Abstract
We provide a new approach for fluorescent probe design termed “PEG-fluorochrome Shielding,” where PEGylation enhances quantum yields while blocking troublesome interactions between fluorochromes and biomolecules. To demonstrate PEG-fluorochrome shielding, fluorochrome-bearing peptide probes were synthesized, three without PEG and three with a 5 kDa PEG functional group. In vitro, PEG blocked the interactions of fluorochrome-labeled peptide probes with each other (absorption spectra, self-quenching) and reduced nonspecific interactions with cells (by FACS). In vivo PEG blocked interactions with biomolecules that lead to probe retention (by surface fluorescence). Integrin targeting in vivo was obtained as the differential uptake of an 111In labeled, fluorochrome shielded, integrin binding RGD and control RAD probes. Using PEG to block fluorochrome mediated interactions, rather than synthesizing de novo fluorochromes, can yield new approaches for the design of actively or passively targeted near infrared fluorescent probes.
Near infrared (NIR) fluorescent, receptor targeted peptides have been used for pre-operative SPECT (or PET) imaging and/or for intraoperative tumor detection in both pre-clinical1 and clinical settings2. Though NIR fluorochromes are desirable because of the tissue penetrating properties of their light and low background, they typically involve multiple unsaturated double bonds linking multiple unsaturated rings. These features lead to self-quenching fluorochrome /fluorochrome interactions, high non-specific binding to many cells, unwanted interactions with proteins and lipids in vivo and enterohepatic circulation rather than renal elimination. Efforts to obtain improved NIR fluorochromes have often followed a “medicinal chemistry” approach of synthesizing low molecular weight fluorochromes and screening them for desirable properties3.
Here we provide an example where a PEG placed on a peptide along with a NIR fluorochrome blocks troublesome interactions between fluorochromes and biological molecules, increasing quantum yield and permitting peptide/target binding. The notion that fluorochromes can be improved, not through fluorochrome redesign, but rather by the association with PEG, can yield new avenues for probe design through PEG-fluorochrome improvement. PEG has been used to extend protein circulation and alter surface chemistry4, but its ability to improve fluorochrome behavior has not been recognized. To demonstrate this “PEG-Fluorochrome Shielding” was compatible with molecular targeting in vivo, we show that a PEGylated, fluorochrome-bearing RGD probe lacks unwanted fluorochrome mediated interactions, yet maintains desirable RGD/integrin interactions.
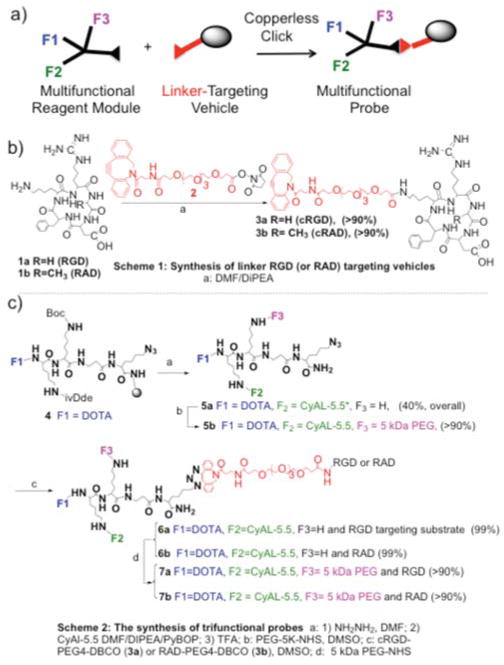
We synthesized PEGylated and unPEGylated, RGD and RAD probes bearing the CyAL5.5 fluorochrome5, using the strategy of synthesizing a multifunctional reagent module to integrin targeted peptides6, see Figure 1. Here three functional groups (F1=DOTA, F2=fluorochrome, F3=5 kDa PEG) were attached to a Lys-Lys-βAla-Lys(N3) peptide scaffold, and the multifunctional reagent module was then reacted with a linker-targeting module bearing a RGD or RAD targeting peptide. DOTA allowed radiometal chelation for biodistribution and/or imaging studies.
