Abstract
Tissue hypoxia occurs in pathologic conditions such as cancer, ischemic heart disease and stroke when oxygen demand is greater than oxygen supply. An imaging method that can differentiate hypoxic versus normoxic tissue could have an immediate impact on therapy choices. In this work, the gadolinium complex of 1,4,7,10-tetraaza-1,4,7,10-tetraacetate (DOTA) having a 2-nitroimidazole attached to one carboxyl group via an amide linkage was prepared, characterized and tested as a hypoxia-sensitive MRI agent. A control complex, Gd(DO3A-monobutylamide), was also prepared in order to test whether the nitroimidazole side-chain alters either the water proton T1 relaxivity or the thermodynamic stability of the complex. The stabilities of these complexes were lower than that of Gd(DOTA)− as expected for mono-amide derivatives. The water proton T1 relaxivity (r1), bound water residence lifetime (τM) and rotational correlation time (τR) of both complexes was determined by relaxivity measurements, variable temperature 17O NMR and proton nuclear magnetic relaxation dispersion (NMRD) studies. The resulting parameters (r1 = 6.38 mm−1s−1 at 20 MHz, τM = 0.71 µs, τR = 141 ps) determined for the nitroimidazole derivative closely parallel those of other Gd(DO3A-monoamide) complexes of similar molecular size. In vitro MR imaging experiments using 9L rat glioma cells maintained under nitrogen (hypoxic) versus oxygen (normoxic) gas showed that both agents enter cells but only the nitroimidazole derivative is trapped in cells maintained under N2 as evidenced by ~2-fold decrease in T1 measured for hypoxic cells versus normoxic cells exposed to this agent. These results suggest that the nitroimidazole derivative may serve as a molecular reporter for discriminating hypoxic versus normoxic tissues by MRI.
Keywords: imaging agents, ligand design, stability, relaxation properties, imaging of hypoxic tissue
Introduction
Tissue hypoxia usually reflects a locally restricted oxygen supply (ischemia) and/or an abnormal increase in oxygen consumption. In clinical practice, hypoxia is related to a variety of pathologic conditions including cancer, ischemic heart disease and stroke. Locally advanced solid tumors often have hypoxic or anoxic regions distributed heterogeneously throughout the tumor as a result from a structurally or functionally abnormal vasculature. Tumor hypoxia is commonly associated with a poor prognosis after radiation and chemotherapy and hypoxia can influence many aspects of tumor proliferation such as growth, apoptosis, angiogenesis and metastasis.[1,2]
In 1955, 2-nitroimidazole was shown to be active against certain anaerobic bacterial infections.[3] Later it was discovered that 2-nitroimidazole analogs could be used to sensitize hypoxic tumors to ionizing radiation.[4,5] Further research on the mechanism of action of this compound revealed that it was capable of accumulating in hypoxic tissue due to selective enzyme-mediated reduction of the nitro group under low oxygen conditions.[6] The diverse bioactivity of nitroimidazoles and related nitroaromatic compounds has the same underlying mechanism; the activities of these compounds depend on the redox potential of the nitro group.[7] The first step of this intracellular process involves the one-electron reduction of the nitro group to a radical anion, which undergoes further reduction in hypoxic cells but is rapidly oxidized back to the nitro compound under normoxic conditions. The reduction potential of the nitro group in nitroimidazoles is sensitive to its position in the imidazole ring and this correlates with hypoxia sensitivity.[8] Given the prospect of nitroimidazole as a platform for hypoxia-homing probes, several radiolabeled nitroimidazole derivatives and related compounds have been tested as imaging probes of hypoxic tissue in vivo.[9] Most prominent among theses have been the 18F labeled fluoromisonidazole (18F-FMISO)[10] for PET imaging and the 99mTc labeled propylene amineoxime (BMS181321)[11] for SPECT imaging. Cyclen (1,4,7,10-tetraazacyclododecane)-based nitroimidazole derivatives tagged with 67GaIII, 153GdIII or 177LuIII have shown only moderate tumor localization.[12–14] Although several multi-fluorinated derivatives of 2-nitroimidazole have been synthesized and used to assess tumor hypoxia by 19F NMR, GdIII-based nitroimidazole derivatives for 1H MRI have not been reported.[15–17] While MRI is considerably less sensitive than nuclear medicine for molecular imaging, it does offer the advantage of wide clinical availability, lack of radiation and high resolution anatomical images. For these reasons, we initiated a program to develop gadolinium-based 2-nitroimidazole derivatives and to evaluate the potential of such agents for detecting hypoxic cells by MRI.
Results and Discussion
Ligand design and synthesis
DOTA (1,4,7,10-tetraaza cyclododecane-1,4,7,10-tetraacetic acid) was selected as the chelate for the conjugation to 2-nitroimidazole because GdIII complexes with DOTA-like ligands have high thermodynamic stability, are kinetically inert and have favorable relaxation properties.[18,19] The 2-nitroimidazole vector was selected based on its one-electron reduction potential, known to fall in the range of −330 to −450 mV, which allows for its entrapment and exclusive reduction under hypoxic conditions as it has been previously reported in the literature by many scientists (vide supra). Since conjugation of DOTA to a targeting vector via a single carboxyl group is relatively straightforward,[20] a 2-nitroimidazole moiety with an extended side-chain containing a primary amino group was chosen for conjugation (Scheme 1). This design affords a neutral GdIII complex which would hopefully help facilitate cellular uptake. It was anticipated that the relatively long linkage between Gd(DO3A-monoamide) unit and the nitroimidazole moiety in 3 would minimize any unwanted interference between the GdIII coordination sphere and the nitroimidazole unit and, at the same time, increase complex lipophilicity. In addition, the linker does not contain groups that can engage in prototropic equilibria at physiological pH and should therefore not affect the redox potential of the nitroimidazole unit.[8,21] The synthesis of the target ligand is outlined in Scheme 1.
