Abstract
Identifying a critical transition and its leading biomolecular network during the initiation and progression of a complex disease is a challenging task, but holds the key to early diagnosis and further elucidation of the essential mechanisms of disease deterioration at the network level. In this study, we developed a novel computational method for identifying early-warning signals of the critical transition and its leading network during a disease progression, based on high-throughput data using a small number of samples. The leading network makes the first move from the normal state toward the disease state during a transition, and thus is causally related with disease-driving genes or networks. Specifically, we first define a state-transition-based local network entropy (SNE), and prove that SNE can serve as a general early-warning indicator of any imminent transitions, regardless of specific differences among systems. The effectiveness of this method was validated by functional analysis and experimental data.
There is usually a sudden health deterioration during gradual progression of many chronic diseases, such as prostate cancer1, asthma attacks2, epileptic seizures3, and others4,5,6,7,8. This critical phenomenon generally results in a drastic or a qualitative transition in the focal system or network from a normal state to a diseased state, which corresponds to a so-called bifurcation point in dynamical systems theory9,10. Clearly, predicting and further elucidating this critical transition at the network level hold the key to understanding the fundamental mechanism of disease development and progression.
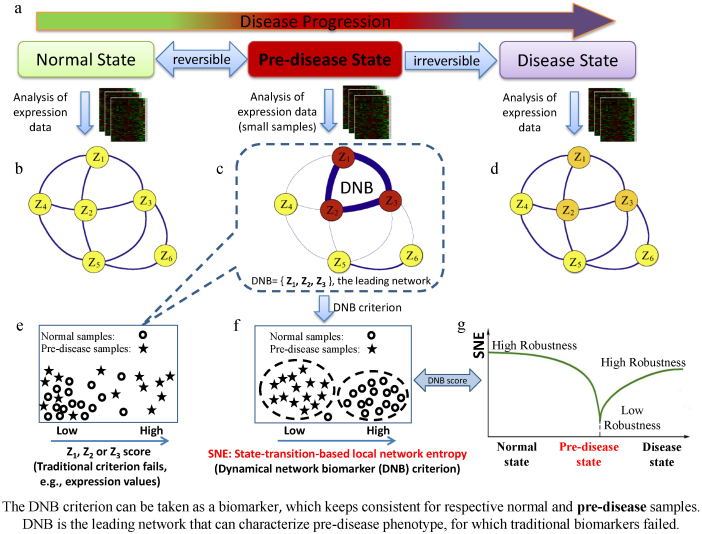
In general, disease progression can be divided into three states11, i.e., a normal state, a pre-disease state (or a critical state), and a disease state (Figs. 1 a–d). In the normal state, a disease is under control or in an incubation period, and dynamically it can be considered to have high resilience and robustness to perturbations (Figs. 1 b and g, also Fig. S1 a). The pre-disease state is defined as the limit of the normal state, which occurs before the imminent phase transition point is reached, but it has low resilience and robustness due to its dynamical structure (Figs. 1 c and g, also Fig. S1 a). At this stage, the system is sensitive to external stimuli but still reversible to the normal state when appropriately interfered with, but a small change in the parameters of the system may suffice to drive the system into collapse through bifurcation, which often implies a phase transition to the disease state. The disease state represents a seriously deteriorated stage possibly with high resilience and robustness (Figs. 1 d and g, also Fig S1 a), where the system usually finds it difficult to recover or return to the normal state even after treatment, which contrasts with the pre-disease state. Therefore, it is crucial to detect the pre-disease state to prevent qualitative deterioration and to further elucidate its molecular mechanism.
Figure 1. Schematics of disease progression and state-transition-based local network entropy: A schematic illustration of dynamical features for disease progression from a normal state to a disease state through a pre-disease state.
(a) Three states during progression of a disease. (b) The normal state is a steady state, where the system generally has high resilience and robustness to perturbations. (c) The pre-disease state is defined as a limit of the normal state and situated before the imminent phase transition point is reached. At this stage, the system is with low resilience and robustness even to small perturbations but still reversible to the normal state when appropriately interfered. {z1, z2, z3} is the dominant group or the DNB. (d) The disease state is the other steady state, at which the system is usually irreversible to the normal state due to its high resilience and robustness. (e) Traditional biomarkers failed to distinguish the pre-disease samples from normal samples. (f) SNE (DNB score) is effective in distinguishing the pre-disease samples. (g) The SNE is the conditional entropy of the previous state, which does not change significantly during the normal state but it drops sharply during the pre-disease state. By contrast, the SNE converges to another steady value during a disease state. The SNE drops drastically whenever the system approaches a critical transition point, so it can provide an effective early-warning signal for identifying the pre-disease state and the leading network that makes the first move toward a disease state.
For many complex diseases, however, it is a difficult task to predict a pre-disease state because the state of the system may change little before the bifurcation point or the critical transition is reached, namely, there may be little difference between the normal and pre-disease states; note that a pre-disease state can be considered as a limit of the normal state but a disease state is different from the normal state. This is also why diagnosis based on traditional biomarkers or static measurements may fail to distinguish a pre-disease state from a normal state (Fig. 1e). Another obstacle that hampers the detection of early-warning signals is the complexity of diseases, which can involve thousands of genetic factors (e.g., SNPs and CNVs) and epigenetic factors (e.g., methylation and acetylation). To overcome those problems, a general early-warning indicator was recently developed based on a new model-free concept, i.e., a dynamical subnetwork of biomarkers, or a dynamical network biomarker (DNB)11, which appears only during a predisease state and satisfies three measurable conditions. A DNB can distinguish a pre-disease state from other states in any disease at least fundamentally with dynamical nature (regardless of their differences) (Figs. 1 c, f and g) and it also has a solid theoretical basis derived from bifurcation theory and center manifold theory12,13. In particular, DNBs have been proven theoretically to be the leading biomolecular networks (or leading networks) in critical transitions, which make the first move from a normal state to a disease state. In other words, the leading network is the first subnetwork that breaks down the limit of a normal state to move into a disease state, which means that they are clearly related to the causal (or driving) genes in a disease network, in contrast to the differential gene expression that results from the disease (as the consequence of the disease). Therefore, identifying the leading network during a critical transition can signal the emergence of a pre-disease state so as to make the early diagnosis on the disease, and also help to elucidate the mechanism of disease initiation and progression at the network level. As shown in Fig. S1, a DNB is a dynamical signal to identify the pre-disease state, rather than the disease state detected by the traditional static biomarkers.
