Abstract
Unraveling the complexity of the adaptive immune system requires the study of T cells in vivo. This protocol describes how populations of T cells specific for a given peptide: Major Histocompatibility Complex (pMHC) epitope can be identified in mice and tracked throughout the course of an immune response. The methodology involves the adoptive transfer of T-cell receptor (TCR) transgenic T cells with defined epitope specificity into histocompatible mice and the subsequent detection of these cells through the use of congenic or clonotypic markers. Alternatively, endogenous epitope-specific T cells can be tracked directly through the use of pMHC tetramers. Using magnetic bead-based enrichment and advanced multi-parameter flow cytometry, populations as small as 5 epitope-specific T cells can be detected from the peripheral lymphoid organs of a mouse. The adoptive transfer procedure can be completed within 3 h, while analysis of epitope-specific cells from mice can be completed within 6 h.
Search Terms: immunology, immune response, T cell, adoptive transfer, TCR transgenic mice, flow cytometry, tetramer, in vivo, magnetic bead enrichment
INTRODUCTION
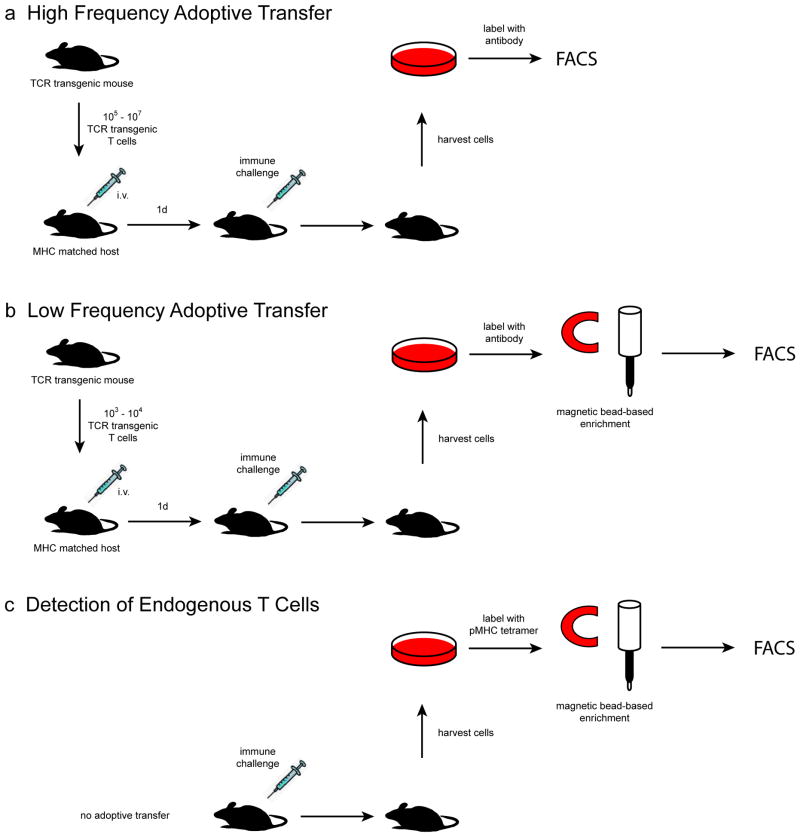
A major goal in the study of adaptive immune responses is to understand the developmental progression of antigen-specific T cells from naive precursors to activated effector cells and long-lived memory cells. The achievement of this goal requires the ability of investigators to track distinct populations of T cells with specificity to a single pMHC epitope throughout the course of an immune response. Over the years, technologies such as flow cytometry, TCR transgenic mice, and pMHC tetramers have enabled the development of powerful in vivo experimental systems that address these needs. In this article, we distill these methods into three related experimental protocols that provide a flexible framework from which to design in vivo studies of pMHC epitope-specific T cells (Fig. 1). Two of these protocols involve the transfer of TCR transgenic T cells into histocompatible host mice while the third protocol involves the detection of endogenous T cell populations via the use of pMHC tetramers. While many techniques are available to obtain quantitative and phenotypic data on these epitope-specific cells throughout the course of an immune response to the relevant epitope, we will focus on the use of flow cytometry in this article.
Figure 1.
Graphical representation of in vivo T cell tracking strategies. (a) A high number of TCR transgenic T cells is transferred into histocompatible recipient mice, and then identified by flow cytometric analysis of cells harvested from the recipient. (b) A low number of TCR transgenic T cells more closely approximating the frequency of an endogenous epitope-specific T cell population is transferred and then tracked with the help of a magnetic bead based enrichment step prior to flow cytometry. (c) In a strategy that does not involve the use of adoptively transferred cells at all, an endogenous population of epitope-specific T cells is tracked directly through the use of a pMHC tetramer and magnetic bead-based enrichment.
TCR transgenic T cell adoptive transfer system
The first two of the three described protocols are based on the T cell receptor (TCR) transgenic T cell adoptive transfer system 1, 2. This approach depends on TCR transgenic mice as a source of monoclonal naive T cells with a single defined epitope specificity. Naive T cells from the secondary lymphoid organs of these donor mice are injected into the bloodsteam of MHC-matched host mice that express an allelic form of a surface protein, usually CD45 or CD90, that is different from the form expressed by the transferred cells. Antibodies that recognize the allelic marker, or alternatively, a unique clonotype-specific epitope on the transgenic TCR itself, are used to distinguish the epitope-specific donor T cells from the background of host T cells with other specificities in tissue samples harvested from the host mouse.
High versus low frequency adoptive transfer
A high frequency transfer experiment involves the injection of 1 × 106 to 5 × 106 TCR transgenic T cells per recipient mouse. Typically, about 10–15% of the transferred population (1 × 105 to 7 × 105 cells) can be found in the secondary lymphoid organs of the recipient a day after the transfer 3–5. The “parked” population is at least 1,000 times larger than the few endogenous naive epitope-specific T cell populations that have been directly measured to date, which number around 20 to 1,200 cells per mouse 6–8. Therefore, although parking a large naïve population by adoptive transfer facilitates detection, it comes at the cost of creating a potentially unphysiologic situation. Indeed, several studies have demonstrated that antigen-induced activation is inefficient when the number of naïve cells is very large, perhaps as a result of increased competition for 4, 9–13 pMHC. Thus, depending on the number of cells transferred and the dose of antigen used, conclusions drawn from adoptive transfer experiments may not accurately reflect the behavior of endogenous T cells.
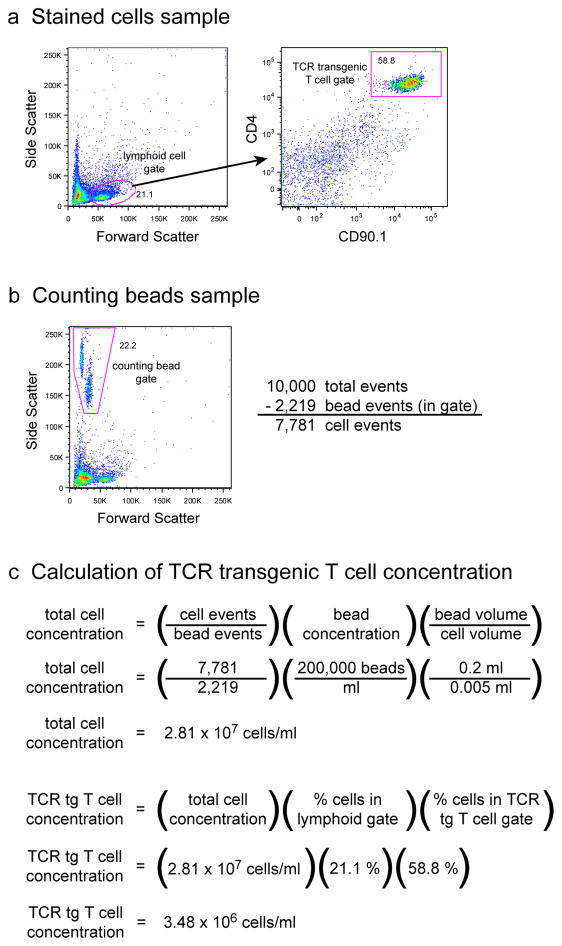
Nevertheless, high frequency transfer experiments are still very useful for applications in which high antigen doses can be used or when no low frequency alternatives are available. These applications may include experiments in which large numbers of epitope-specific cells are needed for biochemical assays, or experiments in which a high frequency of epitope-specific cells in lymphoid tissues are needed to ensure visualization during immunohistochemical or two-photon intravital imaging analysis. It should also be mentioned that only a handful of the estimated 106 endogenous epitope-specific T cell populations in mice have been measured to date 6, 14. Although the largest of these populations was 1,200 cells per mouse, it is possible that much larger populations exist in the normal repertoire 7. High frequency adoptive transfer experiments may be relevant to such populations.
