Abstract
The BCR–ABL fusion kinase is the driving mutation of chronic myelogenous leukemias and is also expressed in a subset of acute lymphoblastic leukemias. Recent advances in elucidating the structure, regulation, and signaling of BCR–ABL have led to the identification of allosteric sites that are distant from the ATP-binding pocket and are critical for BCR–ABL–dependent oncogenic transformation. Here, we review the available data regarding the molecular mechanism of action and the specificity of ATP-competitive tyrosine kinase inhibitors targeting BCR–ABL. In addition, we discuss how targeting of allosteric sites could provide new opportunities to inhibit resistant BCR–ABL mutants, either alone or in combination with conventional ATP-competitive inhibitors.
Introduction
Translocation of the Philadelphia (Ph) chromosome leads to expression of the chimeric BCR–ABL protein, which is a constitutively active tyrosine kinase and a strong driver of different leukemias in humans. Depending on the chromosomal breakpoint, 2 major isoforms of BCR–ABL, p210 and p185, can be expressed. The p210 isoform is the molecular hallmark of chronic myelogenous leukemias (CML), and either p185 or p210 is expressed in a subset of B-cell acute lymphoblastic leukemias (1). The Abl tyrosine kinase inhibitor (TKI) imatinib (Gleevec or Glivec; Novartis) binds to the ATP-binding cleft of the kinase domain and inhibits the kinase activity of BCR–ABL. Administration of imatinib leads to durable remissions in the majority of CML patients when they are treated in the chronic phase and improves the outcome in Ph+ patients with acute lymphoblastic leukemia (ALL). However, the occurrence of point mutations in the BCR–ABL kinase domain that reduce the imatinib sensitivity of BCR–ABL is a leading cause of patient relapse, bearing the risk of disease progression (2). Over the past few years, investigators have developed second- and third-generation TKIs that are active against imatinib-resistant BCR–ABL mutants. Although some of these inhibitors have already received regulatory approval, many more are currently undergoing clinical trials. Still, short-lived responses in patients with advanced-phase CML and Ph+ ALL, general TKI resistance caused by the T315I mutation, compound mutations (2 or more mutations in the same clone), and foreseeable problems with the long-term tolerability of all BCR–ABL inhibitors remain challenging clinical problems (3). Here we present an overview of the mechanisms of action, specificity, and clinical efficacy of BCR–ABL TKIs that bind to the ATP-binding cleft. We also discuss alternative strategies to inhibit BCR–ABL by targeting allosteric regulatory modules of the oncoprotein, which could be used to limit the problems associated with current TKI treatment.
Different Classes of BCR–ABL Inhibitors
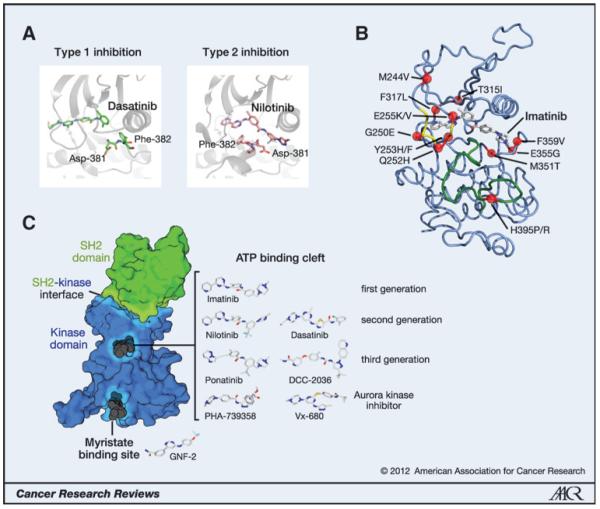
On the basis of their molecular mechanism of action, one can distinguish 2 major classes of TKIs, both of which overlap with the ATP-binding site (Fig. 1A; ref. 4). Type 1 inhibitors target the active conformation of the kinase domain, which is catalytically competent because all elements in the kinase domain are properly arranged for catalysis and able to bind ATP and substrate (Fig. 1A, left). In contrast, type 2 inhibitors target the inactive conformation of the kinase domain. One of the signifying features of type 2 inhibitor binding is the DFG-out conformation, in which the Asp residue of the DFG (Asp-Phe-Gly) sequence motif at the beginning of the activation loop, which is important for the proper positioning of ATP, is rotated out of the active site (Fig. 1A, right). In general, it has been assumed that type 1 inhibitors are less specific than type 2 inhibitors because the active conformation is very similar in most kinases. Recently, however, a systematic survey of the specificity of 72 type 1 and type 2 inhibitors, many of which are in clinical use, showed that this general assumption may be an oversimplification because several highly specific type 1 inhibitors and rather promiscuous type 2 inhibitors exist as well (5). In contrast, allosteric inhibitors do not compete with ATP binding and bind to sites on the kinase domain or other domains in the kinase that are important regulators of kinase activity.