In general, the reliable identification of DNBs and pre-disease stages from many thousands of genes and from many stages using high throughput data is not easy to achieve with noisy data and a small number of samples14, which makes the identification of DNBs and their critical stages inaccurate. Identifying DNBs that satisfy the three conditions among genome-wide scale variables in high-throughput datasets is also a computationally intense task. Therefore, an effective computational method is required to reliably and efficiently identify DNBs to accurately predict pre-disease states and further elucidate the mechanism of disease deterioration. In this study, we used center manifold theory as the basis to develop a novel computational method for identifying the leading networks before critical transitions in complex diseases in an efficient and accurate manner. To achieve this, we derived a new type of entropy, i.e., state-transition-based local network entropy (SNE). The SNE is defined as a state-transition-based Shannon entropy that is conditional on the previous state of a local dynamical network in a Markov process, which is also the entropy rate of the state change in a biomolecular network, where each node represents a gene (or a protein, or a chemical) and each edge represents a regulatory relation between two genes, with the assumption that a Markov process governs the dynamics of each node. Given a biomolecular network, e.g., a protein-protein interaction (PPI) network or a correlation network, we can theoretically prove that the SNE is drastically reduced when the system approaches a pre-disease state, whereas there is no significant change in the SNE at normal and disease states. Thus, the SNE provides a clear signal for detecting the critical transition and its leading network (or DNB) (see Figs. 1 f and g, also Fig. S1). In particular, only local information is required to evaluate the SNE for a specific node, so we can design a very efficient algorithm that avoids the processing of most of the noisy data, which reduces the computational requirements and also improves the reliability and accuracy. In addition, this is a model-free method that can be theoretically applied to any disease or biological system with sudden transition events, while it also requires no parameter tuning due to the nature of the SNE. From a dynamical viewpoint, we show that the SNE can quantify the robustness of a system during the time evolution of disease progression, which can be applied directly to the dynamical and structural analysis of network rewiring during disease progression for many biomedical problems. The entire network can also be naturally decomposed into four layers (i.e., a DNB core, a DNB boundary, a non-DNB boundary, and a non-DNB core) based on the topological structure of the leading network (see Fig. 2a), which reveals the dynamical roles of genes (or proteins, or chemicals) during disease development and progression. To demonstrate the effectiveness and efficiency of the SNE, we applied our method to a simulated dataset and two real disease (i.e., lung injury and liver cancer) datasets, and we successfully identified the critical transitions and the leading networks. The relevance of the identified leading networks to the diseases was also validated by functional enrichment analysis and by using related experimental data. Note that DNB or the leading network in this paper is not for identifying the critical transition phenomenon but for detecting the state just before the critical transition (i.e., a pre-disease state), and therefore, it is of great importance for early diagnosis of complex diseases. Without confusion in this paper, identifying the critical transition means identifying the state just before the critical transition.
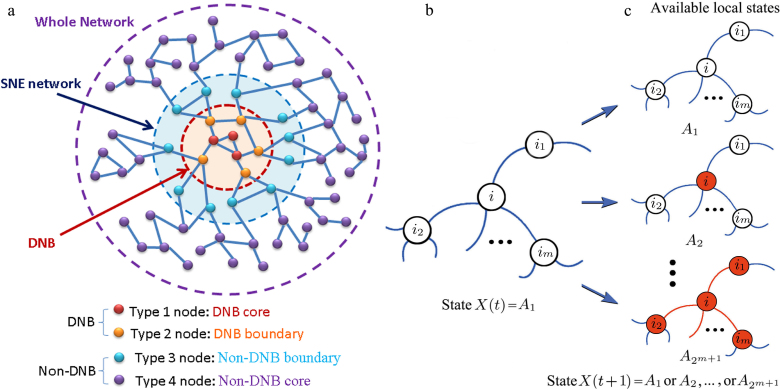
Figure 2. Outline of the SNE network and the state transitions of a local network.
This outline shows the four types of nodes in a general network and the stochastic Markov process of the state transitions between two time points t and t + 1 in a local network centered on node i. (a) The four types of nodes are respectively in four layers. (b) The local network centered on node i with its m linked first-order neighbor nodes i1, i2, …, im, is at transition state X(t) = A1 at time point t. (c) At the next time point t + 1, there are 2m+1 possible transition states 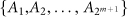 in this local network.
in this local network.
Results
The dynamics of the progression of complex diseases is usually very complicated before and after sudden deterioration, and it may not be fully expressed even by using a very high-dimensional space. However, provided that a system is driven by some known or unknown parameters approaching to a bifurcation point, a system can be generally guaranteed to be eventually constrained to a one- or two-dimensional space (i.e., the center manifold), which can be expressed in a very simple form for any dynamical system regardless of their differences11. This is the theoretical basis for developing a general indicator that detects critical transitions and their leading networks in this study.
State transitions in biomolecular networks
The network entropy was originally proposed for the study of demographic stability in population models15,16 and further extended to analyzing the topology and robustness of protein interaction networks17. Theoretically, the network entropy was derived from the entropy rate18, i.e., a conditional entropy, for measuring network robustness and stability based on a random walk process among the nodes (e.g., genes or proteins) of a network. However, the dynamics of a biological network cannot generally be described simply by individual moves from one gene to another via a random walk, because it is dominated by changes in the system state governed by the dynamical network. In this study, we define a general network entropy based on the state transitions of a local network, rather than the special dynamics of a random walk. This entropy is also a local network-based measurement with state transition variables (not with the original state variables), so it can be exploited to overcome computational difficulties, such as the computational complexity, noisy data, and accuracy, which are encountered during high-throughput data processing.
