Abstract
Nuclear receptors comprise a large family of highly conserved transcription factors that regulate many key processes in normal and neoplastic tissues. Most nuclear receptors share a common, highly conserved domain structure that includes a carboxy-terminal ligand-binding domain (LBD). However, a sub-group of this gene family is known as the orphan nuclear receptors because to date there are no known natural ligands that regulate their activity. Many of the 25 nuclear receptors classified as orphan play critical roles in embryonic development, metabolism, and the regulation of circadian rhythm. Here, we review the emerging role(s) of orphan nuclear receptors in breast cancer, with a particular focus on two of the estrogen-related receptors (ERRα, ERRγ) and several others implicated in clinical outcome and response or resistance to cytotoxic or endocrine therapies, including the COUP-TFs, NGFI-B, DAX-1, LRH-1, and RORα. We also propose that a clearer understanding of the function of orphan nuclear receptors in mammary gland development and normal mammary tissues could significantly improve our ability to diagnose, treat, and prevent breast cancer.
Keywords: orphan nuclear receptor, breast cancer, drug resistance, ERRα, ERRγ, COUP-TF, NGFI-B, DAX-1, LRH-1
What is an orphan nuclear receptor?
Members of the nuclear receptor super-family are some of the most abundant regulators of gene expression in higher eukaryotes. These DNA-binding transcription factors play essential roles in key biological processes from embryonic development to differentiation, and their dysregulation has been widely studied in many different pathologies including cancer (Jeong and Mangelsdorf 2009; Mangelsdorf, et al. 1995; Novac and Heinzel 2004; Robinson-Rechavi, et al. 2003).
Of the 48 members of the human nuclear receptor super-family, 25 are currently considered to be orphan nuclear receptors (Benoit, et al. 2006) (Table 1) because they have no known ligand. Most of these 25 receptors adhere to the classical domain structure that typifies ligand-regulated nuclear receptors (discussed in more detail below), with two notable exceptions. Dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 (DAX-1, NR0B1) and small heterodimerization partner (SHP, NR0B2) lack a classical DNA-binding domain and cannot act alone to bind DNA (Burris, et al. 1996; Seol, et al. 1996). The number of nuclear receptors classified as orphans has decreased over the years as new ligands have been discovered. Two such ‘adopted orphans’ are the retinoid X receptors (RXRs) and the peroxisome proliferator-activated receptors (PPARs), which were initially considered to be orphans but have been firmly in the category of liganded receptors for some time. RXRs and PPARs, along with several other formerly orphaned nuclear receptors, form a group now referred to as natural or nutrient sensors that bind 9-cis-retinoic acid, oleic, and linoleic acids (Benoit, et al. 2004; Francis, et al. 2003).
Table 1.
Orphan Nuclear Receptors in Breast Cancer
DES = diethylstilbestrol; 4HT = 4-hydroxytamoxifen
GSE identification numbers denote publicly available gene expression studies from Gene Expression Omnibus (GEO). For each study, expression of the relevant nuclear receptor is significantly increased in breast cancer vs. normal mammary tissue (p≤0.05, pair-wise t test).
What constitutes an orphan receptor is still the subject of some debate. An example of the lingering controversy is that orphan nuclear receptors like steroidogenic factor-1 (SF1, NR5A1) and hepatocyte nuclear factor-4 (HNF4, NR2A1 and NR2A2) have been shown to bind phospholipids and fatty acids, respectively (Li, et al. 2005; Wisely, et al. 2002). However, because the physiologic and/or functional relevance of these interactions remain unclear, SF1 and HNF4 are still considered to be orphan nuclear receptors.
Nuclear receptor structure and function
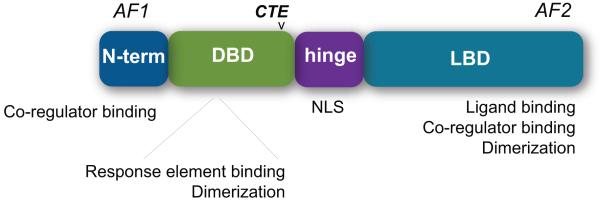
As a group, most nuclear receptors share a common, highly conserved domain structure (Fig. 1). At the amino-terminus, the activation function-1 (AF1) domain is a highly divergent region that assists in regulating the transcriptional activity of nuclear receptors independent from ligand binding (Kumar and Litwack 2009). The AF1 domain is one of the two major sites for the binding of nuclear receptor co-regulators, which include co-activator and co-repressor proteins that can positively or negatively impact transcriptional activity, respectively; it is also an important site of post-translational modification, including phosphorylation and the addition of small ubiquitin-like modifier proteins (SUMOylation) (Cheng, et al. 2007; Garza, et al. 2010; Takimoto, et al. 2003; Tamasi, et al. 2008; Zhang, et al. 2007). Much less is known about the AF1 domain as compared to other regions of nuclear receptors. One key reason is that the AF1 domain has a high level of intrinsic disorder (ID) (Kumar and Litwack 2009), although this is not the only region of these receptors that is disordered (Krasowski, et al. 2008). ID regions are characterized by amino acid sequences that are low in hydrophobicity and highly charged, leading to flexible, highly variable tertiary and quaternary protein structures. In general, all transcription factors are enriched in ID regions (Minezaki, et al. 2006), and these appear to be critical for the regulation of protein-protein interactions (Dunker, et al. 2005). In addition, the distribution of nuclear receptor co-activator proteins that can bind to the AF1 domain and regulate receptor function is tissue- and cell-type specific. It is now apparent that the differential expression and function of the entire group of nuclear receptor co-regulators (co-activators and co-repressors) in normal vs. cancer tissue is a fundamental component of nuclear receptor regulation (Hall and McDonnell 2005; O’Malley and Kumar 2009).
Fig. 1. Nuclear receptor domain structure.
AF1, activation function-1; DBD, DNA-binding domain; CTE, carboxy-terminal extension; NLS, nuclear localization sequence; LBD, ligand-binding domain; AF2, activation function-2.
The DNA-binding domain (DBD) of the nuclear receptor super-family is defined by two cysteine-rich zinc finger motifs that permit binding of the receptor to DNA (Freedman, et al. 1988). This region is also important in mediating the homo- and heterodimerization of nuclear receptors (Claessens and Gewirth 2004). Proximal to the DBD is the flexible hinge region of the nuclear receptor, which typically contains the nuclear localization sequence (NLS) (Aschrafi, et al. 2006; Carrigan, et al. 2007; Claessens, et al. 2001). The hinge region is also a key site for post-translational modifications (Chen, et al. 2006; Hwang, et al. 2009; Sentis, et al. 2005).
Nuclear receptor DBDs contain a short stretch of amino acids downstream of the two zinc fingers known as the carboxy-terminal extension (CTE) (Claessens and Gewirth 2004). The CTE is present in ligand-regulated nuclear receptors like the estrogen receptors (Schultz, et al. 2002), androgen receptor (Schoenmakers, et al. 1999), and the vitamin D receptor (Hsieh, et al. 1999). However, orphan nuclear receptors such as estrogen-related receptor beta (ERRβ, ESRRB, NR3B1) that bind a single half-site rely heavily on the A box of the CTE (which contains a conserved Glycine-Arginine motif) to permit DNA binding in the minor groove (Gearhart, et al. 2003). In addition, residues C-terminal to the A box form intramolecular interactions with the rest of the DBD; together, these interactions serve to stabilize the binding of ERRβ and several other orphan nuclear receptors to DNA.
The carboxy-terminal ligand-binding domain (LBD) and the activation function-2 (AF2) domain are essential for the regulation of nuclear receptor transcriptional activity by mediating ligand-receptor interactions and co-regulator binding; in some cases, these regions also participate in receptor dimerization (Chandra, et al. 2008). Upon the engagement of natural or synthetic ligand, nuclear receptor LBDs undergo a significant conformational change that alters the orientation of several α–helices and β–sheets, most notably the repositioning of helix 12 (H12) that comprises the AF2 domain (Wurtz, et al. 1996). H12 repositioning uncovers a hydrophobic binding groove or charge clamp that recruits co-regulator proteins containing an LXXLL motif (Westin, et al. 1998), and the sum of these changes serves to significantly enhance nuclear receptor transcriptional activity.
In contrast to ligand-regulated nuclear receptors, the orphan nuclear receptors typically display constitutive transcriptional activity. While crystal structures for many orphan LBDs appear to be ligand-filled, some are still modulated by synthetic agonists and antagonists (Table 1). Thus, it remains possible that some of these receptors have natural ligands that have not yet been discovered. Even in the absence of ligand, H12 is often pre-positioned for maximal activation, promoting interactions between the orphan nuclear receptor and its coactivators (Flaig, et al. 2005; Greschik, et al. 2002). It has also become increasingly clear that orphan nuclear receptors are particularly sensitive to the binding of coactivator proteins. Differential co-regulator binding can directly affect which DNA response elements are bound and activated (Gaillard, et al. 2007), and modified coactivators that are selective for particular orphans can be designed to further explore the biology of these receptors (Gaillard, et al. 2006; Stein, et al. 2008).
Benoit et al. have recently published a comprehensive review of all nuclear receptors currently classified as orphaned, summarizing what is known about their expression patterns, co-regulatory molecules, validated transcriptional target genes, and any phenotypes associated with their perturbation in mice (Benoit et al. 2006). Therefore, we will specifically focus on a review of the evidence supporting a role for orphan nuclear receptors in breast cancer pathogenesis and therapeutic response.
Introduction to breast cancer
The American Cancer Society estimates that in 2009, 178,000 women were diagnosed with breast cancer, and over 40,000 women died of breast cancer (Jemal, et al. 2009), making it the second-most common cause of cancer-related death in women. Breast cancer is not a single disease; it is classified into multiple histologic and molecular subtypes with diverse clinicopathological features and survival outcomes (Olopade, et al. 2008; Orlando and Brown 2009). The heterogeneity of breast cancer presents a major challenge for basic, translational, and clinical research. This is most often addressed by studying a combination of model systems, including: tumor-derived cell culture models in vitro; xenograft models in immune-deficient mice; and genetically modified rodent models of mammary gland development and cancer. All of these approaches are required because no single model is a perfect reflection of the diversity of human breast cancer (Clarke 1996). Recently, expression profiling of breast cancer clinical specimens has sought to uncover gene signatures that can classify tumors into various subgroups. The most widely utilized of these classification systems was developed by Sorlie et al., in which tumors are grouped into several classes, including: Luminal A, Luminal B, ERBB2, basal-like, and normal-like (Sorlie, et al. 2001; Sorlie, et al. 2003). While signatures like these may ultimately prove useful in predicting outcome or response to specific therapies, they will require further refinement as additional subdivisions of these breast cancer subtypes (such as claudin low) are identified (Prat and Perou 2010).