Figure 1.
Mutations, inhibitors, and targeting sites on BCR–ABL. A, type 1 versus type 2 kinase-inhibitor complexes. Residues Asp-381 and Phe-382 of the DFG motif are shown in stick representation. Note the dramatically different position of the Asp and Phe side chains, rotated by 180° in the type 1 and 2 inhibitor complexes. B, structure of the Abl kinase domain bound to imatinib [Protein Data Base (PDB) entry 1OPJ]. The activation loop is shown in green, the Gly-rich loop is shown in yellow, and positions of resistance mutation are shown as red balls. C, surface representation of the SH2-kinase domain unit of BCR–ABL. The kinase domain is shown in blue and the SH2 domain bound to the N-lobe of the kinase domain is shown in green (PDB entry 1OPL chain B). The primary drug-binding site in the ATP-binding cleft, the allosteric myristate-binding pocket, and the SH2-kinase interface are highlighted. Prototypic binders of the ATP and myristate pocket are indicated in dark gray. The conformation of the TKIs from the superimposed structures of the respective Abl kinase domain-drug complexes is indicated as a stick model. The orientation of the drugs is uniformly rotated 90° clockwise around the y axis with respect to the orientation of the SH2-kinase domain structure on the left and translated on the xy plane for graphical convenience. The following PDB entries of the Abl kinase domain were used: imatinib (1OPJ), nilotinib (3CS9), dasatinib (2GQG), ponatinib (3IK3), DCC-2036 (3QRJ), PHA-739358 (2V7A), VX-680 (2F4J), and GNF-2 (3K5V). Structural data for bosutinib or SGX393 are not publically available.
Imatinib: The “Magic Bullet” in the Treatment of CML
Imatinib has revolutionized targeted cancer therapy, being the first TKI to be approved for the treatment of human disease (Fig. 1C; ref. 6). As a type 2 inhibitor, it inhibits ABL kinase activity with moderate affinity in the 100-nmol/L range. Imatinib is remarkably specific and inhibits only a few enzymes in addition to the ABL kinases, including the receptor tyrosine kinases KIT, platelet-derived growth factor receptor A/B (PDGFRA/B), Discoidin domain receptor 1/2 (DDR1/2), and the oxidoreductase NQO2 (7). BCR–ABL mutations associated with imatinib resistance cluster in distinct parts of the kinase domain (Fig. 1B): G250E, Q252H, Y253H/F, and E255K/V in the Gly-rich loop (now commonly but incorrectly termed the P-loop); T315I (the gatekeeper residue); H395P/R in the activation loop; and M244V, F317L, M351T, E355G, and F359V in other regions of the kinase domain (8, 9). For most of these mutants, the degree of TKI resistance is relatively mild, increasing the IC50 value for imatinib by only ≤10-fold. However, due to plasma/serum concentrations of imatinib of ~2 μmol/L in treated patients, this mild resistance is sufficient to restore BCR–ABL kinase activity in CML cells carrying a kinase domain mutation (10).