First, we define the network state (or original variables) and the transition states for a dynamical network in a Markov process. For an n-node network, let Z(t) = (z1(t), …, zn(t)) represent the network state at t, where zi(t) denotes the expression value of node (i.e., a gene or protein) i. Then, xi(t) ∈ {0, 1} is defined to measure whether or not node i has a large change at the sampling time point t, i.e., if |zi(t)−zi(t−1)| is sufficiently large such as |zi(t)−zi(t−1)| > di, then xi(t) = 1, otherwise xi(t) = 0 (see Supplementary Information A for detailed definition). Thus, X(t) = (x1(t), …, xn(t)) is the transition state for the network at t.
Next, we define a local network structure that is centered on each node, which is the basis for deriving the network entropy, i.e., the SNE. We assume that node i has m linked first-order neighbor nodes i1, …, im (see Fig. 2b), which form a local network centered on node i with local transition state 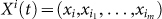 at t. Clearly, based on the current state Xi(t) = A1 at time t, there is a total of 2m+1 possible state transitions (or possible transition states) Xi(t+1), which are denoted as
at t. Clearly, based on the current state Xi(t) = A1 at time t, there is a total of 2m+1 possible state transitions (or possible transition states) Xi(t+1), which are denoted as 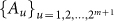 , for this local network at the next time point t + 1 (see Supplementary Information B for details). To simplify the notation, we omit i and denote Xi(t) as X(t), while we also denote the transition state simply as the state.
, for this local network at the next time point t + 1 (see Supplementary Information B for details). To simplify the notation, we omit i and denote Xi(t) as X(t), while we also denote the transition state simply as the state.
Based on the network structure, we can derive the Markov matrix P = (pu,v), where pu,v describes the transition rate from state v to state u as follows:
where u, v ∈ {1, 2, …, 2m+1} and 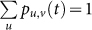 (see Supplementary Information A for detailed definition and derivation). Thus, we derive the following stochastic Markov process for X(t):
(see Supplementary Information A for detailed definition and derivation). Thus, we derive the following stochastic Markov process for X(t):
with X(t + i) = Au, u ∈ {1, 2, …, 2m+1}.
SNE and robustness
We assume a stationary Markov process for each local network centered on node i with its m linked first-order neighbor nodes i1, i2, …, im, and we define the SNE at node i as
where 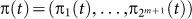 is stationary distribution that satisfies
is stationary distribution that satisfies 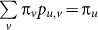 . We can easily prove that Hi(t) = H(X(t)|X(t − 1)), i.e., it is a conditional entropy. Moreover, we can prove (see18,19):
. We can easily prove that Hi(t) = H(X(t)|X(t − 1)), i.e., it is a conditional entropy. Moreover, we can prove (see18,19):
where H(X(t), X(t + 1), …, X(t + T)) is the entropy of {X(t), …, X(t + T)}. In other words, Hi(t) is actually an entropy rate. The detailed derivation is given in Supplementary Information B.
Thus, for a network with n nodes, the average SNE at t is given by
A significant feature is that the SNE can be proven to be positively correlated with the robustness or resilience of the network, which is defined as its capacity to withstand random changes (See Supplementary Information B for detailed descriptions). Thus, a higher Hi(t) is correlated with a more robust local system. At the critical transition, the system undergoes a qualitative structural change with the lowest Hi(t) with Hi(t) → 0. Thus, the SNE can quantitatively measure the structural stability and robustness of the system and detect the critical transition. It is worth noting that the sharp decrease of network robustness coincides with the critical decline of system resilience20,21 when the system approaches a tipping point, a generic dynamical phenomenon known as “critical slowing down”22,23.
Clearly, the SNE has two major differences from other entropy definitions. (a) First, it is a state-transition-based entropy, i.e., it depends on the state transitions of a dynamical network, rather than the special dynamics of a random walk among nodes. (b) Second, it is a local-network-based entropy, i.e., it depends on the local structure of a dynamical network. Based on these two features, we can evaluate the system robustness in an accurate (a) and efficient (b) manner, even with noisy data and a small number of samples. We also adopt the transition state variables in the SNE, rather than the original variables, which characterize the main differences between normal and pre-disease states.
Identifying critical transitions and their leading networks using SNE
Next, we describe our main theoretical results related to the identification of critical transitions and their leading networks during disease progression using the SNE. Note that identifying a critical transition in this paper means identifying the state (or early signal) just before the critical transition, rather than the critical transition phenomenon.
It can be proved that there is a group of molecules (i.e., genes or proteins), known as the dominant group11, that satisfy all of the following conditions in terms of state transition variables (see the proof in Theorem 2 in Supplementary Information A) for any two samples (i.e., at time t and time t + T).
- When the system is in the normal state, the following result holds.
- For any two nodes i and j (including i = j) in the network, xi(t+T) is statistically independent of xj(t).
- When the system approaches a critical transition point, the following result holds.
- If both i and j are in the dominant group or the DNB, there is a strong correlation between xi(t + T) and xj(t);
- If neither i nor j (including i = j) is in the dominant group, xi(t + T) is statistically independent of xj(t).
The network containing this dominant group is known as the DNB11 or the leading network of the critical transition, which makes the first move from the normal state toward the disease state during the critical transition.
Based on the DNB, we can decompose all of the genes in a network into four layers (see Fig. 2a), i.e., DNB core genes, DNB boundary genes, non-DNB boundary genes, and non-DNB core genes. Calculating the SNE using Eq.(3), we can show that the following results hold when a system approaches a critical transition (see Methods and Supplementary Information C for detailed analysis):
for DNB core genes (type-1, red nodes in Fig. 2a), the SNE drastically decreases;
for DNB boundary genes (type-2, orange nodes in Fig. 2a), the SNE decreases;
for non-DNB boundary genes (type-3, blue nodes in Fig. 2a), the SNE decreases;
for non-DNB core genes (type-4, purple nodes in Fig. 2a), the SNE remains almost constant.
Note that the DNB or the leading network is composed of DNB core and DNB boundary genes, whereas non-DNB boundary genes are first-order neighbors of the DNB. The DNB core, DNB boundary, and non-DNB boundary genes form an SNE network, although most genes among the whole biological system are generally expected to be non-DNB core genes.
