Abstract
Droplet digital PCR (ddPCR) is an emerging nucleic acid detection method that provides absolute quantitations of target sequences without relying on the use of standard curves. The ability of ddPCR to detect and quantitate total HIV-1 DNA and 2-LTR circles from a panel of patients on and off antiviral therapy was evaluated compared to established real-time (RT)-PCR methods. To calculate the dynamic range of ddPCR for HIV-1 DNA and 2-LTR circles, serial dilutions of DNA amplicons or episomes were determined by ddPCR as well as with RT-PCR. HIV-1 DNA from 3 viremic patients and 4 patients on suppressive antiretroviral therapy, and 2-LTR circles from 3 patients with low-level viremia was also quantitated. Copy numbers determined by ddPCR of serial dilutions of HIV-1 or human CCR5 DNA amplicon standards were comparable to nominal input copy number. The sensitivity of ddPCR to detect HIV-1 or CCR5 DNA was similar to that of RT-PCR. Low levels of 2-LTR circles were detected in samples from all 3 patients by both ddPCR and RT-PCR. ddPCR is a promising novel technology for the study of HIV-1 reservoirs and persistence, but further optimization of this novel technology would enhance the detection of very low-level viral genetic targets.
Keywords: droplet digital PCR, ddPCR, real-time PCR, HIV-1, HIV DNA, viral quantitation, 2-LTR circles
1. Introduction
Droplet digital PCR (ddPCR) is an emerging nucleic acid detection method that provides absolute quantitation of target sequences without relying on the use of standard curves. To perform ddCPR, the DNA target, fluorescently-labeled probe and the ingredients for a PCR reaction are partitioned into an emulsion of approximately 20,000 droplets, each of which ideally contains 1 or less copies of the target DNA. Using 96-well plates, 2 million PCR reactions can be performed simultaneously (Hindson et al., 2011). Following PCR amplification, enumeration of both fluorescing and non-fluorescing droplets is performed, allowing the absolute quantitation of target molecules present in the original sample. To date, ddPCR has been validated primarily for measurement of germline copy number variation, detection and quantitation of rare alleles, and absolute quantitation of circulating fetal and maternal DNA (Hindson et al., 2011; Pinheiro et al., 2012).
The large-scale partitioning involved with ddPCR leads to greater precision and sensitivity for these applications when compared with real-time PCR (RT-PCR) (Hindson et al., 2011; Pinheiro et al., 2012). Partitioning reactions in picoliter wells may allow ddPCR to handle relatively large amounts of input DNA with little interference from PCR inhibitors. As result, ddPCR is a promising platform for investigation of very low-levels of viral genetic material, and there has been considerable interest in this technology for the sudy of HIV-1 reservoirs. Data are emerging that support ddPCR for the quantitation of low-levels of plasma HIV-1 RNA (Anderson et al., 2012), but the performance and applicability to the study of cell-associated HIV-1 DNA have yet to be fully described. Natural history studies of HIV-1 infection in the setting of antiretroviral therapy show that HIV-1 can persist indefinitely (Siliciano et al., 2003) because the viral genome integrates into host cell chromosomes. HIV-1 cDNA can also enter the cell nucleus, but fail to integrate, resulting in circularized molecules containing one or two copies of the long terminal repeat (LTR) region of HIV(Brussel, Delelis, and Sonigo, 2005; Butler, Johnson, and Bushman, 2002; Morlese et al., 2003). The development of novel methods to detect and quantitate low-levels of HIV-1 DNA is important to advance the study of viral persistence and eradication strategies. We evaluated the dynamic range of ddPCR and tested the ability of ddPCR to detect and quantitate HIV-1 DNA and 2-LTR circles from a panel of patients on and off antiviral therapy compared with traditional RT-PCR methods.