Orphan nuclear receptors in rodent mammary gland biology and development
Understanding mammary gland development and tumorigenesis in rats and mice has been instrumental in advancing the study of human breast cancer (Allred and Medina 2008; Cardiff, et al. 2000; Marcotte and Muller 2008). However, little is known about the role(s) of specific orphan nuclear receptors in the developmental biology of the rodent mammary gland. This may be due to the fact that orphan nuclear receptor deletion in mice often has serious detrimental effects in multiple tissues that are manifested during embryogenesis or early in life, long before mammary glands develop beyond the rudimentary epithelial tree (Chen, et al. 1994; Collins, et al. 2004; Moral, et al. 2008; Pereira, et al. 1999). The creation of mammary gland-specific knockout or transgenic mice for key orphans will be required to explore their function in these tissues. Two orphan nuclear receptors have specifically been studied in the context of mouse mammary epithelial cell differentiation.
DAX-1
DAX-1 is an atypical member of the nuclear receptor super-family because it lacks a DBD and relies on heterodimerization with other transcription factors that can bind DNA. These binding partners include a second orphan nuclear receptor, SF1, and it has been shown that DAX-1 can repress SF1-mediated stimulation of genes regulating steroid synthesis (Wang, et al. 2001); DAX-1 can also repress the activity of estrogen receptors (Zhang, et al. 2000). DAX-1 is strongly upregulated in HC11 mouse mammary epithelial cells that have been induced to differentiate in vitro by withdrawing serum and growth factors prior to treatment with the lactogenic hormones insulin, dexamethasone, and prolactin (Faulds, et al. 2004). It is the withdrawal of epidermal growth factor (EGF), and specifically the resulting decrease in mitogen-activated protein kinase (MAPK) activity, which leads to DAX-1 induction (Helguero, et al. 2006). Once induced, DAX-1 is enriched in the nucleus and inhibits the transcriptional activity of estrogen receptors alpha (ERα) and beta (ERβ), as well as the proliferation of HC11 cells treated with ERα- and ERβ-specific agonists. Finally, the expression of DAX-1 is significantly induced in the mammary glands of pseudo-pregnant and lactating 3 month-old C57BL/6 female mice as compared to virgin animals. Together these data suggest that the lack of sensitivity of pregnant and lactating mammary glands to estrogen stimulation may be due to increased expression of the ER co-repressor DAX-1 (Helguero et al. 2006).
SHP
SHP, the other orphan nuclear receptor lacking a DBD, is also strongly induced during HC11 cell differentiation (Faulds et al. 2004). While SHP can serve as an ER co-repressor (Johansson, et al. 1999), its role in mammary epithelial cell differentiation remains to be elucidated.
Orphan nuclear receptors in breast cancer
While mRNA for all 25 orphan nuclear receptors is significantly over-represented in breast cancer vs. normal breast tissue in multiple gene expression microarray experiments in ONCOMINE and the Gene Expression Omnibus (Table 1) (Barrett, et al. 2009; Rhodes, et al. 2004), the significance of only a handful of these receptors has been studied in breast cancer.
COUP-TFs, EAR2
Chicken ovalbumin upstream promoter-transciption factor-1 (COUP-TFI, NR2F1), -2 (COUP-TFII, NR2F2), and V-erbA related protein 2 (EAR2, NR2F6) are Type III nuclear receptors that preferentially bind to direct DNA repeats as homodimers, although COUP-TFII and EAR2 can also heterodimerize (Avram, et al. 1999). COUP-TFs can interact with DNA responsive elements that are also shared with ERα and ERβ; the consensus estrogen response element (ERE) is a perfect inverted repeat (GGTCA-nnn-TGACC), but there are many genes with imperfect ERE’s or multiple ERE half-sites upstream of their transcriptional start sites. Both COUP-TFs have a high affinity for these ERE half-sites, while ER preferentially binds consensus EREs (Klinge, et al. 1997), and COUP-TFs can inhibit estradiol (E2)-induced, ER− mediated transcriptional activity by physically interacting with ER and disrupting ER/DNA binding. The COUP-TF/ER physical interaction is mediated by amino acid residues within the DBD and LBD of ER, and conformational changes in these domains induced by ER/DNA binding reduce COUP-TF/ER interactions (Klinge 1999). COUP-TFI has a higher affinity for ERE half-sites in the presence of estradiol and ER, and COUP-TFI inhibits E2-induced ERE activity. These data suggest that specific ligands and the specific sequence(s) of EREs influence the degree to which COUP-TFs activate gene transcription.
Nakshatri et al. showed that COUP-TFII mRNA expression is increased in ER-positive (ER+) MCF7, T47D, and ZR75-1 and in ER-negative (ER−) MDA-MB-231 breast cancer cells, but reduced in ER− MDA-MB-468 and SkBr3 breast cancer cells, as compared to the non-tumorigenic mammary epithelial cell line MCF10A (Nakshatri, et al. 2000). Overexpression of COUP-TFII inhibits cell proliferation in MDA-MB-435 cells by delaying progression through the G2/M cell cycle transition, which results from its induction of p21 and the subsequent inhibition of cdk2 activity. However, the more recent classification of MDA-MB-435 cells as melanoma (Rae, et al. 2004) raises the issue of whether this mechanism of growth inhibition is maintained in breast cancer cell lines. This is further called into question by Moré et al. who, in contrast, show that COUP-TFII mRNA and protein expression are highest in SkBr3 and lowest in MCF7 cells (Moré, et al. 2003). Furthermore, mitogenic signals such as epidermal growth factor (EGF) significantly induce COUP-TFII expression in MCF7 cells in a MAPK-dependent manner. Anti-proliferative agents such as oncostatin M (OSM) dramatically reduce COUP-TFII expression. However, neither EGF nor OSM have any effect on COUP-TFI expression or function.
DAX-1
DAX-1 is expressed in benign breast disease, carcinoma in situ, and invasive breast cancer (Conde, et al. 2004), and is significantly more highly expressed in invasive lobular carcinoma than in benign tissues. In this study, expression and nuclear localization of DAX-1 is positively correlated with lymph node-positive status, while cytoplasmic and nucear DAX-1 are both positively associated with androgen receptor (AR) expression.
ERRα
Estrogen-related receptor alpha (ERRα, ESRRA, NR3B1) is the most-studied orphan nuclear receptor in the context of breast cancer (Ariazi and Jordan 2006; Horard and Vanacker 2003; Stein Kunder and McDonnell July 2009; Stein and McDonnell 2006; Tremblay and Giguere 2007). Members of the NR3B or ERR family are classified as Type IV nuclear receptors that can bind to the ERE, which it shares with ERα and ERβ, and the SF1 response element (SF1RE), which it shares with SF1 and LRH-1; this latter element is also referred to as the estrogen-related response element (ERRE). The role of ERRα in energy homeostasis and metabolism is well documented (Giguere 2008), and more recently studies suggest that this orphan nuclear receptor is also involved in the regulation of bone mineral density (Delhon, et al. 2009). In breast cancer, ERRα is the most abundantly expressed member of the ERR family. ERRα can induce expression of the estrogen-regulated gene TFF1 (pS2) (Markićević, et al. 2008; Surowiak, et al. 2006), through its binding to an ERRE in the TFF1 promoter (Lu, et al. 2001). Moreover, ER binds to multiple steroid hormone response element half sites (MHREs) in the ERRα promoter, inducing ERRα expression, and this activity is enhanced by the addition of estrogen (Liu, et al. 2003).
ERRα may play distinct roles in ER+ vs. ER− breast cancer. ERRα’s activity at ERE sites is highly dependent on cell context, acting as a transcriptional activator in ER− cells but a repressor in ER+ cells (Kraus, et al. 2002), and ERRα expression is only positively associated with TFF1 expression in ER− breast tumors (Heck, et al. 2009). Post-translational modification of ERRα provides one potential explanation for these context-dependent activities. Phosphorylation of ERRα by the HER2/MAPK/AKT pathway is a critical determinant in the receptor’s ability to recruit co-activator proteins and activate transcription from ERE sites in ER+ cells (Ariazi, et al. 2007). In breast tumors, ERRα mRNA expression is negatively correlated with ER+ status but shows a strong positive correlation with HER2 expression (Ariazi, et al. 2002). Phosphorylation can subsequently lead to SUMOylation of ERRα, which also modulates its activity (Tremblay, et al. 2008; Vu, et al. 2007). Most recently, Wilson et al. have shown that ERRα acetylation by p300 coactivator associated factor (PCAF) inhibits ERRα transcriptional activity, while histone deacetylase 8 (HDAC8) and a homolog of sirtuin 1 (Sirt1) can reverse acetylation and increase receptor binding to DNA (Wilson, et al. 2010).
Discerning the relevant estrogen signaling pathways controlled by ERRα vs. ER is potentially challenging. However, two recent studies have convincingly shown that genes induced by ERRα in breast cancer cells have very little overlap with those transcribed in response to ER activation. First, using a variant of peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1α) engineered to selectively activate ERRs but not ER or other nuclear receptors, Stein et al. found that very few ERRα-regulated genes are related to estrogen signaling (Stein et al. 2008). Instead, ERRα appears primarily to control oxidative stress signaling and aerobic metabolism (particularly key components of electron transport and the tricarboxylic acid cycle) in ER+ MCF7 breast cancer cells; ERRα also induces the expression of vascular endothelial growth factor (VEGF) (Stein, et al. 2009).
More recently, Deblois et al. have uncovered why the gene targets of ERRα and ER in breast cancer cells are so distinct (Deblois, et al. 2009). Using comparative genome-wide chromatin immunoprecipitation, they determined that the binding sites utilized by ERRα and ER, in the vast majority of cases, do not overlap. They did identify a group of genes (n=212, 18% of the total) that are targets of both receptors. However, while the promoter regions of some of these dual targets have distinct EREs and ERREs in close proximity, the majority of these co-regulated genes are driven by an entirely novel DNA response element termed an ERRE/ERE, in which an ERRE (underlined) merges with a classical ERE (bold) (TCAAGGTCANNNTGACCT). These ERRE/ERE sites are only occupied by a single receptor (ERRα or ER) at a time.
In ER− MDA-MB-231 breast cancer cells, siRNA targeted to ERRα does not inhibit proliferation in vitro but does significantly reduce these cells’ migratory ability and delays xenograft tumor growth (Stein et al. 2008). In contrast, Chisamore et al. have shown that inhibiting ERRα signaling using a novel synthetic antagonist of the receptor (compound A; N-[(2Z)-3-(4,5-dihydro-1,3-thiazol-2-yl)-1,3-thiazolidin-2ylidene]-5Hdibenzo[a,d][7] annulen-5-amine), blocks cell proliferation in ER+ (MCF7, T47D) and ER− (BT-20, MDA-MB-231) breast cancer cell lines in vitro (Chisamore, et al. 2009b). Compound A has no effect on ERRα mRNA expression, but abrogates ERRα-mediated transcriptional activity in MCF7 cells and accelerates degradation of the receptor by the ubiquitin-proteasome pathway (Chisamore, et al. 2009a). A different ERRα antagonist, XCT790, also inhibits MCF7 and MDA-MB-231 proliferation in vitro and tumor formation in nude mice (Bianco, et al. 2009). Like compound A, XCT790 does not inhibit ERRα mRNA expression but instead enhances ERRα protein degradation; XCT790 also blocks ERRα’s interaction with its coactivator PGC-1α (Lanvin, et al. 2007). These findings suggest that pharmacological inhibition of ERRα may represent a promising therapeutic approach, particularly in ER− breast cancer.