Concerns related to a high frequency of naïve cells can be allayed by simply transferring a lower number of TCR transgenic T cells that approximates the naive precursor frequencies of known endogenous epitope-specific T cells. Considering a 10–15% cell parking efficiency, the transfer of about 1,000 TCR transgenic T cells will generate a naive population in the range of observed endogenous populations 4, 6, 7. However, the reliable detection of these low frequency cells necessitates a cell enrichment step and more sophisticated flow cytometric techniques 4, unless the cells have undergone extensive proliferation in vivo.
Endogenous T cell enrichment
A major issue concerning the use of the adoptive transfer system is that the transferred population is monoclonal, that is, each cell expresses the same TCR with a fixed affinity for the relevant pMHC. Therefore, issues related to the complexity of an epitope-specific T cell population, such as TCR repertoire diversity, interclonal competition, and immunodominance, may not be suitably addressed by this experimental system. Rigorous investigation of such issues requires the study of epitope-specific populations within a polyclonal repertoire.
This higher experimental standard can be met through the direct study of endogenous populations of epitope-specific T cells using pMHC tetramers coupled with magnetic bead based enrichment techniques 6–8, 15–26. With this experimental strategy, the protocol is modified such that no donor T cells are transferred, and pMHC tetramers are used instead of congenic or clonotypic marker-specific antibodies to detect endogenous epitope-specific T cells. pMHC tetramers are comprised of the actual ligands recognized by the TCRs of epitope-specific T cells, so in theory they should bind all epitope-specific T cells within a tissue sample. Thus, this experimental system can provide a comprehensive analysis of the entire polyclonal repertoire of epitope-specific T cells within a mouse. As with low frequency adoptive transfer, a magnetic bead-based enrichment of epitope-specific T cells is required as well as sophisticated flow cytometric techniques to improve sensitivity to the point where rare naive populations can be detected.
While pMHC tetramer-based detection systems provide the opportunity to study T cells under physiologic conditions, it is important to recognize the limitations of this approach. Of particular note is the possibility that there may not be an exact correspondence between the capacity of a T cell to bind to a pMHC tetramer and its capacity to become activated when confronted with the relevant pMHC on an antigen-presenting cell (APC) in the host 27. Although cells from many TCR transgenic T cell lines can be stained with pMHC tetramers with relevant specificity 6, it has been suggested that certain T cell clones, perhaps those with low affinity or co-receptor-dependent TCRs, are capable of responding to pMHC displayed by APC but bind weakly to the relevant pMHC tetramer 28–35. In this event, pMHC tetramer staining will detect fewer epitope-specific T cells than are capable of responding. In contrast, our experience suggests the opposite possibility. We found that only about half of an epitope-specific population of naive T cells detected by a pMHCII tetramer could become activated in a mouse following stimulation with a saturating dose of the relevant peptide 6. Although the basis for the incomplete response is unknown, it is possible that some T cells expressed TCRs with high enough affinity to bind the pMHCII tetramer but too low affinity to respond to pMHC displayed by APC. Until these uncertainties are resolved, it is best to keep in mind that pMHC staining may identify a wider or narrower range of epitope-specific T cells that can respond to the relevant pMHC epitope.
In this article, we detail the procedures involved in the tracking of epitope-specific T cells in vivo using high frequency adoptive transfer, low frequency adoptive transfer, or endogenous T cell enrichment strategies (Fig. 1). While there is a considerable degree of overlap between these sytems, each one has its unique set of advantages and drawbacks (Table I), which should be carefully considered during the design of experiments. The successful use of these protocols will also require users to familiarize themselves with relevant experimental parameters of these systems, which we will discuss below.
Table I.
Comparison of Epitope-specific T cell Tracking Protocols
| High Frequency Adoptive Transfer | Low Frequency Adoptive Transfer | Endogenous T cell Enrichment | |
|---|---|---|---|
| Advantages | Easy detection of cells with basic flow cytometric techniques | Near-physiologic naive T cell precursor frequency | T cell population is non- transgenic |
| Compatibility with CFSE dye tracking | Compatibility with CFSE dye tracking | T cell population is polyclonal | |
| Ability to limit genetic deficiencies to transferred T cells | Ability to limit genetic deficiencies to transferred T cells | Physiologic naive precursor frequency | |
| No enrichment required | Capacity to study many different pMHC-specific populations | ||
| Abililty to recover large numbers of epitope-specific T cells | Tetramer staining intensity reveals TCR binding affinity | ||
| Capacity to study pMHC-specific populations in humans | |||
| Disadvantages | T cell population is monoclonal | T cell population is monoclonal | Limited availability of pMHC tetramers |
| Unphysiologic naive precursor frequencies | Enrichment required | Enrichment required | |
| Need for TCR transgenic mice | Elaborate flow cytometric techniques required for analysis | Elaborate flow cytometric techniques required for analysis | |
| Need for TCR transgenic mice | Incompatibility with CFSE dye tracking | ||
| Some epitope-specific populations are exceedingly small | |||
| Best Uses | Cell division analysis | Cell division analysis | Study of epitopes for which TCR transgenic mice do not exist |
| Applications requiring large numbers of cells (biochemical analysis, mRNA isolation, immunohistochemistry, intravital imaging, etc.) | Studies of immune memory | Studies of immune memory | |
| Repertoire analysis | |||
| Immunodominance studies | |||
| References | 1 | 4 | 6 |
TCR transgenic T cells
To date, several dozen TCR transgenic mouse strains have been generated, offering a wide selection of epitope-specific CD4+ and CD8+ T cell clones for the design of adoptive transfer experiments (http://www.jamequist.umn.edu/TCRtable050908.htm). Many of these transgenic strains were derived from or extensively backcrossed to defined inbred strains, ensuring that T cells from these mice will not be rejected when transferred into recipient mice of the same background strain. The sex of the donor and recipient mice should also be matched to prevent any issues of cell rejection based on sex-linked minor antigens 36.
Because allelic exclusion of the TCRα locus is often incomplete, rearrangement and expression of an endogenous TCRα chain can occur in TCR transgenic mice, leading to the pairing of the transgenic TCRβ chain to a second TCRα chain 37. The result is that some of the T cells in a TCR transgenic mouse express TCRs with unknown pMHC specificity. This uncertainty can be avoided by crossing the TCR transgenes into mice with null mutations in either the recombination activating gene-1 (RAG-1) or recombination activating gene-2 (RAG-2) locus, the products of which are required for TCR gene rearrangement. T cells from RAG-1 or RAG-2 deficient TCR transgenic mice are pure with respect to pMHC specificity, and therefore are highly recommended as a source of donor TCR transgenic T cells.
For most studies, it is not necessary to purify TCR transgenic T cells from other non-T cells prior to their transfer into recipient mice. If desired, most of the non-T cells can be removed from harvested donor cells by performing magnetic bead based CD4+ or CD8+ T cell isolation via commercially available kits. To minimize stress on the TCR transgenic T cells, it is advisable to use an isolation kit that is based on negative selection, that is, the non-T cells are labeled with antibody and removed from the target T cells, which are left unlabeled.
Antigen
Antigen containing the peptide of the relevant pMHC epitope may take on many different forms. The simplest form of antigen is the purified peptide itself, or whole protein containing the peptide epitope sequence that can then be produced by antigen processing. Antigenic peptide or protein can be delivered in purified form or as an expressed part of a pathogenic microbe, tumor cell, or transplanted tissue. Often a model peptide epitope will be embedded within the sequence of a protein native to a particular pathogen to act as a surrogate antigen when no endogenous epitopes are known.
There are many different ways of introducing the relevant antigen into the body. We assume that investigators using this protocol to track epitope-specific T cells already have a well-characterized antigen delivery system in mind. The protocols described below recommend analysis of all the secondary lymphoid organs to ensure that all of the relevant epitope-specific T cells are included. The investigator may wish to modify this aspect of the protocol if the response of interest is likely to occur in a subset of the secondary lymphoid organs, for example, the lymph nodes draining a subcutaneous injection site.
Donor cell markers
Discrimination of donor T cells from the recipient’s T cells relies on the expression of a surface marker that is unique to the donor T cells, but does not provide a target for immune rejection by the recipient mouse. A number of allelic variations of surface receptors have been bred onto TCR transgenic mice for this purpose, the most common being CD45.1 (also known as Ly5.2 or Ly5b) and CD90.1 (also known as Thy1.1). For example, OT-I TCR transgenic T cells expressing the CD45.1 allele can be adoptively transferred into C57BL/6 mice, which express the CD45.2 allele 38. Commercially available antibodies specific to the different allelic forms of these markers can be used to identify the donor or host T cell populations. In adoptive transfer systems involving the commonly used C57BL/6 strain as the host, the use of CD45 or CD90 as congenic markers does not appear to elicit immune rejection of the transferred cells. However, the same may not be true for other strains of mice and care should be taken in choosing an appropriate congenic marker when designing adoptive transfer experiments.