Scheme 1.
Synthetic route to ligand 3 (Y.: 28%).
The protected DO3A-monoamide derivative 1 was obtained by alkylating DO3A-tris(tert-butyl ester) with methyl chloroacetate and reacting the resulting methyl ester with excess diaminobutane (putrescine) following a slight modification of a published procedure (ESI).[22] Nitroimidazoyl hexanoic acid (2) was synthesized by alkylation of 2-nitroimidazole with ethyl bromohexanoate followed by the cleavage of the ethyl group in concentrated HCl as previously reported.23 The imidazoyl acid 2 and macrocyclic amine 1 where then coupled using standard HBTU/HOBt chemistry.24,25 The tert-butyl protecting groups of the intermediate ester were cleaved using trifluoroacetic acid (TFA) and the crude product was purified by HPLC to give pure 3 (28% yield). Gd(3) was obtained in quantitative yield by reaction of 3 with GdCl3 at pH 6.
Ligand protonation and complex stability constants
The coordination environment (four macrocyclic N donor atoms, three carboxylate O donor atoms and one amide O donor atom) provided by ligand 3 is typical of a DOTA- and DO3A-bioconjugate in which the chelator is attached to the biomolecule through one of the acetate pendant arms of the ligand.[20] Despite the widespread use of these ligands to chelate Gd3+ for MRI and various radioactive lanthanide ions for radiopharmaceutical applications, only a limited number of papers have appeared on the thermodynamic stability, formation and dissociation kinetics of DO3A-monoamides.[26,27] Here, we report the thermodynamic stabilities of the Ce(3), Gd(3) and Yb(3) and the results of 1H and 17O-NMR relaxometric studies of Gd(3). The results are also compared to corresponding data for Ce(6), Gd(6) and Yb(6).
pH-potentiometry is considered the gold standard technique for determining the protonation constants of polyaminopolycarboxylate ligands and thermodynamic stability constants of complexes formed with various metal ions. The protonation constants obtained from fitting the pH-potentiometric data for ligands 3 and 6 are presented in Table 1 along with the literature data for DOTA (4) and two amide derivatives, DO3A-monopropylamide (5) and DOTA-tetramethylamide (7). The trends found for the protonation constants of these ligands are similar, except for DOTA- tetra- methylamide (7) (Chart 1). For DOTA and DO3A-monoamide derivatives (3 and 6), the first two protonation constants (which have been traced to protonation at ring nitrogen atoms positioned trans- to one another) are relatively high, while the remaining log KiH values (reflecting protonation of the carboxylates) are much lower. For the monoamide ligands studied here, 3 and 6, five protonation steps were observed in the pH range of 2–12. The total basicity of these ligands (expressed as the sum of the protonation constants, log β4) is lower than that of DOTA, reflecting the lower basicity of the macrocyclic N-atoms.[28,29] The protonation constants of 6 as determined previously by pH-potentiometry in 0.1 M Me4NCl (log β4 = 27.94) were found to be somewhat higher than the values measured here in 1.0 M KCl (log β4 = 26.54).[30] This can be attributed to the formation of a weak potassium complex in the presence of 1.0 M KI ions (a similar decrease in basicity was observed for DOTA when the protonation constants were measured in the presence of KCl versus Me4NCl).[31] Interestingly, the first protonation constant of 3 is slightly lower (0.4 log K units) than that of 6, presumably reflecting the elongated side arm on the amide. A similar decrease in the basicity was also observed for some alkyl-EDTA derivatives where an increase in the length of the alkyl substituent had a noticeable effect on the first protonation constant (see Table S1, ESI).
Table 1.
Protonation constants[a] of 3 and some ligands of similar structure, 1 M KCl, 25 °C.
| log K | 3[b] | 4 | 5[e] | 6[b] | 7[h] |
|---|---|---|---|---|---|
| log K1H | 9.78(2) | 12.09[c]; 12.6[d] |
9.6 | 10.17(1), 12.22[f], 10.73[g] |
9.56 |
| log K2H | 9.05(3) | 9.67[c]; 9.70[d] |
9.2 | 9.02(1), 8.90[f], 9.05[g] |
5.95 |
| log K3H | 4.53(4) | 4.55[c]; 4.50[d] |
4.4 | 4.41(1), 4.34[f], 4.53[g] |
– |
| log K4H | 3.17(5) | 4.09[c]; 4.14[d] |
1.7 | 2.94(1), 2.49[f], 3.32[g] |
– |
| log K5H | 2.19(6) | 2.32[d] | – | 1.99(1), 1.47[f], 2.25[g] |
– |
| log β4 | 26.53 | 30.40[c]; 30.94[d] |
24.9 | 26.54, 27.94[f], 27.63[g] |
15.51[i] |
Standard deviations shown in parentheses.
This work, in 1.0 M KCl and at 25 °C.