Figure 1. Synthesis of PEG-Fluorochrome Shielded Integrin Targeted Probes.
(a) A multifunctional reagent module was first synthesized and attached to a linker-targeting vehicle module via a copperless click reaction, to yield a multifunctional probe. (b) Synthesis of the linker-targeting vehicles bearing RGD or RAD peptides. (c) Synthesis of the multifunctional probes with functional groups of F1= DOTA, F2= CyAL5.5 fluorochrome and F3= 5 kDa PEG. Reaction conditions for a: 1) NH2NH2, DMF; 2) CyAL5.5 Acid/ PyBOP/ DMF/ DIPEA; 3) TFA; b: PEG-5K-NHS, DMSO; c: 5a, cRGD-PEG4-DBCO (3a) or RAD-PEG4-DBCO (3b), DMSO; d: PEG-5K-NHS. For the need of the linker arm for RGD/integrin interactions, see Figure S4. Compounds were greater than 90% pure by RP-HPLC and FPLC (Figure 2d).
Though differing in a single methyl group (glycine versus alanine), RGD and RAD peptides have profoundly different strengths of interactions with integrins, see7, with further support provided in Figure–4a. A copperless click reaction joined the terminal DBCO (dibenzylcyclooctane) of the linker-targeting vehicle to the azide of trifunctional reagent, avoiding the possibility of a copper reaction with the DOTA. DBCO and azide moieties were found to be highly stable and readily underwent efficient click reactions when combined. We term as “PEGylated probes” those bearing the F3 5 kDa PEG functional group, which consists of 115 polyethyleneglycol units rather than the short linker-PEG, which consists of just 4 such units. Complete structures of functional groups and targeting peptide structures are given in the Supplement, Figure S1. The molecular weights, along with a summary of our results, are provided in Table 1.
Table 1.
Physical properties and behavior in biological systems of multifunctional reagent modules (5a, 5b), and unPEGylated (6a, 6b), and PEGylated Probes (7a, 7b). Bold = 5 kDa PEG.
| F3: 5 kDa PEG | Target Pept. | MW (Da) | MS Obs. (Da) | Quant. Yield, Fig. 2b | Fluor. Life-time (ns) Fig. 2c | Volume By FPLC (kDa) Fig. 2e | Cell Fluor. (a.u.) Fig. 3b | IM Inject. Fig. 3c, 3d | |
|---|---|---|---|---|---|---|---|---|---|
| 5a | No | None | 1667.0 | 1666.2 | 0.061±0.006 | 0.31±0.04 | 0.4 | 79.7±4.3 | |
| 5b | Yes | None | 6805.1 | 6850 (peak) | 0.17±0.008 | 0.66±0.02 | 20 | 3.1±0.2 | |
| 6a | No | RGD | 2848.3 | 2848.3 | 0.082±0.004 | 0.52±0.05 | 0.4 | 26.7±0.6 | |
