Abstract
The endoplasmic reticulum (ER) stress sensor, IRE1, contains a kinase domain and a ribonuclease domain. Ribonuclease mediates the unconventional splicing of mRNA encoding the transcription factor AtbZIP60 in Arabidopsis, or OsbZIP50 in rice, and thereby transduces signals from stressed ER. Here, we demonstrate the additional roles of plant IRE1 using genetically modified rice plants. Using a gene targeting system based on homologous recombination, genomic IRE1 was replaced with two types of missense alleles, leading to a defect in the kinase or ribonuclease activity of IRE1. Genetic analysis of these alleles demonstrated that the kinase activity of IRE1 plays a vital role independent of ribonuclease activity. Furthermore, the existence of ribonuclease substrates other than OsbZIP50 mRNA is demonstrated for the first time. This study provides new insights into higher plant signalling using a gene targeting approach.
Endoplasmic reticulum (ER) stress is triggered by the accumulation of unfolded proteins in the ER lumen. ER stress activates intracellular signal transduction (called ER stress response or unfolded protein response) pathways that contribute to relief of the stress1. In many eukaryotes, including plants, ER stress is sensed by the transmembrane protein IRE1. The N-terminal portion of IRE1 resides in the ER lumen, and the C-terminal portion resides in the cytosol and contains a serine/threonine kinase domain and an endo-ribonuclease (RNase) domain2,3. Accumulation of unfolded proteins in the ER lumen leads to the clustering of IRE1, autophosphorylation of the kinase domain, and consequent activation of the RNase domain2. The RNase activity mediates an unconventional splicing of the mRNA encoding a key transcription factor, HAC1 (yeast), XBP1 (animals), AtbZIP60 (Arabidopsis) or OsbZIP50 (rice); the spliced forms of these mRNAs are translated as active forms4,5,6,7,8. In the case of rice, the spliced form of OsbZIP50 is involved in the expression of genes that encode ER quality control-related factors such as the ER chaperone, BiP8. The expression of some of these genes, such as OsBiP4, are almost completely dependent on the spliced form of OsbZIP50, indicating that the IRE1-OsbZIP50 pathway is particularly important for the ER stress response in rice8,9.
In animals, IRE1 plays multiple roles other than the cleavage of XBP1 mRNA and serves as a branch point for ER stress response signalling2. Therefore, plant IRE1 orthologues are also expected to play multiple roles10. However, such roles have not been clarified in plants. In our previous study8, transgenic plants were generated in which OsbZIP50 or rice IRE1 (OsIRE1) were knocked down by RNA interference. The OsIRE1 knockdown (KD) lines exhibited severe defects in growth and so were difficult to maintain. In contrast, OsbZIP50 KD lines did not exhibit a similar phenotype even though OsbZIP50 expression was almost completely suppressed. This difference suggests that OsIRE1 plays roles other than the cleavage of OsbZIP50 mRNA.
One strategy that can be employed to clarify the additional roles of OsIRE1 is the analysis of rice plants that are defective in the kinase or RNase activity of OsIRE1. However, such mutants have not been isolated, and the creation of intended plants by T-DNA insertion has not been possible because a disruption mutant of OsIRE1 is not available. To overcome this problem, an alternative plan for obtaining such mutants was considered.
Gene targeting (GT) based on homologous recombination (HR) enables the modification of desired genomic DNA sequences. This technique has greatly contributed to the functional analysis of many genes in various organisms and has been established as an indispensable tool for functional genomics. In plant species, the GT technique is routinely used in moss11. However, practical examples of the use of such a technique in higher plants, which includes most agricultural crops, have been extremely limited due to technical difficulties. Therefore, gene modification in higher plants has generally depended on the occurrence of incidental mutations or random insertions of designed T-DNA.
Recently, experimental techniques for performing Agrobacterium-mediated GT based on HR were improved, and some GT rice plants have become available12,13. Thus, using this improved GT system, we attempted to generate rice plants that are selectively defective in the kinase or RNase activities of OsIRE1 and investigated whether OsIRE1 plays additional roles in plant signalling.
