Abstract
Genetic clustering algorithms require a certain amount of data to produce informative results. In the common situation that individuals are sampled at several locations, we show how sample group information can be used to achieve better results when the amount of data is limited. New models are developed for the structure program, both for the cases of admixture and no admixture. These models work by modifying the prior distribution for each individual’s population assignment. The new prior distributions allow the proportion of individuals assigned to a particular cluster to vary by location. The models are tested on simulated data, and illustrated using microsatellite data from the CEPH Human Genome Diversity Panel. We demonstrate that the new models allow structure to be detected at lower levels of divergence, or with less data, than the original structure models or principal components methods, and that they are not biased towards detecting structure when it is not present. These models are implemented in a new version of structure which is freely available online at http://pritch.bsd.uchicago.edu/structure.html.
Keywords: admixture, divergence, population structure, prior distribution
Introduction
Clustering algorithms for genetic data have become an important tool in a number of fields including conservation and population genetics (Dawson & Belkhir 2001; Corander et al. 2003; Purcell & Sham 2004; Corander & Marttinen 2006;Francois et al. 2006; Patterson et al. 2006).Such methods are often used to understand the structure of populations, as well as to identify migrant or admixed individuals. They are also used to detect cryptic population structure, as undetected structure may lead to false positives when searching for disease-associated markers in case-control studies.
structure is a Bayesian, model-based algorithm that is widely used for clustering genetic data (Pritchard et al. 2000; Falush et al. 2003; Falush et al. 2007). Given the number of clusters (K) and assuming Hardy–Weinberg and linkage equilibrium within clusters, structure estimates allele frequencies in each cluster and population memberships for every individual. In the simplest, ‘no-admixture’ model, it assumes that each individual belongs to a single cluster, whereas in the more general ‘admixture model’, it estimates admixture proportions for each individual. It uses Markov chain Monte Carlo (MCMC) to integrate over the parameter space and make cluster assignments. Although the value of K must be provided to the algorithm, a heuristic method for selecting K is often used, which is based on comparing penalized log likelihoods over independent runs with differing numbers of clusters.
When the data contain relatively little information about population structure, structure sometimes produces results that are difficult to interpret. For example, the samples may have come from several distinct populations, and perhaps FST values calculated between the samples from some pairs of the labelled populations are significantly different from zero, and yet the results indicate no evidence of structure. Or, the population assignments made by the algorithm may hint that there is indeed structure, and yet the highest penalized log likelihood is provided by the model with just one cluster. When such situations arise, it is unclear whether one should conclude that the data are homogeneous after all, or that the amount of data collected is insufficient to make a convincing case for structure.
Although such results may be discouraging, it is worth noting that in a sense, structure aims to solve a rather difficult problem. There is an enormous number of ways that N individuals can be partitioned into K populations. The basic structure models assume that all partitions of the N individuals into K populations are equally likely, a priori. This means that any particular clustering solution is highly unlikely, a priori, and it takes a considerable amount of statistical evidence to provide strong support for any particular partition. This explains why there can be data sets with significant FST values between samples of individuals collected at different locations, and yet structure does not provide a clear indication of population structure.
In this paper, we extend the basic models to allow structure to make use of information about sampling locations, when the data indicate that this information would be helpful. In effect, we place much more prior weight on clustering outcomes that are correlated with the sampling locations. The new models allow much better performance on some data sets where there are too few loci or individuals, or not enough divergence, for the standard structure models to perform well. Our approach could also be used in settings where individuals can be classified into discrete groups on the basis of a phenotypic characteristic. The new models have the desirable properties that (i) they do not tend to find structure when none is present; (ii) they are able to ignore the sampling information when the ancestry of individuals is uncorrelated with sampling locations; and (iii) the old and new models give essentially the same answers when the signal of population structure is very strong. Hence, we recommend using the new models in most situations where the amount of available data is limited, especially when the standard structure models do not provide a clear signal of structure.
The idea of using sampling locations to help infer population structure has also been considered elsewhere. One approach was taken by Corander et al. (2003), and implemented in the program baps. baps allows the user to pre-specify a set of sample groups; all individuals in the same sample group are assumed to have the same ancestry. The authors have shown that the use of sample group information can greatly improve power to detect structure when the amount of data is limited (Corander et al. 2003; Corander & Marttinen 2006). Once the allele frequencies are estimated, migrants and admixture events can be detected in an additional step that does not take the sampling groups into account. By contrast, the methods that we develop here allow for a more flexible relationship between sample groups and ancestry, allowing for the possibility that sample group information might be partially (or even not at all) informative about genetic population structure, and providing simultaneous estimation of allele frequencies and ancestry.
A second type of approach to using location information makes use of spatially explicit models. For example, Wasser et al. (2004) used elephant samples from known locations across Africa to estimate the geographical origin of poached ivory. Their method, implemented in scat, assumes that allele frequencies vary smoothly across the region of study. Another type of approach has been implemented in the program geneland (Francois et al. 2006; Guillot et al. 2008), and in a recent version of baps (Corander et al. 2008). The methodologies of the two programs are somewhat different, but they both use a coloured tessellation to model the distribution of the population clusters across space. These spatially explicit methods differ from the models discussed here in that we do not consider the specific geographical coordinates for each individual, but instead simply group together individuals collected at the same sampling location. This allows us to make fewer assumptions about the geographical structure of populations, while still offering improved performance in the common scenario that individuals are sampled at a modest number of distinct locations.