| 6b | No | RAD | 2862.4 | 2862.2 | 0.081±0.004 | 0.50±0.02 | 0.4 | 20.4±0.4 | Retained |
| 7a | Yes | RGD | 7987.4 | 7980 (peak) | 0.20±0.01 | 0.81±0.02 | 25 | 9.7±0.3 | |
| 7b | Yes | RAD | 8001.4 | 8000 (peak) | 0.20±0.01 | 0.80±0.03 | 25 | 3.2±0.2 | Elim. |
| CyAL5.5 | No | None | 800.01 | 799.31 | 0.08±0.004 | 0.40±0.03 | N.O. | N.O |
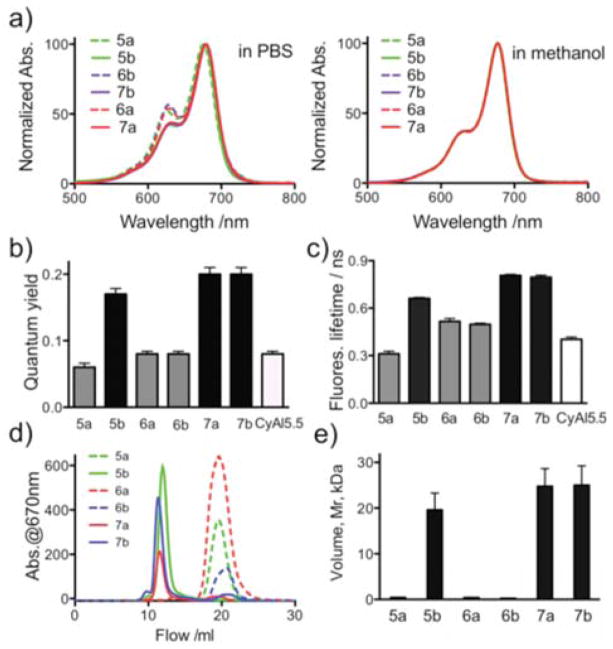
We examined the interactions between the 5 kDa PEG and the CyAL5.5 fluorochrome using the three PEGylated and three unPEGylated compounds from Figure 1c as shown in Figure 2. In PBS, our three unPEGylated peptides (5a, 6a, 6b) exhibited “an aggregation peak” at about 630 nm, typical of stacked fluorochromes, while three PEGylated peptides (5b, 7a, 7b) had reduced aggregation peaks8. Remarkably, in methanol the three PEGylated and three unPEGylated had identical absorption spectra, and those were characteristic of unaggregated fluorochromes.
Figure 2. Physical properties of six compounds from Figure 1c.
(a) Effects of 5 kDa PEG on probe absorption spectra in PBS and methanol. (b) Quantum yields. (c) Fluorescent lifetimes. (d) FPLC chromatograms. (e) Mv’s of PEGylated and unPEGylated probes. For the method of determining volume, see Figure S5.
We examined the effects of the F3 5 kDa PEG functional group on quantum yields (Figure 2b), and fluorescent lifetimes (Figure 2c) using the six fluorochrome-bearing compounds from Figure 1c. PEGylation produced significant (p<0.0001, see Table S1) increases in quantum yield (Figure 2b, 5a vs. 5b; 6a vs. 7a; 6b vs. 7b). PEGylation also produced significant increases in fluorescence lifetime (Figure 2c and Figure S3, 5a vs. 5b; 6a vs. 7a; 6b vs. 7b), with p < 0.0001 in all cases. PEG yielded probes with globular protein equivalent volumes (Mv’s) of 20 to 25 kDa, (5b,7a,7b, Figures 2d, 2e), similar to small proteins. Our three unPEGylated probes (5a,6a,6b) had volumes less than 1 kDa. Thus PEGylation not only blocked fluorochrome-fluorochrome interactions, it dramatically increased probe volume. To determine whether PEG-fluorochrome shielding was occurred biological systems, we examined the effects of PEGylation on the binding of probes to cells and on the elimination of probes after injection.
Figure 5.
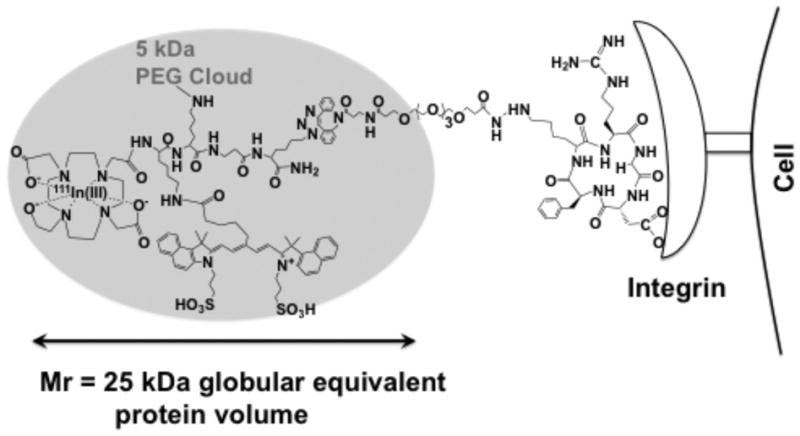
Model of a PEG-Fluorochrome shielded, probe (7a) and its binding to an integrin.
