Abstract
Technological limitations pose a major challenge to acquisition of myocardial fiber orientations for patient-specific modeling of cardiac (dys)function and assessment of therapy. The objective of this project was to develop a methodology to estimate cardiac fiber orientations from in vivo images of patient heart geometries. An accurate representation of ventricular geometry and fiber orientations was reconstructed, respectively, from high-resolution ex vivo structural magnetic resonance (MR) and diffusion tensor (DT) MR images of a normal human heart, referred to as the atlas. Ventricular geometry of a patient heart was extracted, via semiautomatic segmentation, from an in vivo computed tomography (CT) image. Using image transformation algorithms, the atlas ventricular geometry was deformed to match that of the patient. Finally, the deformation field was applied to the atlas fiber orientations to obtain an estimate of patient fiber orientations. The accuracy of the fiber estimates was assessed using six normal and three failing canine hearts. The mean absolute difference between inclination angles of acquired and estimated fiber orientations was 15.4°. Computational simulations of ventricular activation maps and pseudo-ECGs in sinus rhythm and ventricular tachycardia indicated that there are no significant differences between estimated and acquired fiber orientations at a clinically observable level.
Index Terms: Biomedical image processing, cardiomyocyte, electrophysiology, magnetic resonance imaging (MRI), patient-specific modeling, simulation
I. Introduction
The iterative interaction between experimentation and simulation has long played a central role in the advancement of biological sciences. Among computational models of various biological systems, the heart is the most highly advanced example of a virtual organ, capable of integrating data at multiple scales, from genes all the way through the whole organ [1]. State-of-the-art whole-heart models of electrophysiology and electromechanics are currently being used to study a wide range of phenomena, such as normal ventricular propagation, arrhythmia, defibrillation, electromechanical coupling, and cardiac resynchronization [2].
Recent years have witnessed revolutionary advances in imaging, including ex vivo structural and diffusion tensor (DT) magnetic resonance imaging (MRI), that facilitate acquisition of the intact structure of explanted hearts with hitherto unavailable resolution. Leveraging on these advances, a new generation of whole-heart models with unprecedented detail have emerged [3]–[7]. Such ex vivo models are being used in basic research to uncover specific mechanisms of heart dysfunction in diseases such as myocardial infarction and heart failure.
Translation of this ex vivo approach to the in vivo setting of the clinic can yield far reaching consequences. Ultimately, doctors will be able to use simulation tools to predict outcomes of various electrophysiological interventions, and select the optimal cardiac therapy specific to a patient. To this end however, patient-specific ventricular fiber orientation data are necessary, as these orientations determine directions of electrical propagation and strain distributions in the heart. While ventricular geometry and structural remodeling data can now be routinely acquired in vivo with imaging techniques such as late gadolinium enhanced cardiac MR [8], physicians do not currently have access to a method to acquire myocardial orientations data in vivo.
Myocardial fiber orientation data necessary for computational studies of cardiac electrophysiology and mechanics have been predominantly acquired using histological techniques; examples include the canine [9] and swine [10] ventricular models developed at Auckland University, and the rabbit ventricular model developed at the University of California, San Diego [11]. While histological sectioning has traditionally been the gold standard for fiber data acquisition, this technique is exceedingly time consuming, prone to tissue deformation errors introduced due to cutting, restricted to 2-D measurements, and not applicable in vivo. Recently, DTMRI has emerged as a powerful alternative to histology, and DTMRI cardiac fiber data have been acquired in many species [3], [5], [12]. DTMRI-acquired fiber orientations have been shown to match histological data [13], and have begun to be used in whole-heart computational electrophysiology and electromechanics [3], [5]–[7], [14], [15]. Unlike histology, DTMRI offers faster data acquisition, 3-D data, and nondestructive imaging. However, there are several drawbacks to the DTMRI technology, most importantly those that limit its application in the in vivo setting, where the bulk motion of the heart interferes with diffusion measurements.
Attempts to translate the DTMRI technology to the in vivo setting were first made by Edelman et al. [16], but it was later found that their diffusion encoding interacted with myocardial strain, and therefore, a method to correct for strain effects was proposed [17], [18]. But this correction was achieved at the cost of increased acquisition noise and complexity. Although the strain correction method in combination with interpolation has recently been used to perform 3-D tractography of ventricular fiber orientations [19], the data prior to interpolation was very sparse, scan time was about 40 min, and breathholding, a difficult task for patients which typically causes inter-slice misalignments, was necessary. Tseng et al. [20] and Dou et al. [21] have proposed techniques that do not require strain correction, but the former is applicable to only those phases of the cardiac cycle called sweet spots, where the strain effect is negligible, whereas the signal-to-noise ratio of the latter is limited by the gradient intensity available in clinical scanners. Recently, a spin-echo technique to measure cardiac diffusion in vivo was proposed, and the utility of this method in measuring 2-D in-plane diffusion along short axis slices demonstrated [22]. Toussaint et al. [23] have used this method in combination with curvilinear interpolation to perform 3-D tractography of ventricular fiber orientations, but their acquisition was limited to the equatorial part of LV, scan time was about 1.5 h, and breath-hold correction, a very challenging postprocessing step, was necessary. All in all, despite the above mentioned efforts over almost two decades, in vivo translation of cardiac DTMRI remains challenging due to technological inadequacies, e.g., in gradient strength, whole-heart imaging, radio-frequency, and navigators [24].
Previously, there have been a number of efforts to estimate fiber orientations in the ventricles. Among these, rule-based methods have gained some popularity; they assign fiber orientations to the ventricular walls such that the inclination angles vary smoothly in the transmural direction, as a mathematical function of distance from the endocardium [4], [5], [25], [26]. Though there have been two preliminary studies comparing fiber orientations derived using rule-based methods with those obtained from DTMRI [26], [27], how well the idealized rules of myocyte orientation quantitatively correlate with true anatomy in normal and diseased hearts remains largely unknown. A different category of estimation methods utilizes geometric correspondences between two hearts. Panfilov’s group assigned canine heart fiber orientations to the ventricular surfaces of a human heart by computing a landmark-based surface mapping between the human heart and a canine heart [28]. The intramural fiber orientations of the human heart, however, were derived from the acquired surface directions and a rule-based scheme. Sundar et al. [29] presented a technique to estimate fiber orientations via an elastic image transformation of an atlas heart. They tested the methodology using normal and failing canine hearts, and showed that estimated fiber orientations correlate with DTMRI data better than fiber orientations synthesized using a rule-based method. But the elastic mapping employed by them is not guaranteed to be diffeomorphic, and therefore may not preserve the topology of anatomical structures. More importantly, Sundar et al. did not measure the effect of error in their estimated fiber orientations on the results of simulations of cardiac electrophysiology, or any other functions of heart.
We seek to address the need for a new methodology to acquire myocardial fiber orientations for patient-specific simulations of cardiac function. Importantly, we hypothesize that ventricular fiber orientations of a normal or a failing heart can be estimated, given the geometry of the heart and an atlas, where the atlas is a heart whose geometry and fiber orientations are already available. Should our hypothesis be proved, it would pave the way for the use of estimated fiber orientations in patient-specific models of cardiac function.
In this study, we use state of the art techniques to develop the methodology for estimation of cardiac fiber orientations in vivo, and test the hypothesis in normal and failing ventricles. We quantify the estimation error in each heart, and then assess the effect of this error on simulations of cardiac electrophysiology in the specific heart, by computationally simulating local electrical activation maps as well as pseudo-electrocardiograms (pseudo-ECGs).
