Abstract
Rationale
We previously identified conserved G/C Repressor elements in the promoters of most SMC marker genes, and demonstrated that mutation of this element within the SM22α promoter nearly abrogated repression of this transgene following vascular wire injury or within lesions of ApoE−/− mice. However, the mechanisms regulating the activity of the G/C Repressor are unknown, although we have previously shown that phenotypic switching of cultured SMC is dependent on Krupple-Like Factor 4.
Objective
The goals of the present studies were to: 1) injury-induced repression of SM22α gene following vascular injury is mediated through KLF4 binding to the G/C Repressor element; and 2) the transcriptional repressor activity of KLF4 on SMC marker genes is dependent on cooperative binding with pELK-1 and subsequent recruitment of HDAC2 which mediates epigenetic gene silencing.
Methods and Results
Chromatin immunoprecipitation (ChIP) assays were performed on chromatin derived from carotid arteries of mice having either a wildtype or G/C Repressor mutant SM22α promoter-LacZ transgene. KLF4 and pELK-1 binding to the SM22α promoter was markedly increased following vascular injury and was G/C Repressor dependent. Sequential ChIP assays and proximity ligation analyses in cultured SMC treated with PDGF BB or oxidized phospholipids showed formation of a KLF4, pELK-1, and HDAC2 multi-protein complex dependent upon the SM22α G/C Repressor element.
Conclusions
Silencing of SMC marker genes during phenotypic switching is partially mediated by sequential binding of pELK-1, and KLF4 to G/C Repressor elements. The pELK-1-KLF4 complex in turn recruits HDAC2 leading to reduced histone acetylation and epigenetic silencing.
Keywords: KLF4, pELK-1, HDAC2, Smooth muscle cells, acetylation, smooth muscle, gene transcription, transcription factors, vascular disease
INTRODUCTION
Smooth muscle cells (SMC) are remarkably plastic and transition from a quiescent contractile state to a proliferative-migratory state during vascular injury and development of atherosclerosis1. Collectively this process is termed “phenotypic switching”2, and is characterized by the coordinate down-regulation of markers of differentiated SMCs including SM22α, smooth muscle myosin heavy chain (SM-MHC), and SM α–actin, gene products required for SMC contraction1. SMC phenotypic plasticity likely evolved for optimization of vascular repair following injury3, although it is also widely accepted that SMC phenotypic switching plays a key role in development and progression of atherosclerotic lesions4, and regulation of plaque stability.
Our lab5–14 and many others15–28 have studied molecular mechanisms and factors that repress SMC differentiation marker gene expression as a means to elucidate processes involved in mediating SMC phenotypic switching. Importantly, these studies have clearly established that SMC phenotypic switching is actively regulated [reviewed in Owens et al, Physiol Revs2], and is mediated through complex processes including ERK-dependent phosphorylation of ELK-1, loss of SRF-myocardin (MRTF) binding to CArG boxes in SMC marker gene promoters15, 16, 29–32, HERP, and epigenetic silencing processes29–31, 33, 34. Moreover, we have presented multiple lines of evidence that phenotypic switching of cultured SMC in response to PDGF-BB, PDGF-DD, and oxidized phospholipids is dependent on the embryonic stem cell (ESC) pluripotency factor KLF4 including: 1) expression of KLF4 was increased after treatment of cultured SMC with these factors8, 14, 35–38; 2) siRNA induced suppression of KLF4 inhibited suppression of SMC marker genes8, 38–40; and 3) over-expression of KLF4 in cultured SMC was associated with coordinate down-regulation of SMC marker genes and the SMC specific SRF co-activator myocardin39, 40, and epigenetic silencing of SMC marker gene loci30, 31, 33, 34. However, the preceding studies were conducted almost exclusively in cultured SMCs that have already undergone extensive phenotypic switching. Moreover, it is clear that these simple in vitro models of SMC phenotypic switching fail to recapitulate complex environmental cues that mediate SMC phenotypic switching in vivo. As such, very little is known regarding mechanisms and factors that regulate SMC phenotypic switching in vivo following vascular injury or in disease models, including atherosclerosis.
A major advance in understanding the mechanisms that regulate SMC phenotypic switching in vivo were studies from our lab showing that suppression of the SMC marker gene SM22α following vascular injury or within atherosclerotic lesions of ApoE−/− mice was dependent on a G/C Repressor element located in proximity to SM22α 5′ CArG boxes41,42. Notably, mutation of the conserved G/C Repressor element did not alter developmental expression of this gene in transgenic mice41, but nearly completely abrogated down regulation of the gene following carotid wire injury41 or within atherosclerotic lesions42. However, studies failed to identify the transcription factors and mechanisms that regulate the activity of the G/C Repressor element, a regulatory element found within promoters of nearly all CArG-dependent SMC marker genes.
There are a number of transcription factor families capable of binding to G/C rich elements, including Sp1 and Kruppel-like zinc finger transcription factors43. The Owens laboratory first tested Sp1 and Sp3 as potential G/C Repressor binding factors since they are expressed in SMCs and are induced during PDGF-BB phenotypic switching of cultured SMCs41, 42. Interestingly, Sp1 can bind to the SM22α G/C Repressor in electrophorectic mobility shift assays (EMSA), and siRNA suppression of Sp1 inhibited phenotypic switching in cultured SMCs in response to PDGF-BB. However, we were unable to demonstrate direct binding of Sp1 to SMC marker gene promoters within intact chromatin by ChIP assays following PDGF-BB treatment or in vivo following vascular ligation injury41,42. Moreover, we subsequently showed that Sp1 dependence of SMC phenotypic switching in cultured SMC was mediated by Sp1-dependent activation of KLF4 whose promoter contains three conserved Sp1 binding sites45. KLF4 is an attractive alternative candidate given that we have previously shown that it is required for SMC phenotypic switching of cultured SMC8, 30–32, 38–40, 45. Moreover, we demonstrated increased KLF4 binding to SMC promoters following carotid ligation injury in vivo, and that global conditional KLF4 knockout mice showed a transient delay in SMC phenotypic switching following carotid ligation injury in vivo32. However, we have been unable to show specific binding of KLF4 to the SM22α G/C repressor based on EMSA. Furthermore, the effects of global knockout of KLF4 on SMCs may be mediated indirectly through loss of KLF4 in: 1) macrophages where it mediates monocyte to macrophage differentiation46, 47, 48; and/or 2) in endothelial cells where it has pro-inflammatory effects including mediating activation of leukocyte adhesion molecules49, 50. Thus, at present, there is no direct evidence indicating that the activity of the SM22α G/C Repressor in vivo during vascular injury is dependent on KLF4 or mediated through direct binding of KLF4 to the G/C repressor element. The present studies test the hypothesis that repression of SM22α expression during SMC phenotypic switching in vivo following carotid ligation is mediated through binding of pELK-1 and KLF4 to the G/C Repressor element. We also hypothesize that the pELK-1-KLF4 complex in turn recruits HDACs to this gene locus and mediates epigenetic silencing. Finally, given evidence that many SMC marker genes contain G/C Repressor elements, we postulate that these mechanisms contribute to coordinate suppression of multiple SMC genes during SMC phenotypic switching.
MATERIALS AND METHODS
Animal protocol models were approved by the University of Virginia Animal Care and Use Committee. An expanded Materials and Methods section is available online at http://circres.ahajournals.org.
