear plasmid which is associated with infectiousness (34). In other strains of B. burgdorferi, some of the genes of lp36 may be distributed among other replicons (42). Given this variability between strains, we investigated the bbk07 and bbk12 loci in the strains B31, SH2, and TBO2, which alone or in combination elicited high levels of antibodies to both BBK07 and BBK12 (Fig. 3), as well as in the strains HB19, 27577, and WQR, which did not. In strain B31, bbk07 and bbk12 are paralogous but are oppositely oriented and separated by ~2 kb on the plasmid. The near-identity of BBK07 and BBK12 in their N-terminal regions is sufficient for antigenic cross-reactivity (29).
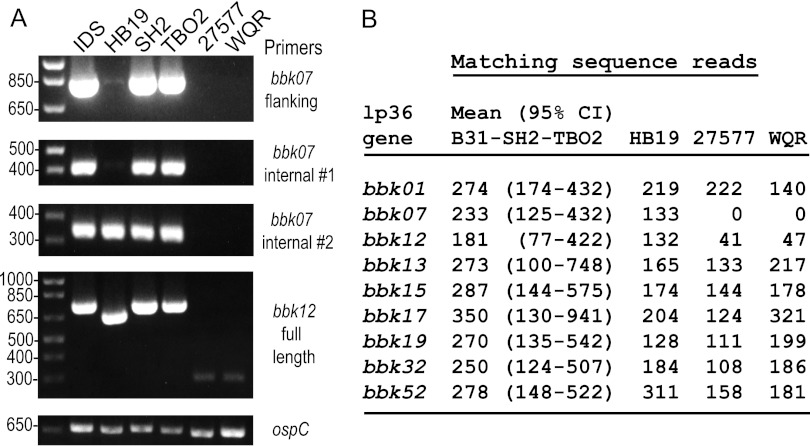
We first used PCR with primers from within and outside the two genes (Fig. 6, left; also, see Table S1 in the supplemental material). With primers for positions that flank bbk07 in strain B31 (see Table S1), there was the expected PCR product of ~800 bp in IDS and also in strains SH2 and TBO2. No products were observed with total DNA from HB19, 27577, and WQR. Similar results were obtained with internal primers that spanned positions 1 to 418 of bbk07. With a forward primer starting from position 66, we observed a product with HB19 as well, but not with 27577 or WQR. PCR with primers comprising the 5′ and 3′ ends of the bbk12 gene revealed the expected products for full-length genes in the IDS, SH2, and TBO2 samples, a truncated product from HB19, and only faint bands at ~280 bp with the 27577 and WQR samples.
FIG 6.
Analysis by PCR and genome-wide sequencing of bbk07 and bbk12 loci in different strains of B. burgdorferi. (Left) Ethidium bromide-stained agarose (1%) electrophoresis gels of PCR products with 3 primer sets for the bbk07 locus, one set for the bbk12 locus, and one set for the ospC locus of isolate IDS and strains HB19, SH2, TBO2, 27577, and WQR. Size markers (in base pairs) are shown on the left of the gels. (Right) Normalized counts of matching sequence reads for selected genes of the lp36 plasmid. Means with 95% confidence intervals (CI) for 3 strains (B31, SH2 and TBO2) with full-length bbk07 and bbk12 loci are compared with the individual counts for HB19, 27577, and WQR, which lacked full-length PCR products for bbk07 and bbk12.
Enumeration of matching reads from genome sequencing indicated that the IDS preparation and the 5 strains of the other samples had genomes with complete or near-complete sequences for the bbk01, bbk13, bbk15, bbk17, bbk19, bbk32, and bbk52 genes of the lp36 plasmid (Fig. 6; also, see Table S2 in the supplemental material), but in strains 27577 and WQR there was not a detectable bbk07 gene sequence and there were significantly fewer reads matching the bbk12 gene (Fig. 6, right). When sequence reads were mapped to strain B31’s lp36 plasmid as a reference, we obtained the full-length lp36 with the sample of B31 that was resequenced (see Fig. S2 in the supplemental material). The assemblies of SH2 and TBO2 reads mapped to the reference were only ~25 kb, but the B31 lp36 plasmid’s left end with its oppositely oriented bbk07 and bbk12 genes was largely preserved in these strains. There was evidence of rearrangements or translocations at both the 5′ and 3′ ends of what remained of the lp36 replicon in HB19 and of deletions in the region containing bbk07 and bbk12 in 27577 and WQR.