ERRα appears to integrate well with known breast tumor molecular subtypes (Sorlie et al. 2001). ERRα target genes are specifically enriched in the ERBB2 or HER2 cluster (Deblois et al. 2009). >80 ERRα targets have prognostic value in several independent datasets, and a subset of these has independent prognostic value beyond ER and ERBB2 status in multivariate analyses. Other ERRα target genes identified by Deblois et al., including GRB7 and ERBB2, are already incorporated into the OncotypeDX diagnostic test (Paik, et al. 2004). Finally, gene expression analysis of distinct murine brain and bone metastases of a breast tumor line established from a patient with advanced breast cancer found that ERRα, its coactivators PGC-1α and β, and several known ERR target genes that control oxidative phosphorylation and the tricarboxylic acid cycle were selectively enriched in brain metastases.
ERRγ
Estrogen-related receptor gamma (ERRγ, ESRRG, NR3B3) shares 90% sequence identity with the DBD, and ~60% sequence identity with the LBD, of ERRα (Horard and Vanacker 2003). Like ERRα, ERRγ is implicated in metabolism and cancer (Ariazi and Jordan 2006; Giguere 2008; Riggins October 2009). However, the role of ERRγ in the etiology of breast cancer remains unclear.
One of the more puzzling aspects of the relationship between ERRα and ERRγ is that, at first glance, they appear to have opposite prognostic value in breast cancer. The different associations arise from a study in which 38 unselected breast tumors were compared to 9 different populations of mammary epithelial cells (Ariazi et al. 2002). Within the tumor samples, ERRα mRNA expression correlates significantly with ER− and progesterone receptor-negative (PR−) status, and with HER2+ status. ER−/PR−/HER2+ tumors are more aggressive than ER+/PR+ tumors, leading to the conclusion that ERRα is a marker of poor clinical outcome. The ability of ERRα (Suzuki, et al. 2004) and its target genes (Deblois et al. 2009) to function independently as poor prognostic factors have been subsequently confirmed. In direct contrast, breast tumors expressing ERRγ are significantly more likely to be ER+ and PR+ (Ariazi et al. 2002). ER+/PR+ status is, overall, a marker of good outcome. However, unlike ERRα, ERRγ has not been shown to have independent prognostic value, positive or negative, in any study. ERRγ’s association with good outcome remains linked to its over-representation in ER+/PR+ tumors, not all of which have a good prognosis (see section on endocrine therapy below).
LRH-1
Liver receptor homolog-1 (LRH-1, NR5A2) is a Type IV nuclear receptor that binds as a monomer to the SF1RE DNA response element (Fayard, et al. 2004). L R H-1 is an important regulator of bile acid homeostasis, reverse cholesterol transport from peripheral tissues to the liver, and steroidogenesis; LRH-1 can bind C16 and C18 phospholipids such as phosphatidylglycerol and phosphatidylethanolamine (Ortlund, et al. 2005). LRH-1 mRNA and protein expression is significantly elevated in breast tumors and their surrounding adipose tissue as compared to normal breast (Miki, et al. 2006; Zhou, et al. 2005). LRH-1 is positively associated with ER+, PR+, and androgen receptor-positive (AR+) status, but negatively correlated with increased tumor stage, grade, and HER2+ status. Interestingly, LRH-1 positive status is associated with improved survival in women with PR+ breast cancer, but worse survival in women with PR− breast cancer. The establishment of LRH-1 as an ER target gene (Annicotte, et al. 2005) has additional implications for endocrine therapy responsiveness in ER+ breast cancer.
NGFI-B, NURR1, NOR1
Nerve growth factor induced-B (NGFI-B, NR4A1; also known as Nur77 or TR3), nuclear receptor related 1 (NURR1, NR4A2), and neuron-derived orphan receptor 1 (NOR1, NR4A3) make up a group of closely related Type IV nuclear receptors. A key feature of all three receptors is their apparent lack of a true ligand-binding pocket (Wang, et al. 2003), although various diindolylmethanes (DIMs) have been shown to have agonist activity toward NGFI-B (Dae Cho, et al. 2010). NGFI-B plays an interesting dual role in cell fate, functioning as a pro-survival factor when found in the nucleus but strongly inducing apoptosis when it translocates to the mitochondria, binds Bcl-2, and promotes cytochrome c release (Ferri and Kroemer 2001; Moll, et al. 2006). MCF7 cells express NGFI-B, and DIM treatment of this cell line inhibits cell proliferation and induces apoptosis (Chintharlapalli, et al. 2005); NOR1 is also strongly induced early in the apoptotic process of MCF7 cells (Ohkubo, et al. 2000).
Rev-Erbα
V-erbA-related protein 1 (Rev-Erbα, NR1D1; also known as EAR1) is a Type IV nuclear receptor that regulates metabolism and circadian rhythm (Duez and Staels 2009). Despite its orphan status Rev-Erbα and the related Rev-Erbβ (NR1D2) are known to bind heme (Burris 2008). Rev-Erbα is localized to chromosome 17q21, a region that is frequently amplified in breast cancer; this region also harbors ERBB2 and Rev-Erbα is often co-amplified and co-expressed with ERBB2 in breast tumors (Chin, et al. 2006; Dressman, et al. 2003). More recently, Davis et al. have reported that amplification of Rev-Erbα together with two other genes (SMARCE1 and BIRC5) functions as an accurate prognostic index independent of patient age or tumor stage in ER−/PR− breast tumors (Davis, et al. 2007).
RORα
Retinoic acid-related orphan receptor alpha (RORα, NR1F1) is a monomeric Type IV nuclear receptor that can, under some circumstances, bind to cholesterol or cholesterol sulfate (Kallen, et al. 2004). RORα is better known for its ability to regulate cerebellar development, the immune response, circadian rhythm, and resistance to atherosclerosis (Jetten 2004). However, several splice variants of this receptor are expressed in ER+ (MCF-7 and T47D) and ER− (BT-20, MDA-MB-231) breast cancer cells (Dai, et al. 2001). RORα is also localized to a well-known site of genomic instability (15q22.2), its expression is reduced in breast and other hormonally regulated tumors, and overexpression of RORα in MCF12F cells significantly inhibits their proliferation (Zhu, et al. 2006).
TR2 and TR4
Human testicular receptor-2 (TR2, NR2C1) and -4 (TR4, NR2C2) are classified as Type III nuclear receptors, although they do not exclusively form homodimers (Lee, et al. 2002). Both TRs are generally considered to be transcriptional repressors; TR2 is implicated in pre-adipocyte proliferation (Gupta, et al. 2007) and erythroid cell differentiation (Tanabe, et al. 2007), while TR4 plays an important role in gluconeogenesis (Liu, et al. 2007) and promyelocyte proliferation (Koritschoner, et al. 2001). Both TR4 (Shyr, et al. 2002) and TR2 (Hu, et al. 2002) can repress ER transcriptional activity, and radiation-stimulated p53 induction in MCF-7 cells downregulates TR2 expression (Lin and Chang 1996).
Orphan nuclear receptors and response to chemotherapy
Several cytotoxic chemotherapies play a key role in the clinical management of locally advanced and/or metastatic breast cancer (Shajahan, et al. 2008). Anthracyclines (e.g. doxorubicin) induce DNA damage by intercalating into DNA while also inhibiting the activity of topoisomerase II and inducing reactive oxygen species (ROS). Alkylating agents (e.g. cisplatin) also induce DNA damage, but do so by forming DNA adducts that block DNA synthesis. In contrast, antimetabolites (e.g. 5-fluorouracil, 5-FU) inhibit the enzyme thymidylate synthase, which normally produces thymidine 5′-monophosphate (dTMP) that is used in the synthesis of DNA. 5-FU can also affect mRNA translation or inhibit rRNA processing (Burger, et al. 2010; Kudo, et al. 2010). Finally, two sub-classes of anti-microtubule drugs are key inhibitors of tumor cell growth and inducers of tumor cell death by apoptosis and other mechanisms; the taxanes (docetaxel, paclitaxel) stabilize, while the Vinca alkaloids (vincristine, vinblastine) promote the destruction of the cellular microtubule network.
ERRα
There is some evidence that supports a role for ERRα in response to 5-FU-based chemotherapy. Uridine phosphorylase (UPase) is one of the essential enzymes required for metabolizing 5-FU (Maring, et al. 2005). UPase mRNA expression is positively regulated by ERRα and the nuclear receptor coactivator PGC-1α (Kong, et al. 2009b). Specifically, PGC-1α/ERRα complexes bind to the UPase promoter, and the ability of PGC-1α to induce UPase transcription in MCF7 cells is abrogated by the ERRα-specific small molecule inhibitor XCT790. Moreover, PGC-1α overexpression sensitizes SkBr3 breast cancer cells to cell death induced by 5-FU, and this sensitization is also reversed by XCT790. These data suggest that the co-expression of PGC-1α and ERRα in breast tumors may indicate enhanced sensitivity to 5-FU-containing treatment regimens.
In contrast, this same group has shown that in multi-drug resistant HepG2 cells overexpressing the MDR1 drug transporter (R-HepG2), XCT790 alone can inhibit growth, alter mitochondrial membrane potential, and stimulate the production of ROS that ultimately induce caspase-dependent apoptosis. (Wu, et al. 2009). R-HepG2 cells are resistant to both doxorubicin and paclitaxel; however, while XCT790 can synergistically restore paclitaxel sensitivity, it has neither an additive nor synergistic effect on doxorubicin response. The authors propose that XCT790’s ability to induce ROS is better complemented by paclitaxel’s stabilization of the microtubule network (thus leading to synergistic inhibition of cell growth). Since doxorubicin already can induce ROS, additive or synergistic interactions are not seen because XCT790 and doxorubicin affect similar downstream pathways. It is not immediately clear whether XCT790 might have similar effects on doxorubicin and/or paclitaxel response in multi-drug resistant breast cancer cells.
NURR1
One of the NR4 orphan nuclear receptors, NURR1, has been implicated in resistance to to doxorubicin. Overexpression of NURR1 significantly decreases expression of a pro-apoptotic member of the BCL2 gene family (BAX) while protecting cells in culture from apoptosis following doxorubicin treatment (Zhang, et al. 2009). The DBD of NURR1 was subsequently found to interact with the C-terminus of the tumor suppressor p53. NURR1/p53 interaction prevents oligomerization of p53, attenuating the induction of p53 target genes (which include BAX). While these studies were not performed in breast cancer cells, BAX is important in doxorubicin sensitivity in MCF7 breast cancer cells (Kong, et al. 2009a) and human breast tumors (Chintamani, et al. 2004; Parton, et al. 2002), suggesting that NURR1 could play an important role in this context.
Orphan nuclear receptors and response to endocrine or hormonal therapy
Estrogen receptor alpha-positive (ER+) breast tumors comprise ~70% of annually diagnosed breast cancer cases (Jemal et al. 2009). Adjuvant or neoadjuvant endocrine therapy is among the least toxic, best tolerated, and most effective therapies available to patients with ER+ breast cancer. There are currently three major classes of endocrine therapy used in the clinic: selective estrogen receptor modulators (SERMs), selective estrogen receptor downregulators (SERDs), and aromatase inhibitors (AIs) (Crago, et al. 2010).