RESULTS
Suppression of SM22α gene expression following carotid ligation is G/C Repressor dependent
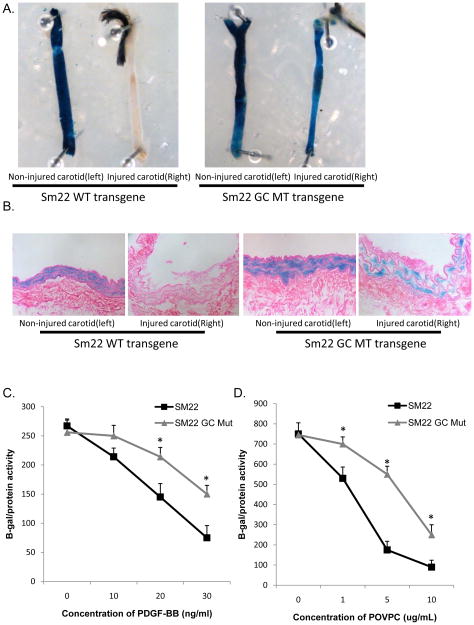
We previously demonstrated that mutation of the SM22α G/C Repressor element inhibited repression of this gene during SMC phenotypic switching following carotid wire injury41 or within atherosclerotic lesions of ApoE−/− Western diet fed mice42. Given evidence that SMC phenotypic switching following carotid ligation is KLF4-dependent32, we first determined if suppression of SM22α in this model is also G/C Repressor dependent. Carotid ligations were done in SM22α wild-type and SM22α G/C Repressor mutant LacZ transgenic mice41. Xgal staining was examined 3-days post ligation, a time point we previously demonstrated that there is significant loss of expression of the endogenous SM22α gene in this model32. Results showed that mutation of the G/C Repressor nearly abolished down-regulation of SM22α following ligation injury [Figs. 1A and 1B]. The un-ligated left carotid of both SM22α wild-type and SM22α G/C Repressor mutant LacZ transgenic mice showed no repression and exhibited SMC restricted expression, indicating that the mutation had no discernible effects on expression in differentiated (non-phenotypically modulated) SMC, consistent with our previous observations showing normal developmental expression of the mutant transgene41. Taken together, these results, and those of our previous studies demonstrate that the G/C Repressor element is required for down-regulation of SM22α gene expression in ALL models of SMC phenotypic switching examined to date. Given the diversity of these models, it is thus likely that G/C Repressor dependent SMC phenotypic switching represents a common transcriptional regulatory pathway for SMC phenotypic switching across highly divergent stimuli. To further test this possibility, we determined the importance of the G/C Repressor element in two in vitro models of SMC phenotypic switching, treatment with PDGF BB9, 10, 14, 39, and the pro-atherogenic oxidized phospholipid POVPC8, 38. The SM22α G/C Repressor mutant promoter reporter showed attenuated PDGF-BB and POVPC induced repression as compared to the WT SM22α promoter reporter, although both showed some repression consistent with previous reports that there are also G/C Repressor independent mechanisms operative within these in vitro models41,42, although it remains to be shown they are functional in vivo.
Figure 1. The SM22 G/C Repressor mutant LacZ transgene fails to down-regulate following vascular ligation injury in vivo.
A). SM22 and SM22 G/C mut LacZ mice were subjected to right common carotid ligation injury. The right and left carotids were harvested 3 days following injury, fixed, Xgal stained, and a gross morphological image was taken. B). Carotid arteries from part A were embedded in paraffin, sectioned and stained as previously described41. Each picture is a representative image from N=5 animals. C). Rat aortic smooth muscle cells were plated and then transiently transfected with LT-Mirus 24 and 300 ng of alpha-actin or SM22 or SM22 with a mutant GC repressor element. 24 hrs after transfection cells were treated with PDGF-BB. 24 hrs after treatment cells were harvested and LacZ activity and protein activity were measured. * indicates significant down-regulation of the wild-type versus the G/C Repressor mutant as mentioned in Materials and methods. D). Cells were treated as mentioned in C. 24 hrs after transfection cells were treated with POVPC. Cells were harvested and assayed as mentioned in 1C.
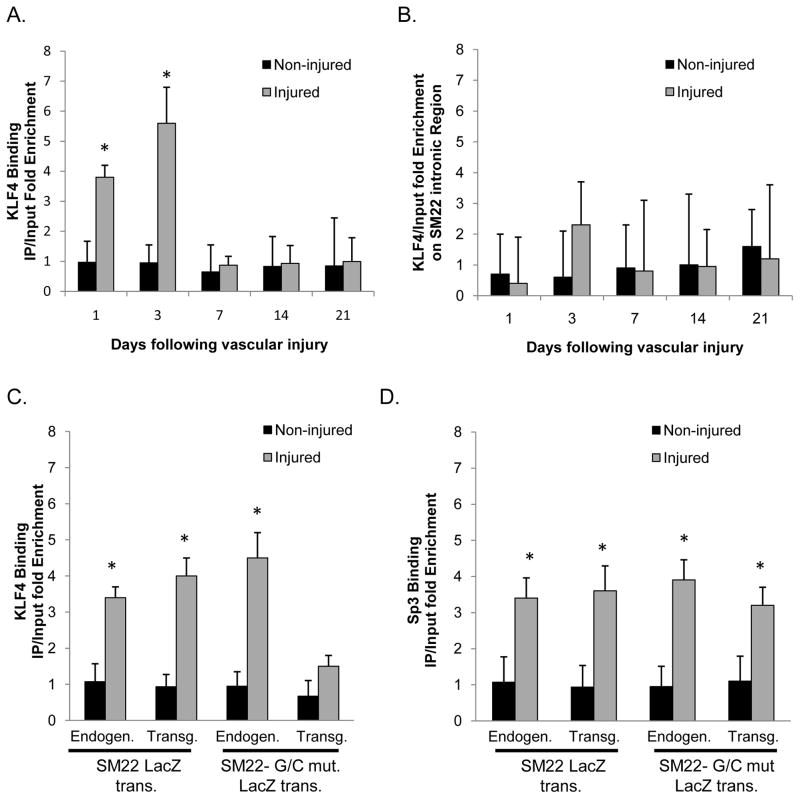
KLF4 binds the G/C Repressor element in vivo following vascular ligation injury
We have previously shown enriched binding of KLF4 to SM22α and other SMC promoters following carotid ligation in vivo32. However, there is no direct evidence that this binding was G/C Repressor dependent. To test if KLF4 binding in vivo is G/C Repressor dependent, we performed ChIP assays following carotid ligation in SM22α wild-type and SM22α G/C Repressor mutant LacZ transgenic mice. Given limited availability of transgenic mice, and the requirement of 10 mouse carotids for each in vivo ChIP data point, we first determined the kinetics of KLF4 binding to the endogenous SM22α promoter region using C57/B6 control mice. Ligated right carotids, and control left carotid arteries were harvested at 1, 3, 7, 14 and 21 days following vascular ligation injury. Consistent with our previous results32, we observed enhanced binding of KLF4 to the SM22α promoter region 1 and 3 days following vascular injury [Fig. 2A]. Binding was selective since KLF4 was not bound to an intronic sequence located within the SM22α promoter [Fig. 2B]. Further ChIP analyses on wild-type and G/C Repressor mutant mice 3 days after vascular ligation injury were completed to determine if KLF4 binding is G/C Repressor element dependent. As a control, KLF4 ChIP assays used PCR primers that distinguished the endogenous mouse SM22α promoter versus the rat SM22α promoter-LacZ transgenes [Online Figure I]. These experiments are important since we originally cloned KLF4 using a yeast one hybrid method based on its ability to bind to another G/C rich cis element located within 200 base pairs of the G/C Repressor element (the TCE element)40, 45. The TCE element is located proximal to the first CArG element while the G/C Repressor element is located proximal to the second CArG element [See Online Fig IVA], and we have shown that mutation of these elements have profoundly different effects39,41. That is, mutation of the TCE element completely abolished transgene expression in vivo in mice, whereas mutation of the G/C repressor had no effect on transgene expression during development and maturation but abrogated repression during SMC phenotypic switching.