Sequencing of products of PCR with the flanking or internal primers for bbk07 confirmed the presence of full-length bbk07 genes in SH2 and TBO2 (see Table S6 in the supplemental material). These alleles were identical in sequence to that of B31. As Fig 6 indicates, bbk07 of HB19 lacked the 5′ end of the gene; it was identical in sequence to that of WI91-23. Analysis of PCR products for the bbk12 gene of SH2 revealed a sequence that was identical to that in strain 156a; in both, the first of several stop codons began at positions 481 to 483 (43). In strain TBO2 the homologous sequence also had a stop codon at positions 481 to 483 but differed as well at other positions from other sequences in the database. The nucleotide sequence of HB19’s shortened bbk12 gene matched that of WI91-23 (see Table S6 in the supplemental material). Both sequences featured (i) an insertion of 7 nucleotides at position 112, which produced a frameshift and then a stop codon at positions 121 to 123, and (ii) a 99-nucleotide deletion in the middle of what otherwise was the open reading frame. In strains 27577 and WQR, the only detectable bbk07 or bbk12 sequence was a region of 231 nucleotides that was identical to part of the bbk12 ortholog in strain 156a. The length of this remnant of the gene was consistent with the PCR findings (Fig. 6).
Under the assumption that strain BL348 is in the same lineage as strain N40, we note that the lower antibody response to the BBK07 and BBK12 proteins for that tick-transmitted infection (Fig. 4) may be explained by 4 differences in the bbk07 locus of strain N40 from B31: (i) an extra A in the area of the −10 element of the σ70-type promoter, (ii) a nonsense mutation at position 77 of the gene, and (iii) a nonsense mutation at position 640.
DISCUSSION
This study of experimental infection extends understanding of the biology of B. burgdorferi in P. leucopus, one of the major reservoirs of this bacterium in North America. While all the strains infected this rodent species and disseminated from the inoculation site to other tissues, the strains differed in the extents of infection, as defined by burdens of spirochetes in selected tissues, and in the antibody responses to certain antigens, notably OspC, VlsE, and the BBK07 and BBK12 proteins. Some traits for strains clustered according their prevalences in ticks in the geographic area from which they were drawn. For instance, in comparison to strains with lower prevalence, higher-prevalence strains tended to have higher tissue burdens overall and were unique in having antibodies to BBK07 and BBK12 proteins. The latter result was unexpected, as was the decidedly strain-specific response to a VlsE protein, and these findings led to further characterization of the bbk07, bbk12, and vlsE loci of these strains.
The study had limitations. (i) The bacterial inocula were obtained from infected animals, not from culture medium, but they were delivered by needle, not by tick. This led us to confirm some of the findings with sera from P. leucopus mice experimentally infected by tick bite. (ii) The animals were reared in captivity, restricted by sex to female, and free from the traumas, nutritional deficiencies, and parasites that wild animals are exposed to. On the other hand, the outbred character of the study’s animals was probably more representative of the diversity expected for host species than inbred mouse strains provide. (iii) The animals were all young adults, and the blood and tissues were sampled at one point, 5 weeks into infection. But these choices were probably not far from the average age at which P. leucopus first acquires the infection and the usual duration of infection at the height of transmission to larval I. scapularis (3). (iv) We did not initially intend to use a mixture of strains along with clonal populations. The mixed character of the IDS isolate was discovered during the course of the study. But we carried on with the isolate because coinfection by two or more different strains in ticks and P. leucopus is common in areas of high prevalence (27, 44, 45) and because both members of the mixture, strains B31 and TBO2, had been characterized.
Acknowledging these limits, we focus here on the findings that were not particularly dependent on the experimental conditions or predictable from previous reports. These findings include the strain-specific antibody response to the VlsE protein and the absent or low levels of antibodies to the BBK07 and BBK12 proteins in sera that were otherwise seropositive. Before discussing these results, we consider the antibody responses to the OspC proteins. Our findings confirm that there are antibodies to conserved regions (30), in particular the N-terminal region, which includes the first alpha-helix. This is consistent with the established sufficiency of a single type of OspC protein for sensitive immunoassays for sera from either patients with Lyme disease (46) or P. leucopus mice captured at field sites where several strains occur (4). But we also observed in the immunoblots and the microarrays evidence of the strain-specific antibody responses to OspC proteins that have been reported in vaccine studies in mice (47–49). On the basis of the array values for the different recombinant OspC proteins and the conserved first helix (see Table S5 in the supplemental material), we estimate that type-specific antibodies constituted at least half of the antibodies binding to a given OspC. This provides scope, in our opinion, for assays comprising multiple OspC types for discerning the strain that an animal in the field or a patient in a clinic was infected with.
In both our previous study and that of Magnarelli et al. of sera from field-captured P. leucopus mice, half or more of the animals in the samples had detectable antibody to a single type of VlsE protein (29, 31). These findings, along with the established diagnostic utility of immunoassays based on all or parts of a single VlsE (33), led us to anticipate that all or most of the P. leucopus mice infected with different strains would have antibodies to strain B31’s VlsE. This could occur equivalently across strains or, more distinctly, from a combination of homotypic (specific) and heterotypic (cross-reactive) responses (50). While interstrain differences in VlsE protein sequences had been noted previously (51), this diversity did not prevent successful application of assays based on a single full-length VlsE or the conserved C6 peptide of VlsE for laboratory diagnosis B. burgdorferi infection (33, 52, 41). Cross-reactivity in VlsE binding by antibodies in human sera between different Borrelia species of LD agents has also been reported (53). The sera from infected M. musculus mice bore out this prediction: mice infected with the SH2 strain had cross-reactive antibodies to the VlsE of strain B31. But, to our surprise, cross-reactive antibodies to the B31 VlsE were not observed in either the immunoblots or the microarrays with sera from P. leucopus mice infected with strains other than B31.