2. Methods
2.1. Patient samples, DNA extraction and preparation
Approval for the use of human material by the Brigham and Women's Hospital/Partners Healthcare Institutional Review Board was obtained prior to study initiation. In order to compare the performance of ddPCR and traditional RT-PCR for HIV-1 DNA, 5 million peripheral blood mononuclear cells (PBMCs) were obtained from 3 HIV-1-infected viremic patients and 4 patients on suppressive antiretroviral therapy. The PBMCs were washed in Hanks Balanced Salt Solution (HBSS) and pelleted. Following resuspension in 200 μL of phosphate-buffered saline, DNA was extracted using the QiaAMP Blood DNA Mini kit (Qiagen, Valencia, CA) as per the manufacturer's protocol. The DNA was eluted in 200 μL of elution buffer AE (10 mM Tris-Cl, 0.5 mM EDTA; pH 9.0), aliquoted and frozen at −80°C. Prior to PCR quantitation, the extracted genomic DNA was digested with MscI (New England Biolabs, Ipswitch, MA) because DNA strands longer than several hundred base-pairs do not package efficiently into the picoliter droplets. The MscI enzyme cuts at the 5′-TGGCCA-3″ sequence, which is absent from the proximal portion of HIV-1 gag and is thus unlikely to interfere with the PCR amplification of the HIV-1 DNA regions targeted in this study. A 50- μ l sample of the eluted DNA was digested in a total reaction volume of 60 μl at 37°C for 1 hour, followed by heat inactivation at 65°C for 20 minutes. Digested genomic DNA was then purified using the StrataClean resin (Agilent Technologies, Santa Clara, CA) as per the manufacturer's protocol.
To amplify 2-LTR circles, episomal DNA was extracted from 5 million PBMCs using the QIAprep Spin Miniprep Kit (Qiagen) following the instructions for the isolation of low-copy plasmids and eluted in 50 μL of nuclease-free water.
2.2. Real-time PCR quantitation of HIV-1 DNA and episomal 2-LTR circles
Following extraction, restriction enzyme digestion and resin purification, HIV-1 DNA was quantified using a validated Taqman real-time PCR method (Malnati et al., 2008). This assay allows for the precise quantitation of HIV-1 DNA down to 1 copy of an HIV-1 standard that comprises a conserved region in the LTR/gag common to nearly all group-M HIV-1 sequences (Malnati et al., 2008). Briefly, 10 μL of genomic DNA, 12.5 μL of Universal Taqman mastermix (ABI), 0.75 μL of 10 μM forward primer (5′-TACTGACGCTCTCGCACC), 0.75 μL of 10 μM reverse priemer (5″-TCTCGACGCAGGACTCG), and 1.0 μL of 5 μM FAM-MGB labeled probe (5′-FAM-CTCTCTCCTTCTAGCCTC) were added to each reaction well. HIV-1 standards were constructed by amplifying a cDNA region with primers that flank the region specified above from the HIV-1 reference strain HXB2 using the forward primer 5′-GGCTCACTATGCTGCCGCCC and the reverse primer 5′- TGACAAGCAGCGGCAGGACC. The exact number of DNA copies was quantified using a Nanodrop 2000c photospectrometer. Serial dilutions of the DNA standard ranging from 3 to 600,000 copies were run in addition to samples, no-template controls and genomic DNA from uninfected PBMCs. Amplifications were performed using the following PCR conditions: 95°C for 15 min; 42 cycles of 95°C for 15 s, 60°C for 1 min. HIV-1 DNA copy numbers were determined by comparing Ct values with those from the standard curves.