Based on the above results, although the average SNE (or the summed SNE) of the entire network decreases as the system approaches a critical transition, it is not an efficient strategy for identifying the critical transition and the leading network because of noisy data and measurement errors when using all of the genes. To obtain a clear early-warning signal, we can intentionally select those genes with the SNEs that decrease or drastically decrease, and then calculate the average SNE using Eq.(5). In this manner, we can reduce the effects of noise and data errors (see Supplementary Information D), and greatly improve the sensitivity for detecting the early warning signal. Close to the critical transition, the average SNE of these genes will decrease drastically, thereby providing a clear early warning signal. It should be noted that genes with the decreasing SNEs are DNB genes (DNB core genes and DNB boundary genes) and the first-order neighbors of the DNB (non-DNB boundary genes), which form an SNE network that covers the leading network (see Fig. 2a). The SNE network provides sufficient information for studying the leading network, but further clustering of these genes based on the SNEs and the three conditions of the DNB can identify accurate gene sets for the leading network or the DNB-core network (see Supplementary Information F). To summarize the above theoretical result on the early warning signal by SNE, we have the following statement.
Drastic decrease of the average SNE is the early warning signal of the critical transition.
In other words, the drastic decrease of the average SNE on the DNB implies the emergence of the critical transition or the pre-disease state. Finally, it should be noted that in this paper we aim to identify the leading network that first moves into the disease state driven by known or unknown factors.
Numerical simulation
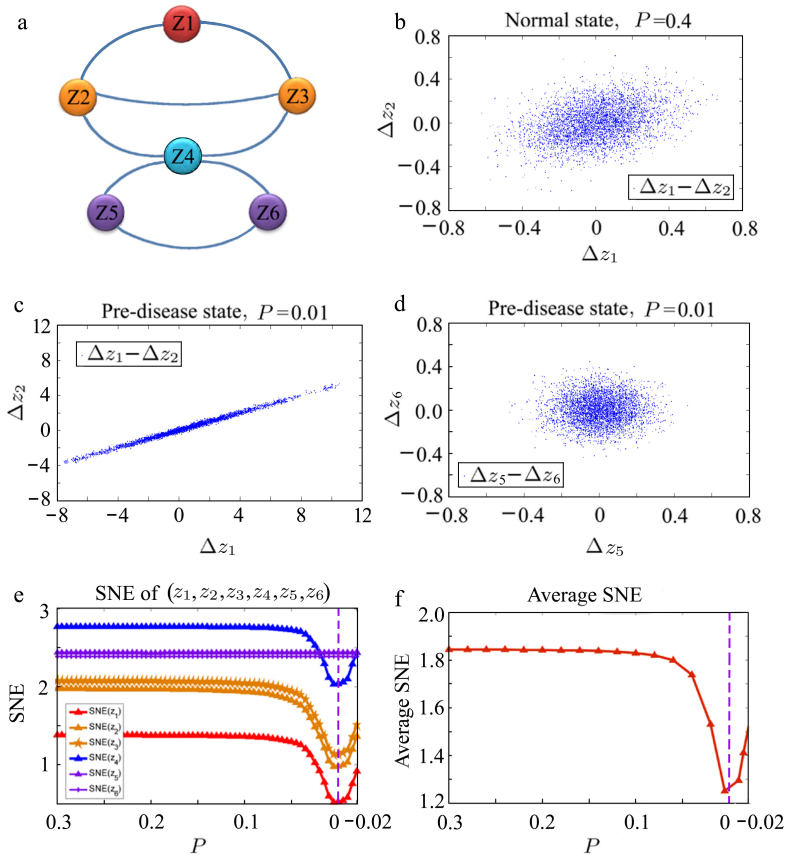
To demonstrate the effectiveness of the SNE, we used a six-node gene regulatory network (shown in Fig. 3a) to show the SNE for each gene and the average SNE. Detailed descriptions of the network represented by a set of stochastic differential equations are provided in Supporting Information E, and numerical simulations are provided in Fig. 3. The numerical simulation shows that a drastic change (or sharp decrease) in the SNE indicates the emergence of a critical transition, which validates that the SNE can serve as a general indicator by detecting an abrupt catastrophic change in the system and the leading network (z1, z2, z3).
Figure 3. Numerical validation of theoretical results.
(a) A six-gene model with the DNB or the leading network (z1, z2, z3), where z1 is the DNB core gene and (z2, z3) are the DNB boundary genes. z4 is the non-DNB boundary or the first-order neighbor of the DNB, and (z5, z6) are the non-DNB core genes. The network model and detailed background are given in Supporting Information E. The critical transition is at parameter P = 0 in the theoretical model, where the system undergoes a critical transition driven by z1, z2 and z3. (b)–(d) When the system approaches the critical transition (P = 0), z1, z2, and z3 (DNB) become closely correlated with increasingly strong deviations from (b) P = 0.4 (uncorrelated in the normal state) to (c) P = 0.01 (strongly correlated in the pre-disease state), whereas z4, z5, and z6 (non-DNB) remain statistically independent of each other from P = 0.4 to P = 0.01 (d), where Δzi(t) = zi(t) − zi(t − 1). (e) The SNE for each gene versus P as the system approaches the critical transition, where the SNEs of z1, z2 and z3 decrease drastically. (f) The average SNE curve shows the critical tendency of the network near the critical transition, which provides a general indicator for detecting the imminent transition and the leading network.