Our new methods are also substantially different from the 'Model with prior population information’ introduced in the original structure paper (Pritchard et al. 2000). That earlier model was designed for the situation in which there is both strong evidence of population structure and in which the sampling locations correspond almost exactly to the inferred clusters. That model allows a user to test whether a small number of individuals might be migrants from a different location than where they were sampled and is only useful for highly informative data. In contrast, the new models presented in this paper help to provide useful inference in settings where the data are not highly informative, and in this case it will usually not be possible to identify migrants with any confidence.
Methods
We present both a no-admixture model and an admixture model that allow the individuals' sampling locations to inform cluster assignments. In order to understand how these models work, it is useful first to review the original model. We provide a brief description here, and Table 1 provides a brief summary of the key model parameters. For the complete details, see Pritchard et al. (2000) and Falushet al. (2003).
Table 1.
Summary of structure parameters
| structure parameters |
|---|
| K: number of clusters |
| N: number of individuals |
| L: number of loci |
| qij: admixture proportion of individual i in cluster j |
| zilm: cluster of origin for locus l, individual i, copy m |
| (α1, … , αK): parameters to Dirichlet distribution which forms a prior for qi |
| pklj: frequency of allele j in locus l, cluster k |
| λ: parameter to Dirichlet distribution which forms a prior for pkl. |
| Fk: the amount of drift from ancestral population to cluster k in the model of correlated allele frequencies |
| New model parameters |
| S: number of sampling locations |
| r: parameter which estimates the informativeness of the sampling location data |
| (η1, … , ηK): for the no-admixture model, these parameters reflect the relative proportion of individuals assigned to each cluster |
| (γs1, … , γsK): for the no-admixture model, these parameters reflect the relative proportion of individuals from location s assigned to each cluster |
| (, …, ): for the admixture model, these parameters reflect the relative levels of admixture from each cluster over all individuals |
| (αs1, … , αsK): for the admixture model, these parameters reflect the relative levels of admixture from each cluster for an individual from location s |
Overview of the structure algorithm
Consider a data set consisting of genotypes for N individuals at L loci. We assume that the sampled individuals have ancestry in K discrete clusters, where the clusters correspond to unobserved populations. K is fixed by the user. Each cluster is characterized by a set of allele frequencies at each locus. The three-dimensional vector P contains the allele frequencies in each cluster for each allele at every locus; the allele frequencies are typically unknown in advance. In the no-admixture model, the algorithm assigns each individual to one of the K clusters. The vector Z records these cluster assignments. In the admixture model, each individual is allowed to have partial ancestry in each of the K clusters. The vector Q describes the proportion of each sampled individual's genome that comes from each cluster. As detailed in Table 1, we use the convention that elements within the vectors P, Q and Z are indexed by lower-case ‘p’, ‘q’, and ‘z’ with appropriate subscripts. The likelihood of an individual's genotype is determined as the roduct of the relevant frequencies of the individual's alleles across all loci (the loci are assumed to be independent given cluster memberships). Our goal is to estimate P, Q and Z from the data.
structure uses MCMC to sample from the posterior distribution of the parameters P, Q, and Z. To estimate the appropriate number of clusters (K), the algorithm is usually run many times independently, varying the value for K. Although there is some debate as to the best method for choosing K (e.g. Evanno et al. 2005), here we use the method suggested in the original structure paper, which involves comparing mean log likelihoods penalized by one-half of their variance (Pritchard et al. 2000). Although a model of linked loci has been developed (Falush et al. 2003), the methods in this paper are most useful when there is a scarcity of data. We assume that when only a small number of loci are genotyped, they are likely to be unlinked, and we will not address the linkage model in this paper.
No-admixture model with sample group information
In the original version of structure, an individual is a priori assumed to be equally likely to come from any of the K clusters. In the no-admixture model, the prior probability that individual icomes from population k (that is, zi = k) is simply given by:
The idea, then, is to modify this prior to take sampling locations into account. We do this by saying that the probability that an individual is assigned to each cluster may vary among the locations:
Here γlk is the prior probability that an individual from location l will be assigned to cluster k, and li denotes the location where individual i was sampled. The γlk values are estimated from the data, and these parameterize the extent to which each sampling location is informative about ancestry. If the γlk are all ~1/K, then the location information is relatively uninformative, and this model is similar to the original structure model. In contrast if, for each location, one value of γlk is estimated to be ~1 and the rest ~0, then the location information will strongly influence the estimated ancestry.
Therefore, while the γlk might help us to improve inference, it is important that they do not overstate the amount of information contained in the location information. To achieve this, we place the following prior structure on γ:
where
and
Here, η is a vector of positive real numbers that, roughly speaking, estimates the overall proportion of individuals from each of the K clusters in the entire data set. Then, r parameterizes the extent to which the ancestry proportions at individual locations can deviate from the overall proportions. rMAX is an upper bound for r, preset by the user. If r is large (>>1), then all the locations have essentially the same prior ancestry proportions (i.e. approximately equal to η). In contrast, if r is ~1 or smaller, then the values of γl· may vary substantially across locations, implying that the location data are informative about ancestry. These priors are chosen so that if either there is no evidence for population structure, or the locations are uncorrelated with ancestry, then r will tend to be large, and we will not be misled by the location information.
For the analyses presented here, we set rMAX = 1000. This choice of rMAX puts considerable prior mass on large values of r, corresponding to the situation where the locations are uninformative. In some circumstances (e.g. with very small data sets, and good prior information that the locations are likely to be informative), a smaller value of rMAX would probably be preferable. We also found that the algorithm converged best if we started r at a small value (rINIT = 1 in our simulations). Appendix I gives details about the MCMC updates for the parameters in this model.