An alternative approach to the use of congenic markers is the use of antibodies specific for a clonotypic TCR. For example, DO11.10 TCR transgenic T cells can be tracked after adoptive transfer into BALB/c mice with the use of the KJ1-26 antibody, which specifically recognizes the DO11.10 TCR 39, 40.
The availability of multiple congenic and clonotypic markers enables the design of experiments involving the adoptive transfer of multiple distinct TCR transgenic T cell populations. For example, CD45.1+ OT-I T cells can be co-transferred with CD90.1+ OT-II T cells into C57BL/6 mice, which are CD45.2+CD90.2+. The two donor cell populations can then be individually tracked based on their respective markers, allowing study of the interaction between CD4+ and CD8+ T cells 41. In some unusual cases, congenic or clonotypic markers may not be sufficient to clearly identify donor T cell populations. In such cases, antibodies to the specific TCR Vα or Vβ segments used by the donor transgenic T cells can be used in addition to congenic or clonotypic antibodies to further verify their identity.
CFSE
The use of carboxyfluorescein diacetate succinimidyl ester (CFSE) to monitor the proliferation history of T cells adds a powerful dimension to the adoptive transfer system 42. TCR transgenic T cells can be labelled with the fluorescent CFSE dye prior to adoptive transfer. At specific timepoints following adoptive transfer, the relative CFSE fluorescence intensity of the cells can be used to indicate the number of divisions they have undergone. More information about the use of CFSE can be obtained from another Nature Protocols article specifically devoted to this topic 43. An added benefit of CFSE labeling adoptively transferred T cells is that it can provide another parameter to distinguish donor from recipient T cells in cases where the donor cells have not divided extensively.
pMHC tetramers
Multimers of pMHCI or pMHCII ligands can be used to detect epitope-specific CD8+ and CD4+ T cell populations, respectively 44, 45. The most commonly used form of pMHC multimer is the tetramer, consisting of four identical biotinylated pMHC complexes bound to streptavidin in a tetrahedral configuration.
The use of phycoerythrin (PE)- or allophycocyanin (APC)-conjugated pMHC tetramers is recommended as these reagents provide strong signals with low background. Other common fluorochromes have not been as effective 46. Moreover, magnetic beads coupled to antibodies specific for these fluorochromes are commercially available, making these fluorochromes more practical for the pMHC tetramer-based enrichment of epitope-specific T cells.
Staining with pMHCI tetramers is generally analogous to staining with antibodies; a 30 minute incubation at 4°C is usually sufficient. However, temperature can be a major factor in the staining of cells with pMHCII tetramers 26, 27, 29, 47. For pMHCII tetramers we recommend a standardized staining protocol of 1 h at room temperature (~22°C).
A final pMHC tetramer concentration of 10 nM is usually adequate to stain the majority of epitope-specific T cells in a sample. Concentrations of 50 nM or higher can result in greater levels of background staining. As the optimal staining concentration is often dependent on the quality of the tetramer, a careful dose titration should be performed on each production lot of tetramer. TCR transgenic T cells specific for the relevant pMHC, if available, are quite useful for this purpose. Alternatively, T cells harvested from mice primed with the relevant antigen can be used, but an enrichment step may be necessary as the frequency of expanded cells is expected to be low.
Once bound to T cells, pMHC tetramers are stable during most fixation and permeabilization protocols used for intracellular staining. Experiments assessing the expression of FoxP3 or intracellular cytokines in tetramer-stained T cells have been successfully performed by our group and others 48 (JJM, MP, JBM, and MKJ, unpublished observations).
Enrichment
The enrichment of rare epitope-specific T cells in mice is accomplished through a magnetic bead-based procedure that results in about a 100-fold increase in the frequency of the target population 4, 6. Therefore, all of the epitope-specific T cells within the ~2 × 108 total nucleated cells harvested from the peripheral lymph nodes and spleen of a mouse can be concentrated into a sample containing only about 5 × 105–3 × 106 total cells, which can be analyzed in its entirety on a flow cytometer. This allows for the comprehensive sampling of epitope-specific T cells in individual mice. Using TCR transgenic T cells as positive and negative controls, we have determined a limit of detection at about 5 epitope-specific T cells per mouse 4, 6.
For the enrichment of transferred TCR transgenic T cells, the cell sample containing the TCR transgenic T cells is stained with a biotinylated antibody specific for the congenic or clonotypic marker expressed by these cells 4. The antibody-labeled cells are then further labeled with magnetic microbeads conjugated to streptavidin. The cell sample is then passed over a magnetized column, to which the microbead-coated T cells bind. The magnetic field is then removed from the column, and the enriched cells are eluted.
Enrichment can also be performed with fluorochrome-conjugated primary antibodies and anti-fluorochrome conjugated magnetic microbeads as alternative reagents. Indeed, the enrichment of rare endogenous epitope-specific T cells is performed using fluorochrome-conjugated pMHC tetramers as the primary staining reagent, followed by corresponding anti-fluorochrome conjugated magnetic microbeads 6. The use of multiple fluorochromes allows for the simultaneous enrichment of multiple populations of transferred or endogenous epitope-specific T cells from a single sample.
The efficiency of positive selection by the magnetic column decreases with increased numbers of magnetically labeled target cells. Therefore, for samples containing very large numbers of epitope-specific cells, it is advisable to also analyze the unbound fraction. The number of epitope-specific cells calculated in this fraction can be added to the number calculated in the enriched bound fraction to give the total number of cells for the entire sample. Alternatively, the enrichment step may be omitted entirely if the frequency of epitope-specific T cells in the mouse is known to be very large, as may be the case at the peak of an immune response to an antigenic stimulus.
Flow cytometry
When large numbers of TCR transgenic T cells are injected, the transferred cells will be numerous enough in the host’s secondary lymphoid organs to be detected by conventional flow cytometry. The use of just two identifying parameters, usually CD4 (or CD8) and the congenic (or clonotypic) marker, allows for unambiguous detection of the donor T cells from a collection of just 100,000 total events, or about 1/2,000th of the total nucleated cells harvested from the peripheral lymphoid organs of a mouse. Thus, a simple flow cytometer capable of analyzing 2 fluorochromes would suffice for many experiments.
However, a more sophisticated flow cytometric analysis is required when enrichment techniques are used to detect small target populations. During both antibody and pMHC tetramer based cell enrichment schemes, a considerable number of “sticky” non-target cells are also enriched in the final bound fraction of the sample. Many of these cells are highly autofluorescent. In addition, detection of low frequency T cell populations requires that most, if not all, of the roughly 5 × 105–3 × 106 cells in the bound fraction of the sample be analyzed by the flow cytometer. Together, these aspects of cell enrichment result in high levels of background noise that prevent effective donor T cell detection with simple 2 or even 4 color analysis.
To overcome these background issues, analysis should be performed on an advanced flow cytometer such as the BD LSR or LSRII that is capable of analyzing at least 6 different fluorochromes. Our protocol includes multiple levels of inclusion and exclusion gating strategies to minimize background and focus critical evaluation on genuine T cell events. At first, a side scatter area versus width plot is used to identify cell aggregates, which are gated out of the analysis. The remaining single cell events are then analyzed on a plot of non-T cell lineage (so-called dump) markers (B220 for B cells, CD11b and F4/80 for macrophages, and CD11c for dendritic cells) versus CD3. This allows for putative T cell events to be gated away from most other cell types, including the highly autofluorescent cells that reside along the center diagonal of the plot, as well as cells that non-specifically bind to antibodies. The gated CD3+ T cell events are then plotted for CD8 versus CD4 expression, which allows for the selection of the desired subpopulation and also provides another chance to gate cells away from any remaining autofluorescent events along the diagonal of the plot. Finally, CD44 is used as a marker to distinguish antigen-experienced T cells 49. We have outlined a 6-color basic staining panel that has been optimized for use with the three different experimental systems described in this article. Remaining available fluorescent channels may be used for additional phenotypic T cell markers, but any new setup should be tested carefully first for fluorochrome compatibility.
Some of the commercially available antibodies specific for CD45 and CD90 alleles can stain cells with such high intensity that they cause fluorescence spillover that cannot be corrected with compensation settings on the flow cytometer. Therefore, we recommend titrating the dose of these antibodies, as well as fluorochrome-conjugated streptavidin used as a secondary staining reagent, so that positive and negative cell populations can be easily distinguished from each other without excessive amounts of fluorescent signal.