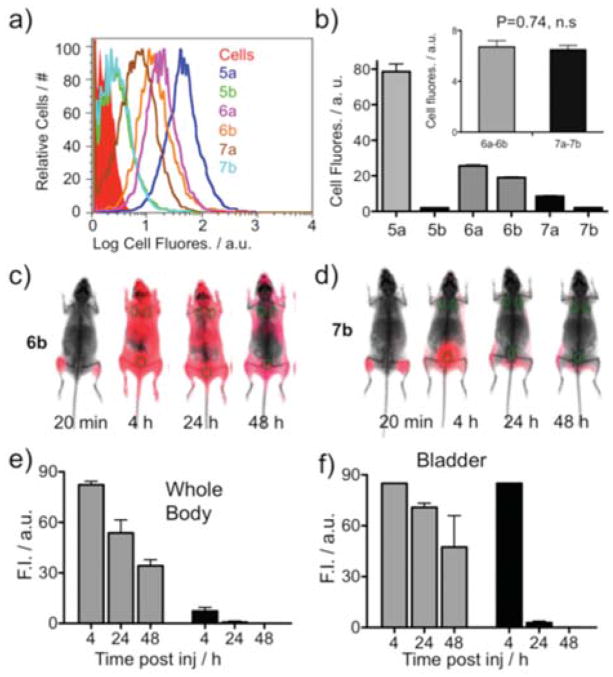
The ability of the F3 5 kDa PEG to reduce the non-specific binding to cultured cells, while maintaining specific binding, is shown in Figures 3a and 3b, with numerical values provided in Table 1. Since BT-20 cells express the RGD binding integrins9, this cell line was used. With the PEGylation of 5a to obtain 5b, cell fluorescence dropped from 79.7 to 3.1 a.u.. PEGylation also reduced the binding of RGD bearing peptides (6a = 26.7 a.u. to 7a = 9.7) and RAD bearing peptides (6b = 20.4 to 7b = 3.2). However, integrin specific binding (see inset for values of 6a–6b, 7a–7b) was not affected by PEGylation (p > 0.05). The binding of the PEGylated RAD probe 7b to cells (3.2±0.2) was similar to that of the PEGylated peptide lacking both RGD and RAD (5b, 3.1±0.2). Therefore our PEGylated RAD probe, 7b, is a valid control for our PEGylated RGD probe, 7a, because of its similar physical properties and because it does not bind integrins.
Figure 3. Effect of PEG on the specific and non-specific binding to cells and on probe elimination.
Cell fluorescence (BT-20 cells) after incubation with the six fluorochrome bearing peptides from Figure 1c was determined by FACS as shown in (a), with peak fluorescence tabulated in (b). (c, d) Whole body surface fluorescence of mice after 6b (RAD control, unPEGylated) or 7b (RAD control, PEGylated) injection at 1 nmole/site. Circles are the regions of interest used to quantify whole body (e) or bladder (f) surface fluorescence. (e) Quantification of whole body surface fluorescence of 7 mice. Gray bars for 6b or black for 7b. (f) Quantification of bladder fluorescence for 7 animals.
A measure of the ability of the 5 kDa PEG to block non-specific binding in vivo was obtained by probe retention/elimination after an IM injection, Figures 3c–f. Mice received dual IM injections of 6b (RAD, unPEGylated) or 7b (RAD, PEGylated), with time dependence of surface fluorescence shown in Figure 3c–d. Two regions of interest (circles near the shoulder) were used to quantify whole body surface fluorescence, while a single region of interest near the bladder was used to quantify bladder fluorescence and renal elimination. Results for whole body and bladder fluorescence are quantified in Figures 3e and 3f, respectively. PEGylated 7b showed pronounced renal elimination at 4h, and a lack of whole body fluorescence. The unPEGylated 6b exhibited strong nonspecific interactions, evident by the retention of whole body fluorescence even at 48h post injection.