The prototypical SERM is the triphenylethylene Tamoxifen, which competes with estrogen (17β-estradiol, E2) for binding to ER and in breast tissue is most often an antagonist; however in bone, brain, and endometrial tissues, Tamoxifen can act as a partial agonist (Clarke, et al. 2001). In contrast, Fulvestrant (ICI 182,780, or Faslodex) functions as a SERD, a pure antagonist of ER in all tissues that competes for E2 binding and accelerates degradation of the receptor via the ubiquitin-proteasome pathway. Steroidal and non-steroidal aromatase inhibitors (Exemestane, Letrozole, Anastrazole) are inhibitors of the cytochrome P450 family member CYP19A1 (aromatase) that catalyzes the conversion of testosterone to estradiol, therefore depriving ER+ breast tumors of growth-stimulatory estrogen signaling (Santen, et al. 2009). Tamoxifen is effective in the treatment of pre-menopausal and post-menopausal breast cancer patients. However, aromatase inhibitors are only indicated for use in post-menopausal women with breast cancer, where the major site of estrogen synthesis has shifted from the ovaries to the adrenal gland and adipose tissue.
As successful as each of these classes of endocrine therapies has been in the treatment of ER+ breast cancer, resistance to Tamoxifen, Fulvestrant, and/or aromatase inhibitors is a significant and widespread clinical problem (Buzdar 2008; Macedo, et al. 2009; Musgrove and Sutherland 2009; Riggins, et al. 2005; Riggins, et al. 2007). The proposed mechanisms of endocrine resistance are varied and can include ER downregulation, silencing, or mutation, ER post-translational modifications, changes in the expression profile(s) of nuclear receptor co-regulators, and altered expression of key networks of growth factor signaling and/or apoptosis. Importantly, several orphan nuclear receptors have demonstrated potential to play important direct or indirect roles in endocrine therapy response and resistance.
COUP-TFs
Like the conflicting effects of the COUP-TFs on cell proliferation (Moré et al. 2003; Nakshatri et al. 2000), results from studies of the role(s) of these receptors in Tamoxifen resistance are also contradictory. A significant decrease in COUP-TFII protein expression is observed in three different MCF7-derived resistance models. Inhibition of COUP-TFII by siRNA in sensitive MCF7 cells prevents Tamoxifen-induced growth inhibition (Riggs, et al. 2006). However, COUP transcription factors have been shown to enhance ER transcriptional activity by forming tight homodimer complexes (Métivier, et al. 2002). These physical complexes increase the affinity of ER for MAPK, resulting in increased ER phosphorylation at Serine 118 and enhanced ER transcriptional activity. Thus, under certain circumstances, overexpression of COUP-TFs might negate the inhibitory effects of Tamoxifen on ER. Moreover, overexpression of COUP-TFI has been shown to stimulate MCF7 cell growth and migration, via an ER− dependent pathway, by selectively upregulating genes involved in cell proliferation; these effects are seen in the presence and absence of estradiol (Le Dily, et al. 2008). Discerning the true role of COUP-TFI and COUP-TFII in ER signaling and endocrine resistance will require further study to clarify their seemingly conflicting activities.
DAX-1
The ability of DAX-1 to function as a co-repressor for ER (Zhang et al. 2000) has potentially significant implications for responsiveness to antiestrogens. However, more is known about the role of DAX-1 in estrogen synthesis through its negative regulation of aromatase mRNA expression in ovarian granulosa cells (Gurates, et al. 2003) and endometrial cells (Gurates, et al. 2002); its family member SHP has also been shown to inhibit aromatase expression (Kovacic, et al. 2004). In contrast, deletion of DAX-1 in male mice significantly upregulates aromatase expression (Wang et al. 2001). To our knowledge there have been no studies of DAX-1 in the peripheral and breast adipose tissues of women with breast cancer. Given that these sites are the major source of estrogen synthesis in post-menopausal women (Macciò, et al. 2009), and that DAX-1 expression is significantly higher in pre-adipocytes than in mature adipose tissue (Kim, et al. 2008), studies of DAX-1 in the context of aromatase inhibitor-responsive and – resistant breast cancer could be informative.
ERRα
Although the expression of ERRα in breast tumors is inversely associated with ER+ status (Ariazi et al. 2002), there are a number of ways in which this orphan nuclear receptor might affect endocrine therapy responsiveness. First, a small subset of ER+ breast cancers also expresses HER2. HER2-dependent phosphorylation allows ERRα to activate transcription from EREs (Ariazi et al. 2007), potentially bypassing the inhibitory effects of SERMs and SERDs. Given that addition of the HER2 inhibitors trastuzumab or gefitinib to endocrine therapy appears to significantly improve disease-free survival in women with ER+/HER2+ breast cancer (Buzdar 2009; Johnston 2009), the contribution of ERRα to endocrine resistance, when it is expressed in ER+/HER2+ tumors, should be explored further. In addition, at least two genes identified as ERRα targets with prognostic value in breast cancer (Deblois et al. 2009) are known effectors of Tamoxifen resistance. GRB7 is a breast cancer antiestrogen resistance (BCAR) gene that can confer estrogen independence and antiestrogen resistance to breast cancer cell lines (van Agthoven, et al. 2009b), and has independent prognostic value in Tamoxifen-resistant breast cancer patients (van Agthoven, et al. 2009a). CCNE1 (cyclin E1), particularly the low molecular weight forms of this cell cycle regulator, has also been implicated in Tamoxifen resistance (Akli, et al. 2004; Dhillon and Mudryj 2002).
Alternatively, the ERRα-specific inhibitor XCT790 has been shown to enhance SERD activity by accelerating Fulvestrant-dependent degradation of ER in MCF7 breast cancer cells (Lanvin et al. 2007). The mechanism by which this occurs is not entirely clear; the extent of ERRα degradation in response to XCT790 is greater in ER− MDA-MB-231 cells than ER+ MCF7 cells, but when ER expression is restored in MDA-MB-231 cells, XCT790’s ability to induce ERRα degradation is unchanged. Furthermore, transfection of MCF7 cells (which express both ER and ERRα) with siRNA specific for each receptor does not lead to enhanced sensitivity of the other receptor to its antagonist. The authors ultimately proposed that ER and ERRα heterodimerize in breast cancer cells in a way that confers protection to each receptor from protein degradation induced by their specific antagonists, and suggest that XCT790 may therefore be useful in combination with Fulvestrant to improve endocrine therapy response in the clinic (Lanvin et al. 2007). However, heterodimerization of ER and ERRα has not been convincingly demonstrated, so this mode of action currently seems somewhat unlikely. Finally, in breast cancer cells ERRα forms a transcriptional complex on aromatase promoters I.3 and II with the nuclear receptor coactivators proline, glutamate, and leucine-rich protein 1 (PELP1) and proline-rich nuclear receptor coactivator 2 (PNRC2), thereby inducing aromatase expression and promoting localized estrogen synthesis (Rajhans, et al. 2008).
ERRγ
As discussed above, the expression of ERRγ in breast tumors is associated with ER+ and PR+ status (Ariazi et al. 2002). This has led to the notion that ERRγ is a marker of good prognosis and response to endocrine therapy. However, 30% of women have ER+/PR+ breast tumors that are intrinsically (de novo) resistant to Tamoxifen, and when initially responsive patients present with recurrent, endocrine-resistant disease, in most cases these recurrent/resistant tumors retain ER expression (Riggins et al. 2005). Therefore, genes that are primarily expressed in ER+ breast tumors are not necessarily good prognostic factors. A clear example is X-box binding protein 1 (XBP1), a key mediator of the unfolded protein response (Feldman, et al. 2005). XBP1 is coexpressed with ER in breast tumors (Lacroix and Leclercq 2004; Tozlu, et al. 2006; Wilson and Giguère 2008). However, XBP1 is an ER coactivator that induces ligand-independent activation of the receptor (Ding, et al. 2003a; Ding, et al. 2003b; Fang, et al. 2004), XBP1 upregulation is evident in some antiestrogen resistant cells (Gu, et al. 2002), and its ectopic overexpression leads to Tamoxifen resistance and estrogen independence in multiple ER+ breast cancer cell lines (Gomez, et al. 2007). The presence of spliced or active XBP1 in breast tumors is also significantly associated with poor clinical response to Tamoxifen (Davies, et al. 2008).
We have shown that ERRγ can play an important functional role in the acquisition of Tamoxifen resistance by breast cancer cell lines derived from invasive lobular carcinoma (Riggins, et al. 2008). The Tamoxifen-resistant variant of the SUM44 breast cancer cell line (Ethier, et al. 1993), LCCTam, expresses significantly more ERRγ mRNA and protein, and knockdown of this orphan nuclear receptor restores Tamoxifen-mediated growth inhibition. In contrast, ectopic overexpression of ERRγ cDNA independently induces Tamoxifen resistance in SUM44 cells and another model of ILC, MDA-MB-134 VI (Reis-Filho, et al. 2006). The mechanism by which ERRγ mediates Tamoxifen resistance is under active investigation. As discussed above, ERRγ can induce gene transcription from ERE and ERRE sites, and one of the more active metabolites of Tamoxifen (4-hydroxytamoxifen, 4HT) inactivates ERRγ with respect to transcription from both of these response elements (Greschik, et al. 2004). However, studies in non-breast cancer cell lines show that in the presence of 4HT, ERRγ can potently activate transcription from activator protein-1 (AP1) sites (Huppunen, et al. 2004). We subsequently demonstrated that the resistant LCCTam cells have significantly higher AP1 activity in the presence of Tamoxifen, and that a peptide inhibitor of AP1 effectively restores Tamoxifen responsiveness (Riggins et al. 2008). Because the full compliment of endogenous ERRγ/AP1 target genes are unknown, current studies are focused on identifying these targets in order to understand how this orphan nuclear receptor contributes to Tamoxifen resistance.
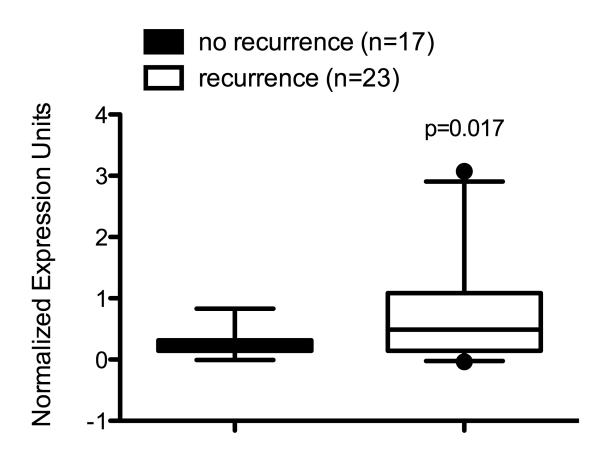
Importantly, ERRγ’s function in Tamoxifen resistance may not be restricted to ILC. We have recently found that ERRγ mRNA expression is significantly increased in the MCF7/RR model (Butler and Fontana 1992) of Tamoxifen resistance (O.Z. Maniya, M.M. Mazzotta, and R.B. Riggins, unpublished observations). This is supported by in silico re-analysis of gene expression microarray data from a study of 60 women diagnosed with ER+ breast cancer who were treated only with Tamoxifen (Ma, et al. 2004). Pre-treatment mRNA levels of ERRγ are significantly elevated in tumors from breast cancer patients who recurred within 5 years of Tamoxifen therapy compared with those from breast cancer patients who did not recur (Fig. 2), and ~80% of this patient population had invasive ductal carcinoma (IDC). Further studies are needed to better understand how ERRγ functions in the full spectrum of Tamoxifen-resistant, ER+ breast cancer.