Application to complex diseases
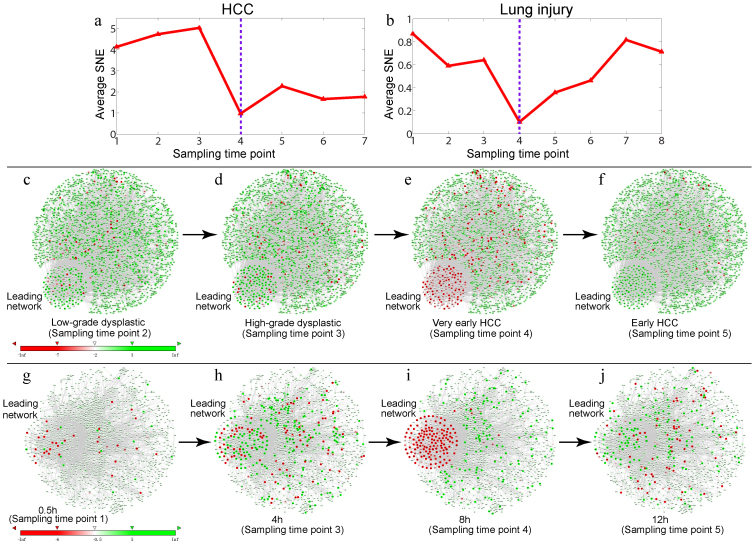
We further analyzed the prediction of two complex diseases using high-throughput experimental data, i.e., microarray data for HCV-induced dysplasia and hepatocellular carcinoma (HCC)24, and lung injury after carbonyl chloride inhalation exposure25. Figure 4 shows the identified pre-transition states and the leading networks just before the critical deteriorations by our SNE-based method, which agreed with the observed biological phenotypes described in the original datasets24,25.
Figure 4. Detecting the critical transitions and leading networks for two complex diseases.
Detection of critical transitions in diseases using two high-throughput experimental datasets for (a) HCC with the whole human molecular network (2291 genes and 6134 edges) and the identified leading network (167 genes), (b) lung injury after carbonyl chloride inhalation exposure with the whole mouse molecular network (1319 genes and 3637 edges) and the identified leading network (178 genes). (c)–(f) show the dynamical evolution of the difference SNE for the PPI network of HCC, where the leading network is indicated. (c) A low-grade dysplastic stage. (d) A high-grade dysplastic stage. (e) A very-early HCC stage. (f) An early HCC stage. (g)–(j) show the dynamical evolution of the difference SNE for the PPI network of acute lung injury, where the leading network is indicated. (g) 0.5 h. (h) 4 h. (i) 8 h. (j) 12 h. Both cases detected strong and significant early-warning signals before the diseases were critically deteriorated, i.e., the SNEs of the identified leading networks decreased drastically at the critical transition points for HCC (the very-early HCC stage) and lung injury (8 h), respectively. The heatmaps of the SNEs for the selected genes in the two diseases are provided in Supplementary Information Fig. S11.
Figures 4a–b show the SNE-based prediction results, i.e., the minima of the average SNEs indicate the critical transition points (i.e., sampling time point 4 for HCC and sampling time point 4 for lung injury). For HCC, Figs. 4 c–f show the dynamics of the difference SNE for the whole human molecular network and the leading network (or DNB) with their functional interactions (protein-protein interactions and TF-target regulations), which clearly shows that the sudden deterioration was near the “very early HCC” stage at which the average SNE of the identified leading network sharply decreases to the minimum (also see the whole network for other periods in Supplementary Information Fig. S9). Clearly, the members of the DNB (or the leading network) behaved in a significantly different way from other genes, but only near the “very early HCC” stage (e), which indicates imminent deterioration (e.g., metastasis). Interestingly, members of the leading network behaved similarly to other genes after the system moved to the deteriorated state, i.e., the early HCC stage (f). Thus, the leading network is not a disease biomarker but a pre-disease (or pre-deterioration) biomarker because it only appeared during the “pre-disease” stage. Figures 4 g-j show the dynamics of the difference SNE in the whole mouse molecular network during the evolution of lung injury, which clearly shows the significance of the leading network (DNB) in terms of expression variations and network structures near the critical state (8 h), i.e., the average SNE of the identified leading network sharply decreases to the minimum at 8 h (also see the whole network for other periods in Supplementary Information Fig. S10). Prior to the disease state, there was no significant differences between the DNB (the leading network) members and other genes during all periods (g and h), with the exception of 8 h when the DNB members behaved very differently in terms of their SNEs, i.e., they decreased sharply. However, after the system was driven into the disease state (j), interestingly the DNB members appeared to behave in a manner similar to other genes again. This was consistent with our previous results11 and the observed biological phenotypes (see the experimental descriptions provided in Supplementary Information F). The results in the two diseases show the effectiveness of our method in detecting the early-signal of critical transitions and their leading networks on the basis of small samples.
The detailed algorithm and data description are provided in Supplementary Information E and F, respectively. To explore the biological implication of the leading network where the SNEs decrease sharply, we conducted the functional analysis for these identified genes (or proteins) in the two datasets respectively (See Methods and Supplementary Information H). In particular, we used HCC liver cancer as an concrete example to explain the calculation procedure in details (see Supplementary Information F).
To verify the biological significance of the identified leading network, we also carried out bootstrap and cross validation analysis of the two diseases individually (described in Supplementary Information G). For phosgene inhalation lung injury (acute lung injury), and HCV-induced dysplasia and HCC liver cancer, some enriched gene ontology (GO) functions and dysfunctional pathways underlying the leading networks are listed in Table 1. Some of the identified members of the leading network are also shown in Table 1 (see Supplementary Table ‘Identified extended leading network’ for complete lists). The detailed descriptions are presented in Supplementary Information H.
Table 1. Functional enrichment of GO biological processes and KEGG pathway enrichment analysis in the identified leading networks of two diseases. Parts of the genes of the leading networks are shown in this table (see Supplementary Table ‘Identified extended leading network’ and Supplementary Information H for complete lists). Note that the identified leading networks are in the respective pre-disease states, rather than in the disease states.