Admixture model with sample group information
The new admixture model works similarly, by modifying the prior distribution for Q. In the original version of structure, the prior distribution for qi, the ancestry of individual i, is given by a Dirichlet distribution with parameters α1,… ,αK. Usually, the α parameters are set equal to each other (α: = α1 = α2 = … = αK), and are estimated during the MCMC. Small values of α (i.e. near 0) indicate that most individuals have little admixture, whereas large values indicate that most individuals have substantial ancestry from multiple clusters.
In order to modify the prior for Q, we now infer a different vector of α's for each location. This is similar in spirit to the new no-admixture model, in that it allows the distribution of cluster assignments to vary by location. If individual i comes from location l, then:
As for the no-admixture model, it is important to prevent the model from over-fitting the location data when the locations are not truly informative. For this reason, we place the following prior structure on the α values, which has the effect of pulling them towards a set of global values unless the locations are genuinely informative. That is, we define a set of global α values:
where denotes the global value of α for the ith cluster. Then the local α values for the lth location are distributed as where
In this model, the global values, α(g), can be thought of as estimating the overall distribution of ancestry. Each is (roughly) proportional to the overall amount of ancestry in cluster i. As in the standard structure model, the mean of α(g) measures the amount of admixture. The distribution of the local a values is constructed so that each αli has mean α(g) and variance . Hence, large values of r imply that the local values of αli are very similar to the global values, and the location information has little impact on the model. Conversely, small values of r allow the local values of αli to differ substantially from the global values, implying that the location information is potentially very informative. As in the no-admixture model, the simulations presented here used rMAX = 1000, although again we note that smaller values would be appropriate for data sets with strong prior reason to expect structure.
Simulations without admixture
Data were simulated with in-house software using a model of correlated allele frequencies (Nicholson et al. 2002) with either two or five populations. It was assumed that each population corresponds perfectly to a sampling location. All simulated data sets were composed of 100 biallelic loci, to model single nucleotide polymorphisms (SNPs). Each individual had an equal probability of being assigned to each of the populations, and the data sets had 100 and 250 diploid individuals for two and five populations, respectively. FST was varied in intervals of 0.005, with 50 independent repetitions for each value of FST. Allele frequencies PR for the root population were simulated from a beta distribution with parameters α = 0.8, β = 0.8. With two populations, the root population was used as population 1, and otherwise a star-like phylogeny of populations was assumed. The allele frequencies for non-root populations were simulated as beta random variables with parameters α = pR(1−FST)/FST, β=(1−pR)(1−FST)/FST, as suggested by Balding & Nichols (1995).
Simulations with admixture
Data were simulated using a model of independent allele frequencies for K = 3, with 100 individuals and a varying number of loci. Each individual had an equal chance of being sampled from each of four locations. The admixture proportions for an individual were drawn from Dirichlet distributions with parameters (10, 0.5, 0.5), (0.5, 10, 0.5), (0.5, 0.5,10), (0.5,0.5, 0.5) for locations 1, 2, 3, and 4, respectively. FST for these simulated data sets was approximately 0.20. An additional set of simulations was performed to demonstrate the behaviour of the admixture model with a large number of sampling locations. Data sets were simulated for K = 5 with 100 individuals and 10 microsatellites, for a range of values of FST. Each individual was assigned to one of 25 sampling locations, and population assignments for each individual were highly determined by the sampling location. Specifically, each location was randomly assigned to one of the five clusters, and admixture proportions were drawn from a Dirichlet distribution with parameter 1 for the main cluster, and 0.01 for each other cluster. For example, if a location was assigned to cluster 3, then every individual from that location would have admixture proportions drawn from a Dirichlet distribution with parameters (0.01, 0.01, 1.0, 0.01, 0.01). The microsatellite data were simulated using the correlated allele frequencies model of Falush et al. (2003). We assumed that all microsatellites had four possible alleles, and the ancestral allele frequencies were simulated from a Dirichlet distribution with parameters (0.8, 0.8, 0.8, 0.8). For this data set, each structure run was repeated four times to ensure proper convergence.
Finally, to illustrate how the results depend on the strength of correlation between location data and population structure, we performed a series of simulations in which we reassigned locations randomly for a fraction f of individuals and re-analysed the data using the new models. This was done for each of the 50 data sets simulated without admixture, assuming five sampling locations, K = 5, and F = 0.03, for values of f in 0,0.04, … ,1.0.
For all the above data sets, structure was run with each value of K ranging from 1 to KT + 1, where KT is the true value of K used in the simulation. The estimate for K was then taken as the K with the highest penalized log likelihood as reported by structure, which calculates the mean log likelihood minus half of its variance. The model of independent allele frequencies was used for the simulations with admixture in which the number of loci was varied. All other runs used the model of correlated allele frequencies, and estimated a separate FST for each population. For all runs using the original admixture model, a separate value of α was estimated for each population as well. All runs consisted of 20 000 burn-in steps followed by 10 000 MCMC steps.