Fig. 2. Elevated ERRγ expression in Tamoxifen-resistant breast tumors.
Re-analysis of gene expression microarray data from Ma et al. shows that pre-treatment ERRγ expression is significantly increased in the tumors of breast cancer patients who recurred within 5 years of Tamoxifen therapy (p=0.017, Mann-Whitney rank sum test).
LRH-1
LRH-1 strongly induces aromatase expression from promoter II in pre-adipocytes (Clyne, et al. 2002) and breast adipose tissue (Clyne, et al. 2004), and its ability to do so can be inhibited by SHP (Kovacic et al. 2004). The presence of higher levels of LRH-1 mRNA in breast tumors and their surrounding adipose tissue, as compared to normal breast, mirrors the expression of aromatase in these tissues (Zhou et al. 2005). More recent studies show that cooperation between LRH-1 and GATA transcription factors (Bouchard, et al. 2005) or PGC-1α (Safi, et al. 2005) strongly induces aromatase expression.
As stated above, LRH-1 is an ER target gene (Annicotte et al. 2005), and tumoral LRH-1 expression is associated with improved survival in women with PR+ breast cancer but worse survival in women with PR− breast cancer (Miki et al. 2006). Together with its demonstrated ability to induce transcription of aromatase, these data suggest that LRH-1 may be a marker of intact aromatase/estrogen/ER signaling that indicates improved sensitivity to endocrine therapies such as Tamoxifen or an aromatase inhibitor. Two clinical studies provide some support for this. Using specimens from the P025 trial in which Letrozole was compared to Tamoxifen, women with PR+ breast tumors that also expressed aromatase trended towards having improved time to progression when treated with Letrozole (Lykkesfeldt, et al. 2009). In the P024 trial that also compared Letrozole to Tamoxifen, increased aromatase expression at baseline was significantly associated with improved relapse-free survival in multivariate analyses (Ellis, et al. 2009).
RORα
RORα has also been shown to induce aromatase expression by enhancing its transcription from promoter I.4, and can significantly increase aromatase activity in ER+ T47D and MCF7 breast cancer cells (Odawara, et al. 2009). This study also showed that RORα and aromatase expression are positively correlated in breast cancer clinical specimens. RORα deletion in mice suggests an even broader role for this gene in regulating multiple components of steroidogenesis other than aromatase, including several sulfotransferases and hydroxysteroid dehydrogenases (Kang, et al. 2007). However, a different study reports that RORα expression is reduced in breast cancer compared with normal tissue (Zhu et al. 2006), so it is still unclear how this orphan nuclear receptor functions in breast tumors or modulates endocrine therapy response.
Summary and Perspectives
The field of breast cancer research has much to gain from the continued study of orphan nuclear receptors. Many of these unique transcription factors have the potential to transform our understanding of how tumorigenesis is integrated with fundamental physiological processes such as metabolism, regulation of circadian rhythm, and obesity (Froy 2010; Sahar and Sassone-Corsi 2009). We also have a great deal to learn about how orphan nuclear receptors function in normal mammary tissue in mice and humans. Exploring these unresolved areas of orphan nuclear receptor biology could ultimately lead to discoveries that will improve our ability to diagnose, treat, and prevent breast cancer.
Acknowledgements
We sincerely thank all members of the Clarke and Riggins labs, as well as the broader Lombardi Comprehensive Cancer Center community, for their insight and helpful comments.
Funding: This work was generously supported by grants from Susan G. Komen for the Cure (KG090187 to RBR; KG090245 to RC), the Department of Defense Breast Cancer Research Program (BC073977 to RC), and the Public Health Service (R03-CA142009 to RBR; R01-CA131465 and R21-CA139246 to RC). This project has also been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E to RC. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Declaration of interest: The authors declare that they have no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contribution statement: RBR and RC developed the original idea and outline, while all authors made significant contributions to the writing and editing of this manuscript.
References
- Akli S, Zheng P, Multani A, Wingate H, Pathak S, Zhang N, Tucker S, Chang S, Keyomarsi K. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 2004;64:3198–3208. doi: 10.1158/0008-5472.can-03-3672. [DOI] [PubMed] [Google Scholar]
- Allred D, Medina D. The relevance of mouse models to understanding the development and progression of human breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:279–288. doi: 10.1007/s10911-008-9093-5. [DOI] [PubMed] [Google Scholar]
- Annicotte J, Chavey C, Servant N, Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F, Maudelonde T, et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–6518. [PubMed] [Google Scholar]
- Ariazi EA, Jordan VC. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr.Top.Med.Chem. 2006;6:203–215. doi: 10.2174/1568026610606030203. [DOI] [PubMed] [Google Scholar]
- Ariazi EA, Kraus RJ, Farrell ML, Jordan VC, Mertz JE. Estrogen-Related Receptor {alpha}1 Transcriptional Activities Are Regulated in Part via the ErbB2/HER2 Signaling Pathway. Mol.Cancer Res. 2007;5:71–85. doi: 10.1158/1541-7786.MCR-06-0227. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Meindl N, Firla B, Brandes R, Steinhilber D. Intracellular localization of RORalpha is isoform and cell line-dependent. Biochim Biophys Acta. 2006;1763:805–814. doi: 10.1016/j.bbamcr.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Avram D, Ishmael J, Nevrivy D, Peterson V, Lee S, Dowell P, Leid M. Heterodimeric interactions between chicken ovalbumin upstream promoter-transcription factor family members ARP1 and ear2. J Biol Chem. 1999;274:14331–14336. doi: 10.1074/jbc.274.20.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup D, Wilhite S, Ledoux P, Rudnev D, Evangelista C, Kim I, Soboleva A, Tomashevsky M, Marshall K, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, Perlmann T, Renaud J, Schwabe J, Sladek F, et al. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev. 2006;58:798–836. doi: 10.1124/pr.58.4.10. [DOI] [PubMed] [Google Scholar]
- Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14:369–376. doi: 10.1016/j.tcb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Bianco S, Lanvin O, Tribollet V, Macari C, North S, Vanacker J. Modulating estrogen receptor-related receptor-alpha activity inhibits cell proliferation. J Biol Chem. 2009;284:23286–23292. doi: 10.1074/jbc.M109.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Taniguchi H, Viger R. Protein kinase A-dependent synergism between GATA factors and the nuclear receptor, liver receptor homolog-1, regulates human aromatase (CYP19) PII promoter activity in breast cancer cells. Endocrinology. 2005;146:4905–4916. doi: 10.1210/en.2005-0187. [DOI] [PubMed] [Google Scholar]
- Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Hölzel M, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010 doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris T. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–1520. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris T, Guo W, McCabe E. The gene responsible for adrenal hypoplasia congenita, DAX-1, encodes a nuclear hormone receptor that defines a new class within the superfamily. Recent Prog Horm Res. 1996;51:241–259. discussion 259-260. [PubMed] [Google Scholar]
- Butler WB, Fontana JA. Responses to retinoic acid of tamoxifen-sensitive and - resistant sublines of human breast cancer cell line MCF-7. Cancer Research. 1992;52:6164–6167. [PubMed] [Google Scholar]
- Buzdar A. Fulvestrant--a novel estrogen receptor antagonist for the treatment of advanced breast cancer. Drugs Today (Barc) 2008;44:679–692. doi: 10.1358/dot.2008.44.9.1256862. [DOI] [PubMed] [Google Scholar]
- Buzdar A. Role of biologic therapy and chemotherapy in hormone receptor- and HER2-positive breast cancer. Ann Oncol. 2009;20:993–999. doi: 10.1093/annonc/mdn739. [DOI] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wakefield LM, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting [see comments] Oncogene. 2000;19:968–988. doi: 10.1038/sj.onc.1203277. [DOI] [PubMed] [Google Scholar]
- Carrigan A, Walther R, Salem H, Wu D, Atlas E, Lefebvre Y, Haché R. An active nuclear retention signal in the glucocorticoid receptor functions as a strong inducer of transcriptional activation. J Biol Chem. 2007;282:10963–10971. doi: 10.1074/jbc.M602931200. [DOI] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris T, Rastinejad F. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature. 2008:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Bradley M, Beaven S, Tontonoz P. Phosphorylation of the liver X receptors. FEBS Lett. 2006;580:4835–4841. doi: 10.1016/j.febslet.2006.07.074. [DOI] [PubMed] [Google Scholar]
- Chen W, Manova K, Weinstein D, Duncan S, Plump A, Prezioso V, Bachvarova R, Darnell JJ. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Cheng J, Zhang C, Shapiro DJ. A functional serine 118 phosphorylation site in estrogen receptor-alpha is required for down-regulation of gene expression by 17beta-estradiol and 4-hydroxytamoxifen. Endocrinology. 2007;148:4634–4641. doi: 10.1210/en.2007-0148. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Chintamani, Singhal V, Singh J, Lyall A, Saxena S, Bansal A. Is drug-induced toxicity a good predictor of response to neo-adjuvant chemotherapy in patients with breast cancer?--a prospective clinical study. BMC Cancer. 2004;4:48. doi: 10.1186/1471-2407-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintharlapalli S, Burghardt R, Papineni S, Ramaiah S, Yoon K, Safe S. Activation of Nur77 by selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J Biol Chem. 2005;280:24903–24914. doi: 10.1074/jbc.M500107200. [DOI] [PubMed] [Google Scholar]
- Chisamore M, Cunningham M, Flores O, Wilkinson H, Chen J. Characterization of a novel small molecule subtype specific estrogen-related receptor alpha antagonist in MCF-7 breast cancer cells. PLoS One. 2009a;4:e5624. doi: 10.1371/journal.pone.0005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisamore M, Wilkinson H, Flores O, Chen J. Estrogen-related receptor-alpha antagonist inhibits both estrogen receptor-positive and estrogen receptor-negative breast tumor growth in mouse xenografts. Mol Cancer Ther. 2009b;8:672–681. doi: 10.1158/1535-7163.MCT-08-1028. [DOI] [PubMed] [Google Scholar]