II. Methods
A. Fiber Orientations Estimation
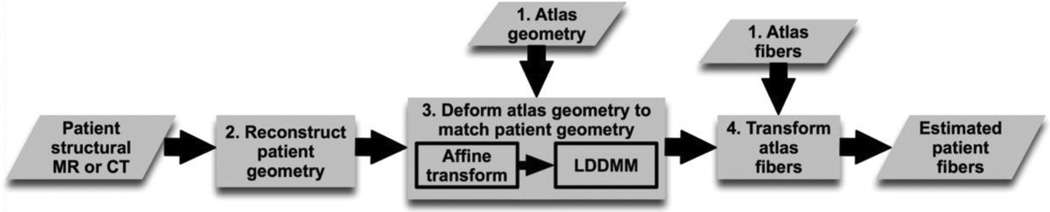
The central idea of our fiber estimation methodology is to exploit similarities in fiber orientations, relative to geometry, between different hearts in order to approximate the fiber structure of a (target) heart for which only the geometry information is available. Fig. 1 illustrates the processing pipeline of our methodology. In the following subsections, we describe the various components of the pipeline by demonstrating how the estimation was performed for an example patient who was scanned using in vivo computed tomography (CT). The numbers inside some of the blocks in Fig. 1 refer to the corresponding subsections below.
Fig. 1.
Our processing pipeline for estimating ventricular fiber orientations in vivo.
1) Atlas Reconstruction
Geometry and fiber orientations of the atlas were extracted from raw data acquired in a previous study [30] using high resolution (0.4297 × 0.4297 × 1 mm3) ex vivo structural MRI and DTMRI, respectively, of a normal human heart in diastole. From the acquired structural image, only the ventricular myocardium of the atlas was extracted by fitting, for each short-axis slice, closed splines through a set of landmark points placed semi-automatically along the epicardial and endocardial boundaries in the slice [Fig. 2(A)]. The placement of landmark points was performed manually for a number of slices that were evenly distributed, with an inter-slice spacing of about 10 mm in the image. The landmark points for the remaining slices were obtained automatically by linearly interpolating the manually identified points. A generic mathematical formulation of this interpolation technique has been reported elsewhere [3], where the technique was used for identifying the atrio-ventricular boundary in an ex-vivo structural MR heart image. Fig. 2 (B) and (C) shows the 3-D geometry and fiber orientations of the reconstructed atlas ventricles; the fibers forming a left-handed helix on the epicardium are clearly visible.
Fig. 2.
Geometry and fiber orientations of the atlas ventricles. (A) Epicardial (red) and endocardial (green and magenta) splines, and corresponding landmarks (yellow) overlaid on an example slice of the atlas image. (B) Atlas ventricles in 3-D. (C) Atlas fiber orientations.
2) Patient Geometry Reconstruction
Ventricular geometry of the patient heart in diastole was reconstructed from an in vivo CT image using a segmentation method that is similar to the one used for the atlas. The patient image was re-sampled prior to reconstruction such that the in-plane resolution was 0.4297 × 0.4297 mm3 as in the atlas. Similarly, the number of slices for which landmarks were manually picked, and the interval of out-of-plane interpolation was adjusted so that the segmented patient heart image had a slice thickness of 1 mm. Fig. 3(A) and (B) illustrates the patient ventricular geometry reconstruction.
Fig. 3.
Application of the fiber orientation estimation methodology to an example patient heart image. (A) Epicardial (red) and endocardial (green and magenta) splines, and corresponding landmarks (yellow) overlaid on an image slice. (B) Patient ventricles in 3-D. (C) Superimposition of ventricles of atlas [magenta, see Fig. 2(B)] and patient. (D) Patient ventricles and the affine transformed atlas ventricles. (E) Patient ventricles and LDDMM-transformed atlas ventricles. (F) Estimated patient ventricular fiber orientations.
3) Atlas Geometry Deformation
Following the reconstruction of the patient ventricles, the atlas ventricular image was deformed using the tools of computational anatomy [31] to match the patient geometry image. The deformation was performed in two steps. The first was an affine transformation based on a set of thirteen landmarks points: the left ventricular (LV) apex, the two right ventricular (RV) insertion points at the base, the two RV insertion points midway between base and apex, and four sets of two points that evenly divide RV and LV epicardial contours at base, and midway between base and apex. Fig. 3 (C) and (D) illustrates the affine transformation of the atlas to match the patient ventricles. In the second step of atlas deformation, the affine-transformed atlas ventricles were further deformed to match the patient geometry, using large deformation diffeomorphic metric mapping (LDDMM).
LDDMM is an image registration algorithm that computes diffeomorphic (invertible, smooth, and with a smooth inverse) transformations between images [32]. Given an atlas image Ia : Ω → R and a patient image Ip : Ω → R, LDDMM computes a flow of diffeomorphisms to transform Ia to match Ip, where Ω ⊂ R3 is the 3-D cube in which the image data are defined, t ∈ [0,1], and υ is a smooth, compactly supported, time-dependent velocity vector field such that
The initial diffeomorphism ϕ0 is the identity transformation. The final diffeomorphism φ = ϕ1 is calculated by integrating the optimal vector field given by
The norm ‖ · ‖V is defined as ‖f‖V = ‖Lf‖L2 to enforce smoothness on the vector fields υ ∈ V, where L = (−α∇2 + γ)2 I3×3 is a differential operator of the Cauchy–Navier type. The parameters α and γ control the elasticity of the transformation, and ‖ · ‖ L2 is the L2 norm of square integrable functions defined on Ω. The optimal solution is computed by means of a gradient descent search, with the gradient
where and |Df| is the determinant of the Jacobian matrix. The diffeomorphic property of LDDMM guarantees that the atlas does not “fold over” itself during deformation, thereby preserving the integrity of anatomical structures. The above equations also ensure that the computed transformations ϕt define a geodesic path in the space of diffeomorphisms, thereby providing a metric quantification of the difference between the atlas and patient. While geodicity is not essential for fiber orientations estimation, from the standpoint of our long-term goals in patient-specific cardiac image analysis, it is a desirable property. For detailed mathematical descriptions of the LDDMM algorithm and implementation, the reader is referred to previous publications [31], [33]. Fig. 3(E) illustrates the LDDMM transformation of the atlas to match the patient geometry; the match is remarkable. Note that the registration was performed on 3-D binary images, and not surfaces.
4) Atlas Fiber Orientations Deformation
In the final phase of the processing pipeline, the transformation matrix of the affine matching and the deformation field of the LDDMM transformation were applied in sequence to morph the DTMRI image of the atlas to obtain an estimate of the patient ventricular fiber orientations. The morphing of the atlas DTMRI image consisted of spatial repositioning of image voxels in accordance with the spatial transformation of the geometry images, and reorientation of the DTs. For the reorientation of the DTs, we considered two methods, namely the preservation of principal directions (PPD) and finite strain (FS) [34]. The FS method models transformations as rigid rotations, and reorients DTs accordingly. On the other hand, the PPD method models the deformation field at an image voxel as an affine transformation, and reorients the DT at that voxel such that the primary eigenvector of the new DT is , where e1 is the primary eigenvector of the original DT and is the affine transformation [34]. The secondary eigenvector of the new DT is orthogonal to e1 and lies in the plane spanned by and Fe2, where e2 is the secondary eigenvector of the original DT. Thus, the PPD method incorporates, in addition to rotations, deformations such as shearing and nonuniform scaling. This method preserves not only the principal DT direction, but also the plane spanned by the two largest eigenvectors. Therefore, in this study, we employed the PDD method. We also performed some quantitative comparisons of the two methods to ensure that the PPD method performs better.
The estimate of the patient fiber orientations was obtained from the morphed atlas DTMRI image by computing the primary eigenvector of the DTs [13]. Fig. 3(F) illustrates a streamlined visualization of the estimated fiber orientations in the patient ventricles. The fiber orientations vary smoothly across the myocardial wall, form a left-handed helix on the epicardium, and appear as a right-handed helix on the endocardium [30].
B. Measurement of Estimation Error and Its Effects
Tests of efficacy of the proposed methodology were performed on canine hearts due to the unavailability of human hearts. Ventricles segmented from a total of six normal and three failing canine hearts, the fiber orientations of all of which were acquired in diastole with ex vivo DTMRI at a resolution of 312.5 × 312.5 × 800 µm3 were used. The datasets have been employed in previous studies [3], [12], [35], [36], where the reader can find details of the acquisition. In brief, the experimental model of cardiac dyssynchrony and heart failure was generated in canines via radio-frequency ablation of the left bundle-branch block followed by three weeks of tachypacing (210 min−1). The animals were then euthanized and hearts explanted. Images of the explanted hearts were acquired and the ventricles segmented similarly to the human atlas heart, as described under Section III-A1. In the following, ventricles segmented from normal canine hearts are referred to as hearts 1–6, and those segmented from failing canine hearts as hearts 7–9.