In 0.1 M KCl.[32]
In 1.0 M Me4NCl.[33]
In 0.1 M NaCl.[26]
In 0.1 M Me4NCl.[30]
The ionic strength was not controlled (25 °C).[34]
In 0.1 M KCl.[35]
The value refers to log β2, since this ligand has only two protonation constants.
Chart 1.
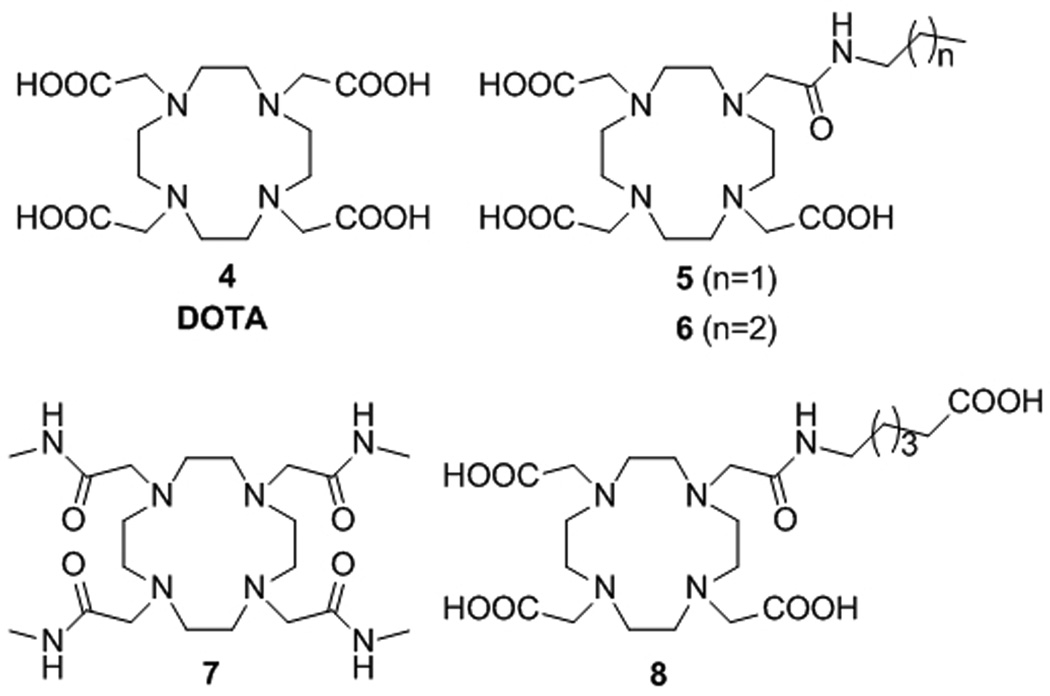
The structures of DOTA (4), a DOTA-tetramethylamide (7) and some DO3A-monoamide derivatives (5, 6 and 8) discussed in this work.
The formation equilibria between these macrocyclic ligands and the LnIII ions cannot be studied by direct pH-potentiometric titration because of slow formation kinetics. Hence, the stability constants were determined by use of the “out-of-cell” technique using separate samples for each titration point (total of 16 points). The stability constants of some Ln(3) and Ln(6) complexes, assuming formation of only 1:1 complexes, are listed in Table 2. The standard deviations associated with these constants significantly decreased when the formation of monoprotonated species (LnLH) was also taken into consideration in the calculations. This suggests that protonation of the complexes also occurs under these conditions. The Ce(6) system was an exception, as the inclusion of the protonated species in the equilibrium model for this complex resulted in a significant increase in the standard deviation of fitted parameters.
Table 2.
Thermodynamic stability constants and protonation constants of lanthanide complexes of ligands 3–7, 25°C.
| LnIII | eq. | 3[a] | 4[c] | 5[f] | 6[a] | 7[h] |
|---|---|---|---|---|---|---|
| CeIII | log KLn•L | 19.2(0.1) | 23.39, 22.86[d] |
– | 19.26(4) | 12.68 |
| log KLnL•H[b] |
– | 2.5[d] | – | 2.65(5) | – | |
| GdIII | log KLn•L | 20.93(9) | 24.67, 25.3[e] |
20.1 | 21.29(1) | 13.54 |
| log KLnL•H | 2.93(4) | 2.3 | – | 2.57(1) | – | |
| LuIII | log KLn•L | 21.51(9) | 25.41, 25.0[e] |
– | 21.83(3)[g] | 13.91 |
| log KLnL•H | 2.44(9) | – | – | 2.33(4) | – |
As expected, the stabilities of all LnIII complexes increase with decreasing lanthanide ion size, CeIII < GdIII < LuIII (Table 2).37 The stabilities of all Ln(3) complexes were similar to the corresponding Ln(6) complexes, as expected for similar DO3A-monoamide structures. This indicates that incorporation of the elongated nitroimidazole-containing pendant arm does little to alter complex stability. However, these complexes are about four orders of magnitude less stable than the corresponding Ln(DOTA)− complexes due to the substitution of a carboxylate in DOTA for the weaker amide donor atom in 3 and 6. This is also reflected in the total basicity of 3 and 6 compared to DOTA. Further substitution of acetate arms for amides results in even lower stabilities as illustrated by the stability of the LnIII complexes of 7. It is worth noting that the stability constant values measured in the presence of KCl represent a lower limit since these calculations assume no significant interaction between the ligands and KI ions, likely an invalid assumption. These results indicate that DO3A-monobutylamide (6) serves as a good model for the nitroimidazole ligand, 3, and other similar DOTA- or DO3A-bioconjugate systems.