Figure 2. KLF4 binds the SM22α promoter in vivo three days following vascular injury.
A&B) C57B6 mice were subjected to right common carotid ligation injury and then ten mice each were harvested at 1, 3, 7, 14 and 21 days following vascular injury and then subjected to ChIP analysis for KLF4 (A) or to an intronic region of the SM22 gene (B). C&D) SM22 and SM22 G/C mut LacZ mice were subjected to right common carotid injury and harvested 3 days after vascular ligation injury, tissues from ten mice were pooled, and then subjected to ChIP assay for KLF4 (C) or Sp3 (D) . In each ChIP IP, qPCR analysis was conducted on both the Endogenous and LacZ transgene as indicated in the Figure. * indicates significant binding as mentioned in Materials and methods.
Results of SM22α in vivo ChIP analyses demonstrated enriched binding of KLF4 to the WT but not the G/C repressor mutant SM22α LacZ transgene and the endogenous SM22α promoter in both transgenic strains [Fig 2C]. In contrast, Sp3 binding to the SM22α promoter was enhanced following carotid ligation but binding was not altered on the G/C Repressor mutation [Fig 2D and Online Fig III]. As such, it is interesting to speculate that Sp3 might represent an alternate G/C Repressor independent repressor pathway in cultured SMC [Fig. 1C-D), or in vivo. These in vivo ChIP assays provide clear evidence that enhanced binding of KLF4 to the SM22α promoter following carotid ligation is G/C Repressor dependent. The G/C Repressor is also required for suppression of SM22α during phenotypic switching of SMC following ligation injury [Fig. 1], wire injury41, and within ApoE−/− atherosclerotic lesions42. Taken together these results provide strong evidence that KLF4 plays an integral role in SMC phenotypic switching and functions through G/C Repressor dependent mechanisms. Significantly, the preceding results are the first to provide direct evidence that the functional effects of mutating the G/C Repressor element (i.e. virtually abrogating down-regulation of SM22α in response to vascular injury or within atherosclerotic lesions), are causally linked to G/C Repressor dependent binding of KLF4.
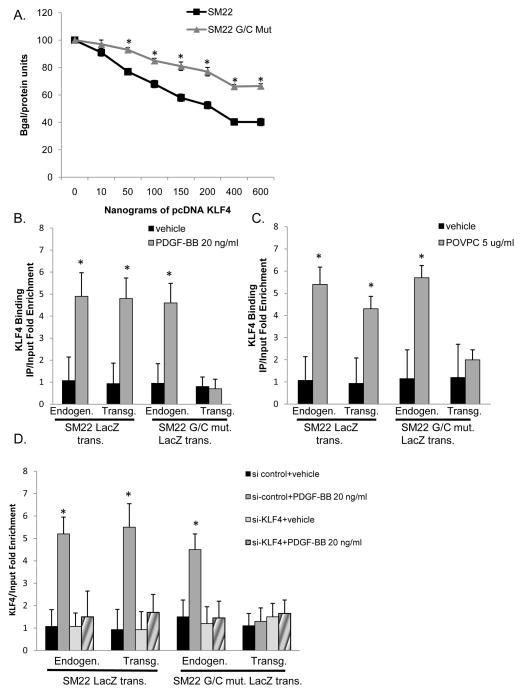
Previously we showed that KLF4 repressed SMC marker gene expression in cultured SMC through multiple mechanisms including: 1) disruption of SRF and myocardin binding to SMC promoters within intact chromatin39, 40, 45; and 2) induction of histone modifications associated with formation of heterochromatin and transcriptional silencing30, 31, 33, 34. However, we have not previously determined if these effects are G/C Repressor dependent which is critical if we are to establish a causal relationship between KLF4 binding, epigenetic changes, and our observations that mutation of the G/C Repressor abrogated SM22α gene suppression in vivo during SMC phenotypic switching. We first tested if KLF4-induced repression of SM22α in cultured SMCs is G/C Repressor dependent via transient transfection assays using a KLF4 over-expression plasmid plus the wild-type or G/C Repressor mutant SM22α promoter LacZ. KLF4-induced repression of the SM22α promoter was markedly attenuated by mutation of the G/C Repressor [Fig. 3A]. We also performed ChIP assays in rat aortic SMCs stably transfected with the wild-type or G/C Repressor mutant SM22α promoter LacZ transgenes [Online Fig. II]. KLF4 binding to the SM22α promoter was enhanced 12-hours following treatment of cultured SMC with either PDGF-BB [Fig 3B] or POVPC [Fig 3C] and binding was G/C Repressor dependent. Significantly, we showed that the G/C Repressor mutation itself did not result in decreased SRF binding [Online Fig. IVB] and thereby contribute to transcriptional repression, consistent with our observations that expression of the G/C Repressor mutant LacZ transgene is normal in differentiated SMCs in vivo [Fig. 1 and Regan et al41]. However, SRF binding to the WT SM22α LacZ promoter and the endogenous SM22α promoter, but not the G/C Repressor mutant, was dramatically reduced in cultured SMCs treated with PDGF BB or POVPC [Online Fig. IVB] commensurate with increased G/C Repressor dependent KLF4 binding [Online Fig. IVC]. Finally, as an additional control for specificity of the KLF4 ChIP assays, we demonstrated that treatment of cultured SMC with a KLF4 siRNA resulted in markedly reduced KLF4 binding to the endogenous SM22α promoters, as well as the WT SM22α promoter-LacZ transgene +/− PDGF-BB treatment [Fig. 3D]. Similar results were obtained in SMC treated with POVPC [data not shown]. In contrast, KLF4 was found to be enriched on both the WT and the mutant TCE SM22α promoters following PDGF-BB [Online Fig. IVC] and POVPC treatment [data not shown] demonstrating that KLF4 binding is G/C Repressor specific. These results extend our previous studies showing that PDGF-BB- and POVPC-induced phenotypic switching of cultured SMC is dependent on KLF439, 40, 45 by demonstrating that effects are mediated through binding to the G/C Repressor thereby establishing for the first time evidence of a direct causal relationship between KLF4 binding to SMC promoters and the functional effects of mutation of the G/C Repressor preventing down-regulation of SM22α during SMC phenotypic switching both in vivo and in vitro. Moreover results validate our in vitro PDGF and POVPC models to further explore mechanisms by which KLF4 directly regulates the G/C Repressor.
Figure 3. KLF4 binds to the SM22α LacZ transgene in vitro and binding is attenuated by mutation of the G/C Repressor element after PDGF-BB or POVPC treatment in rat aortic smooth muscle cells.