To determine if the fiber estimation is accurate in the structurally normal ventricles, five different estimates of ventricular fiber orientations of heart 1 were obtained by using each of hearts 2–6 as an atlas. To test the accuracy of ourmethodology in the structurally remodeled ventricles, fiber orientations for each of the failing ventricles were estimated using heart 1 as the atlas. The purpose of using multiple atlases and a single target in the normal case was twofold. Firstly, it allowed us to confirm that the performance of the proposed methodology was independent of the choice of the atlas. Secondly, it made averaging of ventricular distributions of error across different estimates easier.
Estimation error was computed by comparing inclination angles of estimated and acquired fiber orientations. The effects of this error on outcomes of computational electrophysiological simulations, specifically activation times and ECGs, were examined. Since our estimation methodology was developed for the purpose of modeling, the performance analysis was focused on simulation outcomes.
1) Assessment of Estimation Error
Fiber orientations are typically characterized using inclination angles [13] following the tradition of histology, where angular measurements are performed on tissue sections that are cut parallel to the epicardial surface. Inclination angle of a fiber direction is the angle between the circumferencial direction and the projection of the fiber direction on the epicardial tangent plane. Since the angle between the fiber direction and epicardial tangent plane is generally small [37], [38], the information loss in describing a fiber direction entirely using its inclination angle is insignificant. We used the technique presented in Scollan et al. [13] to calculate inclination angles directly from image data. Briefly, each fiber direction was projected onto the tangential plane at the nearest epicardial point, and the angle between the projection and the circumferential direction measured. The tangential plane at an epicardial point was computed as the plane perpendicular to the short-axis imaging plane, and tangential to the short-axis epicardial contour passing through that point. The circumferential direction was calculated as the vector cross product between the normal to the short-axis imaging plane that points toward the base of the heart, and the normal to the tangential plane that points away from the heart. At each voxel that belongs to the myocardium of each target heart, we computed the estimation error as |θe − θa|, where θe and θa are the inclination angles of estimated and acquired fiber orientations, respectively. Note that inclination angles have values between −9° and +90°, and therefore, the estimation error ranges between 0° and 180°. To demonstrate that the inclination angles characterize the fiber orientations well, we also computed the acute angle between estimated and acquired fiber directions in 3-D by means of the vector dot product.
2) Quantification of the Effects of Estimation Error
We quantified the effect of estimation error on the outcome of computational cardiac electrophysiology simulations by comparing ventricular activation maps and pseudo-ECGs calculated using estimated fiber orientations with those using acquired fiber orientations.
From heart 1, six models, one with the DTMRI-acquired fiber orientations of heart 1 (referred to as model 1), and five with the five estimated fiber orientations datasets (models 2–6), were constructed. For each of the three failing heart geometries, two ventricular models, one with the DTMRI-acquired fiber orientations and the other with the estimated fiber orientations were also constructed. The heart failure models with DTMRI-acquired fibers were denoted as models 7–9, and those with estimated fibers as models 10–12. Computational meshes were generated for each model. Our methodology for mesh generation, given segmented geometry and fiber orientations, involved discretization of the computational domain into finite elements using the software Tarantula,1 and assignment of fiber orientations to the elements using interpolation. Details of the mesh generation procedure have been reported in a previous publication [3].
Mathematical description of cardiac tissue was based on the monodomain representation, which is a reaction-diffusion system composed of a partial differential equation coupled to system of ordinary differential equations [39]. The governing equations are
where σi is the intracellular conductivity tensor; Vm is the transmembrane potential; Cm is the membrane specific capacitance; and Iion is the density of the transmembrane current, which in turn depends on Vm and a set of state variables μ describing the dynamics of ionic fluxes across the membrane. For Cm, we have used a value of 1 µF/cm2. For σi in normal canine heart models, we used longitudinal and transverse conductivity values of 0.34 S/m and 0.06 S/m, respectively, as described by Roberts et al. [40]. Iion was represented by the Greenstein et al. [41] ionic model of the canine ventricular myocyte. This model consisted of canine specific representations of membrane ion channels and intracellular calcium cycling. For the failing canine heart, we used the modifications in canine myocyte membrane dynamics as described by Winslow et al. [42]. Electrical conductivities in canine heart failure ventricular models were also decreased by 30%, consistent with experimental data on electrical propagation in failing hearts [43]. For more details on our cardiac tissue representation including the various parameters, the reader is referred to the relevant publications [40]–[43].
Sinus rhythm was simulated by replicating activation originating from the Purkinje network, The ventricles were activated at six locations on the endocardium: one on the RV free wall, three on the LV septum and two on the LV free wall, as described in [44]. Appropriate timings of the stimuli were chosen such that the resultant 3-D electrical propagation matched with experimental data [45], [46]. Since failing hearts are arrhythmogenic, in addition to sinus rhythm, reentrant ventricular tachycardia (VT) was induced in the six failing models to assess the utility of the fiber estimation methodology in models of cardiac arrhythmias. An S1–S2 pacing protocol [28] was used to induce VT. The timing between S1 and S2 was chosen such that VT activity was sustained for 2 s after S2 delivery. If VT was not induced for any S1–S2 timing, the conductivities were further decreased by up to 70% so that VT was induced, as done in experimental protocols that utilize pharmacological intervention to decrease conduction velocity and thus increase the likelihood of inducing VT [43].
To assess the effects of estimation error on clinically observable electrophysiological data, pseudo-ECGs were calculated by taking the difference of extracellular potentials between two points in an isotropic bath surrounding the hearts. The two points were placed near the base of the heart separated by 18 cm, and the line connecting themwas perpendicular to the base-apex axis. The extracellular potentials at these points were approximated using the integral equation introduced by Gima et al. [47]. To quantify difference between two ECG waveforms, the so-called mean absolute deviation (MAD) metric was computed as
where X and Y are waveforms of length n [48]. MAD varies between 0% and 100%, corresponding to identical and completely different waveforms, respectively. The MAD metric was suitable because it has been utilized in clinical studies, to compare ECGs of reentrant activity and paced propagation for localization of the organizing centers of reentrant circuits [48]. A MAD score of less than 12% implies that the two underlying propagation patterns are clinically equivalent. All simulations were performed using the software package CARP (CardioSolv, LLC); the numerical methods are described in previous publications [49], [50].