2-LTR circles were quantified by RT-PCR using a validated method (Butler, Johnson, and Bushman, 2002). Briefly, 10 μL of episomal PBMCs DNA were added to each PCR reaction in a master mix containing 25 μL of Taqman universal master mix, 11.5 μL of nuclease-free water, and 1.5 μL and 0.5 μL of 10 μM forward/reverse primer and FAM-MGB labeled probes, respectively(Butler, Johnson, and Bushman, 2002). In addition, a 149-bp sequence of the highly conserved human mitochondrial gene ND4 (Mishmar et al., 2003) was quantified from each sample using house primers MitoND4F 5′- ACCACTGACATGACTTTCCA and MitoND4R 5′-GTTAGGGGGTCGGAGGAA, and FAM-MGB labeled probe 5′- CAACCACCCACAGCCTAATT. DNA standards and negative controls were run in parallel. The co-plasmid standard was constructed by ligating the 185-bp sequence HIV-1 LTR R-U5-U3 and the ND4 sequence as internal control using T4 DNA Ligase (Life Technologies, Grand Island, NY) followed by insertion into the pCR4-Topo TA plasmid (Life Technologies) following manufacturer protocol. The vector was amplified by transfection into Oneshot Top-10 electrocompetent cells (Life Technologies) followed by episomal DNA recovery by the Qiaprep Miniprep protocol (Qiagen).
2.3. ddPCR quantitation of HIV-1 DNA and 2-LTR circles
Five to 7.5 μL of MscI-digested, resin-purified genomic DNA was added to 10 μL of ddPCR 2× PCR master mix (Quantalife, Applied Biosystems), 0.75 μL each of 10 μM forward and reverse primers and 1.0 μL of 5 μM probe; the same primer and probes were used for real-time and ddPCR. Twnety microliters of the ddPCR master mix with genomic DNA was added to the droplet generator strips in addition to 60 μL of generator oil and placed in the automated droplet generator. The resulting picoliter droplet emulsion was transferred to a 96-well PCR plate taking care not to disrupt the droplets and the plates were sealed. PCR was then performed using the following program: 95°C for 10 min; 42 cycles of 94°C for 30 s, 60°C for 1 min; and 98°C for 15 min; the final high-temperature cycle cures the droplets. The plates were then placed in the ddPCR reader for enumeration of the number of positive and negative droplets based on fluorescence. The PCR probes have sequence specificity with the target DNA and fluoresce upon target amplification. The number of template molecules per microliter of starting material was then estimated by the Quantalife ddPCR using an internal Poisson algorithm. Dilutions of the HIV-1 DNA standard were run in parallel with the genomic samples using the same protocol.
The 2-LTR circles and plasmid standards incorporating the 2-LTR and mitochondrial DNA sequences were quantified using similar master mix concentrations and reaction conditions and procedure above for genomic DNA with the exception of 40 PCR cycles prior to droplet enumeration. The probe and primer sequences were identical to those used in real-time PCR.
2.4 Statistical methods
GraphPad was used to generate standard curves from real-time PCR and ddPCR results and for comparison of HIV-1 DNA to sample standards and for cross-comparisons of ddPCR and real-time PCR standards. SPSS vs. 20 was used to calculate Pearson correlation coefficients and standard error measurements for comparison of patient sample quantitation.
3. Results
3.1. Real-time PCR and ddPCR quantitation of HIV-1 DNA
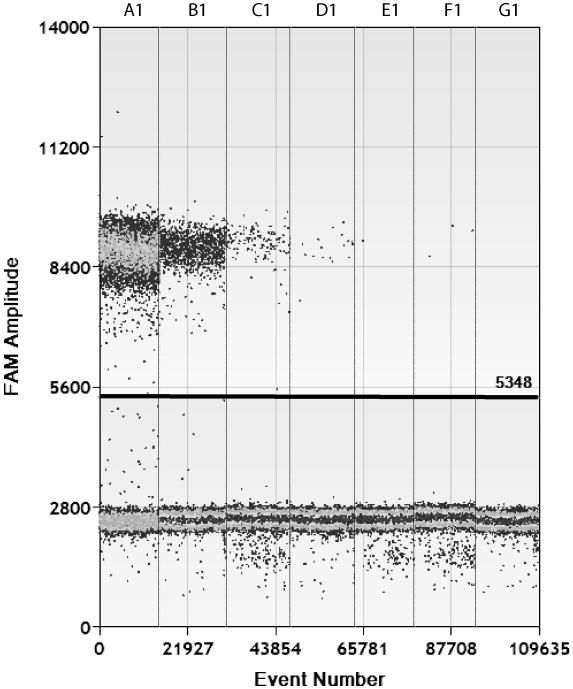
Copy numbers determined by ddPCR of serial dilutions of HIV-1 or CCR5 DNA standards are shown in Table 1. Linear regression correlation coefficients (R2) for the log-transformed copy number between ddPCR results and expected copy numbers were all >0.98 and slopes were all > 0.92. Expected copy numbers measured by ddPCR were comparable to input standard values with the exception of the HIV-1 DNA amplicon standard; ddPCR detected approximately 60% fewer HIV-1 DNA copies than expected by the input standard copy number. However, the sensitivity of ddPCR to detect HIV-1 or CCR5 DNA was similar to that of RT-PCR; ddPCR and RT-PCR both obtained positive signals in 2 of 3 wells for the 3 copy standard. As expected, linearity of ddPCR decreased significantly when greater than 60,000 copies were added into each reaction well (data not shown). Of note, the ddPCR droplet generator led to inadequate droplet formation in approximately 1 out of every 16 wells, leading to loss of sample as noted in Table 1. An example of the visual readout of positive and negative individual ddPCR droplet reactions and the user-determined positive-negative threshold for HIV-1 DNA standard dilutions are shown in Figure 1.