CEPH Human Genome Diversity Panel (HGDP) microsatellite analysis
A microsatellite data set consisting of 377 loci genotyped in 1056 individuals from 52 human populations (Rosenberg et al. 2002) was downloaded from http://rosenberglab.bioinformatics.med.umich.edu/data/rosenbergEtAl2002/diversitydata.stru. We chose one population from each continent for analysis (Surui from South America, Han from Asia, Basque from Europe, Melanesian from Oceania, and Mandenka from Africa), resulting in a data set with 126 individuals. FST among populations from different continents is about 7% in this data set (Rosenberg et al. 2002). All structure analyses were done using the model of correlated allele frequencies, and every run was repeated five times to obtain the run with the highest penalized log-likelihood score. The analysis was repeated 50 times on random subsets of the data for a range of different numbers of loci. Each random subset was created by choosing loci without replacement.
Principal components analysis methods
To provide an additional, and rather different, type of algorithm against which to compare our new methods, we also analysed the simulated data using principal components analysis (PCA). It has been shown (Patterson et al. 2006) that the resolution of principal components methods and structure are quite similar in many cases. The software package eigensoft was downloaded from http://genepath.med.harvard.edu/~reich/Software.htm and the program smartpca (Patterson et al. 2006) was used to analyse the simulated and real data sets. The number of clusters inferred by smartpca was taken as one plus the number of eigenvalues with p-value ≤ 0.05. To get cluster assignments, the k-means algorithm (Hartigan & Wong 1979) was applied to the top K-1 eigenvectors.
Similarity score
To measure the similarity between the true and estimated population assignments, we used an adaptation of the standard Brier similarity score. That is, let qik be the true fraction of ancestry of individual i in population k and let q̂ik be the corresponding estimate of qik. Then, we define a score S as
where N is the number of individuals. Note that S will be zero when Q̂ = Q, and can be as large as 2 if there is a complete mismatch between Q and Q̂. In practice, the labelling of clusters identified by structure is arbitrary, and thus, we computed S for each of the K! possible permutations of the cluster labels, and recorded the minimum of S across permutations (call this S’). When the data are completely uninformative, a clustering solution Q̂* that places a fraction 1/K of each individual into each cluster would receive a smaller score (call this S*) than a solution that puts all individuals into a single cluster (provided that true ancestry is not highly skewed towards particular clusters). Finally, to obtain a similarity score which is equal to one when Q̂ = Q, and zero for any q that performs as poorly as Q̂*, we recorded the similarity score as 1 — min(S’,S*)/S*
Results
To evaluate the performance of the new models, we tested them on simulated and real data under a variety of conditions. Together, the examples illustrate the performance of the methods as a function of the amount of divergence among populations and as a function of the number of loci; as well as under a variety of different conditions: variable numbers of loci; variable levels of information in the location data; discrete populations and admixture; and SNPs and microsatellites. The parameter values for the simulations were chosen because they illustrate the differences between the new and original models; for larger or more informative data sets, the differences between the new and old models tend to be small, and in some contexts, we prefer the original structure models (see below for further discussion).
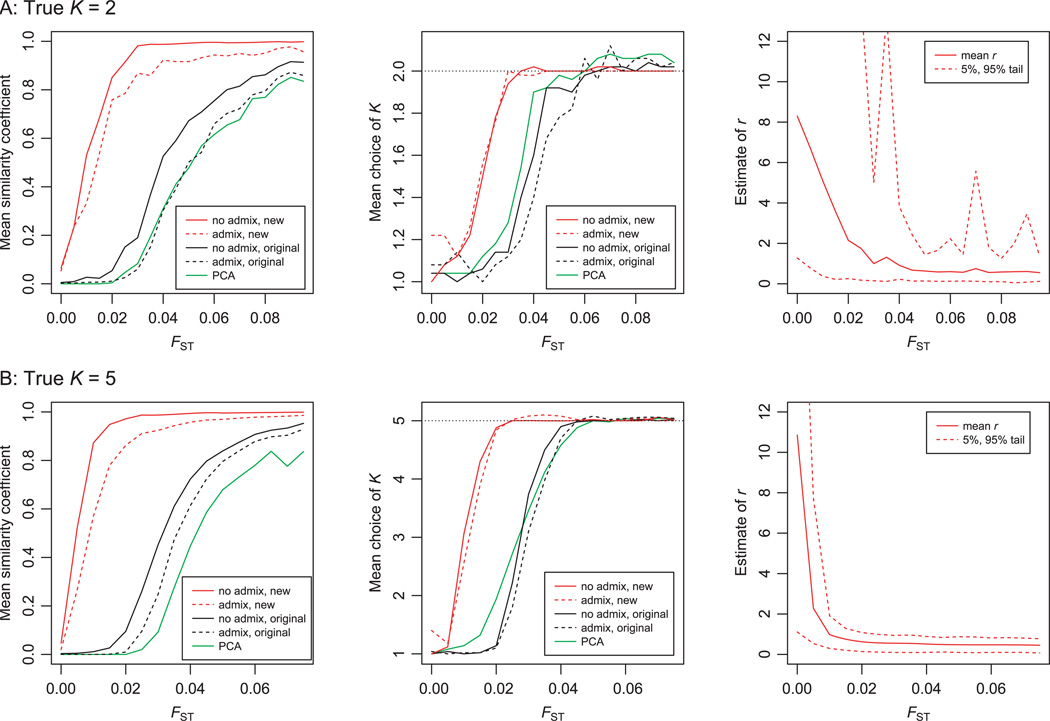
The first set of simulations (Fig. 1) considered a setting in which individuals are sampled from either two or five different sampling locations, and where each sampling location consists of a distinct non-admixed population. As expected, all the methods struggle to assign individuals accurately to populations at low divergence (FST near 0), and provide accurate assignments at high divergence. However, there is a range of FST values for which the new models perform much better than the existing methods: both in terms of making more accurate cluster assignments (similarity coefficient), and in choosing the correct value of K at lower divergence levels. Importantly, all of the models predict just one cluster when FST = 0.0, suggesting that the new models do not bias the algorithm towards finding structure when it is not present.