Nilotinib and Dasatinib: Second-Generation BCR–ABL TKIs
To meet the demand for inhibition of imatinib-resistant BCR–ABL mutants, researchers developed the second-generation TKIs nilotinib (Tasigna; Novartis) and dasatinib [Sprycel Bristol-Myers Squibb (Fig. 1C)]. Dasatinib inhibits BCR–ABL at 10- to 20-fold lower concentrations than nilotinib, which is yet 10- to 20-fold stronger than imatinib (11, 12). All common imatinib resistance mutations, with the exception of the T315I gatekeeper mutant, are efficiently inhibited by dasatinib and nilotinib. In vitro screens for drug resistance clearly showed that the spectrum of emerging resistance mutations is much smaller with second-generation TKIs as compared with imatinib (13, 14). Certain differences in the relative sensitivities for particular mutations in vitro and in cell lines would favor the use of one second-generation TKI over another. For example, mutations in the Gly-rich loop appear to be inhibited more efficiently by dasatinib than nilotinib, whereas the F317L mutation is more sensitive to nilotinib (10). However, because these differences were determined in vitro, the clinical significance of this finding is debatable (15). Currently, there is no strong support from clinical data that would suggest the use of a particular drug depending on a particular BCR–ABL mutation.
In contrast with the similarities observed in targeting imatinib resistance mutations, the mechanism of inhibition and the specificity of nilotinib versus dasatinib differ dramatically. Nilotinib, as a close derivative of imatinib, is a type 2 inhibitor, whereas dasatinib is a type 1 inhibitor (Fig. 1A; ref. 16). Nilotinib exploits an auxiliary binding pocket in the Abl kinase domain, allowing an increase in lipophilic interactions. Together with a decrease in the number of hydrogen bonds with the target, this allows a greater flexibility of the binding interface, explaining the activity of nilotinib for a number of imatinib resistance mutations (11). Despite having a target spectrum very similar to that of imatinib, nilotinib shows higher specificity toward BCR–ABL inhibition as compared with inhibition of Kit and PDGFR (7, 11). In contrast, dasatinib inhibits several other classes of tyrosine kinases, such as the Src, Tec, Csk, and Eph families, as well as several Ser/Thr kinases besides ABL1/2, KIT, PDGFRA/B, and DDR1/2 (7, 17). Many of these additional kinase targets of dasatinib are inhibited with efficiencies comparable to that observed for BCR–ABL, suggesting a broad-range inhibition of tyrosine kinase signaling in dasatinib-treated cells. Therefore, the common designation of dasatinib as a dual Src-Abl inhibitor is misleading and should be avoided. Another major difference is that dasatinib has a much shorter plasma half-life than nilotinib. Initially, to ensure continuous BCR–ABL inhibition in patients with CML, dasatinib was administered twice a day. Surprisingly, a clinical comparison of dosing regimens (once vs. twice daily) showed identical response rates but a significantly lower level of certain severe adverse events, such as pleural effusions and grade 3 to 4 thrombocytopenias, with single daily dosing (18). In line with this, it was shown that a transient inhibition of BCR–ABL with dasatinib, as short as 10 minutes, was sufficient to induce apoptosis in CML cells despite reactivation of BCR–ABL kinase activity after removal of the inhibitor (19).
Both second-generation TKIs are now approved for first-line treatment of CML in chronic phase and CML in chronic and accelerated phases in imatinib-resistant or -intolerant patients. In addition, dasatinib is also approved for imatinibresistant/intolerant patients with blast crisis CML and Ph+ ALL.
Another second-generation TKI is bosutinib (Bosulif/SKI-606; Pfizer). After successful clinical trials (20), the U.S. Food and Drug Administration has approved bosutinib to treat CML patients in all disease stages that show intolerance or resistance to other therapies, including imatinib (21). Bosutinib has characteristics similar to those of dasatinib. Being a potent type 1 inhibitor with a broad-target spectrum, bosutinib also inhibits the majority of imatinib resistance BCR–ABL mutants other than the T315I gatekeeper mutation (22, 23). In contrast with dasatinib, however, bosutinib does not inhibit KIT and PDGFR, and instead has a broad impact on Ser/Thr kinases, including members of the mitogen-activated protein kinase family (23).