A) Rat aortic smooth muscle cells were plated at 1×10^4. 24 hr later cells were transiently transfected using LT-Mirus with 300 ng of SM22α WT or G/C Repressor MT and increasing concentrations of pcDNA-KLF4. 24 hours following transfection, media was removed and replaced and 24 hours later cells were harvested and subjected to Bgal and protein assays. Results are the average of three independent experiments performed in triplicate. B&C) Rat aortic smooth muscle cells stably transfected with either SM22α or G/C Repressor mutant were plated at 1×10^4 and allowed to grow to confluency and then switched to serum free media for three days. Following serum starvation, cells were treated with 20 ng/ml PDGF-BB (B) or 5 ug/ml of POVPC (C) for 12 hours and then subject to ChIP analysis. * indicates significant binding as mentioned in materials and methods. D) Cells were plated and 24 hours following plating cells were treated with either si-control or si-KLF4. 24 hours following siRNA transfection, cells were treated with either vehicle or 20 ng/ml PDGF-BB for 12 hours. Cells were harvested and ChIP analysis was performed as described in part A. * indicates significant binding as mentioned in Fig 2D.
pELK-1 binds via the SM22α G/C Repressor element three days following vascular ligation injury
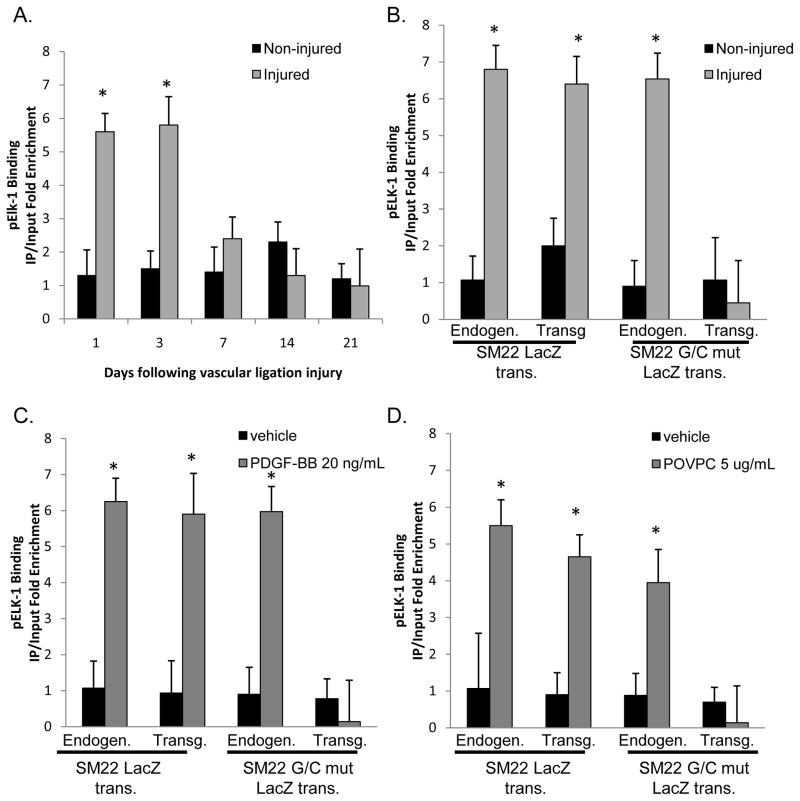
Yoshida et al., and others have demonstrated that pELK-1 binds to the SM22α promoter following PDGF-BB or POVPC treatment and that KLF4 and pELK-1 physically interact based on co-immunoprecipitation assays and sequential ChIP analysis16, 30, 31. pELK-1 also inhibits the interaction between SRF and myocardin by competing for same docking site on SRF as myocardin following PDGF-BB treatment16. The G/C Repressor and ETS binding site overlap by three base pairs within smooth muscle marker genes and we are physically unable to differentiate these sites [See Online Figure IVA]. Therefore, we sought to determine: 1) If there is enhanced binding of pELK-1 to the SM22α promoter following ligation injury in vivo, since previous studies by Wang et al. were done exclusively in cultured cells16; and 2) If pELK-1 binding to the SM22α promoter is altered by mutation of the G/C Repressor. Right carotid ligation injuries were performed and carotid arteries subjected to pELK-1 ChIP analyses at 1, 3, 7, 14 and 21 days after injury [Fig. 4A]. Results showed enhanced pELK-1 binding at 1 and 3 days following carotid ligation – results identical to what we observed for KLF4 [Fig 2A]. To determine if this enhanced pELK-1 binding was G/C Repressor dependent, we performed ChIP analyses in our SM22α WT, and G/C Repressor mutant SM22α promoter-LacZ transgenic mice. Results showed increased pELK-1 binding to the WT but not the G/C Repressor mutant SM22α transgene [Fig 4B], demonstrating that enhanced binding of pELK-1 to the SM22α promoter during SMC phenotypic switching in vivo is also G/C Repressor dependent. PDGF-BB or POVPC treatment resulted in increased pELK-1 binding to the endogenous and the wild-type SM22α LacZ transgene but not the G/C Repressor mutant SM22α promoter in cultured SMC [Fig. 4C, 4D].
Figure 4. pElk-1 binds to the SM22α promoter three days following carotid ligation in vivo.
A) C57B6 mice were subjected to right common carotid injury as mentioned in Fig 2A and then subjected to ChIP analysis for pElk-1. * indicates significant binding compared to non-injured controls. Results are the average of three independent experiments. B) SM22 and SM22 G/C mut LacZ mice were subjected to right common carotid injury and then harvested 3 days after vascular injury, tissues from ten mice were pooled, and then subjected to ChIP assay for pELK-1. In each ChIP IP, qPCR analysis was conducted on both the Endogenous and LacZ transgene as indicated in the Figure. * indicates significant binding as mentioned in Fig 2D. C&D) Stably transfected rat aortic smooth muscle cells were plated at 1×10^4, allowed to grow to confluency and then switched to serum free media for three days. Following serum starvation, cells were treated with 20 ng/ml PDGF-BB (C) or 5 ug/ml of POVPC (D) for 12 hours and then subject to ChIP analysis. * indicates significant binding as mentioned in Fig 2D.
We also performed transient transfection assays to determine if over-expression of pELK-1 inhibited expression of the SM22α promoter [Online Fig. V]. Over-expression of pELK-1 reduced expression of the WT promoter but not the G/C Repressor mutant. Marked reductions in pELK-1 binding to the WT SM22α promoter LacZ transgene and endogenous SM22α promoter was observed following siRNA induced suppression of pELK-1 [Online Fig. XA]. Moreover of major interest, suppression of pELK-1 expression also markedly reduced KLF4 binding, indicating that it is required for binding of KLF4 to the SM22α promoter. Taken together, the preceding data provide direct evidence that mutation of the G/C Repressor attenuates binding of KLF4 and pELK-1, and taken together with previous results16, 30, 31, suggest they act cooperatively to mediate repression of SM22α during SMC phenotypic switching in vivo and in vitro and that pELK-1 binding precedes KLF4 binding on the G/C Repressor element.
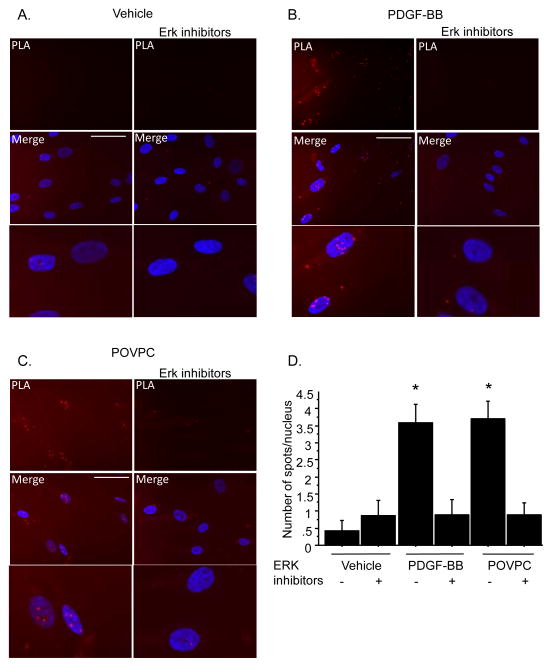
KLF4 and pELK-1 interact based on in situ proximity PLA assays
We previously demonstrated that KLF4 and pELK-1 interact based on co-immunoprecipitation assays on homogenates of cultured SMC treated with POVPC31. To determine if KLF4 and pELK-1 interact within intact cells we performed in situ DuoLink proximity ligation assays which permit detection of proteins located within approximately 40 nm within individual cells based on staining with secondary antibodies containing complementary single stranded DNA molecules (www.olink.com ). Assays were performed in human coronary artery cells due to the availability of higher quality PLA-compatible antibodies for detection of human KLF4 and pELK-1, and since this would allow us to determine if the results seen in our rodent SMC lines also apply to human coronary SMC. Consistent with results of studies in rodent SMC lines, results of ChIP assays in human coronary SMC showed marked enrichment of KLF4 and pELK-1 binding but reduced SRF binding to the SM22α promoter following treatment with PDGF BB [Online Figure VI]. Moreover, results of the PLA assays showed evidence of interaction of KLF4 and pELK-1 following treatment with PDGF-BB [Fig 5B] or POVPC [Fig. 5C]. This increased interaction was blocked by treatment with the ERK inhibitors U1026 and PD8059, indicating that interaction is dependent on ELK-1 phosphorylation, which is critical for physical interaction of ELK-1 with SRF16 and ELK-1 and KLF431. Consistent with reduced SRF binding to the SM22α promoter in ChIP assays, PLA results showed reduced interaction of SRF with myocardin following treatment of cultured human coronary SMC with PDGF-BB or POVPC but was retained in cells treated with ERK inhibitors39, 40 [Online Fig VII, VIII]. To our knowledge, these results are the first to actually show interaction of myocardin and SRF within intact cells, and provide further evidence in support of our hypothesis that cooperative interactions of KLF4 and pELK-1 mediate G/C Repressor dependent transcriptional repression during SMC phenotypic switching.