Results
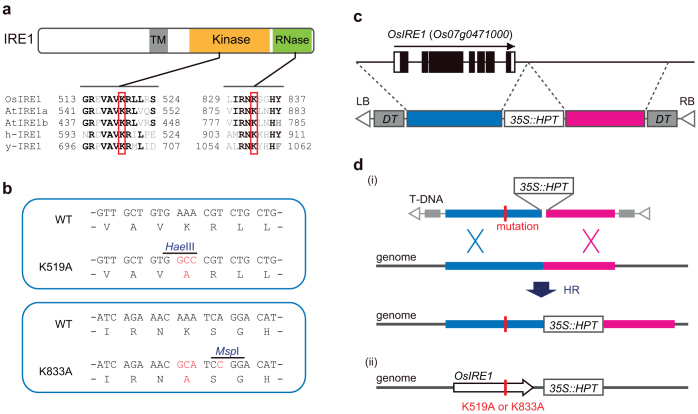
Based on previous data regarding IRE1 homologues in yeast, mammals and plants3,14,15, amino acids required for the enzymatic activities of OsIRE1 were predicted. Multiple alignments of amino acid sequences of IRE1 orthologues indicate that K519 and K833 are essential for the kinase and RNase activities of OsIRE1, respectively (Fig. 1a). Indeed, a mutation of K519A was found to lead to a defect in the autophosphorylation activity of recombinant OsIRE1 proteins produced in Escherichia coli14. The genomic region containing the OsIRE1 locus was cloned, and K519A or K833A mutations were introduced. To easily detect these mutations, restriction enzyme recognition sites were incorporated close to the mutation sites (Fig. 1b). Plasmid vectors for HR-based GT were constructed as shown in Fig. 1c; these vectors were then introduced into rice via Agrobacterium-mediated transformation (Fig. 1c and d). Using PCR and sequencing analysis, we selected GT plants and confirmed that the OsIRE1 loci were modified as designed. At least four T0 plants in which OsIRE1 was replaced with the K519A allele and two T0 plants in which OsIRE1 was replaced with the K833A allele were independently obtained. All of the T0 plants were heterozygous for the alleles of OsIRE1.
Figure 1. Introduction of missense mutations into the OsIRE1 locus by HR-based GT.
(a) Basic structure of IRE1. Transmembrane (TM), kinase and RNase domains are indicated. Partial amino acid sequence alignments of IRE1 orthologues (lower part of figure: At, Arabidopsis; h-, human; y-, yeast). Amino acids identical in at least three sequences are highlighted in boldface type. Red boxes indicate residues that are essential for kinase or RNase activities of IRE1. (b) Missense mutations introduced into OsIRE1. Recognition sites of the restriction enzymes HaeIII or MspI were incorporated into these mutation sites. (c) T-DNA used for GT. White and black boxes indicate untranslated and coding regions of genomic OsIRE1, respectively. A genomic region containing full-length OsIRE1 (blue) and the immediate downstream region (magenta) were cloned into the binary vector for Agrobacterium-mediated GT. K519A or K833A mutations were introduced into the cloned OsIRE1. Genes encoding diphtheria toxin (DT) and hygromycin phosphotransferase (HPT) in the T-DNA were used as negative and positive selection markers, respectively. (d) Schematic representation of GT. The HR event via double crossovers at the HPT-flanking homologous regions in the T-DNA resulted in an insertion of the HPT gene and partial replacements of the homologous regions (i). The successfully modified genomic region containing OsIRE1 is shown (ii).
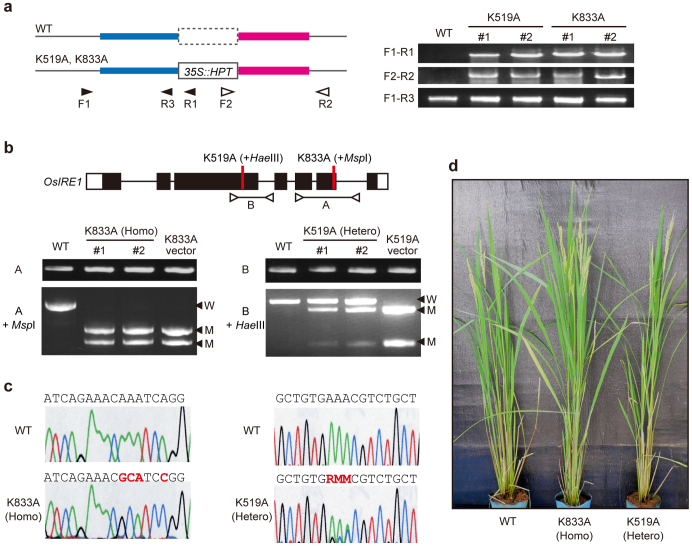
Genotyping showed that 7 of the 39 T1 progeny derived from the K833A heterozygous T0 plants were homozygous for the K833A mutation (Fig. 2a–c). To confirm the deficiency of RNase activity in the K833A homozygous lines (K833A lines), the response of these lines to ER stress was investigated. Dithiothreitol (DTT) treatment, which induces ER stress, led to OsIRE1-mediated splicing of OsbZIP50 mRNA and marked accumulation of OsBiP4 transcripts in wild-type plants, whereas this activity was not detected in the K833A lines (Supplementary Fig. S1). This result indicates that OsIRE1 RNase activity was defective in the K833A lines.