Relaxometric studies
MRI contrast agents are typically characterized by a T1 proton relaxivity (r1p) value. The relaxivity (r1p) of low molecular weight GdL that has rapid water exchange kinetics is dominated by the inner-sphere contribution (r1pis). The Solomon-Bloembergen-Morgan (SBM) theory of relaxivity predicts that r1pis is dependent on several parameters including the number of inner sphere water molecules (q), the longitudinal relaxation time of the protons of the water molecule(s) in the inner coordination sphere (T1MH), the residence time of the inner-sphere water molecule(s) (τM) and the tumbling rate of the paramagnetic complex in solution (τR, rotational correlation time).[38] At clinically relevant fields (1.5 – 3 T), the relaxivity is largely determined by τM and τR. These constants can be obtained by fitting of variable temperature 17O NMR (τM) and 1H nuclear magnetic relaxation dispersion (NMRD) data (τR).[38–40] At 20 MHz, 25°C and pH=7, the relaxivity (r1p) of Gd(3) and Gd(6) were 6.38 ± 0.04 mm−1s−1 and 5.05 ± 0.02 mm−1s−1, respectively, measured at 20 MHz. The r1p value of Gd(3) is about 30% higher than that of Gd(6), reflecting its higher molecular weight (larger size) and consequently longer rotational correlation time (τR). The relaxivity of Gd(6) is nearly the same as that of Gd(8) (r1p = 5.0 mm−1s−1, 25°C, 20 MHz), a Gd-complex of similar molecular weight and size.[41]
Temperature dependence of relaxivity
The relaxivities of Gd(3) and Gd(6) were also measured at 37°C (20 MHz) and found to be only slightly lower (5.25 ± 0.05 and 4.12 ± 0.02 mm−1s−1, respectively) than the values measured at 25°C. The relaxivity of Gd(DOTA)− has a similar temperature dependence at this field, decreasing from 4.7 mm−1s−1 to 3.6 mm−1s−1 as the temperature is increased from 25 to 37°C. This behavior is expected for fast inner-sphere water exchange systems. It has been shown that the relaxivity of low molecular weight Gd-polyaminopolycarboxylate complexes whose bound water lifetime falls into the fast exchange regime (T1M >> τM) decreases exponentially with increasing temperature due to a decrease of the contribution of both r1pis (the residence time of the bound water molecule decreases) and r1pos (the diffusion rate of the water molecules increases on the surface of the complex). This has been observed for the GdIII complexes of DOTA-like ligands that have two inner sphere water molecules (q = 2) (DO3A, PCTA) and consequently fast water exchange rates.[27,41,42] However, as the bound water lifetime approaches the slow exchange condition (T1M ≈ τM), the relaxivity becomes exchange limited and its temperature dependence is less pronounced.[43,44]
Variable temperature 17O NMR
The water exchange dynamics in Gd(3) and Gd(6) were studied by variable temperature 17O NMR measurements. The temperature dependent profiles of the water 17O NMR transverse relaxation rates for Gd(3) and Gd(6) (Figure 1) are similar to those reported previously for GdIII complexes of other small molecular weight complexes such as Gd(7).[41] The maximum in the R2p versus temperature curves for Gd(3) and Gd(6) were observed at about the same temperature as that reported previously for Gd(8) while the maximum observed for Gd(DOTA)− is shifted to lower temperatures, reflecting faster water exchange.[41,42] The residence time (τM) of the inner-sphere water molecule was determined by fitting these data to Swift-Connick theory.[45,44] The value found here for Gd(3) (0.71 µs) (Table 3) is about 50% of the value found for Gd(8) (1.3 µs) and about four times longer than the value found for Gd(DOTA)− (τM = 0.23 µs at 298 K).[38,41] These values are consistent with the empirical observation that substitution of one carboxylate for an amide results in an approximately 3- to 4-fold decrease in the metal bound water exchange rate in Ln-complexes of polyaminopolycarboxylate type ligands.[46] The relatively long water residence times suggest that the water exchange in these complexes likely occur via a dissociative mechanism.[46]
Figure 1.
(Top row) Temperature dependence of the transverse water 17O NMR relaxation rates at 14.1T and pH=7 for 21 mm and 19 mm solutions of Gd(3) (left) and Gd(6) (right), respectively. (Bottom row) NMRD profiles of the 1 mM solutions of Gd(3) (left) and Gd(6) (right) at pH=7. The solid and dashed curves in the lower part represent the inner- and outer-sphere contributions to the overall relaxivity, respectively.
Table 3.
Relaxation parameters calculated from the simultaneous fitting of the temperature dependence of the water 17O transverse relaxation rate and the NMRD profiles of Gd(3) and Gd(6).
| Gd(3) | Gd(6) | Gd(8)[a] | |
|---|---|---|---|
| r1p298 [mm−1s−1] | 6.38 ± 0.04 | 5.05 ± 0.02 | 5.0 |
| q | 1 | 1 | 1 |
| Δ2 [s−2 × 1019] | 1.9 ± 0.1[b] | 1.0 ± 0.1[b] | 1.4 |
| τv298 [ps] | 58 ± 7[b] | 34 ± 4[b] | 40 |
| τR298 [ps] | 141 ± 3[b] | 86 ± 2[b] | 70 |
| τM298 [ps] | 0.71 ± 0.09[c] | 0.66 ± 0.05[c] | 1.3 |
| ΔHM [kJ×mol−1] | 28 ± 4[c] | 40 ± 3[c] | 19 |
| ΔHV [kJ×mol−1] | 25 ± 10[c] | 28 ± 10[c] | 40 |
Magnetic field dependence
The magnetic field dependence of the proton relaxation rates (NMRD profiles) was also measured for Gd(3) and Gd(6) at 25°C and pH 7 (Figure 1). Among other relaxation parameters, the rotational correlation time (τR) can be determined from the magnetic field dependence of the longitudinal relaxation rate of the complex.