Figure 1.
Results for simulations without admixture. Data were simulated for K = 2 and K = 5, as described in the Methods. On the left is plotted the mean similarity coefficient between the true and estimated ancestry, as a function of FST, each averaged over 50 simulated data sets. The middle plots show the average choice of K, with the dotted horizontal lines indicating the true value of K. On the right, the solid line shows the average estimate of r calculated using the new no-admixture model with sampling locations. The dotted lines show the 5% and 95% tails of the distribution.
Figure 1 also plots values of the tuning parameter, r, which measures the amount of information contained in the location information. Recall that r>>1 implies that the location labels are uninformative about ancestry, while small values of r allow the ancestry proportions to vary substantially among locations. Notice that when FST is near 0, the mean estimate of r is considerably larger than 1, consistent with the estimates of K near 1. As the amount of information in the data increases the estimate of r quickly decreases, indicating that the sampling groups are contributing information. At FST = 0, one might have expected that the posterior mean of r should be approximately rMAX/2, since in this case rMAX was set to be very large. The fact that r is much smaller than rMAX/2 suggests that r has not fully explored its posterior range during the course of the MCMC run length used here (recall that r was initialized at 1). However this should not be a serious concern as the model is relatively insensitive to the precise value of r when r is considerably larger than 1, and in practice, we would recommend a smaller value of rMAX for most applications.
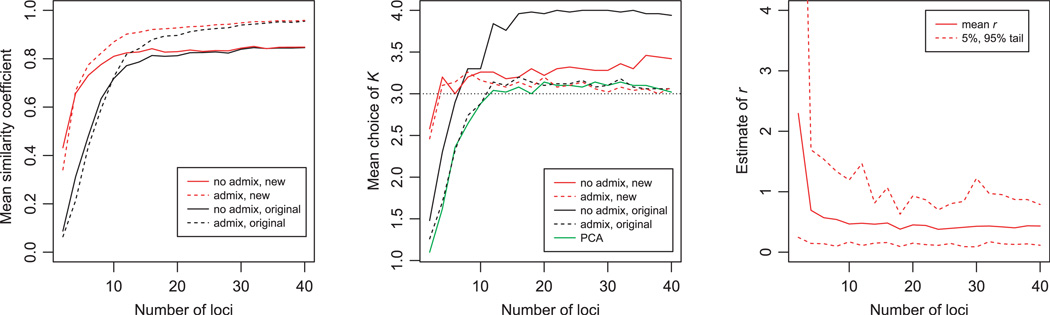
A second set of simulations was performed with admixture (Fig. 2). In this case, we set K = 3 and simulated four sampling locations with different mixtures of ancestry coefficients. We set FST = 0.20 and varied the number of genotyped loci. The plot of similarity coefficients shows that again the new models substantially improve the ancestry estimates when the data sets are small, even providing some information with just one genotyped locus. The old and new models become more similar as the number of genotyped loci increases. We have observed that these new methods tend to improve estimation of admixture coefficients for all the individuals in these data sets, including individuals who are outliers within their sampling group. This indicates that the new methods are not simply working by grouping the individuals in the same location together; instead, the location information also improves the estimation of allele frequencies, leading to more accurate parameter estimation.
Figure 2.
Results for simulations with admixture. Data were simulated with K = 3, as described in the Methods. On the left is the mean similarity coefficient over 50 simulated data sets as a function of the number of loci. In the middle is the mean estimate of K, with the dotted horizontal line indicating the true value of K. The right plot shows the average estimate of r calculated using the new admixture model with sampling locations, with the dotted lines giving the 5% and 95% tails of the distribution.
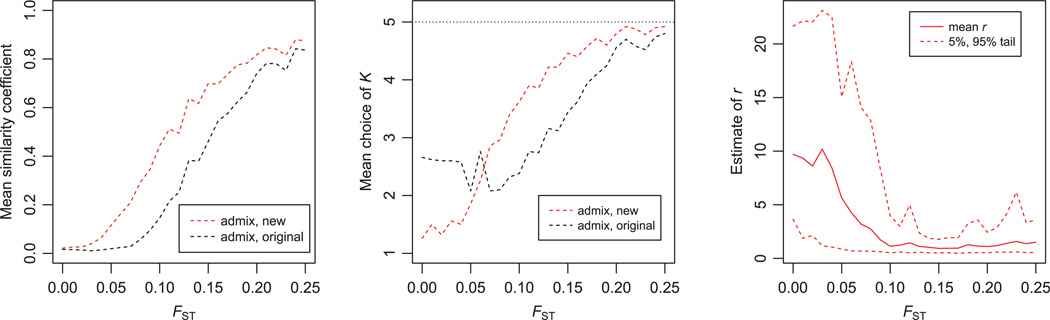
To assess the behaviour of the new model when there are many sampling locations, we also simulated data with 100 individuals sampled from across 25 sampling locations, with K = 5. The simulations were set up so that individuals from the same sampling location generally drew most of their ancestry from the same cluster. Figure 3 shows the performance over a range of values of FST. Even with a relatively small number of individuals per group, the new models still benefit from using the location information, compared to the original models, although the advantage appears to be smaller than when larger numbers of individuals are sampled in each location. We also found that for these data sets, the estimation of K was a little erratic for small values of FST. In particular, both models frequently estimated K > 1 even when FST = 0 (implying that there is no real population structure, so that we would want to estimate K = 1). We believe that structure may be struggling with the relatively small data sets simulated in this case (100 individuals with 10 microsatellites; for example, compare this to Fig. 1A, which includes 100 individuals genotyped at 100 SNPs). In the plot shown in Fig. 3, the new model seems to perform better than the original model at estimating small K when FST = 0, but this does not seem to be a general property of the new model. For example, when we analysed the same data using the ONEFST model in structure, both models overestimated K in the case where FST = 0.