Figure 5. KLF4 and pELK-1 interact following PDGF-BB and POVPC using Proximity Ligation Assay (PLA).
A). Human coronary SMCs were plated and 24 h later switched to serum free medium. B and C). After deprivation with Serum Free Medium for 24h, cells were treated with PDGF-BB (10ng/ml), POVPC (10μg/ml) for 24h. Simultaneously, cells were treated with Erk inhibitors PD98059 (10μM) and U0126 (10μM). PLA amplification corresponding with the interaction of KLF4 and pELK-1 is visualized as red spots localized mainly into the nucleus. Interaction between KLF4 and pELK-1 is induced by PDGF-BB and POVPC treatment. This induction is abolished by the addition of Erk inhibitors. Scale bars, 100μg. D). Quantitation of number of spots per nucleus. * indicates specific binding compared to non-treated serum starved smooth muscle cells with a p-value <0.05.
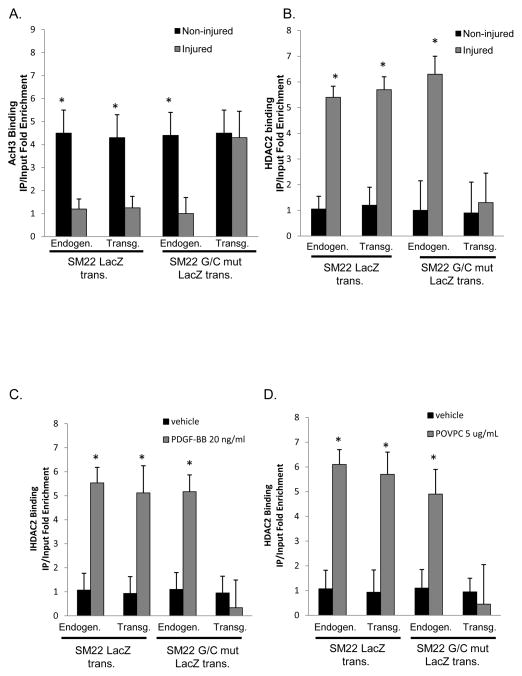
Hypomethylation of the SM22α promoter following carotid ligation is G/C Repressor dependent and mediated at least in part by recruitment of HDAC2
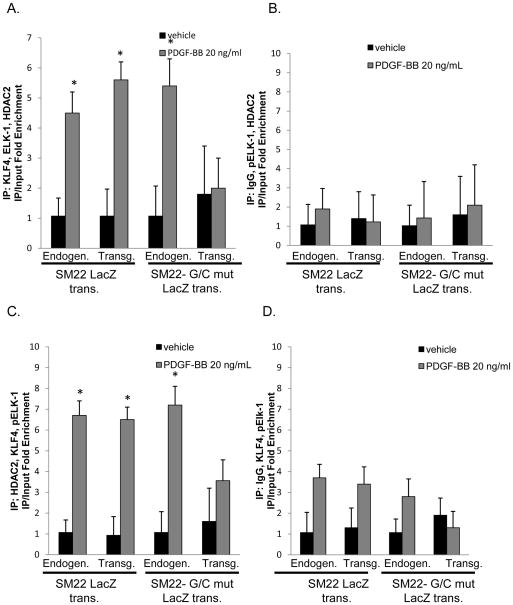
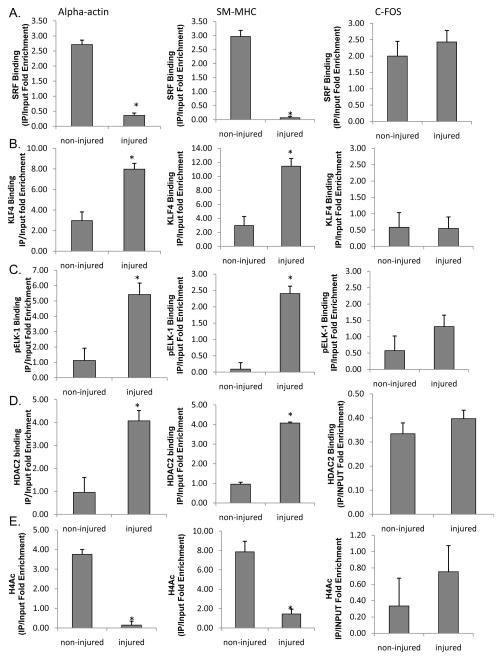
Previously, we demonstrated that PDGF-BB and POVPC-induced phenotypic switching in cultured SMC was mediated in part by KLF4 dependent recruitment of HDACs 2, 4, and 5 and subsequent DNA hypo-acetylation of SMC marker gene promoter regions30–32. To determine if similar mechanisms function in vivo, we performed H3 acetylation ChIP assays [Fig. 6A] in WT and G/C Repressor mutant SM22α promoter LacZ transgenic mice 3 days following carotid ligation. Results showed marked reductions in H3 acetylation of the endogenous SM22α promoter and wild-type LacZ transgene but not the G/C Repressor mutant transgene. There was also marked enrichment of HDAC2 to the WT and endogenous SM22α promoters, but not the G/C Repressor mutant SM22α promoter following ligation [Fig. 6B]. PDGF-BB or POVPC treatment of cultured SMC was associated with increased HDAC2 recruitment to the SM22α promoter that was also G/C Repressor dependent [Fig. 6C and 6D]. HDAC5 also showed increased binding in cultured SMC following PDGF-BB or POVPC treatment or in vivo following carotid ligation injury, but its binding was not affected by the G/C Repressor mutation [Online Fig. XII]. Binding was selective for HDACs 2 and 5 since we saw no evidence for enhanced binding of HDACs 3, 4 or 7 in vitro or in vivo [Online Fig XIII and Salmon and Owens data not shown]. Transient transfection assays of SM22α promoter-reporter genes with HDAC2 demonstrated decreased expression with the WT but not the G/C Repressor mutant [Online Figure V]. We also showed increased HDAC2 binding to the SM22α promoter in human coronary artery SMC following PDGF BB and POVPC treatment [Online Figure VI and data not shown] Furthermore, we were able to demonstrate that HDAC2 binding requires both pELK-1 and KLF4 via siRNA knock-downs and MEK/ERK inhibitor experiments [Online Figs. IX, X, XI]. Finally, to determine if KLF4, pELK-1 and HDAC2 co-localize to the G/C Repressor element, triple sequential ChIP analyses were performed [Fig. 7]. Sequential ChIP analyses demonstrated that KLF4, pELK-1 and HDAC2 were present within the same chromatin fragments consistent with formation of a higher order complex. Significantly we found that the G/C Repressor element was required for binding all three factors [Fig. 7A and 7C]. Finally, we performed additional ChIP analyses on the SM α–actin, SM-MHC and c-Fos promoters in injured and non-injured mouse carotid artery samples three days following ligation injury to determine if pELK-1-KLF4-HDAC2-dependent transcriptional repression is applicable to multiple SMC genes [Fig. 8]. Results showed significant enrichment of KLF4, pELK-1 and HDAC2 binding to the SM α–actin and SM-MHC promoters following ligation injury and indicate that the mechanisms identified using the SM22α promoter as a model system are likely applicable to multiple CArG dependent SMC marker genes. However, direct proof of this would require generation of G/C Repressor mutant promoter reporter transgenic mice for each of these genes.