| Disease | Leading network | GO Term | P-value | Description |
|---|---|---|---|---|
| HCV-induced HCC | {POLRIE; DRD5; SMAD9; CLU; AR; TCERG1; POLRIA; PPARG; STAT1; SNCA; E2F5; JPH2; FGF11; EEF1D; ADSSL1; CYP2B6; XDH; GYS2; FGF4; MAPK10; … } | 0051254 | 5.60E-22 | positive regulation of RNA metabolic process |
| 0031328 | 2.70E-21 | positive regulation of cellular biosynthetic process | ||
| 0042127 | 8.70E-15 | regulation of cell proliferation | ||
| 0042981 | 8.80E-14 | regulation of apoptosis | ||
| 0051049 | 3.40E-05 | regulation of transport | ||
| 0002252 | 1.30E-04 | immune effector process | ||
| 0030217 | 1.30E-04 | T cell differentiation | ||
| 0009615 | 1.50E-04 | response to virus | ||
| 0055088 | 1.80E-04 | lipid homeostasis | ||
| 0043923 | 2.50E-04 | positive regulation by host of viral transcription | ||
| Phosgene-induced Lung Injury | {JUN; NOTCH2; GPX2; MYC; HMOX1; CDO1; PTGS2; CCL2; PLA2G7; MYD88; GCLC; SMAD1; ARG2; CTPS; ASN; IL1B; ALDH4A1; … } | 0042221 | 4.30E-24 | response to chemical stimulus |
| 0042981 | 7.90E-14 | regulation of apoptosis | ||
| 0045935 | 1.90E-11 | positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | ||
| 0055114 | 7.70E-09 | oxidation reduction | ||
| 0019538 | 1.40E-08 | protein metabolic process | ||
| 0016310 | 4.20E-08 | phosphorylation | ||
| 0006954 | 4.60E-08 | inflammatory response | ||
| 0009611 | 6.10E-08 | response to wounding | ||
| 0009064 | 6.20E-05 | glutamine family amino acid metabolic process | ||
| 0006629 | 9.50E-05 | lipid metabolic process |
The functional analysis shows that the members of the leading network are highly relevant to the corresponding complex diseases, which validates the effectiveness of our method. In the HCC study, we found that many genes included in the identified leading network were consistent with the response to HCV infection in vivo, especially the activation of the immune system and the dysfunctions associated with basic cell metabolism of hosts26,27,28. The results of GO and pathway enrichment analyses are provided in Supplementary Table ‘KEGG enrichment analysis’. These results show that the genes of the leading network played significantly important roles during disease development. In the enrichment analysis, the most enriched functions indicate its significant relationship with disease evolution. At the pathway level, the pathways in cancer and Hepatitis C appear to be significant, which provides clear evidence that most of the genes selected by the SNE are directly related to HCC. Some enriched pathways are related to the dysfunctions in basic cell metabolism, which implies the reproduction and release of HCV. Some pathways show the common characteristics of cancer, especially the signaling pathways involved in cell growth, such as transcriptional misregulation in cancer, purine metabolism, the Wnt signaling pathway, and the TGF-beta signaling pathway. These dysfunctional pathways indicate the cell status when HCV invades the host cells. HCV readily uses the host resources to replicate its genetic material (RNA) for viral replication. At the GO function level, some genes were also related to important biological processes beyond the pathways mentioned above. For example, CLU, IL1B, and TNF lead to an inflammatory response27. The regulations of antiapoptosis and growth are also dysfunctional28. However, in order to reproduce and release, the functional interruption of RNA biosynthetic processes and gene expression are enriched in the genes CD81, POLR1A, POLR1E, TCERG1, and AR, while transport activities are enriched in DRD2, PPARG, JPH2, and SNCA. They are often used for releasing new viruses after their reproduction. We also compared the members of the identified leading network with those reported in a previous study28. We validated that 5 out of the 167 genes (hypergeometric test, p-value < 0.02) had a close correspondence with HCC induced by HCV infection during the dysplastic stage and the very early stage of HCC. A detailed functional analysis of lung injury is given in Supplementary Information H.
Discussion
In this study, we proposed a new method with the SNE for identifying critical transitions and their leading networks during disease progression. From the viewpoints of both theoretical analysis and numerical computation, we demonstrated that the SNE can be used to characterize the dynamical behavior of a system and provide a general indicator for the detection of the DNB when a system reaches a critical transition point (see Fig. 1 and Fig. 3).
By only choosing the genes with SNEs that decreased drastically, we identified the leading network during a critical transition that made the first move toward a disease state, or a deteriorated state from a nonlinear dynamics perspective. The identification of the leading network using a small number of samples is vital for the early diagnosis of diseases and elucidating the essential mechanisms of disease deterioration at the network level. The leading networks are also related to causal genes or networks, so they can provide a new type of biomarkers for the detection of pre-disease states in a dynamical manner (Figs. 1 c and g), which contrasts with traditional gene- or protein-based biomarkers that evaluate systems in a generally static manner. It is notable that SNE networks satisfy the three conditions of DNBs in the vicinity of a critical point (see reference 11 and their derivation in Supplementary Information B), so these results are compatible with the DNB theory11.
The advantages of the SNE are obvious. First, the SNE method is efficient because its calculation requires only local information for a local network centered on a specific node. By focusing on the local structure and listing all of the possible state transitions for each node in a network, we can obtain its SNE. Second, we ignore nodes where the SNE increases or changes little, which facilitates a significant reduction in the effect of noise, data errors, and the computational complexity. Third, the SNE can detect the early-signal of the critical transition and identify its leading network, which facilitates the elucidation of the essential mechanisms during disease development and progression at the network level. Compared with correlationbased methods that can only describe linear dependency relationship between any two nodes, our state-transition based SNE describes nonlinear relationship among nodes and the collective behavioral dynamics of a group of nodes. Fourth, the SNE algorithm is simple to implement and it may be viewed as a DNB-free method, although the theoretical background of the SNE is based on DNB. Finally, the SNE can be also considered as a measurement of resilience and robustness for the corresponding local network, which can be applied directly to the dynamical and structural analysis of network rewiring during disease progression in many biomedical problems. We applied our method to two diseases, i.e., lung injury and liver cancer, which demonstrated its effectiveness and efficiency for identifying critical transitions and their leading networks.
One potential application of DNB in medicine is the early diagnosis for complex diseases. As demonstrated in our previous work and also this paper, it is difficult to distinguish the normal and pre-disease states by one sample (e.g., by only one health examination in one year) although it is possible to detect the disease state. But with a number of the consecutive samples (e.g., with several health examinations in one year), we can clearly detect the pre-disease state before the drastic deterioration, thus open a new way to predictive and preventive medicine (and even personalized medicine). Another potential application is to detect the critical transition of a specific biological process (e.g., cell differentiation or cell proliferation process) in biology since our theoretical result can be essentially applied to identify the early-state before any drastic change of a biological system (or state change).