Table 1. ddPCR Detection and Quantitation of Serial Dilutions of HIV-1 DNA, Human CCR5, HIV-1 2-LTR and Human Mitochondria DNA Standards.
| Input Standard Copy Number per Reaction Wella | ddPCR (copies/reaction) | Standard Error | Wells Detected/Tested | % Level of Quantitation of Input Standard by ddCPR |
|---|---|---|---|---|
| HIV-1 DNAb | ||||
| 60,000 | 37,706.7 | 1366.3 | 3/3 | 62.8 |
| 6,000 | 3,613.3 | 40.7 | 3/3 | 60.2 |
| 600 | 391.9 | 28.6 | 3/3 | 65.3 |
| 60 | 50.4 | 3.2 | 2/2c | 84.0 |
| 6 | 3.3 | - | 1/2c | 55.0 |
| 3 | 6.1 | 2.6 | 2/3 | 203.3 |
| Human CCR5b | ||||
| 60,000 | 68,200.0 | 200.0 | 2/2 | 113.7 |
| 6,000 | 6,740.0 | 180 | 2/2 | 112.3 |
| 600 | 742.0 | 54 | 2/2 | 123.7 |
| 60 | 70.6 | 0.6 | 2/2 | 117.7 |
| HIV-1 2-LTRb | ||||
| 75,000 | 71,100.0 | 1300.0 | 2/2 | 94.8 |
| 7,500 | 7,500.0 | 180.0 | 2/2 | 100.0 |
| 750 | 768.0 | 40.0 | 2/2 | 102.4 |
| 75 | 106.2 | 3.0 | 2/2 | 141.6 |
| 7.5 | 9.5 | 1.5 | 2/2 | 126.7 |
| 0.75 | 1.4 | - | 1/2 | 186.7 |
| ND4 Mitochondriab | ||||
| 75,000 | 64100.0 | 900.0 | 2/2 | 85.5 |
| 7,500 | 5760.0 | - | 1/1 b | 76.8 |
| 750 | 752.0 | 36.0 | 2/2 | 100.3 |
| 75 | 85.2 | 7.2 | 2/2 | 113.6 |
| 7.5 | 8.7 | 6.1 | 2/2 | 116.0 |
Abbreviations: LTR, long terminal repeat
Copy number based on serial dilutions of either linear or episomal DNA standard by DNA concentration and known molecular mass; input copy number = expected copy number for ddPCR quantitation.
Linear regression coefficients (R2) for the HIV-1 DNA, CCR5, 2-LTR and ND4 mitochondria standard curves were all >0.98 by real-time PCR [quantitation threshold (Ct) versus input copy number]
ddPCR droplet generator failed to generate sufficient droplets for quantitation leading to fewer test wells
Figure 1.