Figure 6. H3 acetylation of the SM22α promoter was reduced and was G/C Repressor dependent.
A&B). SM22 and SM22 G/C mut LacZ mice were subjected to right common carotid injury and then harvested 3 days after vascular injury, tissues from ten mice were pooled, and then subjected to ChIP assay for AcH3 (A) or HDAC2 (B). In each ChIP IP, qPCR analysis was conducted on both the Endogenous and LacZ transgene as indicated in the Figure. * indicates significant binding as mentioned in Fig 2D. C&D) Rat aortic smooth muscle cells were plated at 1×10^4, allowed to grow to confluency and then switched to serum free media for three days. Following serum starvation, cells were treated with 20 ng/ml PDGF-BB (C) or 5 ug/ml of POVPC (D) for 12 hours and then subject to ChIP analysis. * indicates significant binding as mentioned in Fig 2D.
Figure 7. Triple Sequential ChIP assays demonstrate that KLF4, pELK-1 and HDAC2 occupy the same piece of chromatin and their binding is attenuated with the G/C Repressor mutation.
A) Rat aortic smooth muscle cells were prepared as mentioned previously. Following serum starvation, cells were treated with 20 ng/ml PDGF-BB for 12 hours and then subject to ChIP analysis. Immunoprecipitations were performed in the sequence mentioned in the y-axis. B) IgG was used as a negative control within the sequence of triple immunoprecipitations. Reciprocal immunoprecipitation were performed but are not pictured. * indicates significant binding as mentioned in Fig 2D. C) Triple sequential ChIP analyses were performed as mentioned in part A with a modification to the sequence of the pull-down as indicated on the y axis. D) IgG was used as a negative control during the sequence of the pull-down as mentioned previously in part B.
Figure 8. KLF4, pELK-1 and HDAC2 bind α–actin, SM-MHC but not c-Fos in vivo following caortid ligation injury.
A) mice were ligated as mentioned in Fig 2. following ligation, right and left carotid were harvested and subject to ChIP analysis for (A) SRF, (B) KLF4, (C) pELK-1, (D) HDAC2, and (E) H4ac binding at the α-actin, SM-MHC, and c-Fos promoters. * denotes significant decreases in binding over vehicle treated controls via students t-test with p-value <0.05.
DISCUSSION
Herein we provide direct evidence that KLF4 mediates the effects of mutating the G/C Repressor element in suppression of SM22α during SMC phenotypic switching in vivo in response to vascular injury [Regan et al.41 and Fig.1] and atherogenesis42. Moreover, we provide evidence for a model wherein pELK-1 binds to the G/C Repressor region of the SM22α promoter and in turn recruits KLF4 and HDAC2 ultimately leading to epigenetic silencing of the gene locus mediated at least in part through histone de-acetylation [Online Fig. IXV]. Although the present studies focused primarily on a single SMC marker gene, SM22α we believe it is highly likely that similar mechanisms contribute to coordinate suppression of multiple SMC marker genes during SMC phenotypic switching both in vivo and in vitro given the following observations. First, we have previously shown that KLF4 over-expression markedly suppresses expression of all SMC marker genes examined to date40. Second, most SMC marker genes, but particularly CArG-SRF dependent genes including not only SM22α but also SM-MHC and SM α-actin, contain conserved G/C Repressor and/or ETS elements 41,42,61 (Online Fig. IVA). However, thus far they have only been shown to be functionally important for SM22α in vivo (Fig. 1 and Regan et al.,41 and Wamhoff et al.,42) and in vitro41,42 and SM-MHC in vitro61. Third, inhibition of ELK-1 phosphorylation with MEK or ERK inhibitors has been shown to inhibit SMC phenotypic switching in cultured SMC by many labs16,30,31 although until the results presented herein there was a lack of clear evidence regarding the contribution of this pathway in vivo. Fourth, we have shown that conditional KLF4 knockout mice show delayed SMC phenotypic switching in vivo following ligation injury32, although it remains to be determined if this is a direct function of loss of KLF4 in SMC versus loss in other cell types including macrophages and endothelial cells where it has been shown to regulate transitions in phenotype46–50. Taken together, results are the first, and to date, the only published studies to our knowledge to identify a direct specific molecular mechanism that mediates SMC phenotypic switching in vivo, although further studies including SMC-specific conditional knockout of KLF4 will be required to conclusively show that cell autonomous KLF4 is required for SMC phenotypic switching in vivo.
An additional unresolved issue is that it was not possible for us to distinguish the relative contributions of the ETS domain versus the G/C Repressor element given the partial overlap of these elements, and the fact that it has been impossible to show binding of the higher order pELK-1-KLF4-HDAC2 complex except within intact chromatin or based on PLA assays as demonstrated herein. However, this may be irrelevant since, of course, mother nature did not dictate that these are indeed functionally distinct, and most importantly our studies provide what we believe is compelling evidence that the G/C Repressor mutation was remarkably specific in its effects on promoter function. For example, the mutation had no discernible effect on expression during development and maturation with identical expression patterns to both the WT SM22α promoter-LacZ transgene and the endogenous gene in our previous studies41 and herein in adult SMC tissues [Fig. 1]. Consistent with these findings, we saw no effect of the G/C Repressor mutation on SRF binding, but it dramatically reduced KLF4, pELK-1, and HDAC2 binding following ligation injury in vivo, or POVPC- or PDGF-BB induced phenotypic switching in cultured vascular SMC from mice, rats, and human. Clearly further studies will be required to determine the structural determinants of this promoter region that mediate recruitment of the pELK-1-KLF4 multi-protein complex.
A key unresolved question is the mechanism of activation of KLF4 expression in SMC given this gene is epigenetically silenced in almost all differentiated somatic cells other than epithelial cells51, 52. The KLF4 promoter contains a number of conserved regulatory elements for AP-1, GATA-1, Sp1, NFκB, and HLH factors53 but as yet no studies have directly assessed if these factors regulate KLF4 expression with the exception of our evidence showing that Sp1 bound the KLF4 promoter via ChIP assays45. Oct4 and other ESC factors may play a role given evidence that these factors reciprocally activate one another in ESCs54. There are a number of unresolved questions regarding mechanisms responsible for activation of KLF4 in SMC. First, what are the mechanisms and factors that activate expression expression of KLF4 in vivo during vascular injury or disease? Remarkably, despite widespread interest in this gene because of its involvement in production of iPS cells, as yet, no studies have identified sufficient regions of the promoter necessary to drive expression of the gene in vivo in transgenic mice, a pre-requisite for studies elucidating mechanisms that induce it with injury-inflammation. Second, what mechanisms are responsible for reversing the stable epigenetic silencing of KLF4 during SMC phenotypic switching? Third, are other ESC factors involved in controlling SMC phenotypic switching? Fourth, do post-translational modifications such as differential mRNA splicing, or chemical modifications such as sumoylation or acetylation regulate effects of KLF4? The latter is of interest since we have shown that TGFβ induced expression of SM α-actin in cultured SMC is mediated in part through inactivation of KLF4 through protein sumoylation35. In summary, further studies are needed to determine if these factors regulate activation of KLF4 in SMC in vivo and if post-translational modifications regulate KLF4 function.