We investigated whether the strains that did not elicit an anti-VlsE response had vlsE-like sequences and, if so, the extent to which these sequences differed from the B31 sequence. Identification and characterization of vlsE gene sequences are hampered by the telomeric location of the full-length expressed gene and the long tandem arrays of partial variant versions of the sequence (54). Through an iterative process of first counting the number of reads that mapped to each of a set of known vlsE sequences and then assembling the reads on a scaffold of the highest-ranking reference vlsE sequence, we identified vlsE sequences in all the strains. This approach was validated by resequencing of strain B31 and subsequent searching by the same method, as well as by concordances of the SH2 vlsE sequence with that of strain 297 and of the HB19 vlsE sequence with that of WI91-23.
Interstrain diversity between the assembled vlsE sequences was high. In general, distances between vlsE sequences of some strains were as great as those for their ospC alleles. The lack of reaction to the VlsE of strain B31 of antibodies from P. leucopus infected with other strains is plausibly attributable to the distances between the VlsE proteins of these different strains. Although much of the work on VlsE proteins has justifiably emphasized the role of antigenic variation of VlsE once an infection is under way in an individual animal (32, 39), it is not obvious how this would account for such marked interstrain diversity, particularly when the heterogeneity is most marked in protein regions not involved in antigenic variation. The results here suggest that in addition to intrahost selection for antigenic variants, there may be balancing selection on VlsE proteins in a B. burgdorferi population comprising several strains. If this selection is carried out largely by adaptive immune systems, an animal with antibodies to a particular type VlsE protein, e.g., like that of B31, as the consequence of a prior exposure may be immune on that basis to infection by strains with the same or a similar type of VlsE. But, other things, like anti-OspC immunity, being equal, the animal may remain at risk of infection with strains with another type of VlsE protein, e.g., such as SH2’s.
This account might also explain the more specific anti-VlsE antibody responses observed in P. leucopus in comparison to the more broadly cross-reactive responses that have been noted in humans, dogs, and M. musculus infected with B. burgdorferi. That is, in the coevolution between parasite, B. burgdorferi, and host, P. leucopus, there plausibly was selection against VlsE types that to the P. leucopus immune system were recognizably similar enough to extant VlsE types for there to be cross-protection. By this account, for species, such as humans, that are largely or entirely irrelevant for the parasite’s maintenance in nature, whether or not their anti-VlsE antibodies functionally cross-neutralize strains with other VlsE proteins is of little or no consequence for B. burgdorferi evolution.
We consider a different possible outcome of selection for the case of the paralogous genes bbk07 and bbk12. The deduced proteins BBK07 and BBK12 of strain B31 have an overall sequence identity of 87%, differing only in the last third. There is evidence that one or both proteins are surface-exposed lipoproteins, expressed mainly in mice rather than ticks (35), and elicit antibodies during infections of humans, P. leucopus, and M. musculus (29, 35), but their functions are unknown. All or parts of the lp36 plasmid reportedly are associated with virulence and infectivity (34, 55), and there are DNA rearrangements or deletions involving this extrachromosomal replicon among different strains (see Fig. S2 in the supplemental material) (54–56). The genetic differences in the bbk07 and bbk12 loci between strains of this study plausibly explain the differences in the antibody reactivity in the sera of infected P. leucopus that we observed. Either the genes were undetectable or much truncated, e.g., as in WQR, or there were indels and/or nonsense mutations at the 5′ end of the gene that presumably would diminish protein expression, e.g., in HB19.
BBK07 and BBK12 are not essential for B. burgdorferi’s viability or growth in culture medium (37) and, as shown here, were not necessary for infection of either P. leucopus or immunodeficient M. musculus. But there was a tendency for the low-prevalence strains with missing or mutant bbk07/bbk12 loci to have lower tissue burdens during infection of P. leucopus. While the role of either of these proteins in dissemination or other aspects of virulence remains to be established, this study documents the occurrence of presence/absence polymorphisms in these loci among the B. burgdorferi strains. This may be analogous to the presence/absence phenomenon reported for Streptococcus pneumoniae at a pilus locus under pressure of wide application of a pneumococcal vaccine (57). We propose that immune selection over time has yielded lineages of B. burgdorferi that can infect vertebrate hosts that are otherwise wholly or partially immune to anti-BBK07/12 antibodies. Exploitation of this niche, however, may be at a fitness cost, such as lower densities in the host’s skin when ticks are feeding. This in turn could result in the lower overall prevalences that were observed in the B. burgdorferi population.
MATERIALS AND METHODS
Bacterial strains.