III. Results
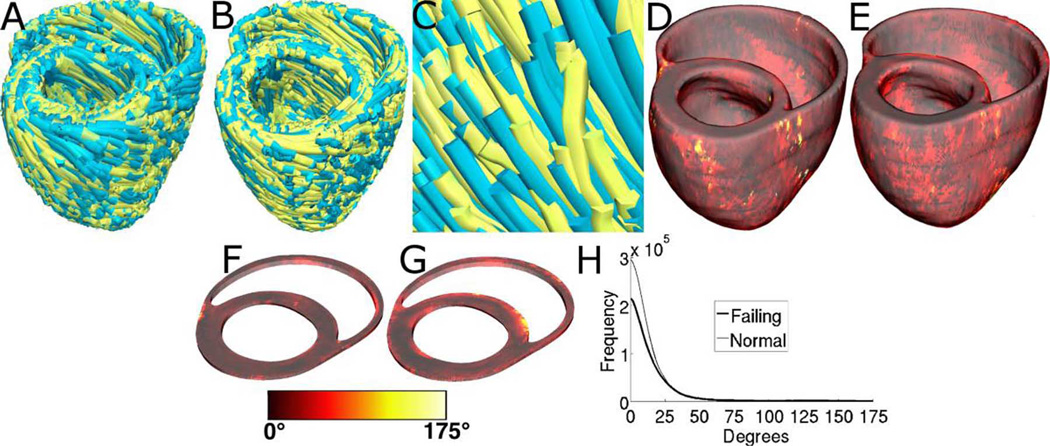
Fig. 4(A)–(C) displays streamlined visualizations of estimated as well as DTMRI-derived fiber orientations in normal and failing hearts. Qualitative examination shows that estimated fiber orientations align well with DTMRI-derived ones. Fig. 4(D) illustrates, overlaid on the geometry of heart 1, the distribution of error in normal hearts’ inclination angles, averaged across all five estimates. Fig. 4(E) shows the mean distribution of error in failing hearts’ inclination angles, overlaid on the geometry of heart 1. Fig. 4(F) and (G) presents sections of tissue from the distributions in Fig. 4(D) and (E), respectively. In Fig. 4(E), the errors calculated in the ventricles of each of the three failing hearts were mapped onto the geometry of heart 1 based on the point-to-point correspondences obtained from affine and LDDMM transformations; the mapped errors were then averaged. Fig. 4(F) and (G) highlights the transmural variation of error. The histograms of errors in Fig. 4(H) suggest that most myocardial voxels have small error values. About 80% and 75% of the voxels have errors less than 20 in normal and failing ventricles, respectively. It was found that the mean error, averaged across all estimated datasets, and all image voxels that belonged to the myocardium, were 14.4° and 16.9° in normal and failing ventricles, respectively. The mean error in the entire myocardium, in normal and failing cases combined, was 15.4°. The mean 3-D acute angle between estimated and acquired fiber directions were 17.5° and 18.8° in normal and failing ventricles, respectively. The 3-D angles are comparable to the estimation errors, demonstrating that the information loss in describing fiber orientations by means of inclination angles is insignificant. The standard deviation of error across the five different estimates of the fiber orientations of heart 1 was only 1.9° indicating that the variation in estimation quality from one atlas to another was small.
Fig. 4.
Validation of the fiber orientation estimation methodology by comparing estimated fiber orientations with DTMRI-derived orientations. (A) Superimposition of DTMRI-acquired fiber orientations (greenish yellow) and one set of estimated fiber orientations (cyan) of heart 1. (B) Acquired and estimated fiber orientations of heart 7. (C) Enlarged portion of (B) showing alignment between acquired and estimated fiber orientations. Note that the streamlines were generated at random locations within the myocardium for visualization purposes only, and so their exact positions are irrelevant. (D) Distribution of mean estimation error in normal ventricles. (E) Distribution of mean estimation error in failing ventricles. (F) Section of tissue extracted from (D). (G) Section of tissue extracted from (E). Colorbar applies to (D)–(G). (H) Histograms of errors in normal and failing ventricles. Frequency denotes the number of voxels having a given error.
To compare the performance of the PDD and FS reorientation strategies in our methodology, the processing pipeline was modified by replacing the PPD method with the FS technique in the affine transformation step. The modified pipeline was then applied to estimate fiber orientations in failing ventricles, and errors were recalculated. The new mean error in the failing ventricles was 18°, which is higher than the error that corresponds to the PPD method. This result indicates that the PPD method outperforms the FS method in predicting fiber orientations in failing ventricles.
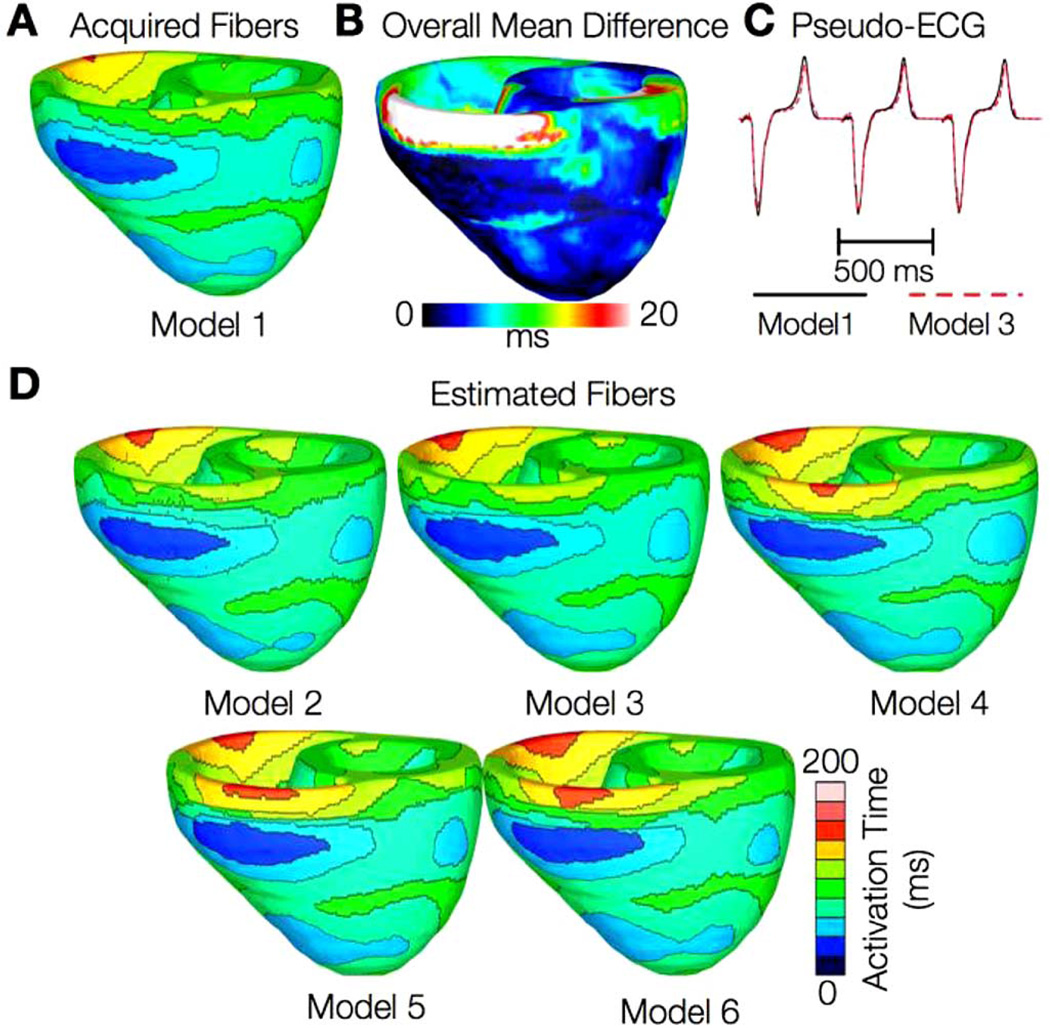
In the following, we describe our findings regarding the effect of estimation error on the results of electrophysiological simulations. Figs. 5 and 6 present the simulated activation maps of one beat of sinus rhythm activation in normal and failing ventricular models, respectively. In both normal and failing ventricles, the activation maps in models with acquired fiber orientations match well experimental activation maps [44], with the earliest epicardial activation at the anterior base, anterior apex, and LV free wall. Models with estimated fiber orientations produce activation maps very similar to those of models with acquired orientations; the earliest epicardial activations occur at the same sites, and the directions of propagation match as well. Among the normal ventricles, the total activation time of model 1 was 151 ms, while for models 2–6, the mean total activation time was 154 ms. The overall mean difference in total activation times between the acquired and estimated fiber orientation cases in the normal ventricular models was 5.7 ms, which is a small fraction (3.7%) of the total activation time. Fig. 5(B) indicates that regions with largest differences in local activation times in the normal ventricles are near the base, especially near the RV outflow tract, where the wall is very thin. We believe that this wall thinness caused imperfect registration and large estimation errors, which in turn led to large differences in local activation times. In models 5 and 6 in Fig. 5(D), the large estimation errors have caused significant conduction slowdown near the RV outflow tract. These errors, however, remained local, and did not have a significant bearing on the global differences in activation times. Fig. 5(C) demonstrates that pseudo-ECGs obtained for sinus rhythm simulations with models 1 and 3 have identical morphologies. The MAD score between these two waveforms was 4.14%. On average, the MAD score between sinus rhythm pseudo-ECGs with each of models 2–6 and model 1 was 10.9%.