Ponatinib and Other Third-Generation BCR–ABL TKIs: Efficient Targeting of BCR–ABL T315I
Despite the high number of preclinical candidates that have been presented over the past 10 years, it is only recently that the first 2 third-generation TKIs targeting the pan-resistant BCR–ABL T315I gatekeeper mutant have entered clinical evaluation. Previous approaches to target BCR–ABL T315I focused on the use of kinase inhibitors that initially were developed against other targets. For example, some available inhibitors against the Aurora family of Ser/Thr kinases (e.g., Vx-680 and PHA-739358), which have a bulky gatekeeper residue, also proved to be potent inhibitors of BCR–ABL T315I (Fig. 1C; refs. 24, 25). However, clinical assessment indicated a lack of effectivity and/or intolerable toxicity of these compounds. Later, it was concluded that the proapoptotic activity of these drugs in CML cells is primarily associated with inhibition of Aurora kinases and not with primary targeting of BCR–ABL (26).
Among the best-studied early inhibitors against BCR–ABL T315I was SGX393, which strongly inhibited BCR–ABL T315I and, in combination with nilotinib or dasatinib, preempted the outgrowth of resistant clones in an in vitro resistance screen (27).
In 2009, the type 2 inhibitor ponatinib (AP24534; Ariad Pharmaceuticals) was shown to inhibit both wild-type and imatinib-resistant BCR–ABL mutants, including T315I, with similar potency (Fig. 1C; ref. 28). Additional ponatinib targets were shown to be the SRC kinases and a number of receptor tyrosine kinases (KIT, RET, PDGFR, VEGF receptor, DDR, EPH, TRK, and FGFR family members), indicating mediumrange specificity (i.e., less specific than imatinib/nilotinib but more specific than dasatinib/bosutinib). An ethinyl linkage (C-C triple bond) in ponatinib allows binding of the compound to the kinase domain despite the presence of the bulky isoleucine residue in the T315I gatekeeper mutation, which blocks the binding of imatinib and second-generation Bcr–Abl TKIs. Many other binding features of ponatinib are remarkably similar to those observed for nilotinib, such as the methylphenyl and the trifluormethylphenyl rings that occupy almost identical positions when bound to the Abl kinase domain (Fig. 1C; ref. 28). A phase II clinical trial of ponatinib showed promising results in patients with the T315I mutation (29).
Recently, another inhibitor of BCR–ABL, T315I was reported to show inhibitory characteristics similar to those of ponatinib (30). DCC-2036 (Deciphera Pharmaceuticals) uses a carboxamide-substituted pyridine ring to engage the hinge region and a well-positioned fluor-phenyl ring to evade clashes with the gatekeeper residue (Fig. 1C). In addition, DCC-2036 binding leads to a displacement of the αC-helix and stabilization of an inactive conformation of the kinase domain (30). In similarity to ponatinib, DCC-2036 has medium specificity. In addition to BCR–ABL, it inhibits SRC kinases and a few receptor tyrosine kinases, including FLT3, TIE2, DDR2, and MER, as well as EPH, TRK, and PDGFR family members, but not KIT. The combination of low concentrations of DCC-2036 with either nilotinib or dasatinib abrogated the emergence of drug-resistant colonies in cell-based mutagenesis screens (31).
In summary, since the approval of imatinib in 2001, investigators have made enormous and rapid progress in developing an arsenal of BCR–ABL TKIs. However, because all current BCR–ABL TKIs target the ATP-binding site, it is likely that still other resistance mechanisms will develop. Thus, strategies to target regulatory modules in BCR–ABL different from the kinase active site may represent an attractive alternative for the treatment of CML.