Figure 3.
Results for simulations with admixture, using 25 sampling locations with an average of 4 individuals per location, and K = 5. See Figs 2 and 3 for descriptions of the plots. Each data point is an average over 50 simulated data sets for a given value of FST.
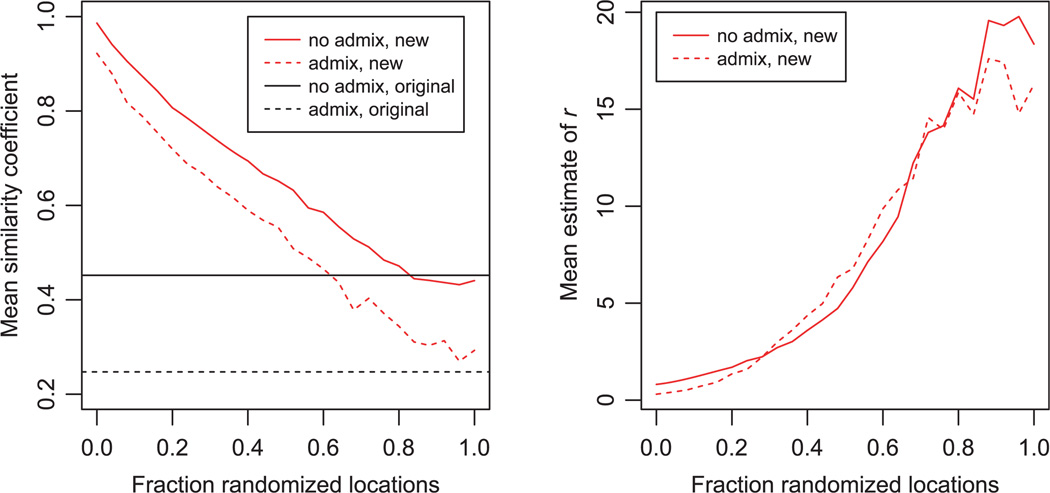
We also investigated the performance of the new models as the correlation between locations and clusters changes. The left plot in Fig. 4 shows the effect of similarity coefficients as the fraction of individuals with randomly assigned locations is increased. The horizontal lines show the average performance of the original structure models on the same data. As expected, the performance of the new models is best when the locations correspond perfectly to the underlying structure. However, even when the locations are completely random, the new models perform almost identically to the old models. This implies that there is little cost to using the new models, even when the location data are potentially uninformative. The right plot in Fig. 4 shows that the value of r estimated by structure seems to be a good indicator of the usefulness of the location data.
Figure 4.
Effect of varying the amount of information contained in the location data. The simulations assumed 250 individuals, five sampling locations, K = 5, FST = 0.03, and no admixture. The x-axis shows the fraction of individuals whose location data were randomized. For all other individuals, the location number matched the true population number.
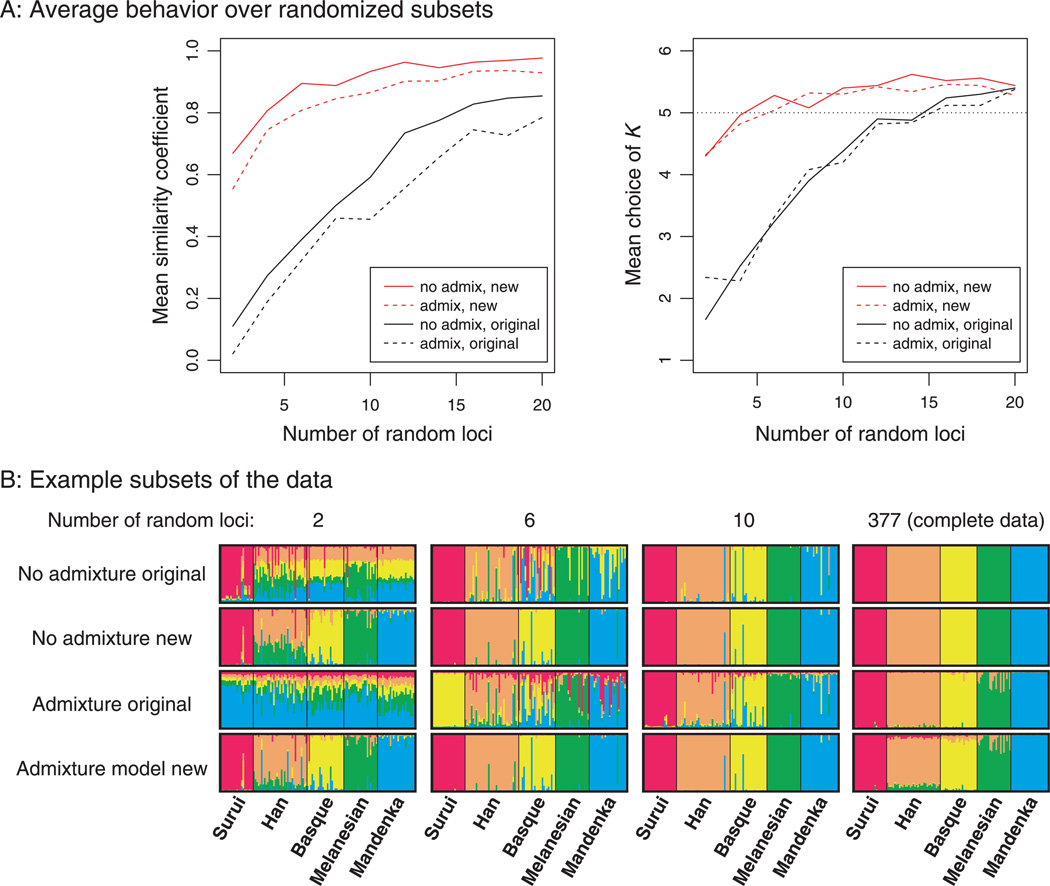
Finally, we illustrate the new methods with a simple application to microsatellite data from the Human Genome Diversity Panel (Rosenberg et al. 2002). We selected a set of 126 individuals representing five populations on five different continents. Figure 5A shows the average results of choosing subsets of the microsatellites at random. We see that the new models almost always estimate K = 5 with as few as 6 random loci, whereas 16 or more loci are required to make the same estimate when the sampling location data are not used. Also, the new models substantially improve the accuracy of the estimated admixture proportions, when the 'true' ancestry proportions are estimated using all 377 microsatellites. Figure 5B shows some example results, using the first 2,6, and 10 microsatellites, respectively, from the data set (in a single random order), compared to the complete data set. It is clear that with 2 and 6 microsatellites, the new models have much more success at separating the continental groups than do the original models.
Figure 5.
Analysis of five populations from the Human Genome Diversity Panel microsatellite data set. In Fig. 5A, the mean similarity coefficient and choice of K are plotted, averaged over 50 runs using a number of randomly chosen microsatellites, shown on the x-axis. Figure 5B shows Structure results for the first 2, 6, and 10 loci, as well as the entire data set.
Once the data set increases to 10 microsatellites or more, the differences among the results become quite subtle. However, for the complete data set of 377 loci, there is a slight but noteworthy difference between results from the new and original admixture models (Fig. 4B). Unlike the original admixture model, the new admixture model estimates that all the Han Chinese individuals contain a small amount of ancestry from both the Melanesians and the Surui. Since it is implausible that there has been recent gene flow of this magnitude from Native Americans and Oceanians into the Chinese population, this argues that the new prior model is subtly shifting the performance of the method on this highly informative data set.
Discussion