A summary of the physical properties of our compounds and their behavior in biological systems in vitro and in vivo is provided in Table 1. In vitro the F3, 5 kDa PEG functional group reduced the interactions of fluorochromes with each other (absorption spectra, quantum yield, Figure 2a, 2b), increased probe volume (Figure 2e), reduced the nonspecific interactions with cells (Figure 3a, 3b), and reduced the interactions with biomolecules that lead to the retention of fluorochrome bearing probes after injection (Figure 3c, 3d).
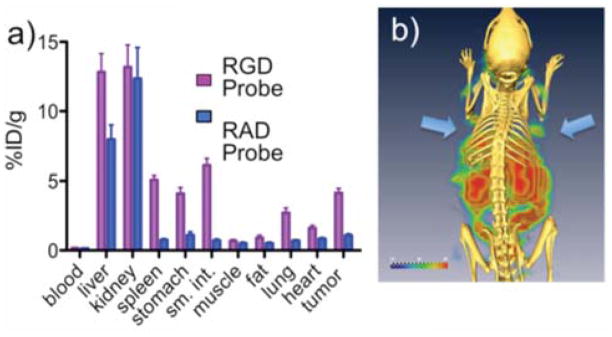
To demonstrate that our PEG-Fluorochrome Shielding allowed RGD/integrin interactions in vivo, the DOTA functional groups of the PEGylated RGD 7a and PEGylated RAD 7b probes were labeled with 111In and their biodistributions determined in a BT-20 xenograft system (Figure 4a, Table S2). Specific RGD binding occurred in the tumor, stomach, small intestine, lung and heart. The differential binding of RGD and RAD probes indicates integrin specific uptake, see above, and provides a method of determining the total RGD/integrin binding of cells (Figure 3) or tissues (Figure 4a). Integrin specific RGD binding to the stomach, spleen and small intestine results from a wide range of RGD integrins on diverse cells and tissues10. A SPECT/CT image of a 7a-injected animal is shown in Figure 4b.
Figure 4.
In vivo integrin targeting using the PEG-Fluorochrome Shielded RGD probe (7a) and RAD (7b) probe. a) Tissue concentrations of 111In labeled 7a and 7b. Numerical data are presented in the Supplement, Table S2. b) SPECT-CT image of 7a. Arrows indicate dual BT-20 tumors. Data are at 24 h post i.v. injection.
A model of the PEG-fluorochrome 7a probe, consistent with our observations and not drawn to scale, is shown in Figure 5. The 5 kDa PEG forms a cloud around the fluorochrome, blocking nonspecific interations (Fig. 2b, 2c) but permiting integrin interactions (Fig. 3b, 4a). PEG provides most of the probe volume (Fig. 2e). However, an essential (Fig. S4), short PEG linker separates the integrin binding RGD targeting peptide from the PEG shielded fluorochrome. With this design PEG shielded the fluorochrome, and blocked fluorochrome mediated interactions, but permitted RGD/integrin interactions.
The ability to improve fluorochromes using covalent linkages between fluorochromes and PEG’s, rather than by de novo fluorochrome synthesis, can yield many new avenues for the design of actively or passively targeted fluorescent probes. When the goal is a receptor binding (actively targeted) probe, the strategy of attaching a PEG and a fluorochrome to a common amino acid or peptide can be employed. Our version of this general strategy employed a modular synthetic route (Figure 1a) using a trifunctional reagent module ((DOTA)Lys(CyAL5.5)-Lys(5 kDa PEG)-βAla-Lys(N3) or 5b). The amino group of an RGD targeting peptide was reacted with a short PEG linker featuring a terminal dibenzylcyclooctyne (DBCO) group, and a copperless click reaction joined the reagent module and linker RGD targeting peptide. For the many intra-operative fluorescent imaging applications11, passively targeted (untargeted), PEG shielded fluorochromes with similar optical properties to ICG, but which have lost their troublesome interactions with biological molecules, might be employed.