- Claessens F, Gewirth DT. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- Claessens F, Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J Steroid Biochem Mol Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- Clarke R. Animal models of breast cancer: their diversity and role in biomedical research. Breast Cancer Research and Treatment. 1996;39:1–6. doi: 10.1007/BF01806073. [DOI] [PubMed] [Google Scholar]
- Clarke R, Leonessa F, Welch JN, Skaar TC. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacological Reviews. 2001;53:25–71. [PubMed] [Google Scholar]
- Clyne C, Kovacic A, Speed C, Zhou J, Pezzi V, Simpson E. Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol Cell Endocrinol. 2004;215:39–44. doi: 10.1016/j.mce.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Clyne C, Speed C, Zhou J, Simpson E. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem. 2002;277:20591–20597. doi: 10.1074/jbc.M201117200. [DOI] [PubMed] [Google Scholar]
- Collins L, Lee Y, Heinlein C, Liu N, Chen Y, Shyr C, Meshul C, Uno H, Platt K, Chang C. Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc Natl Acad Sci U S A. 2004;101:15058–15063. doi: 10.1073/pnas.0405700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde I, Alfaro J, Fraile B, Ruíz A, Paniagua R, Arenas M. DAX-1 expression in human breast cancer: comparison with estrogen receptors ER-alpha, ER-beta and androgen receptor status. Breast Cancer Res. 2004;6:R140–148. doi: 10.1186/bcr766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crago A, Azu M, Tierney S, Morrow M. Randomized clinical trials in breast cancer. Surg Oncol Clin N Am. 2010;19:33–58. doi: 10.1016/j.soc.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Dae Cho S, Lee S, Chintharlapalli S, Abdelrahim M, Khan S, Yoon K, Kamat A, Safe S. Activation of Nerve Growth Factor-Induced B{alpha} by Methylene-Substituted Diindolylmethanes in Bladder Cancer Cells Induces Apoptosis and Inhibits Tumor Growth. Mol Pharmacol. 2010;77:396–404. doi: 10.1124/mol.109.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Ram P, Yuan L, Spriggs L, Hill S. Transcriptional repression of RORalpha activity in human breast cancer cells by melatonin. Mol Cell Endocrinol. 2001;176:111–120. doi: 10.1016/s0303-7207(01)00449-x. [DOI] [PubMed] [Google Scholar]
- Davies M, Barraclough D, Stewart C, Joyce K, Eccles R, Barraclough R, Rudland P, Sibson D. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123:85–88. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]
- Davis L, Harris C, Tang L, Doherty P, Hraber P, Sakai Y, Bocklage T, Doeden K, Hall B, Alsobrook J, et al. Amplification patterns of three genomic regions predict distant recurrence in breast carcinoma. J Mol Diagn. 2007;9:327–336. doi: 10.2353/jmoldx.2007.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblois G, Hall J, Perry M, Laganière J, Ghahremani M, Park M, Hallett M, Giguère V. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- Delhon I, Gutzwiller S, Morvan F, Rangwala S, Wyder L, Evans G, Studer A, Kneissel M, Fournier B. Absence of estrogen receptor-related-alpha increases osteoblastic differentiation and cancellous bone mineral density. Endocrinology. 2009;150:4463–4472. doi: 10.1210/en.2009-0121. [DOI] [PubMed] [Google Scholar]
- Dhillon N, Mudryj M. Ectopic expression of cyclin E in estrogen responsive cells abrogates antiestrogen mediated growth arrest. Oncogene. 2002;21:4626–4634. doi: 10.1038/sj.onc.1205576. [DOI] [PubMed] [Google Scholar]
- Ding L, Yan J, Zhu J, Zhong H, Lu Q, Wang Z, Huang C, Ye Q. Ligand-independent activation of estrogen receptor alpha by XBP-1. Nucleic Acids Res. 2003a;31:5266–5274. doi: 10.1093/nar/gkg731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding LH, Ye QN, Zhu JH, Yan JH, Zhong HJ, Wang ZH, Huang CF. XBP-1 enhances the transcriptional activity of estrogen receptor alpha. Sheng Wu Hua Xue.Yu Sheng Wu Wu Li Xue.Bao.(Shanghai) 2003b;35:829–833. [PubMed] [Google Scholar]
- Dressman MA, Baras A, Malinowski R, Alvis LB, Kwon I, Walz TM, Polymeropoulos MH. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer Res. 2003;63:2194–2199. [PubMed] [Google Scholar]
- Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–1980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Ellis M, Miller W, Tao Y, Evans D, Chaudri Ross H, Miki Y, Suzuki T, Sasano H. Aromatase expression and outcomes in the P024 neoadjuvant endocrine therapy trial. Breast Cancer Res Treat. 2009;116:371–378. doi: 10.1007/s10549-008-0161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL. Differential isolation of normal liminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Research. 1993;53:627–635. [PubMed] [Google Scholar]
- Fang Y, Yan J, Ding L, Liu Y, Zhu J, Huang C, Zhao H, Lu Q, Zhang X, Yang X, et al. XBP-1 increases ERalpha transcriptional activity through regulation of large-scale chromatin unfolding. Biochem Biophys Res Commun. 2004;323:269–274. doi: 10.1016/j.bbrc.2004.08.100. [DOI] [PubMed] [Google Scholar]
- Faulds M, Olsen H, Helguero L, Gustafsson J, Haldosén L. Estrogen receptor functional activity changes during differentiation of mammary epithelial cells. Mol Endocrinol. 2004;18:412–421. doi: 10.1210/me.2003-0290. [DOI] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Chauhan V, Koong AC. The unfolded protein response: a novel component of the hypoxic stress response in tumors. Molecular Cancer Research. 2005;3:597–605. doi: 10.1158/1541-7786.MCR-05-0221. [DOI] [PubMed] [Google Scholar]
- Ferri K, Kroemer G. Mitochondria--the suicide organelles. Bioessays. 2001;23:111–115. doi: 10.1002/1521-1878(200102)23:2<111::AID-BIES1016>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Flaig R, Greschik H, Peluso-Iltis C, Moras D. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem. 2005;280:19250–19258. doi: 10.1074/jbc.M413175200. [DOI] [PubMed] [Google Scholar]
- Francis G, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- Freedman LP, Luisi BF, Korszun ZR, Basavappa R, Sigler PB, Yamamoto KR. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- Froy O. Metabolism and circadian rhythms--implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- Gaillard S, Dwyer M, McDonnell D. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol Endocrinol. 2007;21:62–76. doi: 10.1210/me.2006-0179. [DOI] [PubMed] [Google Scholar]
- Gaillard S, Grasfeder L, Haeffele C, Lobenhofer E, Chu T, Wolfinger R, Kazmin D, Koves T, Muoio D, Chang C, et al. Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol Cell. 2006;24:797–803. doi: 10.1016/j.molcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Garza AM, Khan SH, Kumar R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol. 2010;30:220–230. doi: 10.1128/MCB.00552-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart M, Holmbeck S, Evans R, Dyson H, Wright P. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol. 2003;327:819–832. doi: 10.1016/s0022-2836(03)00183-9. [DOI] [PubMed] [Google Scholar]
- Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr.Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Gomez BP, Riggins RB, Shajahan AN, Klimach U, Wang A, Crawford AC, Zhu Y, Zwart A, Wang M, Clarke R. Human X-box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. The FASEB Journal. 2007;21:4013–4027. doi: 10.1096/fj.06-7990com. [DOI] [PubMed] [Google Scholar]
- Greschik H, Flaig R, Renaud JP, Moras D. Structural basis for the deactivation of the estrogen-related receptor gamma by diethylstilbestrol or 4-hydroxytamoxifen and determinants of selectivity. J.Biol Chem. 2004;279:33639–33646. doi: 10.1074/jbc.M402195200. [DOI] [PubMed] [Google Scholar]
- Greschik H, Wurtz JM, Sanglier S, Bourguet W, van Dorsselaer A, Moras D, Renaud JP. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol.Cell. 2002;9:303–313. doi: 10.1016/s1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- Gu Z, Lee RY, Skaar TC, Bouker KB, Welch JN, Lu J, Liu A, Zhu Y, Davis N, Leonessa F, et al. Association of interferon regulatory factor-1, nucleophosmin, nuclear factor-kappaB, and cyclic AMP response element binding with acquired resistance to faslodex (ICI 182,780) Cancer Research. 2002;62:3428–3437. [PubMed] [Google Scholar]
- Gupta P, Park S, Farooqui M, Wei L. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res. 2007;35:2269–2282. doi: 10.1093/nar/gkl1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurates B, Amsterdam A, Tamura M, Yang S, Zhou J, Fang Z, Amin S, Sebastian S, Bulun S. WT1 and DAX-1 regulate SF-1-mediated human P450arom gene expression in gonadal cells. Mol Cell Endocrinol. 2003;208:61–75. doi: 10.1016/s0303-7207(03)00198-9. [DOI] [PubMed] [Google Scholar]
- Gurates B, Sebastian S, Yang S, Zhou J, Tamura M, Fang Z, Suzuki T, Sasano H, Bulun S. WT1 and DAX-1 inhibit aromatase P450 expression in human endometrial and endometriotic stromal cells. J Clin Endocrinol Metab. 2002;87:4369–4377. doi: 10.1210/jc.2002-020522. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- Heck S, Rom J, Thewes V, Becker N, Blume B, Sinn H, Deuschle U, Sohn C, Schneeweiss A, Lichter P. Estrogen-related receptor alpha expression and function is associated with the transcriptional coregulator AIB1 in breast carcinoma. Cancer Res. 2009;69:5186–5193. doi: 10.1158/0008-5472.CAN-08-3062. [DOI] [PubMed] [Google Scholar]
- Helguero L, Hedengran Faulds M, Förster C, Gustafsson J, Haldosén L. DAX-1 expression is regulated during mammary epithelial cell differentiation. Endocrinology. 2006;147:3249–3259. doi: 10.1210/en.2005-1651. [DOI] [PubMed] [Google Scholar]
- Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J.Mol.Endocrinol. 2003;31:349–357. doi: 10.1677/jme.0.0310349. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Whitfield G, Oza A, Dang H, Price J, Galligan M, Jurutka P, Thompson P, Haussler C, Haussler M. Characterization of unique DNA-binding and transcriptional-activation functions in the carboxyl-terminal extension of the zinc finger region in the human vitamin D receptor. Biochemistry. 1999;38:16347–16358. doi: 10.1021/bi9916574. [DOI] [PubMed] [Google Scholar]
- Hu Y, Shyr C, Che W, Mu X, Kim E, Chang C. Suppression of estrogen receptor-mediated transcription and cell growth by interaction with TR2 orphan receptor. J Biol Chem. 2002;277:33571–33579. doi: 10.1074/jbc.M203531200. [DOI] [PubMed] [Google Scholar]
- Huppunen J, Wohlfahrt G, Aarnisalo P. Requirements for transcriptional regulation by the orphan nuclear receptor ERRgamma. Mol.Cell Endocrinol. 2004;219:151–160. doi: 10.1016/j.mce.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Hwang E, Lee J, Jeong J, Park J, Yang Y, Lim J, Kim J, Baek S, Kim K. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem Biophys Res Commun. 2009;378:513–517. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun M. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Mangelsdorf DJ. Nuclear receptor regulation of stemness and stem cell differentiation. Exp Mol Med. 2009;41:525–537. doi: 10.3858/emm.2009.41.8.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten A. Recent advances in the mechanisms of action and physiological functions of the retinoid-related orphan receptors (RORs) Curr Drug Targets Inflamm Allergy. 2004;3:395–412. doi: 10.2174/1568010042634497. [DOI] [PubMed] [Google Scholar]