Due to the presence of both low and high affinity T cells within most polyclonal T cell populations, pMHC tetramers staining usually produces a wide range of fluorescence intensities 50. Therefore, the discrimination of epitope-specific T cells from background can be more challenging when staining with tetramer than with congenic marker antibodies. The fair placement of gates to identify tetramer+ events should be determined through the use of relevant negative control samples. When analyzing epitope-specific CD4+ T cells with pMHCII tetramers, CD8+ gated events from the same sample can be used as an internal negative control on which to set a tetramer-positive gate. Similarly, CD4+ gated events can be used to help set gates for pMHCI tetramer-positive CD8+ T cells. In addition, T cells from irrelevant TCR transgenic mice can be used as additional negative controls provided that they are processed and stained in the same experiment 6.
MATERIALS
REAGENTS
TCR transgenic mice (see Introduction for more information)
Recipient mice (must be MHC compatible and sex-matched with TCR transgenic mouse)
CAUTION Investigators must follow guidelines for the care and use of laboratory animals and obtain prior approval from their Institutional Animal Care and Use Committee.
complete EHAA media (see REAGENT SETUP)
sorter buffer (see REAGENT SETUP)
Fc block (see REAGENT SETUP)
PBS, pH 7.4
Antigen (see Introduction for more information)
PE- or APC-conjugated pMHC tetramer (produced by the investigator, obtained from the NIH Tetramer Core facility, or purchased from commercial sources)
CRITICAL The stability of pMHC tetramers can vary greatly depending on their design and construction. However, to best avoid any potential issues regarding their stability, they should be stored at 4°C in the dark.
streptavidin-conjugated magnetic microbeads (Miltenyi Biotec 130-048-101)
anti-PE-conjugated magnetic microbeads (Miltenyi Biotec 130-048-801)
anti-APC-conjugated magnetic microbeads (Miltenyi Biotec 130-090-855)
LS columns (Miltenyi Biotec 130-042-401)
counting beads (Caltag-Invitrogen PCB-100)
-
antibodies:
Antibody Clone Vendor T cell markers CD3ε-FITC 145-2C11 eBioscience 11-0031 CD4-Pacific Blue RM4-5 eBioscience 57-0042 CD4-PE RM4-5 eBioscience 12-0042 CD4-PerCP RM4-5 BD-Pharmingen 553052 CD4-AlexaFluor700 RM4-5 eBioscience 56-0041 CD8α-Pacific Orange 5H10 Caltag-Invitrogen MCD0830 Non-T cell lineage (dump) markers B220-Pacific Blue RA3-6B2 eBioscience 57-0452 CD11b-Pacific Blue M1/70.15 eBioscience 57-0112 CD11c-Pacific Blue N418 eBioscience 57-0114 F4/80-Pacific Blue BM8 Caltag-Invitrogen MF48028 Congenic/clonotypic markers CD45.1-biotin A20 eBioscience 13-0453 CD45.2-biotin 104 eBioscience 13-0454 CD90.1-biotin HIS 51 eBioscience 13-0900 CD90.2-biotin 53-2.1 eBioscience 13-0902 KJ1-26-biotin KJ1-26 eBioscience 13-5808 Other CD44-AlexaFluor700 IM-7 eBioscience 56-0441 streptavidin-PE eBioscience 12-4312 antibodies may also be available through other vendors
EQUIPMENT
surgical scissors, tweezers
60 mm Petri dishes
100 μm nylon cell strainers (BD Falcon 352360)
15 ml polypropylene centrifuge tubes with 0.1 ml graduations (Sarstedt 62.554.205)
5 ml polystyrene FACS tubes (BD Falcon 352054)
1.2 ml polypropylene mini-FACS tubes (USA Scientific 1412-1000)
1 cc tuberculin syringes with 25 gauge needles
1 cc tuberculin syringes without needles
MidiMACS or QuadroMACS magnet (Miltenyi Biotec 130-042-302, 130-090-976)
BD LSRII flow cytometer (BD Immunocytometry Systems)
FlowJo flow cytometry analysis software (Tree Star, Inc.)
REAGENT SETUP
EHAA media
EHAA medium (Invitrogen 06-0006DJ) supplemented with 10% heat inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 U/ml streptomycin, 20 μg/ml gentamycin, 2 mM L-glutamine, and 55 μM 2-mercaptoethanol.
Sorter Buffer
PBS pH 7.4 supplemented with 2% heat inactivated FBS and 0.1% sodium azide.
Fc block
Harvest supernatant from 2.4G2 hybridoma (ATCC) cultures grown in hybridoma serum-free media (Invitrogen 12045) and supplement with 2% mouse serum, 2% rat serum, and 0.1% sodium azide.
Counting beads
Counting beads are supplied at a concentration that is specific to each production lot. Dilute the counting beads with sorter buffer to a concentration of 200,000 beads/ml. Verify their concentration with a hemacytometer.
PROCEDURE
There are three basic experimental designs described within this protocol (Fig. 1). Before proceeding, decide which one will be used and navigate accordingly.
| High frequency adoptive transfer | steps 1–25, then 38–50 |
| Low frequency adoptive transfer | steps 1–50 |
| Analysis of endogenous T cells | steps 20–50 |
Isolate TCR transgenic T cells
-
1
Add 2 ml of ice cold EHAA to a 60 mm Petri dish containing a 100 μm nylon cell strainer and place on ice. Work under aseptic conditions until cells are injected into mice.
-
2
Euthanize a TCR transgenic mouse. For simplification purposes, it will be assumed throughout this protocol that the donor TCR transgenic T cells express CD90.1. Make appropriate substitutions if other congenic or clonotypic markers are used.
CAUTION Investigators must follow guidelines for the care and use of laboratory animals and obtain prior approval from their Institutional Animal Care and Use Committee.
-
3
Remove the spleen and as many easily accessible lymph nodes as possible. These should include at least the inguinal, axillary, brachial, cervical, and mesenteric lymph nodes. Place them on top of the cell strainer in the Petri dish.
-
4
Using the plunger end of a 1 ml syringe, mash the lymphoid tissue over the cell strainer to liberate lymphocytes. Transfer cells and media that have passed through the cell strainer into a 15 ml polypropylene centrifuge tube. Rinse the cell strainer with another 2 ml ice cold EHAA and add to the same tube.
-
5
Add ice cold EHAA to a volume of 15 ml and centrifuge the tube at 300g, 4°C for 5 min.
-
6
Aspirate the supernatant and resuspend the cell pellet in 5 ml fresh ice cold EHAA. Keep on ice.
CRITICAL STEP To maximize cell viability, try to minimize the amount of time taken to harvest and analyze donor cells before adoptive transfer. This will result in higher seeding efficiencies in the recipient.
Calculate the concentration of TCR transgenic T cells
-
7
Transfer 98 μl of cells to a 5 ml FACS tube for staining. Transfer another 5 μl of cells to a 5 ml FACS tube containing 200 μl of counting beads (at 200,000 beads/ml) for later use in step 12.
-
8
Add 1 μl of anti-CD4-PE if the TCR transgenic cells are pMHCII-specific or 1 μl of anti-CD8-PE if the TCR transgenic cells are pMHCI-specific, and 1 μl of anti-CD90.1-APC (antibodies from 0.2 mg/ml stocks). Vortex and incubate at 4°C for 30 min.
-
9
Add 5 ml ice cold sorter buffer and centrifuge at 300g, 4°C for 5 min.
-
10
Resuspend the cell pellet in 0.5 ml ice cold sorter buffer.
-
11
Analyze the sample on a flow cytometer, collecting 10,000 total events. Determine the percentage of cells that are TCR transgenic T cells, as identified by lymphoid forward and side scatter profile and co-staining for CD4 (or CD8) and CD90.1 (Fig. 2a).
-
12Vortex the counting beads sample well and immediately analyze on the flow cytometer using the same settings as for the previous stained sample, collecting 10,000 total events (Fig 2b). Determine the concentration of total cells in the sample according to the following equation:
-
13Calculate the concentration of TCR transgenic T cells in the sample by multiplying the total cell concentration (from step 12) by the percentage that are T cells (from step 11).
Figure 2.
Analysis of TCR transgenic T cells prior to adoptive transfer. (a) A sample of cells isolated from the secondary lymphoid tissue of an SM1 RAG1−/− CD90.1+ TCR transgenic mouse is stained for CD4 and the CD90.1 congenic marker to determine the frequency of SM1 TCR transgenic T cells within the sample. (b) A parallel sample of unstained cells is mixed with a known concentration of counting beads to determine the concentration of total cells in the sample. (c) The total cell concentration of the sample is calculated by an equation that incorporates the relative frequencies of the cells and counting beads in the analyzed sample. The concentration of SM1 TCR transgenic T cells in the sample is determined by multiplying the total cell concentration by the percentage of cells that fall within the lymphoid gate and are positive for both CD4 and CD90.1.
Adoptive transfer of TCR transgenic T cells
-
14
If desired, label the cells with carboxyfluorescein diacetate succinimidyl ester (CFSE) to track cell division history (see Introduction).