Allosteric Targeting of the Myristate Pocket
Recently, several allosteric regions in the BCR–ABL molecule have been identified and shown to be potential drug targets. In the following text, we review the most recent developments in this active area of research. A structural analysis of the autoinhibitory mechanisms of ABL revealed a hydrophobic pocket in the C-terminal lobe of the kinase domain that accommodates the N-terminal myristoyl modification of ABL (32, 33). Binding of the myristoyl group to the myristate pocket induces a conformational change in the C-terminal helix of the kinase domain that is necessary for binding of the SH3-SH2 clamp, which keeps the kinase inactive (34). Because BCR–ABL is not myristoylated, it was suggested that this additional pocket could be used to target compounds that mimic myristate binding and push the regulatory interactions toward autoinhibition (31). Indeed, compounds that bind to the myristate pocket, such as GNF-2 (Fig. 1C), were later shown to inhibit BCR–ABL–dependent cell growth and different imatinib resistance mutations, but not the T315I mutant (35). In contrast, the combination of GNF-2 and nilotinib was shown to prolong survival in a BCR–ABL T315I mouse model (36). Therefore, the combination of ATP competitive and myristate pocket inhibitors represents an innovative and rational way to overcome resistance to either agent alone [reviewed in Hantschel (37)].
Targeting Distant Allosteric Modules: Regulatory Roles of the SH2 Domain
Cytoplasmic tyrosine kinases show a conserved domain organization, with the SH2 domain preceding the catalytic domain in most cases (38). Although the canonical function of the SH2 domain is binding to phosphorylated tyrosine residues, a series of experiments with different cytoplasmic tyrosine kinases have revealed additional regulatory roles of the SH2 domain in kinase regulation that are independent of its primary feature.
Results from biochemical and cellular studies suggested that the SH2 domain of the tyrosine kinases FES, SRC, and CSK is necessary for kinase activity (39-41). Likewise, structure-function studies on the Tec-family kinase ITK also suggested a phosphotyrosine-independent regulatory role of the SH2 domain that stimulates catalytic activity of the kinase (42).
SH2-Kinase Interface in Active ABL and BCR-ABL
Direct structural and mechanistic evidence for a stimulatory function of the SH2 domain was obtained from structure-function studies of the kinases FES and ABL (43). In the active conformation of ABL, the SH2 domain binds to the N-terminal lobe of the kinase domain (Fig. 1C). Similar conformations were found in the crystal structure of active SRC, CSK, and FES (44-46). Although superposition of the crystal structures showed that the contact surfaces between the SH2 domain and N-terminal lobe of the kinase domains are not conserved in position or chemical nature among CSK, SRC, FES, or ABL, mutational studies revealed that disruption of the SH2-kinase interface led to a decrease in kinase activity in both FES and ABL (46). Of importance, SH2-domain residues involved in the formation of the interface are not involved in phosphotyrosine binding.
The first evidence that the interaction between the SH2 and kinase domains influences leukemogenesis and can be subjected to pharmacologic interference was recently provided in a study of BCR–ABL (47). A mutational analysis of the involved residues in the SH2 domain showed that disruption of the SH2-kinase interface reduced catalytic activity, whereas mutations that stabilized the interface (initially identified in an imatinib-resistant patient with CML) increased BCR–ABL kinase activity (48). Of note, all of these effects were independent of the phosphotyrosine-binding capability of the BCR–ABL SH2 domain. Therefore, the SH2 domain of BCR–ABL acts as a true allosteric activator of tyrosine kinase activity. Disruption of the SH2-kinase domain interface completely abrogated cellular transformation and leukemia formation in a mouse bone marrow transplant model. This was accompanied by a selective loss of activation of the transcription factor STAT5, a hallmark event that is critical for CML pathogenesis. Disruption of the SH2-kinase interface also increased BCR–ABL’s susceptibility to inhibition by TKIs, and even partly restored sensitivity toward nilotinib in the pan-resistant BCR–ABL T315I mutant (47).