Visual representation of ddPCR results for HIV-1 DNA standards after flow enumeration. Positive droplets (upper cluster) and negative droplets (lower cluster) are shown for 6 serially diluted linear DNA standards (A1 through F1) and negative control (G1). The Y axis represents fluorescent intensity and the X axis is the total number of events for each standard dilution. The manually set threshold for droplet positivity is represented by the horizontal line.
Levels of HIV-1 DNA and the human CCR5 gene measured by RT-PCR and ddPCR for samples from 7 HIV-1 infected individuals (3 off ART with detectable viremia and 4 on suppressive ART) are shown in Table 2. HIV-1 DNA from MscI-digested samples from patients on and off ART ranged from 27 to 4894 copies per million PBMCs by real-time PCR whereas DNA copies by ddPCR ranged from 23 to 2289 per million PBMCs. Pearson correlation coefficients of sample HIV-1 and CCR5 DNA copy number by real-time and ddPCR were 0.98 and 0.85 respectively (all P<0.001). Overall, ddPCR enumerated approximately 10-60% fewer DNA copies compared with RT-PCR, but demonstrated similar detection sensitivity. On average, higher copy numbers of HIV-1 DNA were measured from subjects off ART by both quantitation methods.
Table 2. Cell-Associated HIV-1 and Human CCR5 DNA from Clinical Samples Detected by ddPCR and RT-PCR.
| PBMC HIV-1 DNA (copies/10 μL ± SE) | CCR5 (copies/10 μL) | HIV-1 DNA per 106 PBMCsa | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Sample | Plasma Viral Load | RT-PCR | WD/Tb | ddPCR | WD/Tb | RT-PCR | ddPCR | RT-PCR | ddPCR |
| Off ART | |||||||||
| A | 13683 | 31 ± 1.0 | 3/3 | 15 ± 10.0 | 3/3 | 78116 | 91800 | 794 | 327 |
| B | 119612 | 269 ± 13.8 | 3/3 | 112 ± 34.8 | 3/3 | 115699 | 136800 | 4650 | 1637 |
| C | 7402 | 246 ± 18.7 | 3/3 | 133 ± 15.4 | 3/3 | 100536 | 116200 | 4894 | 2289 |
| On ART | |||||||||
| D | <50 | 32 ± 5.0 | 3/3 | 22 ± 3.9 | 3/3 | 99917 | 99000 | 640 | 444 |
| E | <50 | 26 ± 1.0 | 3/3 | 20 ± 5.7 | 3/3 | 69650 | 77600 | 747 | 515 |
| F | <50 | 1 ± 0.4 | 2/3 | 1 ± 0.9 | 1/3 | 74842 | 86800 | 27 | 23 |
| G | 96 | 19 ± 1.8 | 3/3 | 17 ± 0.2 | 3/3 | 71479 | 84000 | 532 | 404 |
Abbreviations: RT, real-time; SE, standard error; PBMC, peripheral blood mononuclear cell
Total HIV-1 DNA per PBMC number is a function of HIV-1 DNA copies and human genome equivalents (CCR5/2) detected in a set reaction volume (e.g. sample A: 31 HIV-1 DNA copies was quantitated in a reaction volume containing DNA from 39058 PBMCs = 794 copies/106 PBMCs).
WD/T = number of wells detected/tested
3.2. Comparison of RT-PCR and ddPCR quantitation of HIV-1 2-LTR circles
Table 1 shows the correlations between copy numbers determined by ddPCR and the nominal copy number expected from 10-fold serial dilutions of 2-LTR circles and human ND4 mitochondrial co-plasmids. Human mitochondrial DNA levels were used to estimate the efficiency of DNA extraction and processing. Linear regression correlation coefficients (R2) for log-transformed copy number were all >0.98 and slopes ranged from 0.95 to 0.99. The 2-LTR circles from the 0.75 copy co-plasmid standard was detected in 50% of wells by both ddPCR and RT- PCR.