-
15
Centrifuge the cell sample at 300g, 4°C for 5 min.
-
16
Aspirate the supernatant. Resuspend the cell pellet with room temperature PBS (intravenous injection of cold PBS could harm the recipient mouse) to a concentration such that 0.3 ml will contain the desired number of TCR transgenic cells to be transferred. Note that only about 10–15% of the transferred cells will park in the recipient.
-
17
If visible clumps are present, filter the cell suspension through a nylon cell strainer.
CRITICAL STEP If the cells have been labeled with CFSE, an appreciable amount of cell death may occur. Re-analyze the total cell concentration with counting beads to verify the proper concentration of TCR transgenic T cells. It is not necessary to analyze the sample again for CD4 and congenic marker expression. Adjust the concentration of the sample accordingly if necessary.
-
18
Inject 0.3 ml of cell suspension into the tail vein of a histocompatible, CD90.2+ recipient mouse. Although we have not extensively tested different routes of injection, the intravenous route is the most logical choice because recirculating naive cells normally enter secondary lymphoid organs from the blood.
-
19
Prime the mouse with antigen at least 1 day after the adoptive transfer. The transferred naïve cells survive with a half-life of at least 10 days, so it is not necessary to challenge with antigen immediately. A one day interval between transfer and priming maximizes the number of transferred cells while still allowing sufficient time for them to seed the secondary lymphoid organs.
Harvest of epitope-specific T cells
Note – Begin the protocol here for analysis of endogenous T cells in mice that did not receive TCR transgenic T cells.
-
20
For each mouse, set up a 60 mm Petri dish containing a 100 μm nylon cell strainer and 2 ml of ice cold EHAA and place on ice.
-
21
Euthanize each mouse to be analyzed.
-
22
Remove the lymphoid organs of interest. For a comprehensive sampling, harvest the spleen and as many easily accessible lymph nodes as possible. These should include at least the inguinal, axillary, brachial, cervical, and mesenteric lymph nodes. Place organs on top of the cell strainer in the Petri dish.
-
23
Using the plunger end of a 1 ml syringe, rub the spleen and lymph nodes over the cell strainer to release the cells. Transfer cells and media that have passed through the cell strainer into a 15 ml polypropylene centrifuge tube. Rinse the cell strainer with another 1 ml ice cold EHAA and add to the same tube. Repeat the 1 ml rinse two more times to collect as many lymphocytes as possible.
-
24
Add ice cold sorter buffer to a volume of 15 ml and centrifuge the tube at 300g, 4°C for 5 min.
Enrichment of epitope-specific T cells
-
25
If the frequency of epitope-specific T cells is expected to be very high, as in the case of a high frequency T cell adoptive transfer (≥105 TCR transgenic T cells injected per mouse), enrichment of the cell sample is not necessary (perform steps in Option A). For low frequency T cell adoptive transfer or detection of endogenous epitope-specific T cells (no transfer), the cell sample must be enriched for epitope-specific T cells prior to analysis by flow cytometry (perform the steps in Option B).
-
For high frequency adoptive transfer recipients
Aspirate the supernatant from step 24 and resuspend the cell pellet with 5 ml ice cold Fc block.
Proceed to step 38.
-
For low frequency adoptive transfer recipients or mice that did not receive transferred cells
-
Aspirate the supernatant and resuspend the cell pellet with ice cold Fc block in a volume equal to twice the volume of the pellet itself. The spleen and lymph nodes of a naive mouse usually produces a cell pellet of about 100 μl. Therefore, in this case, add 100 μl of Fc block to bring the volume to 200 μl. The graduations on the side of the 15-ml polypropylene tube can be used to adjust the volume to 200 μl. If a large degree of cell clumping has occurred, carefully remove the cell clump at this point with a pipet tip.
CRITICAL STEP When aspirating, be careful not to lose cells from the top of the pellet. Also, be sure to remove all droplets of buffer from the sides of the tube to avoid any unwanted changes in the resuspended volume of cells. Occasionally, cells will stick to the sides of the tube. In this case, a careful aspiration followed by a second wash with ice cold sorter buffer can be performed to get all the cells into the pellet.
Proceed to step 26.
-
-
-
26
Label the epitope-specific T cells. For samples containing adoptively transferred cells, add 2 μg/ml of biotinylated CD90.1 antibody. For endogenous epitope-specific T cells, add PE- or APC-labeled pMHC tetramer to a final concentration of 10 nM (or empirically optimized concentration).
-
27
If labeling with an antibody or a pMHCI tetramer, incubate cells at 4°C for 30 min. If labelling with a pMHCII tetramer, incubate cells at room temperature (~22°C) for 1 h.
-
28
Add ice cold sorter buffer to a volume of 15 ml and centrifuge the tube at 300g, 4°C for 5 min. Keep samples on ice or at 4°C from now on.
-
29
Aspirate and resuspend the cell pellet with ice cold sorter buffer to a volume of 200 μl. For antibody-labeled cells, add 50 μl streptavidin-conjugated magnetic microbeads. For tetramer-labeled cells, add 50 μl of anti PE- or APC-conjugated magnetic microbeads.
-
30
Incubate at 4°C for 30 min.
-
31
Add ice cold sorter buffer to a volume of 15 ml and centrifuge the tube at 300g, 4°C for 5 min.
-
32
During the centrifugation described in step 31, set up a Miltenyi LS column on a MidiMACS magnet and pre-rinse with 3 ml ice cold sorter buffer (discard flow-through). Place a new 15 ml centrifuge tube under the column.
-
33
Resuspend the cell pellet in 3 ml ice cold sorter buffer and add the cells through a nylon cell strainer onto the top of the column, collecting the unbound fraction into the tube placed underneath.
-
34
Rinse the original sample tube with 3 ml ice cold sorter buffer and add to the column, rinsing the cell strainer in the process. Discard the cell strainer. Add two more washes of 3 ml each to the column for a total of three, 3 ml washes. Continue collecting the flow-through into the same collection tube (for unbound material).
-
35
Remove the column from the magnet and place on top of a new 15 ml centrifuge tube. Add 5 ml ice cold sorter buffer and force the buffer through in one smooth motion with the plunger. Discard the column and plunger.
-
36
Centrifuge the bound and unbound collection tubes at 300g, 4°C for 5 min.
-
37
Carefully aspirate all but about 50 μl of buffer from the bound fraction tube. Resuspend the pellet to exactly 95 μl with ice cold Fc block. Vortex the tube after resuspending the pellet to dislodge cells adhering to the sides of the tube near the bottom. Aspirate buffer from the unbound fraction tube and resuspend to 2.0 ml with ice cold Fc block.
CRITICAL STEP The resulting cell pellet from the bound fraction will be very small and extra care should be made to avoid cell loss during the aspiration of the supernatant. When aspirating, be sure to remove all droplets of buffer from the sides of the tube to avoid any unwanted changes in the resuspended volume of cells.
Analysis of epitope-specific T cells by flow cytometry
-
38
For each sample, remove 5 μl and mix with 200 μl counting beads (at 200,000 beads/ml) in a 5 ml FACS tube. Set aside at 4°C for analysis later (step 46).
-
39
Determine the staining strategy for the cells. For optimal background reduction, use a two-stage gating strategy in which CD3+ events are first gated away from a non-T cell lineage panel stain (B220, CD11b, CD11c, F4/80), and then CD4+ events are gated away from CD8+ events (or vice versa). CD44 provides a useful marker for antigen-experienced cells 49. When staining cells previously labeled with pMHC tetramers, avoid the use of biotinylated antibodies, as the structure of pMHC tetramers involves biotin-streptavidin linkages. To minimize compensation issues, avoid the use of tandem dyes when possible. We recommend the following basic 6-fluorochrome setups:
-
For unenriched samples from high frequency adoptive transfer experiments (Fig. 3)
Fluorochrome Antibody Final concentration Pacific Blue dump (B220, CD11b, CD11c, F4/80) 2 μg/ml each Pacific Orange CD8 1 μg/ml FITC CD3 5 μg/ml PE CD90.1 0.4 μg/ml PerCP CD4 2 μg/ml AlexaFluor 700 CD44 2 μg/ml -
For CD90.1 enriched samples from low frequency adoptive transfer experiments (Fig. 4)
Fluorochrome Antibody Final concentration Pacific Blue dump (B220, CD11b, CD11c, F4/80) 2 μg/ml each Pacific Orange CD8 1 μg/ml FITC CD3 5 μg/ml PE streptavidin 1 μg/ml PerCP CD4 2 μg/ml AlexaFluor 700 CD44 2 μg/ml -
For pMHC tetramer enriched samples from endogenous T cell experiments (Fig. 5)
Fluorochrome Antibody Final concentration Pacific Blue dump (B220, CD11b, CD11c, F4/80) 2 μg/ml each Pacific Orange CD8 1 μg/ml FITC CD3 5 μg/ml PE pMHC tetramer (do not add again) PerCP CD4 2 μg/ml APC pMHC tetramer (do not add again) AlexaFluor 700 CD44 2 μg/ml
-
-
40
Stain the cell samples as outlined in step 39. Note that the final concentrations of antibody generally correspond to a 1:100 dilution of the stock concentrations, with the exception of anti-CD90.1-PE and streptavidin-PE, which have been carefully titrated down to doses that will not cause excessive fluorescence intensity that may contribute to difficulties in instrument compensation. For the staining of multiple samples, create an antibody cocktail master mix. For the bound fraction of enriched samples, add one dose of antibody cocktail directly to the whole 90 μl sample and vortex. For unenriched samples or the unbound fraction of enriched samples, transfer 90 μl cells to a 5 ml FACS tube, add one dose of antibody cocktail, and vortex. Incubate at 4°C for 30 min.