Targeting an interdomain association that covers a substantial surface area and has no obvious pockets poses a significant challenge. To target the SH2-kinase domain interface in BCR–ABL, Grebien and colleagues (47) sought to generate a monobody protein that would bind to the SH2 domain with very high affinity and thus prevent the formation of the SH2 kinase interface. Monobodies are small, single-domain proteins based on the fibronectin III scaffold that can be engineered to bind to a bait protein of choice with very high affinity. It was shown that a monobody that targets the ABL SH2 domain could indeed interfere with the formation of the SH2-kinase domain interface in BCR–ABL. This monobody inhibited BCR–ABL kinase activity and induced apoptosis in CML cell lines and primary human CML cells (47). Delivery of the monobody was achieved through lentiviral transduction/transfection in these experiments. Although this manner of delivery may at first glance seem to limit the use of monobody proteins in vivo, other routes of intracellular targeting of monobody proteins should be evaluated for future therapeutic use. The future will show whether the SH2-kinase interface in BCR–ABL and other oncogenic tyrosine kinases can be subjected to pharmacologic inhibition via small molecules that can be applied to treat patients with resistance to TKIs. Although protein–protein interfaces were considered to be undruggable until recently, the clinical use of the BH3-mimetic ABT-737 targeting Bcl-2 family members led investigators to reconsider this old dogma in drug discovery (49, 50).
Socioeconomic Aspects and Outlook
Despite all efforts to inhibit BCR–ABL, TKI therapy is failing to cure CML due to the persistence of treatment-insensitive CML stem cells (51). Therefore, patients with CML must take imatinib or one of its successors continuously for several years. Probably not surprisingly, a significant fraction of patients do not take their imatinib pills regularly. This was shown to be the main reason for a cytogenetic relapse in long-term imatinib therapy (52). Chronic side effects of TKI treatment, such as nausea, diarrhea, and skin rashes, are a contributing factor in this poor compliance. Although these side effects are often mild and occur only transiently, they are clearly bothersome and affect the patients’ quality of life (3). It was concluded that imatinib can be safely discontinued in patients who have been in deep remission for more than 2 years with undetectable BCR–ABL transcripts (53). In addition, it should be mentioned that the annual costs of imatinib, nilotinib, and dasatinib amount to several tens of thousands of dollars per patient. Depending on the health care system in the country of residence, this may represent a large financial burden for the patient. We anticipate that more BCR–ABL TKIs will be entering clinical practice in the coming decade, and possible combinations of these TKIs will be evaluated. Furthermore, imatinib will lose patent protection in 2015, and data on overall survival differences with second-generation TKIs as compared with imatinib will become available. As a result, new guidelines for first-line CML treatment, weighing the benefits and costs of the different TKIs, may be defined.
Despite all these unknowns, we can state with reasonable certainty that the hunt for innovative approaches to BCR–ABL inhibition and CML therapy will continue, keeping the CML research community at the forefront of therapeutic innovation and personalized treatment.
Acknowledgments
The authors thank T. Splettstoesser for help with the preparation of the figure.
Grant Support
The work of O. Hantschel is supported by the ISREC Foundation. The work of F. Grebien was funded by the Austrian Science Fund (FWF), P22282.
Footnotes
Disclosure of Potential Conflicts of Interest
O. Hantschel received speaker honoraria from Bristol-Myers Squibb and was compensated for serving on the scientific advisory board of Novartis. No other potential conflicts of interest were disclosed.