Figure 2. Confirmation of GT plants.
(a) PCR analysis for confirmation of HR events at the target locus. Wild-type and modified (K519A and K833A) genomic regions are shown (left). Blue and magenta bars indicate regions homologous to HPT-flanking regions in the T-DNA used for GT. OsIRE1 is included in the blue region. Arrowheads indicate PCR primers. PCR analysis of genomic DNA extracted from K519A and K833A lines (two independent lines each) is shown (right). (b) Confirmation of genotypes in the progeny of the GT plants. Genotyping of K833A homozygous T1 plants and K519A heterozygous T1 plants (two independent lines each) is shown. K519A homozygous plants were not obtained due to lethality (see Table 1). Open arrowheads and lines (A and B) indicate PCR primers and amplified regions for genotyping (upper panel). Regions A or B were amplified from genomic DNA and were digested with MspI or HaeIII, respectively. Fragments originating from mutant type (M) and wild-type (W) OsIRE1 are indicated by closed arrowheads. (c) Sequencing chromatograms of mutation sites. K833A homozygous plants (left) and K519A heterozygous plants (right). (d) Growth of adult GT plants. Wild-type (left), K833A homozygous (middle) and K519A heterozygous (right) plants germinated at approximately the same time and grown under similar conditions.
In contrast to K833A plants, transgenic plants homozygous for the K519A mutation could not be obtained from the selfed progeny of T0 plants, although the growth conditions were similar to those of the K833A plants (Fig. 2a–c, Table 1). A similar result was also obtained from the progeny of several independent lines, indicating that this phenomenon was not the result of other mutations. These data genetically demonstrate that the deficiency of OsIRE1 kinase activity is lethal and that kinase activity plays a vital role that is independent of RNase activity. The lethality of homozygotes for the K519A mutation is consistent with the growth defect of the OsIRE1 KD line (Supplementary Fig. S2). Additionally, K833A lines did not show a growth phenotype similar to that of the OsIRE1 KD lines (Fig. 2d), again supporting the notion that OsIRE1 plays roles that are independent of RNase activity.
Table 1. Segregation analysis of OsIRE1 K519A mutation.
| Number of plants | ||||||
|---|---|---|---|---|---|---|
| Genotype | Line A | Line B | Line C | Line D | Total | Expected ratio |
| K519A/K519A | 0 | 0 | 0 | 0 | 0 | 1 |
| K519A/WT | 28 | 7 | 5 | 8 | 48 | 2 |
| WT/WT | 14 | 1 | 3 | 3 | 21 | 1 |
Genotypes of OsIRE1 loci in the selfed progeny of four K519A heterozygous plants derived from independent lines (Lines A–D). Genotyping was performed by genomic PCR, followed by restriction digestion with HaeIII.