CRITICAL Antibodies to CD45 and CD90 congenic markers tend to stain cells very brightly and therefore can cause difficulties with compensation settings on the flow cytometer. While the listed concentrations for anti-CD90.1-PE and streptavidin-PE (secondary reagent) offer a guideline for these protocols, these concentrations should be empirically determined by the investigator for each new vial of reagent.
-
41
Prepare single stain compensation controls for calibration of the flow cytometer. For each fluorochrome to be used, set up a 5 ml FACS tube containing 90 μl of fresh mouse spleen or lymph node cells plus 1 μl of anti-CD4 antibody conjugated to that fluorochrome. Set up an unstained cell sample as well. Anti-CD4 antibody is a good choice for compensation samples because many fluorochrome conjugates are available and its staining intensity is equal to or greater than the staining intensity of the actual antibody that will be used in that color. Compensation can also be set on tubes singly stained with each actual antibody used in the experiment. However, this is not always feasible because the target population is sometimes too small to set an accurate compensation gate on.
-
42
Incubate at 4°C for 30 min.
-
43
Add ice cold sorter buffer to a volume of 5 ml for all samples and centrifuge the tubes at 300g, 4°C for 5 min.
-
44
For samples in FACS tubes, decant the supernatant and resuspend the pellet with ice cold sorter buffer to a volume of 1 ml. For bound fraction samples stained in 15 ml centrifuge tubes, carefully aspirate the supernatant, resuspend the pellet in 0.25 ml ice cold sorter buffer, and transfer the cells to a 1.2 ml FACS microtube. Rinse the centrifuge tube with another 0.25 ml ice cold sorter buffer and pool this with the other cells in the FACS microtube. Place the microtube inside a regular 5 ml FACS tube.
-
45
Set up and calibrate the flow cytometer using the single stained compensation controls. Include side scatter-width (SSC-W) as a selected parameter for collection.
-
46
Analyze the samples. For unenriched samples, collect 100,000 total events. For the unbound fractions of enriched samples, collect 500,000 total events. For the bound fractions of enriched samples, collect as many events as possible, up to a maximum of 2,000,000 total events. Try to avoid running the sample completely dry, as air bubbles may become included as part of your saved events. When the sample is almost entirely depleted, you can stop the acquisition, add more sorter buffer to the remaining volume, and restart the acquisition to maximize the collection of cells in the sample. Maintain acquisition rates at about 3,000 events/second or less to avoid high abort rates and altered side scatter profiles.
-
47
Analyze the counting bead samples from step 38. Keeping the same settings from step 46, collect 10,000 total events from each sample.
Figure 3.
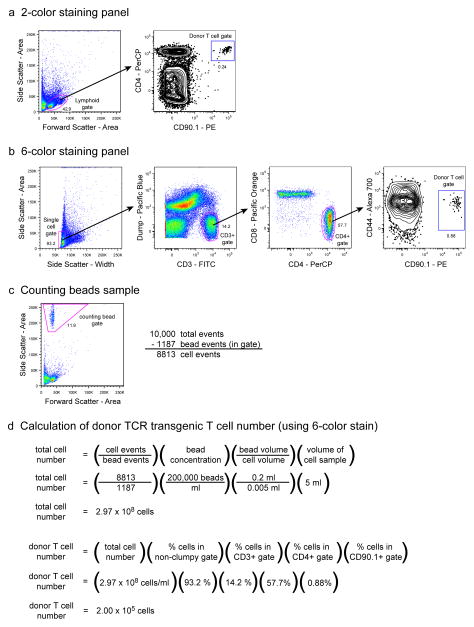
Analysis of donor TCR transgenic T cells from a high frequency adoptive transfer. 106 CD90.1+ SM1 TCR transgenic T cells were adoptively transferred into a C57BL/6 recipient mouse, and one day later cells harvested from the spleen and lymph nodes were analyzed by flow cytometry. Representative results show 100,000 total collected events. (a) Analysis from a simple 2-color staining panel. Cells with a lymphoid forward and side scatter profile are analyzed for CD4 and CD90.1 expression. Double-positive events are identified as donor T cells. (b) Analysis from an extensive 6-color staining panel. Single lymphoid cells are first identified by virtue of their low side scatter area and width intensities. CD3+ events are then gated away from the non-T cell lineage dump panel, and of these, the CD4+CD8− population is evaluated for CD90.1 as well as CD44 expression. Side scatterlow dump− CD3+ CD8− CD4+ CD44lo CD90.1+ events are identified as donor T cells. (c) Analysis of the counting beads sample. 10,000 total events were collected from a mixture of 200 μl of counting beads (200,000/ml) and 5 μl of cells from a 5 ml sample. The number of cells in the sample is equal to the number of total collected events minus the number of events in the counting beads gate. (d) Calculation of the total number of donor TCR transgenic T cells. Values for collected cell events and counting bead events from (c) are entered into an equation that calculates the total number of all cells in the entire cell sample. To calculate the total number of donor TCR transgenic T cells in the entire cell sample, this value is multiplied by the percentage of total events that correspond to these cells in the staining analysis from (a) or (b).
Figure 4.
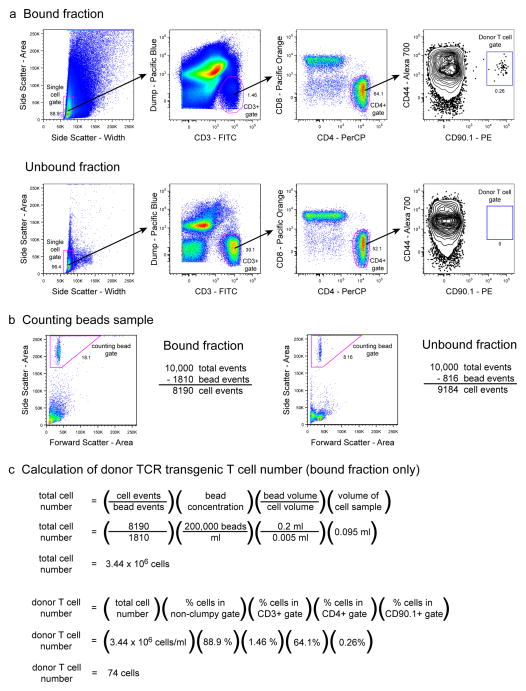
Analysis of donor TCR transgenic T cells from a low frequency adoptive transfer. 103 CD90.1+ SM1 TCR transgenic T cells were adoptively transferred into a C57BL/6 recipient mouse, and one day later cells harvested from the spleen and lymph nodes were analyzed by flow cytometry following CD90.1+ cell enrichment. (a) 6-color staining analysis of the bound and unbound fractions from the CD90.1+ cell enrichment. Cells were gated and evaluated for CD90.1+ donor T cell frequency as described in Figure 3b. Representative results show 2 × 106 total collected events for the bound fraction and 106 total collected events for the unbound fraction. (b) Analysis of the counting beads samples. 10,000 total events were collected from a mixture of 200 μl of counting beads (200,000/ml) and 5 μl of cells from the 95 μl bound or 2 ml unbound fraction samples. The number of cells in the sample is equal to the number of total collected events minus the number of events in the counting beads gate. (c) Calculation of the total number of donor TCR transgenic T cells in the entire cell sample (bound fraction only) was calculated as described in Figure 3d.
Figure 5.