The new models presented in this study are designed to help detect population structure and to produce more accurate ancestry estimates for data sets with low information content. Our simulation studies suggest that the models can help considerably in such cases. As the information content in the data increases, the results become similar to those obtained using the original models. In general, our simulations show that the new models provide an appropriate balance between the potential value of incorporating location information into the inference, while still remaining reasonably robust when there is no population structure. Moreover, the new models are able to ignore the sampling information when there is clear evidence of population structure, but the structure is uncorrelated with sampling locations.
For these reasons, we feel that it will often be beneficial to use the new models for analysing small- or medium-sized data sets, such as are currently typical in studies of molecular ecology or conservation genetics. However, we would still encourage users to run the original models as well, and to check that substantial differences between results from the new and old models seem biologically sensible. We also suggest that the value of r can be a useful indicator of whether the location information is relevant to the model: values of r near or below 1 imply that the ancestry proportions differ substantially between sampling locations.
However, we also caution that the new models are not a panacea. For example, structure sometimes overestimates the number of clusters: for example when there is inbreeding or relatedness among some individuals. Moreover, the number of clusters is not well-defined in settings where the allele frequencies vary smoothly across the landscape (Wasser et al. 2004). The new models are likely to be affected similarly by these issues. Finally, for very informative data sets, the new and old models should provide very similar results. However, in one example (the HGDP data, described above), we noted slight differences between results with the old and new priors. Given this, and the fact that there is now a great deal of accumulated experience with the standard structure models, we recommend that the standard models should continue to be the default for data sets in which the data are highly informative.
Finally, we remind users that the new models serve a very different purpose from an existing model in structure that also uses location information (obtained in the software by setting USEPOPINFO = 1) (Pritchard et al. 2000). That model was designed for identifying migrant individuals in data that are highly informative, in contrast to the goal here of detecting very weak population structure.
The models presented here have been implemented in a forthcoming version of structure, version 2.3. The use of the new models will be described in detail in the next release of the structure manual. The new software and documentation will be available online at http://pritch.bsd.uchicago.edu/structure.html.
Acknowledgements
This work was supported by a National Institutes of Health Genetics and Regulation Training Grant (M.J.H.), a Packard Foundation grant (J.K.P.), and a Science Foundation of Ireland (D.F., grant no. 05/FE1/B882). J.K.P. is an investigator of the Howard Hughes Medical Institute. We thank Jukka Corander, three anonymous reviewers, and the editor, Jared Strasburg, for helpful comments, and Tim Wootton for a conversation that helped to stimulate this project.
Appendix: MCMC updates
No admixture model with sample groups
To sample from Pr(P, Z, r, η, γ|X), the algorithm proceeds as follows:
Sample P(m) from Pr(P | Z(m−1), γ(m−1), η(m−1), r(m−1), X).