Strains of B. burgdorferi and their geographic origins are listed in Table 1, and their sources are given in Text S1 in the supplemental material. The B. hermsii strain was HS1. Spirochetes were cultivated in Barbour-Stoenner-Kelly (BSK) II broth medium containing 6% rabbit serum at 34°C, and cells were counted microscopically with a Petroff-Hausser chamber.
Experimental infections.
Adult female CB-17 SCID mice (Charles Rivers Laboratories, Wilmington, MA) were inoculated intraperitoneally (i.p.) with stocks of infected plasma and on day 21 terminally exsanguinated. Densities of spirochetes in the plasma were determined by quantitative PCR as described (12). Pathogen-free, 8-week-old, female, closed-colony outbred P. leucopus animals (Peromyscus Genetic Stock Center, University of South Carolina) in groups of 5 were inoculated i.p. on day 0 with fresh plasma containing 50 to 100 cells. Female, 6- to 8-week-old C3H/HeN mice (Charles River) were infected with SH2 or IDS in groups of 3 by the same protocol. Uninfected controls were 12 P. leucopus and 5 M. musculus mice. On day 35, plasma, the heart, distal tail, an ear, and a rear ankle were collected aseptically from each P. leucopus mouse and stored at −80°C. Three 1-mm-diameter punch biopsy specimens were taken from each ear. Day 56 sera from laboratory-reared P. leucopus mice infected with either strain BL206 or strain B348 of B. burgdorferi by bites of I. scapularis placed on mice on day 0, as described elsewhere (22), were provided by Lindsay Rollend and Durland Fish (Yale University, New Haven, CT).
DNA extractions.
Cells were harvested from medium by centrifugation at 9,500 × g for 10 min at 20°C, and washed twice with PBS (pH 7.4)-5 mM MgCl2. B. burgdorferi genomic DNA was extracted from culture medium, tissue, or blood with Qiagen’s DNeasy blood and tissue kit with proteinase K and TissueLyser II and QIACube apparatuses (Valencia, CA). The ear punch samples were processed with the Phire animal tissue PCR kit (Thermo, Fisher Scientific, Lafayette, CO). DNA concentrations were determined on a Nanodrop ND-1000 (Thermo Scientific) spectrophotometer.
PCR.
Primers, the corresponding nucleotides positions in the B31 reference genome, and expected product sizes are listed in Table S1 in the supplemental material. Additional primers for amplification of selected vlsE sequences are given in Text S1 in the supplemental material. Reactions were performed with Phusion DNA polymerase (New England Biolabs, Ipswich, MA) in a MyCycler thermal cycler (Bio-Rad, Richmond, CA). A 3-minute denaturation step at 98°C was followed by 35 cycles at 98°C for 30 s, 30 s at the specified annealing temperature (see Table S1 in the supplemental material), and 72°C for 90 s and a final extension at 72°C for 7 min. PCR products were purified using DNA Clean & Concentrator-5 (Zymo Research, Irvine, CA) and analyzed by agarose gel electrophoresis. Quantitative PCR using a Rotorgene 6000 thermal cycler (Qiagen, Valencia, CA) and primers and probe for the 16S rRNA gene was carried out in duplicate as described elsewhere (12, 27). PCR results were normalized for total DNA concentrations of the samples; a positive result was ≥10 gene copies per µg tissue DNA. For tissues from uninfected P. leucopus mice in the same runs, there were <0.1 gene copies.
DNA sequencing and analysis.
Nucleotide sequencing of PCR products was performed by Genewiz (La Jolla, CA). For next-generation sequencing, DNA extracted from culture harvest was first treated with RNase I (Fermentas, Burlington, Ontario, Canada), purified as described above, and then enzymatically sheared with an Ion Express Plus fragment library kit (Life Technologies, Grand Island, NY). Products were size selected by gel purification with the E-Gel system (Invitrogen). Templates were prepared by emulsion PCR on an Ion Torrent OneTouch apparatus (Life Technologies), and these were sequenced on an Ion Torrent personal genome machine with either Ion 314 or 316 chips (Life Technologies). Single reads of ~100 or ~200 bases were assembled into de novo contigs or mapped to reference sequences using CLC Genomics Workbench version 5.1 (CLC bio, Aarhus, Denmark). The coverage was at least 20×. The RNA-Seq algorithm of this software suite was used to count reads mapping to selected reference sequences with criteria of a minimum length fraction of 0.8 and minimum similarity fraction of 0.95. For comparisons of values from reference sequences of different lengths and between different sequencing runs, total reads were first converted to reads per base of the reference sequence, standardized to a fragment size of 750 bases, and then normalized as described in the supplemental material (see Text S1). Other sources of reference sequences are also given in Text S1. De novo sequence contigs were placed on a local BLAST server (version 2.2.6) (http://blast.ncbi.nlm.nih.gov) and interrogated with selected reference sequences. The multiple locus sequence types of strains not previously typed were determined by mapping sequence reads to the strain B31 chromosome. The assembled chromosome sequences were used for BLASTn searches of alleles of 8 chromosomal loci (58), and the deduced individual alleles were submitted to the reference MLST server for B. burgdorferi for assignment of MLST genotype (http://borrelia.mlst.net). The 16S-23S intergenic spacer and the ospC type of each strain were determined as described elsewhere (27).