2-LTR circles were quantified in PBMCs samples from 3 HIV-1 infected individuals, 2 from viremic controllers (viral load from 547 to 780 copies/mL) and one from a patient with persistent low-level viremia on ART (viral load = 203 copies/mL). Low levels of 2-LTR circles were detected in samples from all 3 patients by both ddPCR and RT-PCR (Table 3). However, there was considerable intra-sample variation in copy number by ddCPR for one sample. Given the relatively low volumes of eluted DNA, only two ddPCR wells could be tested per subject. Insufficient sample existed to perform the LTR assay on genomic DNA from patients included in analysis of total HIV-1 DNA.
Table 3. HIV-1 2-LTR Circle and Human Mitochondria Copy Number Detected by ddPCR and RT-PCR for Patients with Low-Level Viremia.
| HIV-1 2-LTR Circles (copies/106 PBMCs ± SE) | ND4 Mitochondrial DNA (copies/106 PBMCs) a | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Sample | Plasma Viral Load | RT-PCR | WD/Tb | ddPCR | WD/Ta | RT-PCR | ddPCR |
| H (PLLV) | 203 | 3.3 ± 0.8 | 2/3 | 3.3 ± 0.2 | 2/2 | 2.9×107 | 2.3×107 |
| I (VC) | 547 | 2.3 ± 2.2 | 2/3 | 63.7 ± 60 | 2/2 | 3.1×107 | 5.4×107 |
| J (VC) | 780 | 1.0 | 1/3 | 9.4 | 1/2 | 5.3×107 | 3.0×107 |
Abbreviations: PLLV, persistent low-level viremia on ART; VC, viremic controller; RT, real-time; SE, standard error
Human mitochondria copy numbers differ between cell and patient and is therefore used to check efficiency of DNA extraction
WD/T = number of wells detected/tested
4. Discussion
Digital droplet PCR is a promising novel technology for the detection and quantitation of viral genetic material. In this study, we demonstrate that the platform is generally comparable to traditional RT-PCR methods for both total HIV-DNA and episomal 2-LTR circles. Despite good correlations of both linear and episomal DNA standards between the two methods, differences in the number of genomic HIV-1 DNA copies and episomal 2-LTR circles were observed. Whereas ddPCR is a direct measure of target DNA concentration, real-time PCR relies on a standard curve to extrapolate the amount of target present. The PCR quantitation threshold cycle (Ct) of an unknown sample is compared to the Ct of known input/expected copy number of a dilutional standard. The copy number of the standard is calculated prior to PCR quantitation by measuring concentrations of a known linear or episomal DNA sequence with a known molecular mass. One potential cause of the discrepancy between the assays could be errors introduced by spectrophotometric determination of the DNA concentration, or subsequent serial dilution of the standards. It is also possible that PCR was less efficient under one set of PCR conditions, or that some of the genomic DNA remained undigested prior to droplet generation. Larger fragments may not have packaged efficiently into picoliter droplets, leading to an underestimation of HIV-1 DNA copies by ddPCR. By contrast, real-time PCR quantitation most likely would not be affected by size differences in genomic DNA strands as all genetic material is included in each reaction without packaging or partitioning.
Discrepancies between the two PCR approaches in measuring the number of 2-LTR circles may have been related to the fact that the input copy numbers were low overall. Despite relatively good linearity at low standard copy numbers for both real-time and ddPCR, a relatively small number of positive ddPCR droplets could dramatically alter the results in low copy-number samples. For example, 63 LTR circles/million PBMCs were quantified by ddPCR for one patient, but only 2.3 copies/million PBMCs by real-time PCR. There was, however, a high degree of variation between the observed copy numbers of the duplicate samples evaluated by ddPCR for this patient (3.7 and 123.6 copies). Furthermore, the maximum volume of input sample in ddPCR is 7.5 μL, whereas a larger sample volume can be included in real-time PCR. As a result, higher concentrations of DNA substrate may be required in ddPCR reaction wells in order to maximize sensitivity for detecting HIV-1 DNA at very low levels.