In a typical NMRD experiment, a field-cycling relaxometer is used to measure the longitudinal relaxation rates of the solvent protons in the presence of the paramagnetic complex over a continuum of magnetic fields that range from 0.01 MHz up to ~70 MHz. Normally, τM is independently determined by variable temperature 17O NMR, while the inner-sphere relaxivity parameters are obtained by fitting the NMRD profiles to the Solomon–Bloembergen–Morgan equations and the outer-sphere parameters are determined by fitting the data to Freed’s equations.[47,48] The solid curves through the experimental data points (Figure 1) represent the best fit calculated values obtained by use of the relaxation parameters summarized in Table 3. The NMRD profile of both complexes show a single dispersion centered near 4 MHz with two plateaus in the low and high magnetic field regions. (The rotational correlation times of the complexes Gd(6) (τR = 86 ps) and Gd(8) (τR =70 ps) are similar, whereas the τR value of the complex Gd(3) (141 ps) is about twice as high as that of Gd(8).[41] Obviously, the long pendant arm in Gd(3) slows molecular rotation in this complex resulting in a higher relaxivity. The inner- and outer sphere contribution to the r1p relaxivity of Gd(3) and Gd(6) could also be estimated from the fitting of the NMRD profile (Figure 1). At 25°C and 20 MHz, the inner-sphere relaxivity (r1pis) of Gd(3) (4.0 mm−1s−1) is almost twice as high as that of Gd(6) (2.4 mm−1s−1) due to its slower tumbling rate. On the other hand, as expected, the contribution of the outer-sphere water molecules to the overall relaxivity (r1pos) is comparable (2.5 mm−1s−1) for both complexes. This value is also in good agreement with the r1p value of Gd(7) (r1p=2.5 mm−1s−1, 25°C, 20 MHz), a complex whose water proton longitudinal relaxation rate is dominated by the outer-sphere relaxation due to the very slow exchange between the inner-sphere water molecule and the bulk water.[49]
MRI of cells
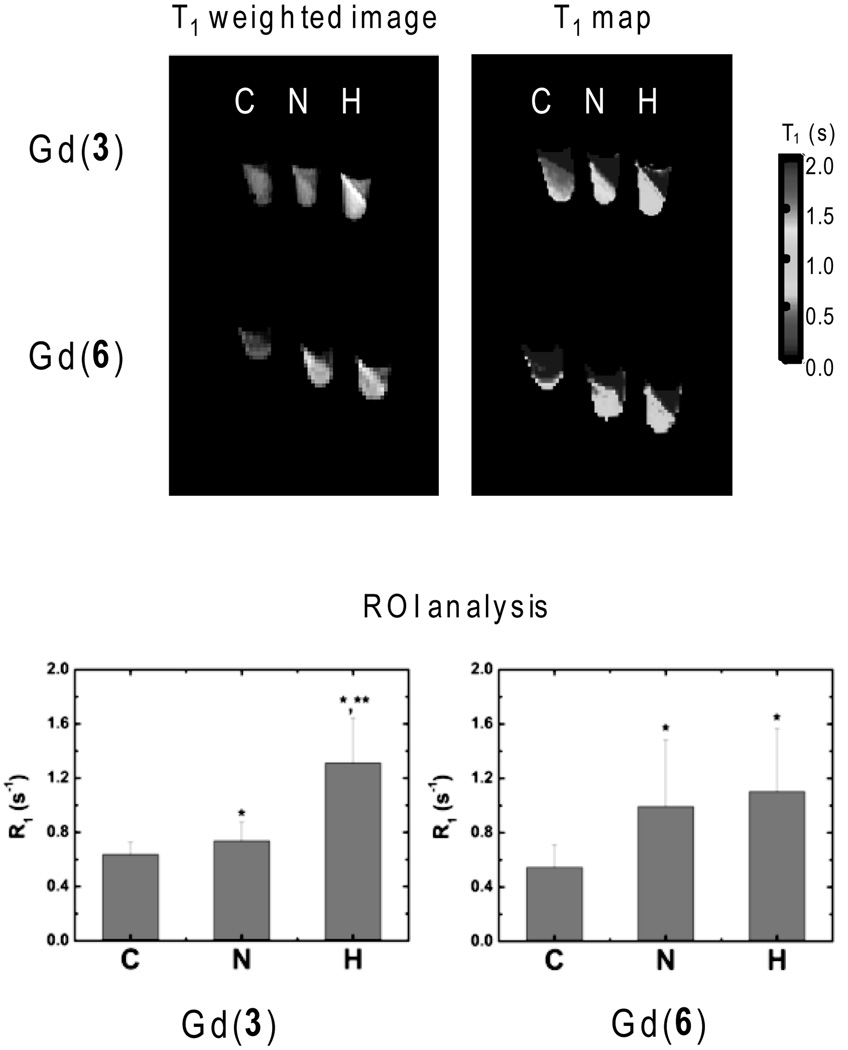
Cellular uptake of Gd(3) (or Gd(6) as control) under hypoxic versus normoxic conditions was first examined using 9L rat glioma cells as an in vitro model. This cell line has been extensively used in the past as a hypoxia model in radiosensitivity studies under low oxygen conditions.[52–54] In our experiments, hypoxia was induced in 9L cells by exposing a plate of cells to nitrogen gas at a rate of 1L/min for two hours at room temperature prior to the addition of the complex. After this induction phase, the cells were incubated with either 5 mm Gd(3) or Gd(6) for two hours under a continuous flow of nitrogen gas. The control consisted of the same cells exposed to the same concentration of each agent but using a constant flow of air rather than nitrogen. In addition, a negative control was prepared in which cells were incubated with PBS buffer alone (no gadolinium). After the incubation period, the samples were washed with PBS, detached from the culture plates using trypsin, pelleted into centrifuge tubes, and imaged immediately at 4.7T. T1-weighted images and T1 maps for three samples 1) negative control (C), 2) normoxic incubation (N) and 3) hypoxic incubation (H) of each complex are shown in Figure 2.