Methods
Identifying critical transitions and their leading networks using SNE
Dynamical networks
The theoretical results were derived by considering the following equations with noise perturbations near the equilibrium 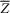 :
:
We assume that {λ1(P), …, λn(P)} are the eigenvalues of the Jacobian matrix of f at 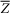 with each |λi| between 0 and 1. Among the eigenvalues, the largest in terms of its modulus, say λ1, approaches 1 when parameter P → Pc. This eigenvalue characterizes the system's rate of change around a fixed point and it is known as the dominant eigenvalue. In the overall network, nodes that are influenced directly by the dominant eigenvalue λ1 are known as the dominant group members, the leading network, or the DNB (see Supplementary Information B), i.e., a group of nodes that make the first move toward the disease state thereby indicating the approach of a sudden deterioration. Thus, in the network shown in Fig. 2a, the nodes or genes can be categorized into four groups based on the topological structure of the DNB, i.e., DNB core genes, DNB boundary genes, non-DNB boundary genes, and non-DNB core genes.
with each |λi| between 0 and 1. Among the eigenvalues, the largest in terms of its modulus, say λ1, approaches 1 when parameter P → Pc. This eigenvalue characterizes the system's rate of change around a fixed point and it is known as the dominant eigenvalue. In the overall network, nodes that are influenced directly by the dominant eigenvalue λ1 are known as the dominant group members, the leading network, or the DNB (see Supplementary Information B), i.e., a group of nodes that make the first move toward the disease state thereby indicating the approach of a sudden deterioration. Thus, in the network shown in Fig. 2a, the nodes or genes can be categorized into four groups based on the topological structure of the DNB, i.e., DNB core genes, DNB boundary genes, non-DNB boundary genes, and non-DNB core genes.
Dynamical evolution of state transitions
We consider the linearized equations of Eq.(6) with the Jacobian matrix 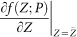 which has different real eigenvalues (λ1(P), …, λn(P))29. After introducing new variables Y (k) = (y1(k), …, yn(k)) and a transformation matrix S, i.e., ΔY (t) = S−1(Z(t)−Z(t − 1)) (Supplementary Information A2), we have
which has different real eigenvalues (λ1(P), …, λn(P))29. After introducing new variables Y (k) = (y1(k), …, yn(k)) and a transformation matrix S, i.e., ΔY (t) = S−1(Z(t)−Z(t − 1)) (Supplementary Information A2), we have
where Λ(P) is the diagonalized matrix of 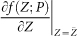 , ζ(k) = (ζ1 (k), …, ζn(k)) are Gaussian noises with zero means and covariances κij = Cov(ζi, ζj), Λ(P) = diag(λ1(P), …, λn(P)), and y1 is the eigenvector of the dominant eigenvalue λ1.
, ζ(k) = (ζ1 (k), …, ζn(k)) are Gaussian noises with zero means and covariances κij = Cov(ζi, ζj), Λ(P) = diag(λ1(P), …, λn(P)), and y1 is the eigenvector of the dominant eigenvalue λ1.
For any integer T > 0, it holds that
where εi(t) is a small white noise (see Supplementary Information A2).
Therefore, given that ΔZ(t) = SΔY (t), we can use Eq.(8) to derive the main results of the dynamical evolution for the original variable ΔZ and the local transition state X (also see the derivations in Supplementary Information A). The detailed derivations of the dynamics for the four types of nodes at different stages are presented in Supplementary Information C.
Data preprocessing and functional analysis
Data processing
Two gene expression profiling datasets were downloaded from the NCBI GEO database (ID: GSE6764, GSE2565) (www.ncbi.nlm.nih.gov/geo). In these datasets, probe sets without corresponding gene symbols were not considered during our analysis. The expression values of probe sets mapped to the same gene were averaged.
In each disease dataset, the expression profiling information was mapped to the integrated networks individually. For each species, we downloaded the biomolecular interaction networks from various databases, including BioGrid (www.thebiogrid.org), TRED (www.rulai.cshl.edu/cgi-bin/TRED/), KEGG (www.genome.jp/kegg), and HPRD (www.hprd.org). First, the available functional linkage information for Mus musculus and Homo sapiens was downloaded from these databases and combined. After removing any redundancy, we obtained 65625 linkages in 11451 human proteins/genes and 37950 linkages in 6683 mouse proteins/genes. Next, the genes evaluated in these microarray datasets were mapped individually to these integrated functional linkage networks. Furthermore, the leading networks (LNs) for two diseases were identified using the proposed SNE algorithm. In total, there were 167 proteins including four transcriptional factors (TFs) in the leading network identified for HCC, and 182 proteins including 16 TFs in the leading network identified for acute lung injury. These networks were then visualized using Cytoscape (www.cytoscape.org).
Functional analysis of the leading networks
In the high throughput gene expression profiling datasets for the two diseases, i.e., HCC induced by HCV infection and acute lung injury induced by phosgene gas, we detected the early-warning signals as well as the leading networks using our SNE algorithm and the corresponding protein interaction networks.
It is reported that TFs with high differential expression of their target genes might be causal factors. Bearing this in mind, we added TFs if their targets were present in the leading networks, as well as proteins or genes adjacent to any members of the leading networks. We treated them as the extended leading networks. In Supplementary Information H, we describe the data processing method in detail and present the results of the functional analysis (g:profiler: http://biit.cs.ut.ee/gprofiler/ and NOA: http://app.aporc.org/NOA/)30,31 of these leading networks for two complex diseases.
Based on the functional analysis, we found close relationship between the members of the leading networks and complex diseases. Several genes that had been verified in other published reports were also identified. Newly identified genes can be treated as novel biomarker candidates.
We performed a functional analysis of genes in the leading networks identified by the SNE in HCC and the acute lung injury. We also applied enriched pathways in the leading networks and the results were validated by bootstrap and cross validation analysis (See Supplementary Information G for details). Functional enrichment showed that the leading networks have significantly strong relationship with the corresponding diseases. Supplementary Table ‘KEGG Enrichment analysis’ shows the results for the two diseases (see Supplementary Information H).