One inherent limitation of ddPCR as compared to RT-PCR is the need to dilute samples with more than 75,000 copies of the target DNA, as overloading the picoliter droplets results in a significant loss of linearity at higher copy numbers. While this problem has less impact on quantitation at the low copy numbers that would be expected for HIV-1 DNA (Richman et al., 2009; Siliciano et al., 2003), human chromosomal and mitochondrial DNA require significant dilution prior to assay.
Whereas ddPCR is a promising platform for the detection of total DNA, episomal DNA and viral RNA, the technology may not be compatible with certain methods used to detect integrated HIV-1 DNA without involving standard curves. For example, Alu-PCR has become a standard technique to quantitate the integrated, proviral HIV-1 DNA fraction (Brussel, Delelis, and Sonigo, 2005; Liszewski, Yu, and O'Doherty, 2009). Alu-PCR relies on two PCR steps: the first amplifies the sequences between the nearest chromosomal Alu element and integrated HIV-1 DNA. However, because the target sequence is randomly inserted between Alu elements, different length strands from first-round PCR are created. The second step involves amplification and real-time PCR quantitation of only the HIV-1 sequence. However, this method requires a PCR standard generated by infection of a cell line containing randomly integrated HIV-1 DNA as input into the first-round PCR (Brussel, Delelis, and Sonigo, 2005; Liszewski, Yu, and O'Doherty, 2009). Although ddPCR could be used to quantitate HIV-1 DNA from the second PCR step, the need persists for an input standard.
Despite these limitations, ddPCR is a promising tool for the study of HIV-1 reservoirs and persistence. ddPCR and real-time PCR have similar dynamic ranges for linear and episomal DNA standards and ddPCR is generally comparable to real-time PCR for the detection and quantitation of HIV-1-DNA and 2-LTR circles. However, PCR inhibition during quantitation of large amounts of DNA may be reduced by the partitioning reactions into droplets, allowing for investigation of very low levels of HIV-1 without the use of repetitive sampling required by standard quantitative PCR methods. Employing statistical methods to define positive-negative cutoffs rather than relying on manual methods may also enhance ddPCR, and further optimization of this novel technology would improve the detection of very low-level viral genetic targets.
Highlights.
ddPCR provides quantitation of sequences without the use of standard curves.
The ability of ddPCR to detect HIV-1 DNA from patient samples was evaluated.
The ability of ddPCR to detecting and quantitate HIV-1 DNA was similar to RT-PCR.
ddPCR is a promising novel technology for the study of HIV-1 reservoirs.
Further optimization would enhance the detection of low-level viral genetic targets.
Acknowledgments
We would like to thank Bio-Rad Laboratories, Inc. for providing trial reagents and equipment. Funding: NIH/NIAID K23AI098480 to T.J.H.
Abbreviations
- ddPCR
droplet digital PCR
- RT-PCR
real-time PCR
- PBMCs
peripheral blood mononuclear cells
- LTR
long terminal repeat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson EM, Wiegand A, Boltz VF, Spindler J, Dueppen P, Troppman R, Maldarelli F, Mellors JW, Kearney MF, Coffin JM. Single-Copy Detection of Plasma HIV-1 RNA Using Droplet Digital PCR Technology (Abstract V-1002). 19th Conference on Retroviruses and Opportunistic Infections; Seattle, WA. 2012. [Google Scholar]
- Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–54. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- Butler SL, Johnson EP, Bushman FD. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J Virol. 2002;76:3739–47. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski MK, Yu JQJ, O'Doherty U. Detecting HIV-1 integration by repetitive-sampling (Alu)under-bar-gag PCR. Methods. 2009;47(4):254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc. 2008;3:1240–8. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci U S A. 2003;100:171–6. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlese J, Teo IA, Choi JW, Gazzard B, Shaunak S. Identification of two mutually exclusive groups after long-term monitoring of HIV DNA 2-LTR circle copy number in patients on HAART. Aids. 2003;17:679–83. doi: 10.1097/00002030-200303280-00005. [DOI] [PubMed] [Google Scholar]
- Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003–11. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–7. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4(+) T cells. Nature Medicine. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]