References
- 1.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–56. [PubMed] [Google Scholar]
- 2.Hochhaus A, O’Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, et al. IRIS Investigators Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–61. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- 3.Schiffer CA. CML: how low can you go? Blood. 2010;116:3686–7. doi: 10.1182/blood-2010-08-302323. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 5.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 6.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–17. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 7.Rix U, Hantschel O, Dürnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–63. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 8.Hochhaus A, Kreil S, Corbin AS, La Rosée P, Müller MC, Lahaye T, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–6. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 9.Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio G, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood. 2011;118:1208–15. doi: 10.1182/blood-2010-12-326405. [DOI] [PubMed] [Google Scholar]
- 10.O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–9. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- 11.Weisberg E, Manley PW, Breitenstein W, Brüggen J, Cowan-Jacob SW, Ray A, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–41. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 13.von Bubnoff N, Manley PW, Mestan J, Sanger J, Peschel C, Duyster J. Bcr-Abl resistance screening predicts a limited spectrum of point mutations to be associated with clinical resistance to the Abl kinase inhibitor nilotinib (AMN107) Blood. 2006;108:1328–33. doi: 10.1182/blood-2005-12-010132. [DOI] [PubMed] [Google Scholar]
- 14.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102:3395–400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laneuville P, Dilea C, Yin OQ, Woodman RC, Mestan J, Manley PW. Comparative In vitro cellular data alone are insufficient to predict clinical responses and guide the choice of BCR-ABL inhibitor for treating imatinib-resistant chronic myeloid leukemia. J Clin Oncol. 2010;28:e169–71. doi: 10.1200/JCO.2009.26.4945. author reply e172. [DOI] [PubMed] [Google Scholar]
- 16.Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW, Grzesiek S, et al. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J Biol Chem. 2008;283:18292–302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 17.Hantschel O, Rix U, Superti-Furga G. Target spectrum of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib. Leuk Lymphoma. 2008;49:615–9. doi: 10.1080/10428190801896103. [DOI] [PubMed] [Google Scholar]
- 18.Shah NP, Kantarjian HM, Kim DW, Réa D, Dorlhiac-Llacer PE, Milone JH, et al. Intermittent target inhibition with dasatinib 100 mg once daily preserves efficacy and improves tolerability in imatinib-resistant and -intolerant chronic-phase chronic myeloid leukemia. J Clin Oncol. 2008;26:3204–12. doi: 10.1200/JCO.2007.14.9260. [DOI] [PubMed] [Google Scholar]
- 19.Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14:485–93. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 20.FDA approved drug products [cited 2012 Sep 19]. Available from: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm318160.htm.
- 21.Cortes JE, Kantarjian HM, Brümmendorf TH, Kim DW, Turkina AG, Shen ZX, et al. Safety and efficacy of bosutinib (SKI-606) in chronic phase Philadelphia chromosome-positive chronic myeloid leukemia patients with resistance or intolerance to imatinib. Blood. 2011;118:4567–76. doi: 10.1182/blood-2011-05-355594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puttini M, Coluccia AM, Boschelli F, Cleris L, Marchesi E, Donella-Deana A, et al. In vitro and in vivo activity of SKI-606, a novel Src-Abl inhibitor, against imatinib-resistant Bcr-Abl+ neoplastic cells. Cancer Res. 2006;66:11314–22. doi: 10.1158/0008-5472.CAN-06-1199. [DOI] [PubMed] [Google Scholar]
- 23.Remsing Rix LL, Rix U, Colinge J, Hantschel O, Bennett KL, Stranzl T, et al. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–85. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 24.Giles FJ, Cortes J, Jones D, Bergstrom D, Kantarjian H, Freedman SJ. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–2. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 25.Modugno M, Casale E, Soncini C, Rosettani P, Colombo R, Lupi R, et al. Crystal structure of the T315I Abl mutant in complex with the aurora kinases inhibitor PHA-739358. Cancer Res. 2007;67:7987–90. doi: 10.1158/0008-5472.CAN-07-1825. [DOI] [PubMed] [Google Scholar]