Fig. 5.
Results from simulations of one beat of sinus rhythm in normal canine ventricular models. (A) Activation map simulated using the model with acquired fiber orientations (model 1). (B) Absolute difference between simulated activation maps obtained from a ventricular model with acquired fiber orientations and that with estimated fiber orientations, averaged over the five estimates. (C) Simulated pseudo-ECGs with models 1 and 3. (D) Simulated activation maps from ventricles with estimated fiber orientations (models 2–6).
Fig. 6.
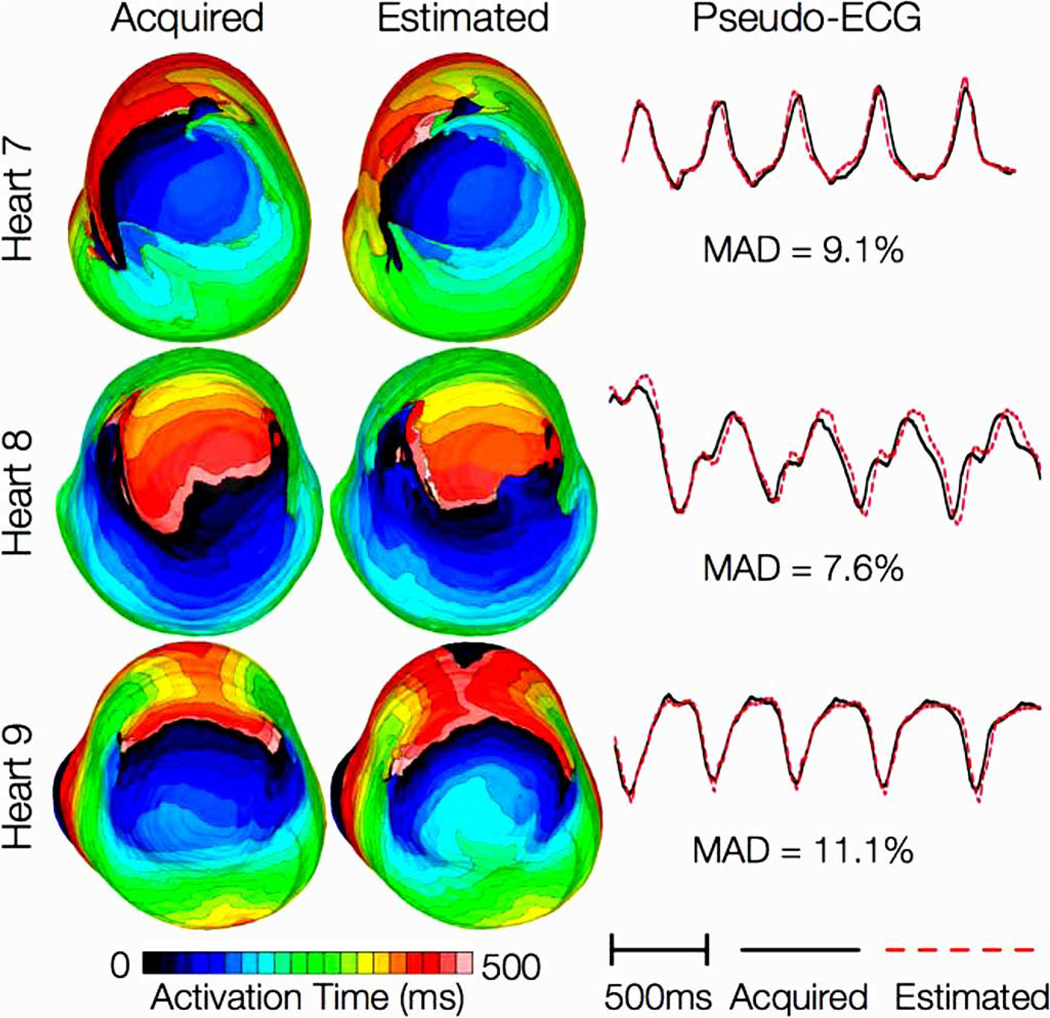
Results from simulations of one beat of sinus rhythm in failing heart models. In the first column, rows 1–3 show activation maps calculated using models 7–9, respectively. In the second column, rows 1–3 display results of simulations with models 10–12, respectively. Rows 1–3 in the third column portray the absolute difference between the activation maps shown in the first and second columns of the corresponding row. Rows in the fourth column display simulated pseudo-ECGs from models in the first and second columns of the corresponding row.
In simulations of sinus rhythm with failing ventricular models, total activation times for models 7–9 were 183, 144, and 166 ms, respectively, while for models 10–12, they were 187, 139, and 162 ms, respectively. The total activation times in the heart failure models are longer than those in normal ventricular models due to the lower tissue conductivity values. The failing ventricles were also on average 29% larger than normal ventricles resulting in longer paths for wavefront propagation and thus longer total activation times. The mean difference in total activation times between heart failure models with acquired and estimated fiber orientations was only 5.2 ms (3.1%), while the mean MAD score was 4.68%. These results indicate that the outcomes of simulation of ventricular activation in sinus rhythm in normal and failing canine ventricular models with fiber orientations estimated with the present methodology closely match those with acquired orientations. In particular, presence of heart failure did not diminish the accuracy of the estimation. Incidentally, we did not average the three ventricular distributions of difference in activation maps displayed in column 3 of Fig. 6 because these distributions are not similar.
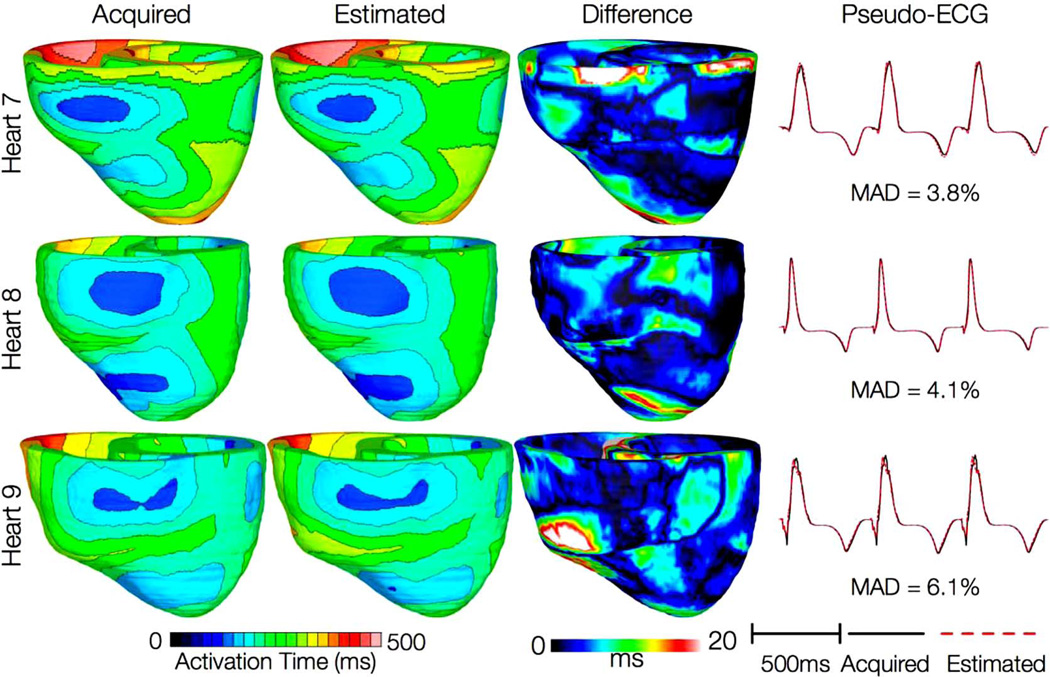
Fig. 7 shows simulated activation maps, in apical views of the ventricles, during one cycle of induced VT in the heart failure models, and corresponding pseudo-ECGs. Simulations with acquired and estimated fiber orientation both exhibit similar figure-of-eight reentrant patterns. The VT cycle lengths for models 7–12 were 392, 393, 436, 391, 408, and 432 ms, respectively. Accordingly, the percentage difference in VT cycle length between models with acquired and estimated fiber orientations, averaged across the three failing hearts, was 1.7%. In hearts 7 and 9, the ECG morphologies corresponding to estimated and acquired fiber orientations were in excellent agreement. In heart 8, the longer VT cycle length in the model with estimated fibers was manifested as a minor phase shift in the ECG, but the overall morphology of the two ECG recordings was still the same. The mean MAD score was 9.3%. These results indicate that canine heart failure models with estimated fiber orientations can closely replicate outcomes of VT simulations performed using acquired fiber orientations.