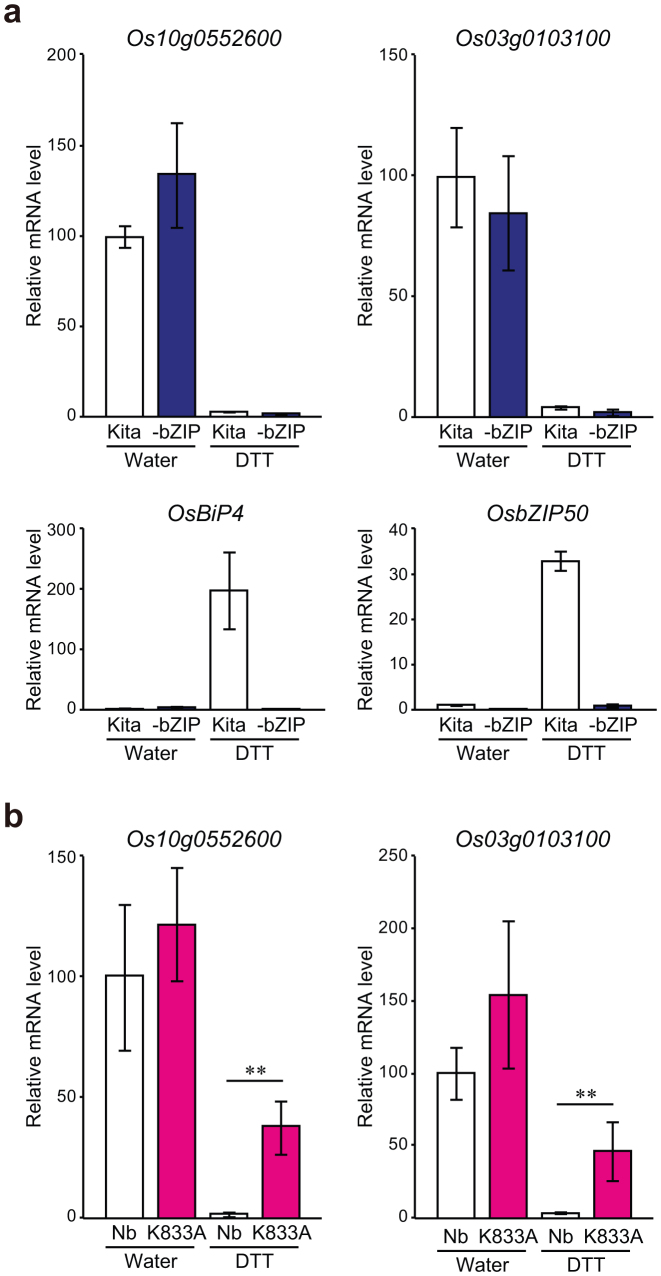
In our previous study8, genes that are up-regulated or down-regulated by OsIRE1 were screened by DNA microarray analysis of OsIRE1 KD plants after treatment with DTT. Detailed analysis demonstrated that, in response to ER stress, OsIRE1 up-regulates the expression of various genes (e.g., OsBiP4) by mediating mRNA splicing of OsbZIP508. In this study, to investigate whether the down-regulation of some genes by OsIRE1 is also dependent on OsbZIP50, the expression levels of these down-regulated genes were analysed in detail. Consistent with the result of the first screening, the mRNA levels of some genes, such as Os10g0552600 and Os03g0103100 (putative lipid transfer proteins), were strikingly reduced following two hours of DTT treatment (Fig. 3a). As shown in Fig. 3a, there was little difference in the level of reduction between the wild-type and OsbZIP50 KD plants, indicating that the down-regulation of these genes is independent of OsbZIP50 function. Next, to investigate whether the down-regulation of these genes is also independent of OsIRE1 RNase activity, we analysed the mRNA levels of these genes in the K833A lines. As shown in Fig. 3b, the reduction of the mRNA levels of these genes was significantly less pronounced in the K833A lines. These results genetically demonstrate that the RNase activity of OsIRE1 is partly implicated in the reduction of mRNA levels of Os10g0552600 and Os03g0103100, independent of OsbZIP50. Furthermore, these results strongly suggest the existence of substrates of OsIRE1 RNase other than OsbZIP50 mRNA, which is currently the only known OsIRE1 substrate.
Figure 3. OsIRE1 RNase activity is involved in the reduction of mRNA levels of some genes independent of OsbZIP50.
(a) mRNA levels, determined by quantitative RT-PCR, of Os10g0552600 and Os03g0103200 in OsbZIP50 KD (-bZIP) and wild-type plants (Kita, Kitaake) after treatment for 2 h with or without DTT. mRNA levels of OsBiP4 and OsbZIP50 reflect the activity of OsbZIP50. Data are expressed as the means ± s.d. of four or six biological replicates. (b) mRNA levels, determined by quantitative RT-PCR, of Os10g0552600 and Os03g0103100 in K833A (K833A) and wild-type plants (Nb, Nipponbare) after treatment for 2 h with or without DTT. Data are expressed as the means ± s.d. of four or five biological replicates. Asterisks indicate significant differences (**, P<0.01; t-test).
Discussion
We generated GT rice plants in which the OsIRE1 gene was replaced with missense alleles, and we genetically demonstrated that OsIRE1 plays multiple roles and serves as a branch point in signalling. As reported previously8, one of the multiple roles of this gene is the cleavage of OsbZIP50 mRNA through RNase activity. This leads to transcription of various genes, including those related to ER quality control. In this study, we demonstrated that RNase plays an additional role in the reduction of the mRNA levels of some genes in an OsbZIP50-independent manner. Previously, OsIRE1 kinase activity was thought to activate RNase activity, and other roles for OsIRE1 kinase had not been identified14. However, we demonstrated that kinase activity plays a vital role that is independent of RNase activity.