Figure 2.
In vitro MR imaging of 9L rat glioma cells after exposure to either Gd(3) or Gd(6) (top). T1-weighted images (TR = 300 msec; TE = 10 msec) and T1 maps of negative control (C), normoxic (N) and hypoxic (H) cells. Relaxation rates (R1) for the packed cell layers (bottom) (* = p <0.05 compared to negative control (C); ** = p<0.05 compared to normoxic sample (N)). Color version of Figure 2 is included in the Electronic Supporting Information.
If the nitroimidazole moiety in Gd(3) undergoes reduction and is trapped in hypoxic cells, then the accumulation of Gd(3) should result in brighter proton images as a result of the shortening of T1 of the bulk water by the intracellular agent. As seen from the T1-weighted images in Figure 2, the hypoxic sample (H) has higher signal intensity when compared to the other two samples, whereas the normoxic and negative control samples are indistinguishable. The T1 maps also show that cells incubated with Gd(3) under normoxic conditions have a somewhat shorter T1 than the negative control cells. The average T1 values for the negative controls, the normoxic incubation samples and the hypoxic incubation samples were 1.6±0.2 s, 1.4±0.1 s, and 0.8±0.1 s, respectively. This illustrates that Gd(3) enters cells by passive diffusion in both cell samples (normoxia and hypoxia) and a small amount of complex likely gets trapped even in normoxic cells. Nevertheless, the hypoxic cell pellet displays the shortest T1 of the three samples. Converting the values to relaxation rates (0.64±0.09, 0.74±0.14 and 1.31±0.33 s−1, respectively) highlights the differences in R1p. Subtraction of the R1 value for the negative control (cells only, no Gd(3)), yields R1p values of 0.10 and 0.67 s−1 for the samples incubated with Gd(3) under normoxic and hypoxic conditions, respectively. Thus, the hypoxic cells accumulate ~2-fold more Gd(3) compared to normoxic cells. In comparison, the distribution of T1 values found for normoxic and hypoxic cells incubated with Gd(6) are significantly different (p<0.05) from the negative control but not from each other. The R1p values of for the samples are 0.45 and 0.56 s−1 under normoxic and hypoxic conditions, respectively. While the two complexes (Gd(3) and Gd(6)) appear to accumulate to about the same extent in hypoxic cells as the nitroimidazole derivative, only Gd(3) shows hypoxia selectivity. The higher uptake of Gd(6) indicates that its uptake and retention is mediated by an oxygen-independent factor other than just passive diffusion. The higher uptake of Gd(6) may reflect its lower molecular weight and/or higher lipophilicity. Whatever the exact reasons for the nonselective uptake of Gd(6), the important fact is that this complex does not show hypoxia selectivity, while Gd(3) exhibits selective hypoxia-mediated retention.
Conclusion
DO3A-monoamide conjugate of 2-nitroimidazole (ligand 3) was synthesized and its complexes with GdIII and other lanthanide ions characterized. As expected, the protonation constants of this ligand and stability of the resulting lanthanide ion complexes are quite comparable to published values for similar compounds. The ligand has a total of five protonation steps in the pH range of 2–12; two of these are assigned to the protonation of two macrocyclic nitrogen atoms. The stability of the Ln(3) complexes ranges from 19.24 to 21.51 for CeIII to LuIII, about four orders of magnitude lower than that of the corresponding Ln(DOTA)− complexes. This is in accordance with the lower total basicity of 3 compared to DOTA. The r1p relaxivity, residence time of the metal bound water molecule (τM) and rotational correlation time (τR) of Gd(3) was determined by relaxivity measurements, variable temperature 17O NMR and proton nuclear magnetic relaxation dispersion (NMRD) studies and the data (r1p = mm−1s−1, 6.38 (τM = 0.71 µs, τR = 141 ps) are in agreement with those obtained for other GdIII complexes of similar size. In vitro MRI experiments with hypoxic 9L rat glioma cells revealed that Gd(3) is selectively trapped in hypoxic cells as evidenced by a nearly 2-fold contrast enhancement in the images of cells compared to normoxic cells. This suggests that this nitroimidazole conjugate may be suitable for assessment of hypoxia in vivo. Preliminary MR imaging of tumor bearing rats indicates that Gd(3) indeed accumulates largely in hypoxic tissues.[55]
Experimental Section
Abbreviations
MRI - magnetic resonance imaging; PET - positron emission tomography; SPECT - single photon emission computed tomography; L - ligand; DOTA - 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid; DO3A - 1,4,7,10-tetraazacyclododecane-1,4,7-triacetic acid; PCTA - pyclen triacetic acid; NMRD- nuclear magnetic resonance dispersion; ROI - region of interest.