- Johansson L, Thomsen J, Damdimopoulos A, Spyrou G, Gustafsson J, Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J Biol Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- Johnston S. Enhancing the efficacy of hormonal agents with selected targeted agents. Clin Breast Cancer. 2009;9(Suppl 1):S28–36. doi: 10.3816/CBC.2009.s.003. [DOI] [PubMed] [Google Scholar]
- Kallen J, Schlaeppi J, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- Kang H, Angers M, Beak J, Wu X, Gimble J, Wada T, Xie W, Collins J, Grissom S, Jetten A. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- Kim G, Lee G, Nedumaran B, Park Y, Kim K, Park S, Lee Y, Kim J, Choi H. The orphan nuclear receptor DAX-1 acts as a novel transcriptional corepressor of PPARgamma. Biochem Biophys Res Commun. 2008;370:264–268. doi: 10.1016/j.bbrc.2008.03.098. [DOI] [PubMed] [Google Scholar]
- Klinge C. Role of estrogen receptor ligand and estrogen response element sequence on interaction with chicken ovalbumin upstream promoter transcription factor (COUP-TF) J Steroid Biochem Mol Biol. 1999;71:1–19. doi: 10.1016/s0960-0760(99)00124-7. [DOI] [PubMed] [Google Scholar]
- Klinge C, Silver B, Driscoll M, Sathya G, Bambara R, Hilf R. Chicken ovalbumin upstream promoter-transcription factor interacts with estrogen receptor, binds to estrogen response elements and half-sites, and inhibits estrogen-induced gene expression. J Biol Chem. 1997;272:31465–31474. doi: 10.1074/jbc.272.50.31465. [DOI] [PubMed] [Google Scholar]
- Kong W, Jiang X, Mercer W. Downregulation of Wip-1 phosphatase expression in MCF-7 breast cancer cells enhances doxorubicin-induced apoptosis through p53-mediated transcriptional activation of Bax. Cancer Biol Ther. 2009a;8 doi: 10.4161/cbt.8.6.7742. [DOI] [PubMed] [Google Scholar]
- Kong X, Fan H, Liu X, Wang R, Liang J, Gupta N, Chen Y, Fang F, Chang Y. Peroxisome proliferator-activated receptor gamma coactivator-1alpha enhances antiproliferative activity of 5′-deoxy-5-fluorouridine in cancer cells through induction of uridine phosphorylase. Mol Pharmacol. 2009b;76:854–860. doi: 10.1124/mol.109.056424. [DOI] [PubMed] [Google Scholar]
- Koritschoner N, Madruga J, Knespel S, Blendinger G, Anzinger B, Otto A, Zenke M, Bartůnĕk P. The nuclear orphan receptor TR4 promotes proliferation of myeloid progenitor cells. Cell Growth Differ. 2001;12:563–572. [PubMed] [Google Scholar]
- Kovacic A, Speed C, Simpson E, Clyne C. Inhibition of aromatase transcription via promoter II by short heterodimer partner in human preadipocytes. Mol Endocrinol. 2004;18:252–259. doi: 10.1210/me.2003-0211. [DOI] [PubMed] [Google Scholar]
- Krasowski MD, Reschly EJ, Ekins S. Intrinsic disorder in nuclear hormone receptors. J Proteome Res. 2008;7:4359–4372. doi: 10.1021/pr8003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus RJ, Ariazi EA, Farrell ML, Mertz JE. Estrogen-related receptor alpha 1 actively antagonizes estrogen receptor-regulated transcription in MCF-7 mammary cells. J.Biol Chem. 2002;277:24826–24834. doi: 10.1074/jbc.M202952200. [DOI] [PubMed] [Google Scholar]
- Kudo K, Xi Y, Wang Y, Song B, Chu E, Ju J, Russo J. Translational control analysis by translationally active RNA capture/microarray analysis (TrIP-Chip) Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Litwack G. Structural and functional relationships of the steroid hormone receptors’ N-terminal transactivation domain. Steroids. 2009;74:877–883. doi: 10.1016/j.steroids.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Lanvin O, Bianco S, Kersual N, Chalbos D, Vanacker J. Potentiation of ICI182,780 (Fulvestrant)-induced estrogen receptor-alpha degradation by the estrogen receptor-related receptor-alpha inverse agonist XCT790. J Biol Chem. 2007;282:28328–28334. doi: 10.1074/jbc.M704295200. [DOI] [PubMed] [Google Scholar]
- Le Dily F, Métivier R, Guéguen M, Le Péron C, Flouriot G, Tas P, Pakdel F. COUP-TFI modulates estrogen signaling and influences proliferation, survival and migration of breast cancer cells. Breast Cancer Res Treat. 2008;110:69–83. doi: 10.1007/s10549-007-9693-6. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lee H, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol. 2002;81:291–308. doi: 10.1016/s0960-0760(02)00118-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham N, Kliewer S, Xu H. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lin D, Chang C. p53 is a mediator for radiation-repressed human TR2 orphan receptor expression in MCF-7 cells, a new pathway from tumor suppressor to member of the steroid receptor superfamily. J Biol Chem. 1996;271:14649–14652. doi: 10.1074/jbc.271.25.14649. [DOI] [PubMed] [Google Scholar]
- Liu D, Zhang Z, Gladwell W, Teng C. Estrogen stimulates estrogen-related receptor alpha gene expression through conserved hormone response elements. Endocrinology. 2003;144:4894–4904. doi: 10.1210/en.2003-0432. [DOI] [PubMed] [Google Scholar]
- Liu N, Lin W, Kim E, Collins L, Lin H, Yu I, Sparks J, Chen L, Lee Y, Chang C. Loss of TR4 orphan nuclear receptor reduces phosphoenolpyruvate carboxykinase-mediated gluconeogenesis. Diabetes. 2007;56:2901–2909. doi: 10.2337/db07-0359. [DOI] [PubMed] [Google Scholar]
- Lu D, Kiriyama Y, Lee K, Giguère V. Transcriptional regulation of the estrogen-inducible pS2 breast cancer marker gene by the ERR family of orphan nuclear receptors. Cancer Res. 2001;61:6755–6761. [PubMed] [Google Scholar]
- Lykkesfeldt A, Henriksen K, Rasmussen B, Sasano H, Evans D, Møller S, Ejlertsen B, Mouridsen H. In situ aromatase expression in primary tumor is associated with estrogen receptor expression but is not predictive of response to endocrine therapy in advanced breast cancer. BMC Cancer. 2009;9:185. doi: 10.1186/1471-2407-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Macciò A, Madeddu C, Mantovani G. Adipose tissue as target organ in the treatment of hormone-dependent breast cancer: new therapeutic perspectives. Obes Rev. 2009;10:660–670. doi: 10.1111/j.1467-789X.2009.00592.x. [DOI] [PubMed] [Google Scholar]
- Macedo L, Sabnis G, Brodie A. Aromatase inhibitors and breast cancer. Ann N Y Acad Sci. 2009;1155:162–173. doi: 10.1111/j.1749-6632.2008.03689.x. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Muller W. Signal transduction in transgenic mouse models of human breast cancer--implications for human breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:323–335. doi: 10.1007/s10911-008-9087-3. [DOI] [PubMed] [Google Scholar]
- Maring J, Groen H, Wachters F, Uges D, de Vries E. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5:226–243. doi: 10.1038/sj.tpj.6500320. [DOI] [PubMed] [Google Scholar]
- Markićević M, Petrović A, Kanjer K, Nesković-Konstantinović Z, Nikolić-Vukosavujević D. Estrogen-regulated cut-off values of pS2 and cathepsin D expression in breast carcinomas. Adv Exp Med Biol. 2008;617:341–348. doi: 10.1007/978-0-387-69080-3_32. [DOI] [PubMed] [Google Scholar]
- Miki Y, Clyne C, Suzuki T, Moriya T, Shibuya R, Nakamura Y, Ishida T, Yabuki N, Kitada K, Hayashi S, et al. Immunolocalization of liver receptor homologue-1 (LRH-1) in human breast carcinoma: possible regulator of insitu steroidogenesis. Cancer Lett. 2006;244:24–33. doi: 10.1016/j.canlet.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Moll U, Marchenko N, Zhang X. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- Moral R, Wang R, Russo IH, Lamartiniere CA, Pereira J, Russo J. Effect of prenatal exposure to the endocrine disruptor bisphenol A on mammary gland morphology and gene expression signature. J.Endocrinol. 2008;196:101–112. doi: 10.1677/JOE-07-0056. [DOI] [PubMed] [Google Scholar]
- Moré E, Fellner T, Doppelmayr H, Hauser-Kronberger C, Dandachi N, Obrist P, Sandhofer F, Paulweber B. Activation of the MAP kinase pathway induces chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) expression in human breast cancer cell lines. J Endocrinol. 2003;176:83–94. doi: 10.1677/joe.0.1760083. [DOI] [PubMed] [Google Scholar]
- Musgrove E, Sutherland R. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- Métivier R, Gay F, Hübner M, Flouriot G, Salbert G, Gannon F, Kah O, Pakdel F. Formation of an hER alpha-COUP-TFI complex enhances hER alpha AF-1 through Ser118 phosphorylation by MAPK. EMBO J. 2002;21:3443–3453. doi: 10.1093/emboj/cdf344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H, Mendonca M, Bhat-Nakshatri P, Patel N, Goulet RJ, Cornetta K. The orphan receptor COUP-TFII regulates G2/M progression of breast cancer cells by modulating the expression/activity of p21(WAF1/CIP1), cyclin D1, and cdk2. Biochem Biophys Res Commun. 2000;270:1144–1153. doi: 10.1006/bbrc.2000.2562. [DOI] [PubMed] [Google Scholar]
- Novac N, Heinzel T. Nuclear receptors: overview and classification. Curr Drug Targets Inflamm Allergy. 2004;3:335–346. doi: 10.2174/1568010042634541. [DOI] [PubMed] [Google Scholar]
- O’Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Res. 2009;69:8217–8222. doi: 10.1158/0008-5472.CAN-09-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odawara H, Iwasaki T, Horiguchi J, Rokutanda N, Hirooka K, Miyazaki W, Koibuchi Y, Shimokawa N, Iino Y, Takeyoshi I, et al. Activation of aromatase expression by retinoic acid receptor-related orphan receptor (ROR) alpha in breast cancer cells: identification of a novel ROR response element. J Biol Chem. 2009;284:17711–17719. doi: 10.1074/jbc.M109.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo T, Ohkura N, Maruyama K, Sasaki K, Nagasaki K, Hanzawa H, Tsukada T, Yamaguchi K. Early induction of the orphan nuclear receptor NOR-1 during cell death of the human breast cancer cell line MCF-7. Mol Cell Endocrinol. 2000;162:151–156. doi: 10.1016/s0303-7207(00)00222-7. [DOI] [PubMed] [Google Scholar]
- Olopade O, Grushko T, Nanda R, Huo D. Advances in breast cancer: pathways to personalized medicine. Clin Cancer Res. 2008;14:7988–7999. doi: 10.1158/1078-0432.CCR-08-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando F, Brown K. Unraveling breast cancer heterogeneity through transcriptomic and epigenomic analysis. Ann Surg Oncol. 2009;16:2270–2279. doi: 10.1245/s10434-009-0500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund E, Lee Y, Solomon I, Hager J, Safi R, Choi Y, Guan Z, Tripathy A, Raetz C, McDonnell D, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of Tamoxifen-treated, node-negative breast cancer. The New England Journal of Medicine. 2004 doi: 10.1056/NEJMoa041588. NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Parton M, Krajewski S, Smith I, Krajewska M, Archer C, Naito M, Ahern R, Reed J, Dowsett M. Coordinate expression of apoptosis-associated proteins in human breast cancer before and during chemotherapy. Clin Cancer Res. 2002;8:2100–2108. [PubMed] [Google Scholar]