In animals, it was proposed that the RNase activity of IRE1 directly degrades various mRNAs16. Additionally, many of the degraded mRNAs encode membrane or secretory proteins that pass through the ER lumen. Os10g0552600 and Os03g0103100, which were analysed in this study, are also predicted to encode membrane proteins. Although it remains undetermined whether the mRNAs of these rice genes are directly catalysed by OsIRE1 RNase, there is no doubt that OsIRE1 RNase has various substrates other than OsbZIP50 mRNA.
IRE1 is encoded by a single gene in rice, whereas Arabidopsis has two IRE1 paralogues3. Additionally, a double disruption mutant of the Arabidopsis IRE1 paralogues did not show lethality7. These differences between rice and Arabidopsis suggest that IRE1 plays species-specific roles in plants. In mammals, adaptor proteins that bind to the cytosolic domain of IRE1 have been isolated and suggest the existence of cellular signalling pathways mediated by the adaptors2. It is possible that OsIRE1 also interacts with such signalling components in a kinase activity-dependent manner. Such a pathway may be revealed in the future by isolating factors that genetically or physically interact with plant IRE1.
Gene disruption in higher plants has generally depended on incidental mutations or random insertions of transgenes such as T-DNA and transposons. Additionally, in planta analysis of modified genes in the absence of wild-type genes requires corresponding disruption mutants. Therefore, studies of gene function in higher plants have been significantly influenced by the availability of disruption mutants. Unfortunately, an OsIRE1 disruption mutant was not available for the current study. However, the HR-based GT approach successfully overcame this problem and readily demonstrated the existence of additional roles of OsIRE1, as described above. Furthermore, the GT technique is advantageous because mutagenesis by gene replacement based on HR enables functional analysis of the mutated gene in plants with the same genetic background as the wild type. Indeed, the mRNA level of the OsIRE1 gene was similar in wild-type and GT plants (Supplementary Fig. S3).
In this study, HR-based GT rice plants were obtained by employing exogenous selection markers and highly efficient Agrobacterium-mediated transformation. Some GT rice plants obtained using fundamentally the same selection system have already been reported. In these studies, the genomic Waxy, Adh2 and OsDRM2 genes were disrupted by the insertion of a positive selection marker gene11,17,18. Additionally, several rice plants modified by HR-based GT systems using endogenous selection markers have also been reported19,20. In systems using endogenous markers, the target genes and the types of mutations that were induced were restricted because specific phenotypes of modified target genes were used for selection. In contrast to previous studies, we introduced desired missense mutations into genomic OsIRE1 to investigate several functions of the OsIRE1 protein. Additionally, our study is the first to provide new insights into higher plant signalling using such an approach. This investigation will encourage the use of similar techniques in other studies of higher plants.
Methods
Oligonucleotides
The oligonucleotides used in this study are listed in Supplementary Table S1.
Plant material and growth conditions
Oryza sativa L. (cv. Nipponbare or Kita-ake) were used in this study. Plants were grown on MS medium (1x Murashige and Skoog salt mix, 3% sucrose, B5 vitamin, 2.5 mM MES [pH 5.8] and 0.25% Gelrite) at 25°C using a 16 h light/8 h dark cycle. For ER stress treatment, seedlings (8-days-old) were incubated in liquid MS medium containing 2 mM DTT. The DTT was substituted with equal volumes of water in the negative controls.
Generation of transgenic rice lines
To generate GT plants, an approximately 7.0 kb DNA fragment containing a promoter (2.0 kb), a cistron (4.5 kb) and a terminator (0.5 kb) of OsIRE1 was cloned by PCR, and mutations that led to a defect in kinase activity (K519A, Fig. 1a and b) or RNase activity (K833A, Fig. 1a and b) were introduced into the fragments. Additionally, 4.5 kb of the region downstream of the OsIRE1 terminator was cloned. As shown in Figure 1c, these fragments were inserted into the binary vector for Agrobacterium-mediated GT12,21. Agrobacterium-mediated transformation and selection of candidate cells (gene-targeted calli) were performed as described by Ozawa et al. 201212. In this GT system, Nipponbare calli were used. To select gene-targeted calli, genomic DNA was extracted from calli growing on selection medium and subjected to PCR using specific primer sets (Fig. 2a, F1 and R1 or F2 and R2). Gene-targeted calli (approximately 0.7% [K519A] and 2.2% [K833A] of transformants) were regenerated on MS regeneration medium. Regenerated plantlets were grown in a closed glasshouse at constant temperature (28°C). Genotyping of the OsIRE1 loci was performed by genomic PCR followed by restriction digestion, as shown in Figure 2b. The absence of undesirable mutations in the OsIRE1 loci was confirmed by sequencing analysis.