Fig. 7.
Results from simulations of VT induction with the failing heart models. Rows 1–3 in the first column show activation maps during one cycle of reentrant activity in simulations with models 7–9, respectively. Rows 1–3 in the second column show activation maps corresponding to models 10–12, respectively. Rows in the third column illustrate pseudo-ECGs from models in the first and second columns of the corresponding row.
IV. Discussion
The goal of this project was to develop a methodology for estimating ventricular fiber orientations from in vivo images of ventricular geometry, and to assess the accuracy of this estimation and its utility in simulations of cardiac electrophysiology. We hypothesized that the fiber orientations can be estimated in normal ventricles and those with structural remodeling, such as heart failure. The results described in the previous section support our hypothesis. They show that inclination angles of predicted fiber orientations are comparable to those acquired by ex vivo DTMRI, the state-of-the-art technique [12], [35]. Furthermore, activation maps and pseudo-ECGs generated by electrophysiological simulations using estimated fiber orientations closely match those using acquired fiber orientations. This research demonstrates quantitatively that, in the absence of DTMRI, myocardial fiber orientations of normal and failing ventricles can be estimated from in vivo images of their geometries for use in simulations of cardiac electrophysiology. The proposed methodology is demonstrated with in vivo CT data, but it is equally applicable to in vivo MR images of ventricular geometry, addressing the lack of ability to directly acquire patient fiber orientations. It is thus an important step towards the development of personalized models of ventricular electrophysiology for clinical applications. The methodology can also be used to estimate the fiber orientations in ex vivo hearts with high resolution. This is especially helpful when acquiring submillimeter resolution DTMRI images is difficult or prohibitively expensive, due to the very long acquisition times, in the order of days [12].
Our results are in agreement with research conducted by Helm et al. [35], who showed that heart failure does not significantly alter fiber orientations relative to geometry. Furthermore, our results demonstrate that the largest errors in estimation occur near the epicardial and endocardial surfaces, and the RV insertion points. The decreased accuracy near the surfaces is due to the partial volume effect in DTMRI [51] which introduces error and imperfect LDDMM registration, while the larger errors near RV insertion points are due to the complex branching of fiber bundles and the difference in fiber orientations between LV and RV.
For the reorientation of DTs, we chose the PPD strategy, as it accounts for shearing and nonuniform scaling, and therefore is more generic than the FS strategy [34]. On the other hand, Peryrat et al. [36] have preferred the FS method because it is consistent with the Log-Euclidian framework that they used to compute statistics on DTs, and is not affected by errors in eigenvector extraction. They also argued that FS method is better at preserving transmural variation of fiber structure, and their quantitative experiments comparing all three eigenvectors of DTs indicated that the FS strategy registers images better than PPD. However, FS outperformed PPD only by a very small margin in their work, and their experiments were limited to normal canine hearts. Form the standpoint of patient-specific modeling of cardiac function, it was important for us to adopt a reorientation strategy that performs well in the presence of structural remodeling such as in heart failure, which involves shape changes that PPD may better account for. Furthermore, only the primary eigenvector was relevant in this study. Indeed, when we replaced PPD with FS as part of our pipeline, the fiber estimation error for failing hearts increased. A more comprehensive comparison of various reorientation strategies may be helpful in the future, but such an analysis is outside the scope of this research.
Our electrophysiological simulations suggested that activation maps were not very sensitive to changes in fiber orientations. More importantly, we demonstrated that effects of fiber estimation errors on gross electrophysiology were insignificant at a clinically observable level by means of the MAD score of the pseudo-ECGs. The MAD scores in our results were less than 12%, implying that the propagation patterns generated using estimated and acquired fiber orientations were clinically equivalent [48]. Note that the similarity of propagation patterns will translate to low differences in mechanical activation patterns as well, as experiments show that local electrical and mechanical activation times during sinus rhythm are highly correlated [52], [53]. In summary, our research will facilitate ventricular simulation studies of any species in health and disease when it is not feasible to acquire fiber orientations using DTMRI. In particular, the proposed methodology paves the way for patient-specific modeling of ventricular whole-heart electrophysiology (and possibly) electromechanics based only on in vivo clinical imaging data. Simulations with such models may ultimately aid physicians to arrive at highly personalized decisions for therapeutic interventions as well as prophylaxis. For example, it has recently been shown that such models can be used to predict the organizing centers of reentrant activity in infarcted hearts, which could aid in guiding ablation therapy for infract-related VT [7]. Also, electromechanical simulations using models of failing hearts have shown promise in noninvasively determining optimal cardiac resynchronization therapy [54].
The current study has a number of limitations. Firstly, human heart image data were not available to us, and therefore the proposed estimation methodology was validated with images of canine hearts. We expect that the methodology will accurately estimate fiber orientations in human hearts as well because, just as in canine hearts, fiber orientations relative to geometry have been shown to be similar between different human hearts [37]. Also, we tested our methodology in normal and failing hearts only. It would be important to test it under conditions such as myocardial infarction and hypertrophy, where fiber disorganizations are known to occur [55], [56]. Furthermore, the present study is a proof-of-concept for estimating ventricular fiber orientations for use in personalized simulations of cardiac electrophysiology. Patient-specific acquisition of other electrophysilogically relevant data such as action potential heterogeneity will also be necessary for the development of personalized models of cardiac electrophysiology; our methodology is only one step towards achieving this goal. Finally, the accuracy of the fiber orientation estimation in simulations of cardiac mechanics will also need to be assessed.
Acknowledgment
The authors would like to thank Dr. R. Winslow, Dr. E. McVeigh, and Dr. P Helm at Johns Hopkins University for providing the ex vivo datasets online.
This work was supported in part by the National Institutes of Health under Grant R01-HL082729 and Grant R01-HL091036, and in part by the National Science Foundation under Grant CBET-0933029.
Footnotes
Contributor Information
Fijoy Vadakkumpadan, Email: fijoy@jhu.edu, Institute for Computational Medicine and the Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21205 USA.
Hermenegild Arevalo, Institute for Computational Medicine and the Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21205 USA.
Can Ceritoglu, Center for Imaging Science, Johns Hopkins University, Baltimore, MD 21218 USA.
Michael Miller, Center for Imaging Science, Institute for Computational Medicine, Baltimore, MD 21218 USA, and also with the Department of Biomedical Engineering, Johns Hopkins University, MD 21205 USA.
Natalia Trayanova, Institute for Computational Medicine and the Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21205 USA.