- Pereira F, Qiu Y, Zhou G, Tsai M, Tsai S. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Perou C. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2010 doi: 10.1016/j.molonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Rae JM, Ramus SJ, Waltham M, Armes JE, Campbell IG, Clarke R, Barndt RJ, Johnson MD, Thompson EW. Common origins of MDA-MB-435 cells from various sources with those shown to have melanoma properties. Clinical and Experimental Metastasis. 2004;21:543–552. doi: 10.1007/s10585-004-3759-1. [DOI] [PubMed] [Google Scholar]
- Rajhans R, Nair H, Nair S, Cortez V, Ikuko K, Kirma N, Zhou D, Holden A, Brann D, Chen S, et al. Modulation of in situ estrogen synthesis by proline-, glutamic acid-, and leucine-rich protein-1: potential estrogen receptor autocrine signaling loop in breast cancer cells. Mol Endocrinol. 2008;22:649–664. doi: 10.1210/me.2007-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas 1 7372. Clin.Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins R. ESRRG (estrogen-related receptor gamma) Atlas Genet Cytogenet Oncol Haematol. 2009 Oct; [Google Scholar]
- Riggins R, Bouton AH, Liu MC, Clarke R. Antiestrogens, aromatase inhibitors, and apoptosis in breast cancer. Vitam Horm. 2005;71:201–237. doi: 10.1016/S0083-6729(05)71007-4. [DOI] [PubMed] [Google Scholar]
- Riggins RB, Lan JP, Zhu Y, Klimach U, Zwart A, Cavalli LR, Haddad BR, Chen L, Gong T, Xuan J, et al. ERR{gamma} Mediates Tamoxifen Resistance in Novel Models of Invasive Lobular Breast Cancer. Cancer Res. 2008;68:8908–8917. doi: 10.1158/0008-5472.CAN-08-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. Pathways to tamoxifen resistance. Cancer Lett. 2007;256:1–24. doi: 10.1016/j.canlet.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs K, Wickramasinghe N, Cochrum R, Watts M, Klinge C. Decreased chicken ovalbumin upstream promoter transcription factor II expression in tamoxifen-resistant breast cancer cells. Cancer Res. 2006;66:10188–10198. doi: 10.1158/0008-5472.CAN-05-3937. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Garcia HE, Laudet V. The nuclear receptor superfamily. J Cell Sci. 2003;116:585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- Safi R, Kovacic A, Gaillard S, Murata Y, Simpson E, McDonnell D, Clyne C. Coactivation of liver receptor homologue-1 by peroxisome proliferator-activated receptor gamma coactivator-1alpha on aromatase promoter II and its inhibition by activated retinoid X receptor suggest a novel target for breast-specific antiestrogen therapy. Cancer Res. 2005;65:11762–11770. doi: 10.1158/0008-5472.CAN-05-2792. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Santen R, Brodie H, Simpson E, Siiteri P, Brodie A. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr Rev. 2009;30:343–375. doi: 10.1210/er.2008-0016. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E, Alen P, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F. Differential DNA binding by the androgen and glucocorticoid receptors involves the second Zn-finger and a C-terminal extension of the DNA-binding domains. Biochem J. 1999;341(Pt 3):515–521. [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Loven M, Melvin V, Edwards D, Nardulli A. Differential modulation of DNA conformation by estrogen receptors alpha and beta. J Biol Chem. 2002;277:8702–8707. doi: 10.1074/jbc.M108491200. [DOI] [PubMed] [Google Scholar]
- Sentis S, Le Romancer M, Bianchin C, Rostan M, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol. 2005;19:2671–2684. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- Seol W, Choi H, Moore D. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- Shajahan A, Riggins R, Clarke R. Apoptosis, cell death, and breast cancer. In: Pasqualini J, editor. Breast Cancer: Prognosis, Treatment, and Prevention. 2nd edition Informa Healthcare; 2008. [Google Scholar]
- Shyr C, Hu Y, Kim E, Chang C. Modulation of estrogen receptor-mediated transactivation by orphan receptor TR4 in MCF-7 cells. J Biol Chem. 2002;277:14622–14628. doi: 10.1074/jbc.M110051200. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Van de RM, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc.Natl.Acad.Sci.U.S.A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc.Natl.Acad.Sci.U.S.A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein Kunder R, McDonnell D. ESRRA (estrogen-related receptor alpha) Atlas Genet Cytogenet Oncol Haematol. 2009 Jul; [Google Scholar]
- Stein R, Chang C, Kazmin D, Way J, Schroeder T, Wergin M, Dewhirst M, McDonnell D. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68:8805–8812. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R, Gaillard S, McDonnell D. Estrogen-related receptor alpha induces the expression of vascular endothelial growth factor in breast cancer cells. J Steroid Biochem Mol Biol. 2009;114:106–112. doi: 10.1016/j.jsbmb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R, McDonnell D. Estrogen-related receptor alpha as a therapeutic target in cancer. Endocr Relat Cancer. 2006;13(Suppl 1):S25–32. doi: 10.1677/erc.1.01292. [DOI] [PubMed] [Google Scholar]
- Surowiak P, Materna V, Györffy B, Matkowski R, Wojnar A, Maciejczyk A, Paluchowski P, Dziegiel P, Pudełko M, Kornafel J, et al. Multivariate analysis of oestrogen receptor alpha, pS2, metallothionein and CD24 expression in invasive breast cancers. Br J Cancer. 2006;95:339–346. doi: 10.1038/sj.bjc.6603254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miki Y, Moriya T, Shimada N, Ishida T, Hirakawa H, Ohuchi N, Sasano H. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64:4670–4676. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- Takimoto GS, Tung L, Abdel-Hafiz H, Abel MG, Sartorius CA, Richer JK, Jacobsen BM, Bain DL, Horwitz KB. Functional properties of the N-terminal region of progesterone receptors and their mechanistic relationship to structure. J Steroid Biochem Mol Biol. 2003;85:209–219. doi: 10.1016/s0960-0760(03)00197-3. [DOI] [PubMed] [Google Scholar]
- Tamasi V, Miller KK, Ripp SL, Vila E, Geoghagen TE, Prough RA. Modulation of receptor phosphorylation contributes to activation of peroxisome proliferator activated receptor alpha by dehydroepiandrosterone and other peroxisome proliferators. Mol Pharmacol. 2008;73:968–976. doi: 10.1124/mol.107.036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe O, Shen Y, Liu Q, Campbell A, Kuroha T, Yamamoto M, Engel J. The TR2 and TR4 orphan nuclear receptors repress Gata1 transcription. Genes Dev. 2007;21:2832–2844. doi: 10.1101/gad.1593307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozlu S, Girault I, Vacher S, Vendrell J, Andrieu C, Spyratos F, Cohen P, Lidereau R, Bieche I. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- Tremblay AM, Giguere V. The NR3B subgroup: an ovERRview. Nucl.Recept.Signal. 2007;5:e009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay AM, Wilson BJ, Yang XJ, Giguere V. Phosphorylation-dependent sumoylation regulates estrogen-related receptor-alpha and -gamma transcriptional activity through a synergy control motif. Mol.Endocrinol. 2008;22:570–584. doi: 10.1210/me.2007-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Agthoven T, Sieuwerts A, Meijer-van Gelder M, Look M, Smid M, Veldscholte J, Sleijfer S, Foekens J, Dorssers L. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009a;27:542–549. doi: 10.1200/JCO.2008.17.1462. [DOI] [PubMed] [Google Scholar]
- van Agthoven T, Veldscholte J, Smid M, van Agthoven T, Vreede L, Broertjes M, de Vries I, de Jong D, Sarwari R, Dorssers L. Functional identification of genes causing estrogen independence of human breast cancer cells. Breast Cancer Res Treat. 2009b;114:23–30. doi: 10.1007/s10549-008-9969-5. [DOI] [PubMed] [Google Scholar]
- Vu EH, Kraus RJ, Mertz JE. Phosphorylation-dependent sumoylation of estrogen-related receptor alpha1. Biochemistry. 2007;46:9795–9804. doi: 10.1021/bi700316g. [DOI] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker N, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jeffs B, Ito M, Achermann J, Yu R, Hales D, Jameson J. Aromatase (Cyp19) expression is up-regulated by targeted disruption of Dax1. Proc Natl Acad Sci U S A. 2001;98:7988–7993. doi: 10.1073/pnas.141543298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, Milburn MV, Rosenfeld MG, Glass CK. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- Wilson B, Giguère V. Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer. 2008;7:49. doi: 10.1186/1476-4598-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B, Tremblay A, Deblois G, Sylvain-Drolet G, Giguère V. An Acetylation Switch Modulates the Transcriptional Activity of Estrogen-Related Receptor {alpha} Mol Endocrinol. 2010 doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisely G, Miller A, Davis R, Thornquest AJ, Johnson R, Spitzer T, Sefler A, Shearer B, Moore J, Willson T, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Wu F, Wang J, Wang Y, Kwok T, Kong S, Wong C. Estrogen-related receptor alpha (ERRalpha) inverse agonist XCT-790 induces cell death in chemotherapeutic resistant cancer cells. Chem Biol Interact. 2009;181:236–242. doi: 10.1016/j.cbi.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Wurtz J, Bourguet W, Renaud J, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- Zhang H, Thomsen J, Johansson L, Gustafsson J, Treuter E. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J Biol Chem. 2000;275:39855–39859. doi: 10.1074/jbc.C000567200. [DOI] [PubMed] [Google Scholar]
- Zhang T, Jia N, Fei E, Wang P, Liao Z, Ding L, Yan M, Nukina N, Zhou J, Wang G. Nurr1 is phosphorylated by ERK2 in vitro and its phosphorylation upregulates tyrosine hydroxylase expression in SH-SY5Y cells. Neurosci.Lett. 2007;423:118–122. doi: 10.1016/j.neulet.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wang P, Ren H, Fan J, Wang G. NGFI-B nuclear orphan receptor Nurr1 interacts with p53 and suppresses its transcriptional activity. Mol Cancer Res. 2009;7:1408–1415. doi: 10.1158/1541-7786.MCR-08-0533. [DOI] [PubMed] [Google Scholar]
- Zhou J, Suzuki T, Kovacic A, Saito R, Miki Y, Ishida T, Moriya T, Simpson E, Sasano H, Clyne C. Interactions between prostaglandin E(2), liver receptor homologue-1, and aromatase in breast cancer. Cancer Res. 2005;65:657–663. [PubMed] [Google Scholar]
- Zhu Y, McAvoy S, Kuhn R, Smith D. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25:2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]