Immunoblot assay.
The Borrelia ViraStripe IgG test kit from Viramed Biotech AG (Planegg, Germany) contained a whole-cell lysate of low-passage-number strain B31 cells with an added stripe of a recombinant VlsE of this strain. The strips were blocked, incubated with serum samples diluted 1:100, washed, and then incubated with alkaline phosphatase-conjugated goat anti-P. leucopus IgG heavy and light chain (Kirkegaard-Perry Laboratory, Gaithersburg, MD) as described elsewhere (4). After strips had been washed with buffer and then distilled water, bound antibodies were detected using 1-Step nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) (Pierce). A serum sample was considered positive if there were detectable bands for at least 3 of the standard antigens (18 kDa, 39 kDa, 41 kDa, and 45 kDa) on the blot, negative if there were no bands on the blot, and indeterminate if there was one band among these or other antigens.
Protein microarray.
Expression and purification of strain B31 recombinant proteins BBA25 (DbpB) and BBK12 were described previously (29). The procedure used to express and purify the following proteins was described by Baum et al. (Baum, E, Randall, A., Baldi, P., and Barbour, A.G. Submitted for publication.): BB0147 (FlaB), BBA15 (OspA), BBK07, BB0383 (BmpA), and VlsE from strain B31; OspC proteins type A, E, F, I, K, N, and T; and Vlp17 outer membrane protein of B. hermsii strain HS1. Lipoproteins (BmpA, BBK07, VlsE, OspA, OspC, and Vlp17) were cloned and expressed without signal peptides. A cell lysate of low-passage-number strain B31 was prepared by sonication in PBS (4). A 63-residue peptide comprising a methionine, then amino acids 20 to 73 of OspC of strain B31, and finally the influenza A hemagglutinin peptide YPYDVPDYA was synthesized by Invitrogen (Carlsbad, CA). Purified proteins were printed in duplicate on Whatman nitrocellulose-coated glass FAST slides (Piscataway, NJ) in 1.5-nanoliter volumes (30 pg) per spot as described elsewhere (29). An anti-His tag antibody (Sigma, St. Louis, MO) confirmed the presence of protein on the array. Negative controls were spots of storage buffer, PBS, and recombinant Burkholderia pseudomallei protein BPSS1525.
Binding of antibodies to array.
Kirkegaard-Perry (Gaithersburg, MD) produced a custom Cy3-conjugated, goat anti-P. leucopus heavy- and light-chain IgG antiserum. M. musculus antibodies were detected with Cy3-conjugated, goat anti-mouse heavy- and light-chain IgG (Jackson ImmunoResearch). Sera were diluted 1:100 in protein array blocking buffer (Whatman, Sanford, ME) that was supplemented with 10% (vol/vol) Escherichia coli lysate (MCLAB, San Francisco, CA). The array slides were incubated with sera, washed, incubated with secondary antisera, and washed again as described elsewhere (29). Slides were scanned in a ScanArray Express HT (PerkinElmer, Waltham, MA), and the bindings to spots were quantified with ProScanArray Express software (PerkinElmer) with spot-specific background correction. The mean pixel values for the two separate replicates for each antigen were calculated; values of ≤0 after background correction were assigned a value of 1. Replicate-to-replicate and pad-to-pad variation are described in the supplemental material (see Text S1). The results from two P. leucopus samples were excluded because of nonuniform exposure of the array pad. The cutoff for a positive interpretation of a value was 4 standard deviations above the mean for the control sera.
Statistics.
For means and OR, 95% confidence intervals are given in parentheses or brackets. Significance tests were two sided. A z score for an assay result was the number of standard deviations above or below the mean for the entire sample of infected animals. The total z score was the sum of scores for each tissue or for each antibody binding value for an individual mouse. Asymptotic, exact, and nonparametric statistical analyses of continuous data were carried out with Stata/MP version 10.1 (StataCorp, College Station, TX) or Systat version 11 (Systat Software, Chicago, IL).
Nucleotide sequence accession numbers.
The VlsE genes identified in TBO2, HB19, SH2, 27577, and WQR were assigned accession numbers JX406867 to JX406871, respectively.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download Text S1 , PDF file, 0.1 MB.
Alignment of partial amino acid sequences of deduced VlsE proteins for selected strains of B. burgdorferi and B. garinii (PLi) with the C6 peptide (35). Also shown is the corresponding peptide sequence for the orthologous Vlp17 protein of Borrelia hermsii. Download Figure S1 , PDF file, 0.1 MB.