References
- 1.Noble D. Modeling the heart—From genes to cells to the whole organ. Science. 2002;vol. 295:1678–1682. doi: 10.1126/science.1069881. [DOI] [PubMed] [Google Scholar]
- 2.Trayanova N. Whole heart modeling: Applications to cardiac electrophysiology and electromechanics. Circulat. Res. 2011;vol. 108:113–128. doi: 10.1161/CIRCRESAHA.110.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vadakkumpadan F, Arevalo H, Prassl AJ, Chen J, Kickinger F, Kohl P, Plank G, Trayanova N. Image-based models of cardiac structure in health and disease. Wiley Interdisciplin. Rev.: Syst. Biol. Med. 2010;vol. 2:489–506. doi: 10.1002/wsbm.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop M, Plank G, Burton R, Schneider J, Gavaghan D, Grau V, Kohl P. Development of an anatomically detailed MRI-derived rabbit ventricular model and assessment of its impact on simulations of electrophysiological function. Am. J. Physiol.: Heart Circulat. Physiol. 2010;vol. 298:H699–H718. doi: 10.1152/ajpheart.00606.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plank G, Burton RA, Hales P, Bishop M, Mansoori T, Bernabeu MO, Garny A, Prassl AJ, Bollensdorff C, Mason F, Mahmood F, Rodriguez B, Grau V, Schneider JE, Gavaghan D, Kohl P. Generation of histo-anatomically representative models of the individual heart: Tools and application. Philos. Trans. Series A, Math., Phys., Eng. Sci. 2009;vol. 367:2257–2292. doi: 10.1098/rsta.2009.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurev V, Lee T, Constantino J, Arevalo H, Trayanova NA. Models of cardiac electromechanics based on individual hearts imaging data: Image-based electromechanical models of the heart. Biomechan. Model. Mechanobiol. 2010;vol. 10:295–306. doi: 10.1007/s10237-010-0235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vigmond E, Vadakkumpadan F, Gurev V, Arevalo H, Deo M, Plank G, Trayanova N. Towards predictive modelling of the electrophysiology of the heart. Exp. Physiol. 2009;vol. 94:563–577. doi: 10.1113/expphysiol.2008.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt A, Azevedo CF, Cheng A, Gupta SN, Bluemke DA, Foo TK, Gerstenblith G, G R, Marbán E, Tomaselli GF, Lima JAC, Wu KC. Infarct tissue heterogeneity by magnetic resonance imaging identifies enhanced cardiac arrhythmia susceptibility in patients with left ventricular dysfunction. Circulation. 2007;vol. 115:2006–2014. doi: 10.1161/CIRCULATIONAHA.106.653568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen PMF, Legrice IJ, Smaill BH, Hunter PJ. Mathematical model of geometry and fibrous structure of the heart. Am. J. Physiol.—Heart Circulat. Physiol. 1991;vol. 260:H1365–H1378. doi: 10.1152/ajpheart.1991.260.4.H1365. [DOI] [PubMed] [Google Scholar]
- 10.Stevens C, Hunter PJ. Sarcomere length changes in a 3-D mathematical model of the pig ventricles. Progress Biophys. Molecular Biol. 2003;vol. 82:229–241. doi: 10.1016/s0079-6107(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 11.Vetter FJ, McCulloch AD. Three-dimensional analysis of regional cardiac function: A model of rabbit ventricular anatomy. Progress Biophys. Molecular Biol. 1998;vol. 69:157–183. doi: 10.1016/s0079-6107(98)00006-6. [DOI] [PubMed] [Google Scholar]
- 12.Helm P, Beg MF, Miller M, Winslow R. Measuring and mapping cardiac fiber and laminar architecture using diffusion tensor MR imaging. Ann. NY Acad. Sci. 2005;vol. 1047:296–307. doi: 10.1196/annals.1341.026. [DOI] [PubMed] [Google Scholar]
- 13.Scollan DF, Holmes A, Winslow R, Forder J. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. Am. J. Physiol.—Heart Circulat. Physiol. 1998;vol. 275:H2308–H2318. doi: 10.1152/ajpheart.1998.275.6.H2308. [DOI] [PubMed] [Google Scholar]
- 14.Vadakkumpadan F, Rantner LJ, Tice B, Boyle P, Prassl AJ, Vigmond E, Plank G, Trayanova N. Image-based models of cardiac structure with applications in arrhythmia and defibrillation studies. J. Electrocardiol. 2009;vol. 42:157.e1–157.e10. doi: 10.1016/j.jelectrocard.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Relan J, Chinchapatnam P, Sermesant M, Rhode K, Delingette H, Razavi R, Ayache N. Coupled personalisation of electrophysiology models for simulation of induced ischemic ventricular tachycardia. Med. Image Comput. Comput.-Assist. Intervent.; Beijing, China. 2010. pp. 420–428. [DOI] [PubMed] [Google Scholar]
- 16.Edelman RR, Gaa J, Wedeen VJ, Loh E, Hare JM, Prasad P, Li W. In vivo measurement of water diffusion in the human heart. Magn. Reson. Med. 1994;vol. 32:423–428. doi: 10.1002/mrm.1910320320. [DOI] [PubMed] [Google Scholar]
- 17.Reese TG, Wedeen VJ, Weisskoff RM. Measuring diffusion in the presence of material strain. J. Magn. Reson. 1996;vol. 112:253–258. doi: 10.1006/jmrb.1996.0139. [DOI] [PubMed] [Google Scholar]
- 18.Reese TG, Weisskoff RM, Smith RN, Rosen BR, Dinsmore RE, Wedeen VJ. Imaging myocardial fiber architecture in vivo with magnetic resonance. Magn. Reson. Med. 1995;vol. 34:786–791. doi: 10.1002/mrm.1910340603. [DOI] [PubMed] [Google Scholar]
- 19.Nielles-Vallespin S, Mekkaoui C, Reese TG, Gatehouse P, Feiweier T, Speier P, Sosnovik DE, Firmin D. Int. Soc. Magn. Reson. Med. Montreal, Canada: 2011. Fiber tracking of the human heart in vivo; p. 3372. [Google Scholar]
- 20.Tseng WY, Reese TG, Weisskoff RM, Wedeen VJ. Cardiac diffusion tensor MRI in vivo without strain correction. Magn. Reson. Med. 1999;vol. 42:393–403. doi: 10.1002/(sici)1522-2594(199908)42:2<393::aid-mrm22>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 21.Dou J, Reese TG, Tseng WY, Wedeen VJ. Cardiac diffusion MRI without motion effects. Magn. Reson. Med. 2002;vol. 48:105–114. doi: 10.1002/mrm.10188. [DOI] [PubMed] [Google Scholar]
- 22.Gamper U, Boesiger P, Kozerke S. Diffusion imaging of the in vivo heart using spin echoes—Considerations on bulk motion sensitivity. Magn. Reson. Med. 2007;vol. 57:331–337. doi: 10.1002/mrm.21127. [DOI] [PubMed] [Google Scholar]
- 23.Toussaint N, Sermesant M, Stoeck CT, Kozerke S, Batchelor PG. In vivo human 3-D cardiac fibre architecture: Reconstruction using curvilinear interpolation of diffusion tensor images. Medical Image Computing and Computer Assisted Intervention. 2011:418–425. doi: 10.1007/978-3-642-15705-9_51. [DOI] [PubMed] [Google Scholar]
- 24.Sosnovik DE, Wang R, Dai G, Reese TG, Wedeen VJ. Diffusion MR tractography of the heart. J. Cardiovas. Magn. Reson. 2009;vol. 11:47–61. doi: 10.1186/1532-429X-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potse M, Dube B, Richer J, Vinet A, Gulrajani RM. A comparison of monodomain and bidomain reaction-diffusion models for action potential propagation in the human heart. IEEE Trans. Biomed. Eng. 2006 Dec.vol. 53(no. 12):2425–2435. doi: 10.1109/TBME.2006.880875. [DOI] [PubMed] [Google Scholar]
- 26.Bishop MJ, Hales P, Plank G, Gavaghan DJ, Scheider J, Grau V. Comparison of rule-based and DTMRI-derived fibre architecture in a whole rat ventricular computational model. Int. Conf. Functional Imag. Model. Heart; Nice, France. 2009. pp. 87–96. [Google Scholar]
- 27.Bayer J. Modeling fiber orientation in unstructured meshes of cardiac tissue using image- and rule-based approaches. presented at the CARP Workshop; Banff, Canada. 2008. [Google Scholar]