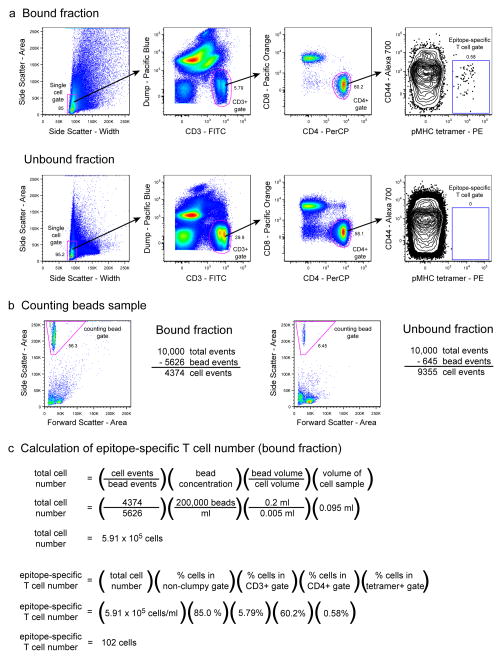
Analysis of endogenous epitope-specific T cells. Cells harvested from the spleen and lymph nodes of a naive C57BL/6 mouse were analyzed by flow cytometry following enrichment with the 2W1S:I-Ab pMHCII tetramer 6, 51. (a) 6-color staining analysis of the bound and unbound fractions from the enrichment. Cells were gated and evaluated for 2W1S:I-Ab-specific CD4+ T cell frequency as described in Figure 3b, with the substitution of 2W1S:I-Ab tetramer for CD90.1 antibody. The tetramer-positive gate was set using the CD8+ gated events as a negative control. Representative results show 2 × 106 total collected events for the bound fraction and 106 total collected events for the unbound fraction. (b) Analysis of the counting beads samples were performed as described in Figure 4b. (c) Calculation of the total number of 2W1S:I-Ab-specific T cells in the entire cell sample (bound fraction only) was calculated as described for donor TCR transgenic T cells in Figure 3d.
Calculation of cell numbers
-
48
Analyze the data from step 46 using FlowJo software (Fig. 3a–c). Use the following gating sequence:
-
Side scatter area vs. side scatter width
Gate on non-clumpy lymphoid events that are low for both parameters.
-
Dump (B220, CD11b, CD11c, F4/80) vs. CD3
Gate on dump- CD3+ events.
-
CD8 vs. CD4
Gate on CD4+CD8− or CD8+CD4− events.
-
CD44 vs. congenic/clonotypic marker or pMHC tetramer
Gate on epitope-specific T cells that are either CD44lo (naive) or CD44hi (activated).
-
-
49Using the data from step 47, calculate the total number of cells in the sample with the following equation:
-
50Calculate the total number of epitope-specific T cells in the sample by multiplying the total number of cells in the sample (step 49) by the percentage of events that correspond to epitope-specific T cells (step 48).
TIMING
It takes approximately 2 h to isolate and quantify TCR transgenic T cells from a mouse. Injection of these cells into recipient mice should require about 15 min for 1–4 mice, but may take longer depending on the technical skill of the operator. Therefore, the entire adoptive transfer procedure should take about 2–3 h.
Adoptively transferred cells are usually given a day to seed the lymphoid organs before the mouse is primed with antigen. Effector cell studies are usually conducted within the first 1–2 weeks after priming, and memory studies cover any period of time after that.
Isolating cells from a comprehensive set of peripheral lymphoid organs takes about 15 min per mouse. Cell enrichment and staining takes about 3–4 h. Flow cytometry takes about 15 min per sample. Therefore, the analysis part of a medium-sized experiment of 4 samples should take about 6 h.
Ideally, all of the steps should be performed continuously and as quickly as possible, since any increases in cell death will lead to concomitant increases in background staining. However, if necessary, the best place to stop would be at a point after the harvested tissues have been processed into single cell suspensions, but before they have been stained with any reagents. Although not advisable, cell suspensions can be kept on ice, preferably in EHAA media, for several hours before analysis. Some of the incubation times (antibodies, magnetic beads) can be extended with minimal effects, but room temperature tetramer incubation times should be adhered to rigidly. As a last resort, stained cells may be fixed with formaldehyde or paraformaldehyde and analyzed on the flow cytometer a day later.
TROUBLESHOOTING
See Table 1 for troubleshooting advice.
ANTICIPATED RESULTS
The total yield of TCR transgenic T cells from a TCR transgenic mouse can vary quite widely depending on the transgenic strain. In our experience, roughly 107 to 108 TCR transgenic T cells can be harvested from the spleen and most lymph nodes of a TCR transgenic mouse. In general, less TCR transgenic T cells are present in TCR transgenic mice that are bred onto a RAG-1 or RAG-2 deficient background. These yields usually range from around 106 to 3 × 107 cells per mouse.
The overall goal of these methods is to provide an assay with the sensitivity to reliably distinguish epitope-specific T cells from a background of other T cells. In the high frequency adoptive transfer method, the frequency of donor T cells in the sample is great enough that the analysis of just 100,000 total events on the flow cytometer yields a readily identifiable population of donor T cell events (Fig. 3). The donor T cells can be seen using a simple 2-color staining analysis (Fig. 3a), but are better resolved from background when a full 6-color staining panel is used (Fig 3b). The expected naive phenotype of the CD90.1+ donor T cells is verified by their low expression of CD44. The total calculated number of donor TCR transgenic T cells in the illustrated example is 2 × 105, which corresponds to a “park” rate of about 20% of the original 106 transferred cells (Fig. 3d). This is at the upper end of the 5–20% park rate seen for most adoptive transfer experiments.
The detection of low frequency adoptively transferred cells following CD90.1 antibody-based enrichment is shown in the example in Figure 4. As expected, CD90.1+ events are seen in the bound, but not unbound fraction of the enrichment (Fig. 4a). During antibody-based enrichment of T cells, about 5 × 105–3 × 106 total cells end up in the bound fraction. In addition to CD90.1+ T cells, this fraction contains many other cell types that have bound to the magnetic column nonspecifically. Compared to the unbound fraction, the bound fraction consists of more highly autofluorescent cells that make the gating of discrete populations more difficult. The total calculated number of donor TCR transgenic T cells in this example is 74, which corresponds to 7.4% of the original 1,000 transferred cells (Fig. 4c). This is at the low end of the expected 5–20% park rate, but due to the extremely low concentration of cells in the transfer inoculum, the accuracy of cell transfer doses in low frequency adoptive transfer experiments is usually less than that of high frequency experiments.
Tetramer-based enrichment analysis (Figure 5) is similar to that of antibody-based enrichment. Tetramer-positive T cell events are readily detected in the bound, but not unbound fraction samples. The expected naive phenotype of the pMHC tetramer-positive T cells is verified by their low expression of CD44. Because the epitope-specific T cells are polyclonal and exhibit a repertoire of TCR with varying affinities for pMHC, there is more variation in the staining intensities of the tetramer-positive cells (Fig. 5a) as compared to CD90.1+ T cells in antibody-enrichment experiments (Fig. 4a). Therefore, the tetramer-positive gate is positioned carefully using CD8+ gated events as an internal negative control. The total calculated number of endogenous epitope-specific T cells in the naive mouse in this example is 102, which is similar to the published expected frequency of 190 cells per mouse 6(Fig. 5c).
Table 2.
Troubleshooting Guide.
| Problem | Possible reason | Solution |
|---|---|---|
| Few or no transferred cells are detected above background. | Low donor T cell viability prior to adoptive transfer | Ensure media is supportive of good cell health. Minimize preparation time of cells. If applicable, reduce concentration of CFSE to minimize toxicity. High background. Include exclusion gate. |
| Donor cells are offscale during flow cytometric analysis | Too much congenic marker antibody is used (especially in case of CD45 or CD90) | Reduce/titrate the amount of congenic marker antibody used to stain cells |
| Inability to compensate flow cytometer | Bright and weak signals are in adjacent fluorochrome channels | Change fluorochrome setup to further separate the two signals |
| Erratic flow rates and cell profiles during flow cytometric acquisition | Cells are clumpy and causing clogs in the flow cytometer | Filter the cells through a nylon strainer and clean the flow cell before resuming |
Acknowledgments
This work was supported by the National Institutes of Health (J.J.M., J.H., A.J.P., M.P., J.B.M., M.K.J.) and the American Heart Association (H.H.C.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Contributor Information
James J. Moon, Email: jjmoon@umn.edu.
H. Hamlet Chu, Email: chuxx080@umn.edu.
Jason Hataye, Email: jhataye@mednet.ucla.edu.
Antonio J. Pagán, Email: pagan017@umn.edu.
Marion Pepper, Email: peppe033@umn.edu.
James B. McLachlan, Email: mclac016@umn.edu.
Traci Zell, Email: traci.zell@ebioscience.com.
Marc K. Jenkins, Email: jenki002@umn.edu.