- 26.Donato NJ, Fang D, Sun H, Giannola D, Peterson LF, Talpaz M. Targets and effectors of the cellular response to aurora kinase inhibitor MK-0457 (VX-680) in imatinib sensitive and resistant chronic myelogenous leukemia. Biochem Pharmacol. 2010;79:688–97. doi: 10.1016/j.bcp.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 27.O’Hare T, Eide CA, Tyner JW, Corbin AS, Wong MJ, Buchanan S, et al. SGX393 inhibits the CML mutant Bcr-AblT315I and preempts in vitro resistance when combined with nilotinib or dasatinib. Proc Natl Acad Sci U S A. 2008;105:5507–12. doi: 10.1073/pnas.0800587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–12. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes JE, Kim D-W, Pinilla-Ibarz J, le Coutre P, Chuah C, Nicolini F, et al. Initial findings from the PACE trial: a pivotal phase 2 study of ponatinib in patients with CML and Ph+ ALL resistant or intolerant to dasatinib or nilotinib, or with the T315I mutation. Blood. 2011;118:109. [Google Scholar]
- 30.Chan WW, Wise SC, Kaufman MD, Ahn YM, Ensinger CL, Haack T, et al. Conformational control inhibition of the BCR-ABL1 tyrosine kinase, including the gatekeeper T315I mutant, by the switch-control inhibitor DCC-2036. Cancer Cell. 2011;19:556–68. doi: 10.1016/j.ccr.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eide CA, Adrian LT, Tyner JW, Mac Partlin M, Anderson DJ, Wise SC, et al. The ABL switch control inhibitor DCC-2036 is active against the chronic myeloid leukemia mutant BCR-ABLT315I and exhibits a narrow resistance profile. Cancer Res. 2011;71:3189–95. doi: 10.1158/0008-5472.CAN-10-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–71. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 33.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–57. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 34.Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 35.Adrián FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Adrián FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–6. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hantschel O. Allosteric BCR-ABL inhibitors in Philadelphia chromosome-positive acute lymphoblastic leukemia: novel opportunities for drug combinations to overcome resistance. Haematologica. 2012;97:157–9. doi: 10.3324/haematol.2012.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr Opin Cell Biol. 2009;21:147–53. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Sadowski I, Stone JC, Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol. 1986;6:4396–408. doi: 10.1128/mcb.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu B, Miller WT. Src homology domains of v-Src stabilize an active conformation of the tyrosine kinase catalytic domain. Mol Cell Biochem. 1996;158:57–63. doi: 10.1007/BF00225883. [DOI] [PubMed] [Google Scholar]
- 41.Mikkola ET, Gahmberg CG. Hydrophobic interaction between the SH2 domain and the kinase domain is required for the activation of Csk. J Mol Biol. 2010;399:618–27. doi: 10.1016/j.jmb.2010.04.045. [DOI] [PubMed] [Google Scholar]
- 42.Joseph RE, Min L, Andreotti AH. The linker between SH2 and kinase domains positively regulates catalysis of the Tec family kinases. Biochemistry. 2007;46:5455–62. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 43.Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, et al. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21:787–98. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa A, Takayama Y, Sakai H, Chong KT, Takeuchi S, Nakagawa A, et al. Structure of the carboxyl-terminal Src kinase, Csk. J Biol Chem. 2002;277:14351–4. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- 45.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–71. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Filippakopoulos P, Kofler M, Hantschel O, Gish GD, Grebien F, Salah E, et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grebien F, Hantschel O, Wojcik J, Kaupe I, Kovacic B, Wyrzucki AM, et al. Targeting the SH2-kinase interface in Bcr-Abl inhibits leukemogenesis. Cell. 2011;147:306–19. doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherbenou DW, Hantschel O, Kaupe I, Willis S, Bumm T, Turaga LP, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116:3278–85. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 50.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 51.Sloma I, Jiang X, Eaves AC, Eaves CJ. Insights into the stem cells of chronic myeloid leukemia. Leukemia. 2010;24:1823–33. doi: 10.1038/leu.2010.159. [DOI] [PubMed] [Google Scholar]
- 52.Marin D, Bazeos A, Mahon F-X, Eliasson L, Milojkovic D, Bua M, et al. Adherence is the critical factor for achieving molecular responses in patients with chronic myeloid leukemia who achieve complete cytogenetic responses on imatinib. J Clin Oncol. 2010;28:2381–8. doi: 10.1200/JCO.2009.26.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahon FX, Réa D, Guilhot J, Guilhot F, Huguet F, Nicolini F, et al. Intergroupe Français des Leucémies Myéloïdes Chroniques. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–35. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]