Dot matrix plots of the lp36 linear plasmid of strain B31 against each of the same strands of assembled contigs of corresponding sequence reads for strains SH2, TBO2, HB19, 27577, and WQR. In the top left panel, an assembly from a resequencing of strain B31 is plotted against the reference B31 sequence. The alignments and dot matrix plots were performed using the MAFFT version 6 program (http://mafft.cbrc.jp/alignment/server/index.html) with the default settings for window size and stringency. The values on both axes are nucleotide positions from the 5′ ends. Highly similar sequences in same orientations on both of the pairwise sequences are indicated by red lines, and highly similar sequences oppositely oriented in the sequences are indicated by blue lines. Regions of the strain B31 lp36 that were not found in the assembled contigs for the other strains are denoted by gray areas. Download Figure S2 , PDF file, 0.2 MB.
PCR primers, annealing temperatures, and expected sizes of products for four strains of Borrelia burgdorferi.
Counts of sequence reads by strain that mapped to selected genes and loci of Borrelia burgdorferi.
Spirochete burdens in Peromyscus leucopus mice infected with different strains of Borrelia burgdorferi by strain prevalence and immunoblot result.
Binding of antibodies in sera of Peromyscus leucopus and Mus musculus mice to selected Borrelia antigens on a microarray.
Binding of antibodies in sera of infected Peromyscus leucopus to different OspC proteins of Borrelia burgdorferi on a protein microarray.
Pairwise percent amino acid identities of ungapped sites for translations of aligned partial vlsE nucleotide sequences (Fig. 6; also, see Text S1 in the supplemental material) of different strains of B. burgdorferi or B. garinii (PLi).
Summary of sequence analysis for PCR products for bbk07 and bbk12 genes.
ACKNOWLEDGMENTS
The work was supported by Public Health Service grants U54 AI-065359 (A.G.B.), R43 AI-072872 (A.G.B.), and F31 AI-078734 (E.B.).
We thank the reviewers for their constructive and insightful reviews of the manuscript, Lindsay Rollend and Durland Fish for providing sera from P. leucopus mice infected by tick bites, and Arash Ghalyanchilangeroudi and Azadeh Shojaee Estabragh for their assistance on an experiment.
Footnotes
Citation Baum E, Hue F, and Barbour AG. 2012. Experimental infections of the reservoir Peromyscus leucopus with diverse strains of Borrelia burgdorferi, a Lyme disease agent. mBio 3(6):e00434-12. doi:10.1128/mBio.00434-12
REFERENCES
- 1. Hoen AG, et al. 2009. Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proc. Natl. Acad. Sci. U. S. A. 106:15013–15018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fikrig E, Kantor FS, Barthold SW, Flavell RA. 1993. Protective immunity in Lyme borreliosis. Parasitol. Today 9:129–131 [DOI] [PubMed] [Google Scholar]
- 3. Tsao JI. 2009. Reviewing molecular adaptations of Lyme borreliosis spirochetes in the context of reproductive fitness in natural transmission cycles. Vet. Res. 40:36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bunikis J, et al. 2004. Borrelia burgdorferi infection in a natural population of Peromyscus leucopus mice: a longitudinal study in an area where Lyme borreliosis is highly endemic. J. Infect. Dis. 189:1515–1523 [DOI] [PubMed] [Google Scholar]
- 5. Tsao JI, et al. 2004. An ecological approach to preventing human infection: vaccinating wild mouse reservoirs intervenes in the Lyme disease cycle. Proc. Natl. Acad. Sci. U. S. A. 101:18159–18164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steppan S, Adkins R, Anderson J. 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 53:533–553 [DOI] [PubMed] [Google Scholar]
- 7. Barthold SW, Cadavid D, Phillip MT. 2010. Animal models of borreliosis. In Radolf JD, Samuels DS, Borrelia: molecular biology, host interaction, and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 8. Moody KD, Terwilliger GA, Hansen GM, Barthold SW. 1994. Experimental Borrelia burgdorferi infection in Peromyscus leucopus. J. Wildl. Dis. 30:155–161 [DOI] [PubMed] [Google Scholar]
- 9. Wright SD, Nielsen SW. 1990. Experimental infection of the white-footed mouse with Borrelia burgdorferi. Am. J. Vet. Res. 51:1980–1987 [PubMed] [Google Scholar]
- 10. Brown RN, Lane RS. 1994. Natural and experimental Borrelia burgdorferi infections in woodrats and deer mice from California. J. Wildl. Dis. 30:389–398 [DOI] [PubMed] [Google Scholar]
- 11. Duray PH, Johnson RC. 1986. The histopathology of experimentally infected hamsters with the Lyme disease spirochete, Borrelia burgdorferi. Proc. Soc. Exp. Biol. Med. 181:263–269 [DOI] [PubMed] [Google Scholar]
- 12. Barbour AG, et al. 2009. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am. J. Trop. Med. Hyg. 81:1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodman JL, Jurkovich P, Kodner C, Johnson RC. 1991. Persistent cardiac and urinary tract infections with Borrelia burgdorferi in experimentally infected Syrian hamsters. J. Clin. Microbiol. 29:894–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwan TG, Burgdorfer W, Schrumpf ME, Karstens RH. 1988. The urinary bladder, a consistent source of Borrelia burgdorferi in experimentally infected white-footed mice (Peromyscus leucopus). J. Clin. Microbiol. 26:893–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwanz LE, Voordouw MJ, Brisson D, Ostfeld RS. 2011. Borrelia burgdorferi has minimal impact on the Lyme disease reservoir host Peromyscus leucopus. Vector Borne Zoonotic Dis. 11:117–124 [DOI] [PubMed] [Google Scholar]