General
Details of all synthetic procedures, characterization of all intermediates, preparation of the GdIII complexes, pH potentiometric titrations of ligands and complexes are reported in ESI.
Relaxivity and NMRD measurements
T1 values were recorded at 20 MHz (0.47 T), 25°C and 37°C using a Stelar Spinmaster spectrometer (Stelar, Italy) and a Maran Ultra relaxometer (Oxford Instruments, United Kingdom) at 23 MHz and 37°C. Longitudinal relaxation times were measured using the inversion-recovery pulse sequence (180° – τ – 90°). The T1 relaxivities were determined by the linear regression analysis of the water proton relaxation rates in solutions ranging in concentration from 0.2 to 21 mm, 50 mm HEPES buffer, in triplicate. The NMRD profiles were recorded on 1 mM samples over a continuum of magnetic field strengths from 0.00024 to 0.47 T (corresponding to 0.01 – 20 MHz proton Larmor Frequency) at 25°C using a Stelar field-cycling relaxometer under computer control with an absolute uncertainty of 1%. Relaxation data covering the range 0.47 T (20 MHz) to 1.7 T (70 MHz) were collected using a Stelar Spinmaster spectrometer operating at variable fields.
Variable temperature 17O NMR measurements
17O NMR linewidth data were collected using a Bruker DRX 600 NMR (14.1 T) spectrometer equipped with a 5 mm probe and a Bruker VT-1000 thermocontroller. NMR data were acquired by using a spectral width of 10,000 Hz, a 90° pulse (7 µs), an acquisition time of 10 ms, 1000 scans and no sample spinning. Water enriched in 17O to 2.6% (Yeda, Israel) was used. The observed transverse relaxation rates (RO2obs) were calculated from the signal width at half-height (Δν1/2): RO2obs = π Δν1/2 The paramagnetic contribution to the water 17O transverse relaxation rate (RO2p) were calculated from the RO2obs values according to the following equation: RO2p = RO2obs − RO2d where RO2d is the water 17O transverse relaxation rate in the absence of a GdL complex. The concentrations of Gd(3) and Gd(6) used for 17O NMR measurements were 21 and 19 mm, respectively.
In vitro cell experiments
Rat 9L-glioma cells were grown to confluence in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 1% L-glutamine and 1% penicillin/streptomycin. The growth medium was replaced with 8 mL of un-supplemented DMEM prior to the uptake experiment. One plate (100 mm i.d.) of cells was made hypoxic by exposing it to nitrogen gas (1 L/min) in an enclosed chamber for two hours at room temperature while two other plates were exposed to room air. 1 mL of phosphate buffer saline (PBS) was then added to one of the two normoxic samples (as negative control), and 1 mL of 45 mm Gd(3) to the remaining two samples (one normoxic and one hypoxic) yielding a final concentration of 5 mm. The same procedure was followed to prepare three other samples (control, normoxic and hypoxic) for assessing cell uptake of Gd(6). These samples were incubated for two hours at room temperature (negative control and reference samples in air, hypoxic sample in nitrogen at 1 L/min), rinsed three times with PBS, trypsinized to release the cells, and centrifuged in 15 mL tubes at 2500 rpm for 5 min. Excess buffer was removed from the pellets, and the samples were transferred to 0.2 mL tubes for a final centrifugation at 14,500 rpm for 5 min.
MRI imaging of cells
The three sample tubes were positioned in a 2 cm volume coil and imaged at 4.7 T. Images were collected using a TE of 12 ms and ten different TR values ranging from (0.1 s – 6 s). A T1 map was computed on a voxel-by-voxel basis from a least-squares fitting of the exponential relaxation curves using Varian software (VNMR 6.1C). Using software written in MATLAB, a region of interest (ROI) analysis was performed to extract voxel T1 values for each pellet for statistical comparisons. The voxel T1 data were imported into Origin 6.1 (OriginLab, Northampton, MA, USA) for student t-test analyses.
Supplementary Material
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (CA-126608, RR-02584, CA-84697, and CA-115531) (ADS), the Robert A. Welch Foundation (AT-584) (ADS), the Hungarian Scientific Research Found (OTKA K84291 (GT), OTKA PD-83253 (KFK)); the TÁMOP 4.2.1./B-09/1/KONV-2010-0007 and TÁMOP-4.2.2./B-10/1-2010-0024 projects implemented through the New Hungary Development Plan, co-financed by the European Social Fund and the European Regional Development Fund and by the János Bolyai Research Scholarship (GT) of the Hungarian Academy of Sciences. This work has been carried out in the frame of the COST CM1006 “EUFEN: European F-Element Network” and COST TD1004 “Theragnostics Imaging and Therapy: An Action to Develop Novel Nanosized Systems for Imaging-Guided Drug Delivery” Actions.
Footnotes
Supporting information for this article is available on the WWW under http://www.chemeurj.org/ or from the author.