Author Contributions
L.C., R.L. and K.A. conceived the research. L.C., R.L. and M.L. designed the numerical simulation and the actual experimental data analysis. R.L. and M.L. performed the numerical experiments. Z.P.L., M.L. and J.W. contributed to the functional analysis. All the authors wrote the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
This work was supported by NSFC under grant Nos. 91029301, 61134013, and 61072149, by the Chief Scientist Program of SIBS of CAS under grant No. 2009CSP002, and by the Knowledge Innovation Program of SIBS of CAS under grant No. 2011KIP203. This work was also supported by the Shanghai Pujiang Program, 973 Program No.2011CB910201, the National Center for Mathematics and Interdisciplinary Sciences of CAS, and the Aihara Project, the FIRST program from JSPS initiated by CSTP. We thank Yoshito Hirata for his comment on cross validation.
Data and materials availability: Data are available in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE2565 and GSE6764.
References
- Hirata Y., Bruchovsky N. & Aihara K. Development of a mathematical model that predicts the outcome of hormone therapy for prostate cancer. J. Theor. Biol. 264, 517–527 (2010). [DOI] [PubMed] [Google Scholar]
- Venegas J. G. et al. Self-organized patchiness in asthma as a prelude to catastrophic shifts. Nature 434, 777–782 (2005). [DOI] [PubMed] [Google Scholar]
- Litt B. et al. Epileptic seizures may begin hours in advance of clinical onset: a report of five patients. Neuron 30, 51–64 (2001). [DOI] [PubMed] [Google Scholar]
- McSharry P. E., Smith L. A. & Tarassenko L. Prediction of epileptic seizures: are nonlinear methods relevant? Nature Med. 9, 241–242 (2003). [DOI] [PubMed] [Google Scholar]
- Roberto P. B., Eliseo G. & Josef C. Transition models for change-point estimation in logistic regression. Statist. Med. 22, 1141–1162 (2003). [DOI] [PubMed] [Google Scholar]
- Paek S., Chung H., Jeong S. & Park C. Hearing preservation after gamma knife stereotactic radiosurgery of vestibular schwannoma. Cancer 104, 580–590 (2005). [DOI] [PubMed] [Google Scholar]
- He D., Liu Z., Honda M., Kaneko S. & Chen L. Coexpression network analysis in chronic hepatitis B and C hepatic lesion reveals distinct patterns of disease progression to hepatocellular carcinoma. Journal of Molecular Cell Biology 4, 140–152 (2012). [DOI] [PubMed] [Google Scholar]
- Liu J. K., Rovit R. L. & Couldwell W. T. Pituitary Apoplexy. Seminars in Neurosurgery 12, 315–320 (2001). [Google Scholar]
- Gilmore R. Catastrophe Theory for Scientists and Engineers, (Dover, 1981). [Google Scholar]
- Murray J. D. Mathematical Biology, (Springer, 1993). [Google Scholar]
- Chen L., Liu R., Liu Z. P., Li M. & Aihara K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2, 342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnol'd V. I. Dynamical systems V: bifurcation theory and catastrophe theory, (Springer, 1994). [Google Scholar]
- Murdock J. Normal forms and unfoldings for local dynamical systems, (Springer, 2003). [Google Scholar]
- Chen L., Wang R., Li C. & Aihara K. Modeling Biomolecular Networks in Cells: Structures and Dynamics, (Springer, New York, 2010). [Google Scholar]
- Demetrius L., Gundlach V. & Ochs G. Complexity and demographic stability in population models. Theoretical Population Biology 65, 211–225 (2004). [DOI] [PubMed] [Google Scholar]
- Demetrius L. & Manke T. Robustness and network evolutionłan entropic principle. Physica A 346, 682–696 (2005). [Google Scholar]
- Manke T., Demetrius L. & Vingron M. An entropic characterization of protein interaction networks and cellular robustness. J. R. Soc. Interface 30, 51–64 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover T. & Thomas J. Elements of information theory, (Wiley, New Jersey, 2005). [Google Scholar]
- Gomez-Gardenes J. & Latora V. Entropy rate of diffusion processes on complex networks. Phy. Rev. E 78, 065102(4) (2008). [DOI] [PubMed] [Google Scholar]
- Gunderson L. H. Ecological resilience - in theory and application. .Ann. Rev. Ecol. Syst. 31, 425–439 (2000). [Google Scholar]
- Walker B., Holling C. S., Carpenter S. R. & Kinzig A. Resilience, adaptablility and transformability in social - ecological systems. Ecology and Society 9, 1–5 (2004). [Google Scholar]
- Strogatz S. H. Nonlinear Dynamics And Chaos: With Applications To Physics, Biology, Chemistry And Engineering, (Addison-Wesley, Reading, MA, 1994). [Google Scholar]
- Van Nes E. H. & Scheffer M. Slow recovery from perturbations as a generic indicator of a nearby catastrophic shift. Am. Nat. 169, 738–747 (2007). [DOI] [PubMed] [Google Scholar]
- Wurmbach E. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45, 938–947 (2007). [DOI] [PubMed] [Google Scholar]
- Sciuto A. M., Phillips C. S. & Orzolek L. D. Genomic analysis of murine pulmonary tissue following carbonyl chloride inhalation. Chem. Res. Toxicol. 18, 1654–1660 (2005). [DOI] [PubMed] [Google Scholar]
- Bruix J., Boix L., Sala M. & Llovet, J,M. Focus on hepatocellular carcinoma. Cancer Cell 5, 215–219 (2004). [DOI] [PubMed] [Google Scholar]
- Farazi P. A. & DePinho R. A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 6, 674–687 (2006). [DOI] [PubMed] [Google Scholar]
- Wurmbach E., Chen Y., Khitrov G. & Zhang W. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45, 938–947 (2007). [DOI] [PubMed] [Google Scholar]
- Scheffer M., Bascompte J., Brock W. & Brovkin V. Early-warning signals for critical transitions. Nature 461, 53–59 (2009). [DOI] [PubMed] [Google Scholar]
- Reimand J., Arak T. & Vilo J. g:Profiler – a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 39, W307–315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Huang Q., Liu Z., Wang Y., Wu L., Chen L. & Zhang X. NOA: a novel Network Ontology Analysis method. Nucleic Acids Res. 39, e87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information