- 28.ten Tusscher KHWJ, Hren R, Panfilov AV. Organization of ventricular fibrillation in the human heart. Circulat. Res. 2007;vol. 100:e87–e101. doi: 10.1161/CIRCRESAHA.107.150730. [DOI] [PubMed] [Google Scholar]
- 29.Sundar H, Shen D, Biros G, Litt H, Davatzikos C. Estimating myocardial fiber orientations by template warping. IEEE Int. Symp. Biomed. Imag.; Arlington, VA. 2006. pp. 73–76. [Google Scholar]
- 30.Rohmer D, Sitek A, Gullberg GT. Reconstruction and visualization of fiber and laminar structure in the normal human heart from ex vivo diffusion tensor magnetic resonance imaging (DTMRI) data. Investigat. Radiol. 2007;vol. 42:777–789. doi: 10.1097/RLI.0b013e3181238330. [DOI] [PubMed] [Google Scholar]
- 31.Beg MF, Helm PA, McVeigh E, Miller MI, Winslow RL. Computational cardiac anatomy using MRI. Magn. Reson. Med. 2004;vol. 52:1167–1174. doi: 10.1002/mrm.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beg MF, Miller MI, Trouve A. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vis. 2005;vol. 61:139–157. [Google Scholar]
- 33.Ceritoglu C, Oishi K, Li X, Chou MC, Younes L, Albert M, Lyketsos C, van Zijl PC, Miller MI, Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. Neuroimage. 2009;vol. 47:618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans. Med. Imag. 2001 Nov.vol. 20(no. 11):1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- 35.Helm PA, Younes L, Beg MF, Ennis DB, Leclercq C, Faris OP, McVeigh E, Kass D, Miller MI, Winslow RL. Evidence of structural remodeling in the dyssynchronous failing heart. Circulat. Res. 2006;vol. 98:125–132. doi: 10.1161/01.RES.0000199396.30688.eb. [DOI] [PubMed] [Google Scholar]
- 36.Peyrat J-M, Sermesant M, Pennec X, Delingette H, Chenyang X, McVeigh ER, Ayache N. A computational framework for the statistical analysis of cardiac diffusion tensors: Application to a small database of canine hearts. IEEE Trans. Med. Imag. 2007 Nov.vol. 26(no. 11):1500–1514. doi: 10.1109/TMI.2007.907286. [DOI] [PubMed] [Google Scholar]
- 37.Lombaert H, Peyrat J-M, Croisille P, Rapacchi S, Fanton L, Clarysse P, Delingette H, Ayache N. Statistical analysis of the human cardiac fiber architecture from DT-MRI. presented at the 6th Int. Conf. Funct. Imag. Model. Heart; New York. 2011. [Google Scholar]
- 38.Streeter DD. Gross Morphology and Fiber Geometry of the Heart. Baltimore, MD: Johns Hopkins Press; 1979. [Google Scholar]
- 39.Plank G, Zhou L, Greenstein JL, Plank G, Zhou L, Greenstein JL, Cortassa S, Winslow RL, O’Rourke B, Trayanova NA. From mitochondrial ion channels to arrhythmias in the heart: Computational techniques to bridge the spatio-temporal scales. Philos. Trans. Series A, Math., Phys., Eng. Sci. 2008;vol. 366:3381–3409. doi: 10.1098/rsta.2008.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts DE, Scher AM. Effect of tissue anisotropy on extra-cellular potential fields in canine myocardium in situ. Circulat. Res. 1982;vol. 50:342–351. doi: 10.1161/01.res.50.3.342. [DOI] [PubMed] [Google Scholar]
- 41.Greenstein J, Wu R, Po S, Tomaselli GF, Winslow RL. Role of the calcium-independent transient outward current I(to1) in shaping action potential morphology and duration. Circulat. Res. 2000;vol. 87:1026–1033. doi: 10.1161/01.res.87.11.1026. [DOI] [PubMed] [Google Scholar]
- 42.Winslow R, Rice J, Jafri S, Marbán E, O’Rourke B. Mechanisms of altered excitation-contraction coupling in canine tachycardia-induced heart failure, II: Model studies. Circulat. Res. 1999;vol. 84:571–586. doi: 10.1161/01.res.84.5.571. [DOI] [PubMed] [Google Scholar]
- 43.Akar F, Nass R, Hahn S, Cingolani E, Shah M, Hesketh G, DiSilvestre D, Tunin R, Kass D, Tomaselli G. Dynamic changes in conduction velocity and gap junction properties during development of pacing-induced heart failure. Am. J. Physiol.—Heart Circulat. Physiol. 2007;vol. 293:H1223–H1230. doi: 10.1152/ajpheart.00079.2007. [DOI] [PubMed] [Google Scholar]
- 44.Gurev V, Constantino J, Rice JJ, Trayanova N. Distribution of electromechanical delay in the ventricles: Insights from a 3-D electromechanical model of the heart. Biophys. J. 2010;vol. 99:745–754. doi: 10.1016/j.bpj.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durrer D, Dam RTV, Freud GE, Janse MJ, Meijler FL, Arzbaecher RC. Total excitation of the isolated human heart. Circulation. 1970;vol. 41:899–912. doi: 10.1161/01.cir.41.6.899. [DOI] [PubMed] [Google Scholar]
- 46.Spach MS, Barr RC. Ventricular intramural and epicardial potential distributions during ventricular activation and repolarization in the intact dog. Circulat. Res. 1975;vol. 37:243–257. doi: 10.1161/01.res.37.2.243. [DOI] [PubMed] [Google Scholar]
- 47.Gima K, Rudy Y. Ionic current basis of electrocardiographic waveforms. Circulat. Res. 2002;vol. 90:889–896. doi: 10.1161/01.res.0000016960.61087.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerstenfeld E, Dixit S, Callans D, Rajawat Y, Rho R, Marchlinski F. Quantitative comparison of spontaneous and paced 12-lead electrocardiogram during right ventricular outflow tract ventricular tachycardia. J. Am. College Cardiol. 2003;vol. 41:2046–2053. doi: 10.1016/s0735-1097(03)00427-3. [DOI] [PubMed] [Google Scholar]
- 49.Vigmond E, Hughes M, Plank G, Leon L. Computational tools for modeling electrical activity in cardiac tissue. J. Electrocardiol. 2003;vol. 36:69–74. doi: 10.1016/j.jelectrocard.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 50.Vigmond E, Santos RWD, Prassl A, MDM, Plank G. Solvers for the cardiac bidomain equations. Progress Biophys. Molecul. Biol. 2007;vol. 96:3–18. doi: 10.1016/j.pbiomolbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexander AL, Hasan KM, Lazar M, Tsuruda JS, Parker DL. Analysis of partial volume effects in diffusion-tensor MRI. Magn. Reson. Med. 2001;vol. 45:770–780. doi: 10.1002/mrm.1105. [DOI] [PubMed] [Google Scholar]
- 52.Faris O, Evans F, Ennis D, Helm P, Taylor J, Chesnick AS, Guttman M, Ozturk C, McVeigh E. Novel technique for cardiac electromechanical mapping with magnetic resonance imaging tagging and an epicardial electrode sock. Ann. Biomed. Eng. 2003;vol. 31:430–440. doi: 10.1114/1.1560618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Provost J, Gurev V, Trayanova N, Konofagou EE. Mapping of cardiac electrical activation with electromechanical wave imaging: An in silico-in vivo reciprocity study. Heart Rhythm. 2010;vol. 8:752–759. doi: 10.1016/j.hrthm.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Constantino J, Gurev V, Trayanova N. Electromechanical modeling applied to cardiac resynchronization therapy. In: Pahlm O, Wagner G, editors. Multimodal Cardiovascular Imaging: Principles and Clinical Applications. New York: McGraw-Hill; 2011. [Google Scholar]
- 55.Chen J, Song S-K, Liu W, McLean M, Allen SJ, Tan J, Wickline SA, Yu X. Remodeling of cardiac fiber structure after infarction in rats quantified with diffusion tensor MRI. Am. J. Physiol.—Heart Circulat. Physiol. 2003;vol. 285:H946–H954. doi: 10.1152/ajpheart.00889.2002. [DOI] [PubMed] [Google Scholar]
- 56.Karlon WJ, Covell JW, Mcculloch AD, Hunter JJ, Omens JH. Automated measurement of myofiber disarray in transgenic mice with ventricular expression. Anatom. Rec. 1998;vol. 252:612–625. doi: 10.1002/(SICI)1097-0185(199812)252:4<612::AID-AR12>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]