In summary, results of the present studies provide direct evidence that KLF4 mediates phenotypic switching of SMCs in vivo during vascular injury and does so through binding to a G/C Repressor element. Moreover, our results support a model wherein there is sequential binding of pELK-1 and KLF4 followed by binding of HDAC2 to epigenetically silence the gene locus. Although the present studies have focused on studies of the SM22α gene, we feel results have broad significance for overall control of SMC phenotypic switching. However, further studies are needed to directly test if KLF4, pELK-1 and HDAC2 play similar roles in SMC phenotypic switching in disease models such as atherosclerosis.
Supplementary Material
Novelty and Significance.
What Is Known?
Mutating the G/C Repressor element within an SM22α promoter-lacZ transgene virtually abolished down-regulation of the transgene following carotid wire injury or within atherosclerotic lesions of ApoE−/− Western diet fed mice. Thus, the G/C Repressor element is required for suppression of this smooth muscle cell (SMC) marker during phenotypic switching of this cell in vivo.
PDGF-BB and oxidized phospholipid-induced phenotypic switching of cultured SMC is KLF4 dependent. KLF4 over-expression induces profound SMC phenotypic switching of cultured SMC. However, there is no direct evidence that KLF4 regulates the activity of the G/C repressor.
There is no evidence that the functional effects of mutating the G/C repressor in vivo are related to KLF4 or to pELK-1, although we have shown that KLF4 and pELK-1 bind SMC marker gene promoters based on chromatin immunoprecipitation (ChIP) assays following carotid ligation injury in vivo and during phenotypic switching of cultured SMC.
What New Information Does This Article Contribute?
We show that KLF4 binding to the SM22α promoter within intact chromatin is markedly elevated following carotid ligation injury in vivo and is dependent on the G/C Repressor element. This findings provides the first direct evidence that the functional effects of the G/C repressor mutations in abrogating suppression of SM22α during SMC phenotypic switching in vivo are mediated by KLF4.
Binding of pELK-1, and HDAC2 to the SM22α promoter following carotid ligation injury in vivo is also G/C repressor dependent. Results of studies in cultured SMC provide evidence that there is sequential binding of pELK-1, KLF4 and HDAC2, with the latter contributing to histone hypo-acetylation, chromatin remodeling, and transcriptional silencing.
Studies provide novel evidence that phenotypic switching of SMC in vivo is mediated, at least in part, by binding of the stem cell pluripotency factor KLF4 to a G/C repressor cis element contained in the promoter of many SMC marker genes. In addition, studies show that KLF4 binds to SMC promoters in conjunction with pELK-1, and HDAC2, and that the latter mediates histone hypo-acetylation and transcriptional silencing through chromatin remodeling-epigenetic mechanisms. These results are significant in that they are the first to define a specific molecular mechanism that contributes to SMC phenotypic switching in vivo, a process believed to play a critical role in post-angioplasty restenosis, and the pathogenesis of atherosclerosis.
Acknowledgments
We would like to thank Mary E. McCanna and Rupa S. Tripathi for their knowledge and technical expertise.
SOURCES OF FUNDING
This work was supported by NIH grants R01 HL57353, R01 HL098538, and R01 HL087867 (to GKO). Dr. Salmon is supported by an American Heart Postdoctoral Fellowship 11POST7760056. Dr. Gomez is supported by an American Heart Postdoctoral Fellowship 11POST7760009. Ms. Shankman is funded by a pre-doctoral American Heart Fellowship 11PRE17008.
Non-standard Abbreviations
- CArG box
binding element for SRF
- ERK
extra cellular-signal related kinases
- ETS binding site
Binding site for the ETS family of transcription factors
- G/C Repressor element
Conserved cis element within the promoter regions of multiple smooth muscle marker genes
- HDAC2
Histone de-acetylase two
- KLF4
Kruppel-like factor four
- MEK
mitogen-activated protein kinase
- pELK-1
down-stream activator of the mitogen-activated protein kinase pathway
- Oct4(octamer-4)
a homeodomain transcription factor of the POU family
- PLA
proximity ligation assay
- PDGF-BB
platelet derived growth factor BB
- POVPC
oxidized phospholipid
- SM22α
smooth muscle 22 alpha
- SMC
vascular smooth muscle cells
- SM-MHC
smooth muscle myosin heavy chain
- SM α–actin
smooth muscle alpha actin
- Sp1
kruppel-like zinc finger transcription factor
- Sp3
kruppel-like zinc finger activator and/or repressor of transcription
- SRF
serum response factor
- TCE element
conserved cis element within the promoter regions of multiple smooth muscle marker genes, activates gene transcription
Footnotes
DISCLOSURES
None.
References
- 1.Chamley-Campbell J, Campbell G, Ross R. The smooth muscle cell in culture. Physiol Reviews. 1979;59:1–6. doi: 10.1152/physrev.1979.59.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Owens GK. Regulation of Differentiation of Vascular Smooth Muscle Cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 4.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic swiching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 5.Adam PJ, Regan CJ, Hautmann MB, Owens GK. Positive and negative acting kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle differentiation marker SM22alpha in vivo. J Biol Chem. 2000;275:37798–806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 6.Blank RS, McQuinn TC, Yin KC, Thompson MM, Takeyasu K, Schwartz RJ, Owens GK. Elements of the smooth muscle alpha-actin promoter required in cis for transcriptional activation in smooth muscle. Evidence for cell type-specific regulation. J Biol Chem. 199;267:984–9. [PubMed] [Google Scholar]
- 7.Blank RS, Owens GK. Platelet-Derived growth factor regulates actin isoform expression and growth state in cultured rat aortic smooth muscle cells. J Cell Physiol. 1990;142:635–42. doi: 10.1002/jcp.1041420325. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanova OA, Pidkovka NA, Sarmento OF, Yoshida T, Gan Q, Adiguzel E, Bendeck MP, Berliner J, Leitinger N, Owens GK. Oxidized Phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ Res. 2009;104:609–18. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dandre F, Owens GK. Platelet derived growth factor BB and Ets-1 negatively regulate transcription of multiple smooth muscle differentiation marker genes. Am J Physiol Heart Circ Physiol. 2004;286:H2042–51. doi: 10.1152/ajpheart.00625.2003. [DOI] [PubMed] [Google Scholar]
- 10.Corjay MH, Blank RS, Owens GK. Platelet-derived growth factor-induced destabilization of smooth muscle alpha actin mRNA. J Cell Physiol. 1990;145:391–7. doi: 10.1002/jcp.1041450302. [DOI] [PubMed] [Google Scholar]
- 11.Hautmann MB, Adam PJ, Owens GK. Similarities and Differences in smooth muscle alpha-actin induction by transforming growth factor beta in smooth muscle versus non-muscle cells. Arterioscler Thromb Vasc Biol. 1999;19:2049–58. doi: 10.1161/01.atv.19.9.2049. [DOI] [PubMed] [Google Scholar]
- 12.Hautmann MB, Thompson MM, Swartz EA, Olson EN, Owens GK. Angiotensin II induced stimulation of smooth muscle alpha-actin expression by serum response factor and the homeodomain factor MHox. Circ Res. 1997;81:600–10. doi: 10.1161/01.res.81.4.600. [DOI] [PubMed] [Google Scholar]
- 13.Hendrix J, Wamhoff BR, McDonald T, Sinha S, Yoshida T, Owens GK. 5′ CArG degeneracy in smooth muscle alpha-actin is required for injury induced expression in vivo. J Clin Invest. 2005;115:418–27. doi: 10.1172/JCI22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holycross BJ, Blank RS, Thompson MM, Peach MJ, Owens GK. Platelet-Derived Growth FActor-BB-Induced Supppression of Smooth Muscle Cell Differentiation. Circ Res. 1992;71:1525–32. doi: 10.1161/01.res.71.6.1525. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional co-activator of serum response factor. Cell. 2001;105:851–62. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Wang D-Z, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–9. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 17.Chang PS, Li L, McAnally J, Olson EN. Muscle spcificity encoded by serum response factor-binding sites. J Biol Chem. 2001;276:17206–12. doi: 10.1074/jbc.M010983200. [DOI] [PubMed] [Google Scholar]