Supplementary Material
Acknowledgments
This work was supported by R01’s from the NIH: EB 011996, EB 009691.
Footnotes
Supporting Information Available: Detailed synthesis of all compounds, methods for cell binding assay, animal model, surface fluorescence imaging, absorption spectra, quantum yield, volume determination, SPECT-CT, fluorescence lifetime, biodistribution, and additional data, including expanded structure of compounds, more absorption and emission spectra, fluorescence lifetime curve, and displacement by direct PEG probe. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Kuil J, Velders AH, van Leeuwen FW. Bioconjugate chem. 2010;21:1709. doi: 10.1021/bc100276j. [DOI] [PubMed] [Google Scholar]; (b) Luo S, Zhang E, Su Y, Cheng T, Shi C. Biomaterials. 2011;32:7127. doi: 10.1016/j.biomaterials.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 2.van Dam GM, Themelis G, Crane LM, Harlaar NJ, Pleijhuis RG, Kelder W, Sarantopoulos A, de Jong JS, Arts HJ, van der Zee AG, Bart J, Low PS, Ntziachristos V. Nat Med. 2011;17:1315. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 3.(a) Choi HS, Nasr K, Alyabyev S, Feith D, Lee JH, Kim SH, Ashitate Y, Hyun H, Patonay G, Strekowski L, Henary M, Frangioni JV. Angew Chem Int Ed Engl. 2011;50:6258. doi: 10.1002/anie.201102459. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Licha K, Riefke B, Ntziachristos V, Becker A, Chance B, Semmler W. Photochem photobiol. 2000;72:392. doi: 10.1562/0031-8655(2000)072<0392:hcdaca>2.0.co;2. [DOI] [PubMed] [Google Scholar]; (c) Licha K, Welker P, Weinhart M, Wegner N, Kern S, Reichert S, Gemeinhardt I, Weissbach C, Ebert B, Haag R, Schirner M. Bioconjugate chem. 2011;22:2453. doi: 10.1021/bc2002727. [DOI] [PubMed] [Google Scholar]
- 4.Pasut G, Veronese FM. Adv Drug Deliv Rev. 2009;61:1177. doi: 10.1016/j.addr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Shao F, Yuan H, Josephson L, Weissleder R, Hilderbrand SA. Dyes Pigm. 2011;90:119. doi: 10.1016/j.dyepig.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Garanger E, Aikawa E, Reynolds F, Weissleder R, Josephson L. Chem Commun (Camb) 2008:4792. doi: 10.1039/b809537j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Garanger E, Blois J, Hilderbrand SA, Shao F, Josephson L. J Comb Chem. 2010;12:57. doi: 10.1021/cc900141b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Mulder WJ, Strijkers GJ, Habets JW, Bleeker EJ, van der Schaft DW, Storm G, Koning GA, Griffioen AW, Nicolay K. FASEB J. 2005;19:2008. doi: 10.1096/fj.05-4145fje. [DOI] [PubMed] [Google Scholar]; (b) Kok RJ, Schraa AJ, Bos EJ, Moorlag HE, Asgeirsdottir SA, Everts M, Meijer DK, Molema G. Bioconjugate chem. 2002;13:128. doi: 10.1021/bc015561+. [DOI] [PubMed] [Google Scholar]
- 8.Galande AK, Hilderbrand SA, Weissleder R, Tung CH. J Med Chem. 2006;49:4715. doi: 10.1021/jm051001a. [DOI] [PubMed] [Google Scholar]
- 9.(a) Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. J Med Chem. 2006;49:6087. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]; (b) Montet X, Montet-Abou K, Reynolds F, Weissleder R, Josephson L. Neoplasia. 2006;8:214. doi: 10.1593/neo.05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphries JD, Byron A, Humphries MJ. J Cell Sci. 2006;119:3901. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sevick-Muraca EM. Annu Rev Med. 2012;63:217. doi: 10.1146/annurev-med-070910-083323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.