- 16. Dykhuizen DE, et al. 2008. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg. 78:806–810 [PMC free article] [PubMed] [Google Scholar]
- 17. Earnhart CG, Buckles EL, Dumler JS, Marconi RT. 2005. Demonstration of OspC type diversity in invasive human Lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC murine antibody response. Infect. Immun. 73:7869–7877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seinost G, et al. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67:3518–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang G, et al. 2002. Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J. Infect. Dis. 186:782–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeidner NS, Schneider BS, Dolan MC, Piesman J. 2001. An analysis of spirochete load, strain, and pathology in a model of tick-transmitted Lyme borreliosis. Vector Borne Zoonotic Dis. 1:35–44 [DOI] [PubMed] [Google Scholar]
- 21. Dolan MC, et al. 2004. Comparison of disseminated and nondisseminated strains of Borrelia burgdorferi sensu stricto in mice naturally infected by tick bite. Infect. Immun. 72:5262–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derdáková M, et al. 2004. Interaction and transmission of two Borrelia burgdorferi sensu stricto strains in a tick-rodent maintenance system. Appl. Environ. Microbiol. 70:6783–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanincová K, et al. 2008. Fitness variation of Borrelia burgdorferi sensu stricto strains in mice. Appl. Environ. Microbiol. 74:153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barbour AG, Travinsky B. 2010. Evolution and distribution of the ospC gene, a transferable serotype determinant of Borrelia burgdorferi. mBio 1:e00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bunikis J, et al. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741–1755 [DOI] [PubMed] [Google Scholar]
- 26. Brisson D, Vandermause MF, Meece JK, Reed KD, Dykhuizen DE. 2010. Evolution of northeastern and midwestern Borrelia burgdorferi, United States. Emerg. Infect. Dis. 16:911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Travinsky B, Bunikis J, Barbour AG. 2010. Geographic differences in genetic locus linkages for Borrelia burgdorferi. Emerg. Infect. Dis. 16:1147–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu WG, et al. 1997. A population genetic study of Borrelia burgdorferi sensu stricto from eastern Long Island, New York, suggested frequency-dependent selection, gene flow and host adaptation. Hereditas 127:203–216 [DOI] [PubMed] [Google Scholar]
- 29. Barbour AG, et al. 2008. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect. Immun. 76:3374–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theisen M, Frederiksen B, Lebech AM, Vuust J, Hansen K. 1993. Polymorphism in ospC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implications for taxonomy and for use of OspC protein as a diagnostic antigen. J. Clin. Microbiol. 31:2570–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magnarelli LA, Stafford KC, III, Ijdo JW, Fikrig E. 2006. Antibodies to whole-cell or recombinant antigens of Borrelia burgdorferi, Anaplasma phagocytophilum, and Babesia microti in white-footed mice. J. Wildl. Dis. 42:732–738 [DOI] [PubMed] [Google Scholar]
- 32. Norris SJ. 2006. Antigenic variation with a twist—the Borrelia story. Mol. Microbiol. 60:1319–1322 [DOI] [PubMed] [Google Scholar]
- 33. Bacon RM, et al. 2003. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J. Infect. Dis. 187:1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jewett MW, et al. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coleman AS, Pal U. 2009. BBK07, a dominant in vivo antigen of Borrelia burgdorferi, is a potential marker for serodiagnosis of Lyme disease. Clin. Vaccine Immunol. 16:1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hojgaard A, Eisen RJ, Piesman J. 2008. Transmission dynamics of Borrelia burgdorferi ss during the key third day of feeding by nymphal Ixodes scapularis (acari: Ixodidae). J. Med. Entomol. 45:732–736 [DOI] [PubMed] [Google Scholar]
- 37. Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 97:13865–13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strother KO, Broadwater A, De Silva A. 2005. Plasmid requirements for infection of ticks by Borrelia burgdorferi. Vector Borne Zoonotic Dis. 5:237–245 [DOI] [PubMed] [Google Scholar]
- 39. Zhang JR, Hardham JM, Barbour AG, Norris SJ. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275–285 [DOI] [PubMed] [Google Scholar]