- 18.Camoretti-Mercado B, Liu HW, Halayko AJ, Forsythe SM, Kyle JW, Li B, Fu Y, McConville J, Kogut P, Vieira JE, Patel NM, Hershenson MB, Fuchs E, Sinha S, Miano JM, Parmacek MS, Burkhardt JK, Solway J. Physiological control of smooth muscle-specific gene expression through regulated nuclear translocation of serum response factor. J Biol Chem. 2000;275:30387–93. doi: 10.1074/jbc.M000840200. [DOI] [PubMed] [Google Scholar]
- 19.Du K, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical SRF co-factor in the transcriptional program regulating smooth muscle differentiation. Mol Cell Biol. 2003;23:2425–37. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–34. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du K, Chen M, Li J, Lepore J, Mericko P, Parmacek MS. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem. 2004;279:17578–86. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program idientifies distinct smooth muscle cell sublineages. Mol Cell Biol. 1997;17:2266–78. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miano JM. Serum response factor; toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–93. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 24.Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain marks exclusively the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–12. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–56. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Miano JM, Mercer B, Olson EN. Expression of the Sm22a promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–59. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Van Putten V, Zarinetchi F, Nicks ME, Thaler S, Heasley LE, Nemenoff RA. Suppression of smooth muscle alpha-actin expression by platelet derived growth factor in vascular smooth muscle cells involves Ras and cytosolic phospholipase A2. Biochem J. 1997;327:709–16. doi: 10.1042/bj3270709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herring BP, Smith AF. Telokin expression is mediated by a smooth muscle cell-specific promoter. Am J Physiol. 1996;270:C1656–65. doi: 10.1152/ajpcell.1996.270.6.C1656. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida T, Owens GK. Molecular Determinants of vascular smooth muscle cell diversity. Circ Res. 2005;96:280–91. doi: 10.1161/01.RES.0000155951.62152.2e. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor BB represses smooth muscle cell marker genes via changes in binding of MKl factors and histone deactylases to their promoters. Am J Physiol Cell Physiol. 2007;292:C886–95. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elf-1 and histone deacetylases cooperatively suppress smooth muscle differentiation markers in reponse to oxidized phospholipids. Am J Physiol Cell Physiol. 2008;295:C1175–82. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida T, Kaestner KH, Owens GK. Conditional Deletion of Kruppel-Like Factor 4 Delays Downregulation of Smooth Muscle Cell Differentiation Markers but Accelerates Neointimal Formation Following Vascular Injury. Circ Res. 2008;102:1548–57. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–41. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 34.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF Binding to CARG-box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawai-Kowase K, Ohshima T, Matsui H, Tanaka T, Shimizu T, Iso T, Arai M, Owens GK, Kurabayashi M. PIAS1 Mediates TGFbeta-Induced SM alpha-Actin Gene Expression Through Inhibition of KLF4 Function-Expression by Protein Sumoylation. Arterioscler Thromb Vasc Biol. 2009;29:99–106. doi: 10.1161/ATVBAHA.108.172700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hautmann MB, Madsen CS, Owens GK. A transforming growth factor beta control element drives TGF-induced stimulation of SM-alpha-actin gene expression in concert with two CArG elements. J Biol Chem. 1997;272:10948–56. doi: 10.1074/jbc.272.16.10948. [DOI] [PubMed] [Google Scholar]
- 37.Kawai-Kowase K, Kumar MS, Hoofnagle MH, Yoshida T, Owens GK. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol Cell Biol. 2005;25:8009–23. doi: 10.1128/MCB.25.18.8009-8023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, Leitinger N, Owens GK. Oxidized Phospholipids induce phenotypic switchin of vascular smooth muscle cells in vitro and in vivo. Circ Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Sinha S, Owens G. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem. 2003;278:48004–11. doi: 10.1074/jbc.M301902200. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Sinha S, McDonald O, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–27. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 41.Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker gene expression after vascular injury. J Clin Invest. 2000;106:1139–47. doi: 10.1172/JCI10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. A G/C element mediates repression of the SM22a promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ Res. 2004;95:981–8. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 43.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 44.Thomas JA, Deaton RA, Hastings NE, Shang Y, Moehle CW, Eriksson U, Topouzis S, Wamhoff BR, Blackman BR, Owens GK. PDGF-DD, A novel mediator of smooth muscle cell phenotypic modulation, is up-regulated in endothelial cells exposed to atherosclerotic-prone flow patterns. Am J Physiol Heart Circ Physiol. 2009;296:H442–52. doi: 10.1152/ajpheart.00165.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am J Physiol Heart Circ Physiol. 2009;296:H1027–37. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, SenBanerjee S, Jain MK. Kruppel-like Factor 4 Is a Mediator of Proinflammatory Signaling in Macrophages. J Biol Chem. 2005;280:38247–58. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 47.Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, Flask CA, Kim J, Doreian BW, Lu KQ, Kaestner KH, Hamik A, Clement K, Jain MK. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–49. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–52. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohnesorge N, Viemann D, Schmidt N, Czymai T, Spiering D, Schmolke M, Ludwig S, Roth J, Goebeler M, Schmidt M. Erk5 Activation Elicits a Vasoprotective Endothelial Phenotype via Induction of Kruppel-like Factor 4 (KLF4) J of Biol Chem. 2010;285:26199–210. doi: 10.1074/jbc.M110.103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–79. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional Profiling of Kruppel-like Factor 4 Reveals a Function in Cell Cycle Regulation and Epithelial Differentiation. J of Mol Biol. 2003;326:665–77. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shields JM, Christy RJ, Yang VW. Identification and Characterization of a Gene Encoding a Gut-enriched Kruppel-like Factor Expressed during Growth Arrest. J of Biol Chem. 1996;271:20009–17. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahatan C, Kaestner KH, Geiman D, Yang VW. Characterization of the structure and regulation of the murine encoding gut-enriched Kruppel-like factor. Nucleic Acid Research. 1999;27:4562–9. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wernig M, Benninger F, Schmandt T, Rade M, Tucker KL, Bussow H, Beck H, Brustle O. Functional Integration of Embryonic Stem Cell-Derived Neurons In Vivo. The Journal of Neuroscience. 2004;24:5258–68. doi: 10.1523/JNEUROSCI.0428-04.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madsen CS, Regan CP, Hungerford JE, White SL, Manabe I, Owens GK. Smooth muscle-specific expression of the smooth muscle myosin heavy chain gene in transgenic mice requires 5′-flanking and first intronic DNA sequence. Circ Res. 1998;82:908–17. doi: 10.1161/01.res.82.8.908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.