OsIRE1 KD and ObZIP50 KD lines (background, Kita-ake) were obtained as described by Hayashi et al. 20128.
RNA extraction and gene expression analysis
Total RNA was extracted from root tissues using an RNeasy® Plant Mini Kit (Qiagen). For RT-PCR analysis, first-strand cDNA was synthesised from 1.0 µg of total RNA using the SuperScript® III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen), which includes both oligo(dT) and random hexamers. Quantitative RT-PCR analysis was performed using 1/80 of the prepared cDNA, SYBR® Premix Ex Taq™ (Takara) and specific primer sets (Supplementary Table S1). The ubiquitin-encoding gene (Os06g0681400) was used as an internal reference.
Author Contributions
FT, YW and SH designed the experiments. KO, YW and SH performed the experiments. YW and SH analysed the data. FT, YW and SH wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Info
Acknowledgments
We thank Y. Ikemoto, K. Miyashita, Y. Suzuki, M. Utsuno and Y. Yajima for technical assistance. This work was supported by a grant from Genomics for Agricultural Innovation (GMC0003) and NIAS Strategic Research Funding from the Ministry of Agriculture Forestry and Fisheries of Japan to F.T.
References
- Walter P. & Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- Hetz C. & Glimcher L. H. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell 35, 551–561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N. et al. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127, 949–962 (2001). [PMC free article] [PubMed] [Google Scholar]
- Mori K., Ogawa N., Kawahara T., Yanagi H. & Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 97, 4660–4665 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T. & Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001). [DOI] [PubMed] [Google Scholar]
- Deng Y. et al. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7247–7252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y. et al. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1, 29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Wakasa Y., Takahashi H., Kawakatsu T. & Takaiwa F. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 69, 946–956 (2012). [DOI] [PubMed] [Google Scholar]
- Wakasa Y., Hayashi S. & Takaiwa F. Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 236, 1519–1527 (2012). [DOI] [PubMed] [Google Scholar]
- Iwata Y. & Koizumi N. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends in Plant Science (2012). doi:10.1016/j.tplants.2012.06.014 [DOI] [PubMed] [Google Scholar]
- Schaefer D. G. A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53, 477–501 (2002). [DOI] [PubMed] [Google Scholar]
- Terada R., Urawa H., Inagaki Y., Tsugane K. & Iida S. Efficient gene targeting by homologous recombination in rice. Nat. Biotechnol. 20, 1030–1034 (2002). [DOI] [PubMed] [Google Scholar]
- Ozawa K. et al. Development of an efficient agrobacterium-mediated gene targeting system for rice and analysis of rice knockouts lacking granule-bound starch synthase (Waxy) and β1,2-xylosyltransferase. Plant Cell Physiol. 53, 755–761 (2012). [DOI] [PubMed] [Google Scholar]
- Okushima Y. et al. Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol. 43, 532–539 (2002). [DOI] [PubMed] [Google Scholar]
- Imagawa Y., Hosoda A., Sasaka S.-I., Tsuru A. & Kohno K. RNase domains determine the functional difference between IRE1alpha and IRE1beta. FEBS Lett. 582, 656–660 (2008). [DOI] [PubMed] [Google Scholar]
- Hollien J. & Weissman J. S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107 (2006). [DOI] [PubMed] [Google Scholar]
- Terada R., Johzuka-Hisatomi Y., Saitoh M., Asao H. & Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 144, 846–856 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S. et al. Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 71, 85–98 (2012). [DOI] [PubMed] [Google Scholar]
- Endo M. et al. Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J. 52, 157–166 (2007). [DOI] [PubMed] [Google Scholar]
- Saika H. et al. Application of gene targeting to designed mutation breeding of high-tryptophan rice. Plant Physiol. 156, 1269–1277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa Y., Ozawa K. & Takaiwa F. Agrobacterium-mediated co-transformation of rice using two selectable marker genes derived from rice genome components. Plant Cell Reports 31, 2075–2084 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Info