- 40. Jewett MW, et al. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liang FT, et al. 1999. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi vlsE. J. Clin. Microbiol. 37:3990–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Casjens S, Eggers CH, Schwartz I. 2010. Borrelia genomics: chromosome, plasmids, bacteriophages and genetic variation. In Radolf JD, Samuels DS, Borrelia: molecular biology, host interaction, and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 43. Schutzer SE, et al. 2011. Whole-genome sequences of thirteen isolates of Borrelia burgdorferi. J. Bacteriol. 193:1018–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anderson JM, Norris DE. 2006. Genetic diversity of Borrelia burgdorferi sensu stricto in Peromyscus leucopus, the primary reservoir of Lyme disease in a region of endemicity in southern Maryland. Appl. Environ. Microbiol. 72:5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Swanson KI, Norris DE. 2008. Presence of multiple variants of Borrelia burgdorferi in the natural reservoir Peromyscus leucopus throughout a transmission season. Vector Borne Zoonotic Dis. 8:397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fung BP, McHugh GL, Leong JM, Steere AC. 1994. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect. Immun. 62:3213–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bockenstedt LK, et al. 1997. Borrelia burgdorferi strain-specific Osp C-mediated immunity in mice. Infect. Immun. 65:4661–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gilmore RD, Jr, Kappel KJ, Dolan MC, Burkot TR, Johnson BJ. 1996. Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational protective epitope in OspC. Infect. Immun. 64:2234–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Probert WS, Crawford M, Cadiz RB, LeFebvre RB. 1997. Immunization with outer surface protein (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J. Infect. Dis. 175:400–405 [DOI] [PubMed] [Google Scholar]
- 50. McDowell JV, Sung SY, Hu LT, Marconi RT. 2002. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with Lyme disease Spirochetes. Infect. Immun. 70:4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang FT, Bowers LC, Philipp MT. 2001. C-terminal invariable domain of VlsE is immunodominant but its antigenicity is scarcely conserved among strains of Lyme disease Spirochetes. Infect. Immun. 69:3224–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liang FT, Jacobson RH, Straubinger RK, Grooters A, Philipp MT. 2000. Characterization of a Borrelia burgdorferi VlsE invariable region useful in canine Lyme disease serodiagnosis by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 38:4160–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goettner G, et al. 2005. Improvement of Lyme borreliosis serodiagnosis by a newly developed recombinant immunoglobulin G (IgG) and IgM line immunoblot assay and addition of VlsE and DbpA homologues. J. Clin. Microbiol. 43:3602–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Casjens SR, et al. 2012. Genome stability of Lyme disease Spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One 7:e33280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu Y, Bruno JF, Luft BJ. 2003. Detection of genetic diversity in linear plasmids 28-3 and 36 in Borrelia burgdorferi sensu stricto isolates by subtractive hybridization. Microb. Pathog. 35:269–278 [DOI] [PubMed] [Google Scholar]
- 56. Casjens S, et al. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 57. Basset A, et al. 2007. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J. Clin. Microbiol. 45:1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Margos G, et al. 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 105:8730–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download Text S1 , PDF file, 0.1 MB.
Alignment of partial amino acid sequences of deduced VlsE proteins for selected strains of B. burgdorferi and B. garinii (PLi) with the C6 peptide (35). Also shown is the corresponding peptide sequence for the orthologous Vlp17 protein of Borrelia hermsii. Download Figure S1 , PDF file, 0.1 MB.
Dot matrix plots of the lp36 linear plasmid of strain B31 against each of the same strands of assembled contigs of corresponding sequence reads for strains SH2, TBO2, HB19, 27577, and WQR. In the top left panel, an assembly from a resequencing of strain B31 is plotted against the reference B31 sequence. The alignments and dot matrix plots were performed using the MAFFT version 6 program (http://mafft.cbrc.jp/alignment/server/index.html) with the default settings for window size and stringency. The values on both axes are nucleotide positions from the 5′ ends. Highly similar sequences in same orientations on both of the pairwise sequences are indicated by red lines, and highly similar sequences oppositely oriented in the sequences are indicated by blue lines. Regions of the strain B31 lp36 that were not found in the assembled contigs for the other strains are denoted by gray areas. Download Figure S2 , PDF file, 0.2 MB.
PCR primers, annealing temperatures, and expected sizes of products for four strains of Borrelia burgdorferi.
Counts of sequence reads by strain that mapped to selected genes and loci of Borrelia burgdorferi.
Spirochete burdens in Peromyscus leucopus mice infected with different strains of Borrelia burgdorferi by strain prevalence and immunoblot result.
Binding of antibodies in sera of Peromyscus leucopus and Mus musculus mice to selected Borrelia antigens on a microarray.
Binding of antibodies in sera of infected Peromyscus leucopus to different OspC proteins of Borrelia burgdorferi on a protein microarray.
Pairwise percent amino acid identities of ungapped sites for translations of aligned partial vlsE nucleotide sequences (Fig. 6; also, see Text S1 in the supplemental material) of different strains of B. burgdorferi or B. garinii (PLi).
Summary of sequence analysis for PCR products for bbk07 and bbk12 genes.