Sample Z(m) from Pr(Z | P(m), γ(m−1), η(m−1), r(m−1), X).
Update r using a Metropolis-Hastings step.
Update η using a Metropolis-Hastings step.
Update γ using a Metropolis-Hastings step.
Because the new models have only modified the prior for Z, Pr(P | Z(m−1), γ(m−1), η(m−1), r(m−1), X) does not depend on γ, η, or r, and step 1 does not need to be modified from the original structure algorithm.
For step 2, we note that since η and r form a prior for γ, Pr(Z | P(m), γ(m−1), η(m−1), r(m−1), X) is equivalent to Pr(Z | P(m), γ(m−1), X). Then, for each individual i from location li we can sample zi based on the distribution:
where Pr(zi = k|γ) = γlik, and Pr(X | P, Zi = k) is a product of allele frequencies in cluster k corresponding to the genotype data. The exact expression is defined in the appendix of Pritchard et al. (2000).
For step 3, r′ is simulated from a uniform distribution in (r(m−1)—rε, r(m−1) + rε). r′ is rejected if it is not in the range (0, rMAX. Otherwise, it is accepted with the probability:
where l = 1 … S indicates the sampling locations, and where f(γl· | r, η) is given by the Dirichlet distribution:
If r′ is accepted, than r(m) is set to r′, otherwise r(m) is set to r(m−1).
In all the analyses in this manuscript, rε was set to 0.1.
For step 4, two clusters, i and j, are chosen at random so that i ≠ j. A random number ε is simulated randomly in the range (0, εMAX). Then, is set to , and is set to . All other elements are set to for k not equal to i or j. The update is rejected if either or is not in the range (0,1). In this way, the elements of the η′ vector are guaranteed to sum to 1, given that the elements of η(m−1) sum to 1. Then, η′ is accepted with the probability:
If η′ is accepted, η(m) is set to η′. Otherwise, η(m) is set to η(m−1). For all analysis in this paper, εMAX was set to 0.025.
For step 5, each vector γl· is updated in turn, for each location l. A .is generated in exactly the same manner as η’, and is rejected if any of the elements are not in the range (0,1). Then, γl·′ is accepted with the probability:
Here, I(li = l) is the indicator function which equals 1 if individual i comes from location l, and zero otherwise, and g(zi | γ) is the probability of observing a particular value of zi, given γ. If is accepted, is set to γ′, otherwise is set to .
Admixture model with sample groups
To sample from Pr(Z, Q, P, α, r | X), the algorithm proceeds as follows:
Sample P(m) from Pr(P | Z(m−1), Q(m−1), α(m−1), r(m−1), X).
Sample Q(m) from Pr(Q | P(m), Z(m−1), α(m−1), r(m−1), X).
Sample Z(m) from Pr(Z | P(m), Q(m), α(m−1), r(m−1), X).
Update r using a Metropolis-Hastings step.
Update α using a Metropolis-Hastings step.
The new admixture model only affects the prior for Q, and therefore steps 1 and 3 do not need to be modified from the original algorithm. To perform step 2, the admixture proportions for individual i from location l have a distribution given by:
where nik is the total number of copies of each locus assigned to population k in individual i.
For step 4, r′ is simulated from a uniform distribution in (r(m−1) − rε, r(m−1) + rε), where rε is the same as in the new no-admixture model. r′ is rejected if it is not in the range (0, rMAX). Otherwise, it is accepted with the probability:
where h(αlk | r,) is given by the Gamma distribution with parameters r, 1/r.
Step 5 is achieved by independently updating every element of the α vector. First each element of α(g) is updated. is simulated from a normal distribution with mean and standard deviation σα, It is rejected if it is outside the range (0, αMAX). Otherwise, it is accepted with the probability:
Finally, to update each element of αlk, an is simulated from a normal distribution with mean and standard deviation σα. It is accepted with the probability:
For all the analysis in this paper, σα was set to 0.025.
References
- Balding DJ, Nichols RA. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica. 1995;96:3–12. doi: 10.1007/BF01441146. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multilocusmolecular markers. Molecular Ecology. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Siren J, Arjas E. Bayesian spatial modeling of genetic population structure. Computational Statistics. 2008;23:111–129. [Google Scholar]
- Corander J, Waldmann P, Sillanpaa MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson KJ, Belkhir K. A Bayesian approach to the identification of panmictic populations and the assignment of individuals. Genetics Research. 2001;78:59–77. doi: 10.1017/s001667230100502x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois O, Ancelet S, Guillot G. Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics. 2006;174:805–816. doi: 10.1534/genetics.106.059923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G, Santos F, Estoup A. Analysing georeferenced population genetics data with geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics. 2008;24:1406–1407. doi: 10.1093/bioinformatics/btn136. [DOI] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. A K-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- Nicholson G, Smith AV, Jónsson F, et al. Assessing population differentiation and isolation from single-nucleotide polymorphism data. Journal of the Royal Statistical Society B. 2002;64:695–715. [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigen analysis. Public Library of Science, Genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Sham P. Properties of structured association approaches to detecting population stratification. Human Heredity. 2004;58:93–107. doi: 10.1159/000083030. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Wasser SK, Shedlock AM, Comstock K, et al. Assigning African elephant DNA to geographic region of origin: applications to the ivory trade. Proceedings of the National Academy of Sciences, USA. 2004;101:14847–14852. doi: 10.1073/pnas.0403170101. [DOI] [PMC free article] [PubMed] [Google Scholar]