References
- 1.Kearney ER, Pape KA, Loh DY, Jenkins MK. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1994;1:327–339. doi: 10.1016/1074-7613(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 2.Pape KA, et al. Use of adoptive transfer of T-cell-antigen-receptor-transgenic T cell for the study of T-cell activation in vivo. Immunol Rev. 1997;156:67–78. doi: 10.1111/j.1600-065x.1997.tb00959.x. [DOI] [PubMed] [Google Scholar]
- 3.Blattman JN, et al. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 5.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotturi MF, et al. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford ML, et al. Antigen-specific precursor frequency impacts T cell proliferation, differentiation, and requirement for costimulation. J Exp Med. 2007;204:299–309. doi: 10.1084/jem.20062319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulds KE, Shen H. Clonal competition inhibits the proliferation and differentiation of adoptively transferred TCR transgenic CD4 T cells in response to infection. J Immunol. 2006;176:3037–3043. doi: 10.4049/jimmunol.176.5.3037. [DOI] [PubMed] [Google Scholar]
- 11.Garcia Z, et al. Competition for antigen determines the stability of T cell-dendritic cell interactions during clonal expansion. Proc Natl Acad Sci U S A. 2007;104:4553–4558. doi: 10.1073/pnas.0610019104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzo AL, et al. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis MM. The alphabeta T cell repertoire comes into focus. Immunity. 2007;27:179–180. doi: 10.1016/j.immuni.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Luxembourg AT, et al. Biomagnetic isolation of antigen-specific CD8+ T cells usable in immunotherapy. Nat Biotechnol. 1998;16:281–285. doi: 10.1038/nbt0398-281. [DOI] [PubMed] [Google Scholar]
- 16.Bodinier M, et al. Efficient detection and immunomagnetic sorting of specific T cells using multimers of MHC class I and peptide with reduced CD8 binding. Nat Med. 2000;6:707–710. doi: 10.1038/76292. [DOI] [PubMed] [Google Scholar]
- 17.Lemaitre F, et al. Detection of low-frequency human antigen-specific CD4(+) T cells using MHC class II multimer bead sorting and immunoscope analysis. Eur J Immunol. 2004;34:2941–2949. doi: 10.1002/eji.200425281. [DOI] [PubMed] [Google Scholar]
- 18.Barnes E, et al. Ultra-sensitive class I tetramer analysis reveals previously undetectable populations of antiviral CD8+ T cells. Eur J Immunol. 2004;34:1570–1577. doi: 10.1002/eji.200424898. [DOI] [PubMed] [Google Scholar]
- 19.Keenan RD, et al. Purification of cytomegalovirus-specific CD8 T cells from peripheral blood using HLA-peptide tetramers. Br J Haematol. 2001;115:428–434. doi: 10.1046/j.1365-2141.2001.03106.x. [DOI] [PubMed] [Google Scholar]
- 20.Lucas M, et al. Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J Virol. 2004;78:7284–7287. doi: 10.1128/JVI.78.13.7284-7287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohn H, et al. MHC class II tetramer guided detection of Mycobacterium tuberculosis-specific CD4+ T cells in peripheral blood from patients with pulmonary tuberculosis. Scand J Immunol. 2007;65:467–478. doi: 10.1111/j.1365-3083.2007.01924.x. [DOI] [PubMed] [Google Scholar]
- 22.Jager E, et al. Peptide-specific CD8+ T-cell evolution in vivo: response to peptide vaccination with Melan-A/MART-1. Int J Cancer. 2002;98:376–388. doi: 10.1002/ijc.10165. [DOI] [PubMed] [Google Scholar]
- 23.Jang MH, Seth NP, Wucherpfennig KW. Ex vivo analysis of thymic CD4 T cells in nonobese diabetic mice with tetramers generated from I-A(g7)/class II-associated invariant chain peptide precursors. J Immunol. 2003;171:4175–4186. doi: 10.4049/jimmunol.171.8.4175. [DOI] [PubMed] [Google Scholar]
- 24.McDermott AB, Spiegel HM, Irsch J, Ogg GS, Nixon DF. A simple and rapid magnetic bead separation technique for the isolation of tetramer-positive virus-specific CD8 T cells. Aids. 2001;15:810–812. doi: 10.1097/00002030-200104130-00024. [DOI] [PubMed] [Google Scholar]
- 25.Day CL, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scriba TJ, et al. Ultrasensitive detection and phenotyping of CD4+ T cells with optimized HLA class II tetramer staining. J Immunol. 2005;175:6334–6343. doi: 10.4049/jimmunol.175.10.6334. [DOI] [PubMed] [Google Scholar]
- 27.Vollers SS, Stern LJ. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology. 2008;123:305–313. doi: 10.1111/j.1365-2567.2007.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falta MT, et al. Class II major histocompatibility complex-peptide tetramer staining in relation to functional avidity and T cell receptor diversity in the mouse CD4(+) T cell response to a rheumatoid arthritis-associated antigen. Arthritis Rheum. 2005;52:1885–1896. doi: 10.1002/art.21098. [DOI] [PubMed] [Google Scholar]
- 29.Reichstetter S, et al. Distinct T cell interactions with HLA class II tetramers characterize a spectrum of TCR affinities in the human antigen-specific T cell response. J Immunol. 2000;165:6994–6998. doi: 10.4049/jimmunol.165.12.6994. [DOI] [PubMed] [Google Scholar]
- 30.Gebe JA, et al. Low-avidity recognition by CD4+ T cells directed to self-antigens. Eur J Immunol. 2003;33:1409–1417. doi: 10.1002/eji.200323871. [DOI] [PubMed] [Google Scholar]
- 31.Mallet-Designe VI, et al. Detection of low-avidity CD4+ T cells using recombinant artificial APC: following the antiovalbumin immune response. J Immunol. 2003;170:123–131. doi: 10.4049/jimmunol.170.1.123. [DOI] [PubMed] [Google Scholar]
- 32.Kerry SE, et al. Interplay between TCR affinity and necessity of coreceptor ligation: high-affinity peptide-MHC/TCR interaction overcomes lack of CD8 engagement. J Immunol. 2003;171:4493–4503. doi: 10.4049/jimmunol.171.9.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniels MA, Jameson SC. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J Exp Med. 2000;191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallone R, et al. Functional avidity directs T-cell fate in autoreactive CD4+ T cells. Blood. 2005;106:2798–2805. doi: 10.1182/blood-2004-12-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laugel B, et al. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J Biol Chem. 2007;282:23799–23810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- 36.Gordon RD, Mathieson BJ, Samelson LE, Boyse EA, Simpson E. The effect of allogeneic presensitization on H-Y graft survival and in vitro cell-mediated responses to H-y antigen. J Exp Med. 1976;144:810–820. doi: 10.1084/jem.144.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinmetz M, Bluthmann H, Ryser S, Uematsu Y. Transgenic mice to study T-cell receptor gene regulation and repertoire formation. Genome. 1989;31:652–655. doi: 10.1139/g89-119. [DOI] [PubMed] [Google Scholar]
- 38.Lefrancois L, et al. The role of beta7 integrins in CD8 T cell trafficking during an antiviral immune response. J Exp Med. 1999;189:1631–1638. doi: 10.1084/jem.189.10.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrack P, Shimonkevitz R, Hannum C, Haskins K, Kappler J. The major histocompatibility complex-restricted antigen receptor on T cells. IV. An antiidiotypic antibody predicts both antigen and I-specificity. J Exp Med. 1983;158:1635–1646. doi: 10.1084/jem.158.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 41.Ehst BD, Ingulli E, Jenkins MK. Development of a novel transgenic mouse for the study of interactions between CD4 and CD8 T cells during graft rejection. Am J Transplant. 2003;3:1355–1362. doi: 10.1046/j.1600-6135.2003.00246.x. [DOI] [PubMed] [Google Scholar]
- 42.Parish CR. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol Cell Biol. 1999;77:499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- 43.Quah BJ, Warren HS, Parish CR. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat Protoc. 2007;2:2049–2056. doi: 10.1038/nprot.2007.296. [DOI] [PubMed] [Google Scholar]
- 44.Bousso P. Generation of MHC-peptide tetramers: a new opportunity for dissecting T-cell immune responses. Microbes Infect. 2000;2:425–429. doi: 10.1016/s1286-4579(00)00324-5. [DOI] [PubMed] [Google Scholar]
- 45.Mallone R, Nepom GT. MHC Class II tetramers and the pursuit of antigen-specific T cells: define, deviate, delete. Clin Immunol. 2004;110:232–242. doi: 10.1016/j.clim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, et al. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin Immunol. 2006;120:21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Cameron TO, Cochran JR, Yassine-Diab B, Sekaly RP, Stern LJ. Cutting edge: detection of antigen-specific CD4+ T cells by HLA-DR1 oligomers is dependent on the T cell activation state. J Immunol. 2001;166:741–745. doi: 10.4049/jimmunol.166.2.741. [DOI] [PubMed] [Google Scholar]
- 48.Burchill MA, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 50.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 51.Rees W, et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci U S A. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]