Contributor Information
Gyula Tircsó, Email: gyula.tircso@science.unideb.hu.
A. Dean Sherry, Email: dean.sherry@utsouthwestern.edu, sherry@utdallas.edu.
References
- 1.Wouters A, Pauwels B, F. Lardon JB, Vermorken JB. Oncologist. 2007;12:690–712. doi: 10.1634/theoncologist.12-6-690. [DOI] [PubMed] [Google Scholar]
- 2.Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KSC, Chapman JD, Eckelman WC, Fyles AW, Giaccia AJ, Hill RP, Koch CJ, Krishna MC, Krohn KA, Lewis JS, Mason RP, Melillo G, Padhani AR, Powis G, Rajendran JG, Reba R, Robinson SP, Semenza GL, Swartz HM, Vaupel P, Yang D, Croft B, Hoffman J, Liu G, Stone H, Sullivan D. Int. J. Radiat. Biol. 2006;82:699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura S. Pharm. Bull. 1955;3:379–383. doi: 10.1248/cpb1953.3.379. [DOI] [PubMed] [Google Scholar]
- 4.Sheldon PW, Foster JL, Fowler JF. Br. J. Cancer. 1974;30:560–565. doi: 10.1038/bjc.1974.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JM. Radiat. Res. 1975;64:633–647. [PubMed] [Google Scholar]
- 6.Edwards DI. J. Antimicrob. Chemother. 1993;31:9–20. doi: 10.1093/jac/31.1.9. [DOI] [PubMed] [Google Scholar]
- 7.Knox RJ, Knight RC, Edwards DI. Br. J. Cancer. 1981;44:741–745. doi: 10.1038/bjc.1981.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams GE, Clarke ED, Flockhart IR, Jacobs RS, Sehmi DS, Stratford IJ, Wardman P, Watts ME, Parrick J, Wallace RG, Smithen CE. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1979;35:133–150. doi: 10.1080/09553007914550151. [DOI] [PubMed] [Google Scholar]
- 9.Rasey JS, Krohn KA, Freauff S. Radiat. Res. 1982;91:542–554. [PubMed] [Google Scholar]
- 10.Grierson JR, Link JM, Mathis CA, Rasey JS, Krohn KA. J. Nucl. Med. 1989;30:343–350. [PubMed] [Google Scholar]
- 11.Ballinger JR, Kee JW, Rauth AM. J. Nucl. Med. 1996;37:1023–1031. [PubMed] [Google Scholar]
- 12.Das T, Chakraborty S, Banerjee S, Mukherjee A, Samuel G, Sarma HD, Nair CKK, Kagiya VT, Venkatesh M. Bioorg. Med. Chem. 2004;12:6077–6084. doi: 10.1016/j.bmc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Norman TJ, Smith FC, Parker D, Harrison A, Royle L, Walker CA. Supramol. Chem. 1995;4:305–308. [Google Scholar]
- 14.Hoigebazar L, Jeong JM, Hong MK, Kim YJ, Lee JY, Shetty, Y.-S. Lee D, Lee, J.-K. Chung DS, Lee MC. Bioorg. Med. Chem. 2011;19:2176–2181. doi: 10.1016/j.bmc.2011.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell RJ, Workman P, Griffiths JR. Int. J. Radiat. Oncol., Biol., Phys. 1989;16:925–929. doi: 10.1016/0360-3016(89)90888-2. [DOI] [PubMed] [Google Scholar]
- 16.Aboagye EO, Lewis AD, Johnson A, Workman P, Tracy M, Huxham IM. Br. J. Cancer. 1995;72:312–318. doi: 10.1038/bjc.1995.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seddon BM, Payne GS, Simmons L, Ruddle R, Grimshaw R, Tan S, Turner A, Raynaud F, Halbert G, Leach MO, Judson I, Workman P. Clin. Cancer Res. 2003;9:5101–5112. [PubMed] [Google Scholar]
- 18.Aime S, Crich SG, Gianolio E, Giovenzana GB, Tei L, Terreno E. Coord. Chem. Rev. 2006;250:1562–1579. [Google Scholar]
- 19.Hylton N. J. Clin. Oncol. 2006;24:3293–3298. doi: 10.1200/JCO.2006.06.8080. [DOI] [PubMed] [Google Scholar]
- 20.De Leon-Rodriguez LM, Kovacs Z. Bioconjugate Chem. 2008;19:391–402. doi: 10.1021/bc700328s. [DOI] [PubMed] [Google Scholar]
- 21.Brown JM, Workman P. Radiat. Res. 1980;82:171–190. [PubMed] [Google Scholar]
- 22.Andre JP, Geraldes CFGC, Martins JA, Merbach AE, Prata MIM, Santos AC, de Lima JJP, Toth E. Chem.--Eur. J. . 2004;10:5804–5816. doi: 10.1002/chem.200400187. [DOI] [PubMed] [Google Scholar]
- 23.Hay MP, Wilson WR, Moselen JW, Palmer BD, Denny WA. J. Med. Chem. 1994;37:381–391. doi: 10.1021/jm00029a010. [DOI] [PubMed] [Google Scholar]
- 24.Dourtoglou V, Gross B, Lambropoulou V, Zioudrou C. Synthesis. 1984:572–574. [Google Scholar]
- 25.Knorr R, Trzeciak A, Bannwarth W, Gillessen D. Tetrahedron Lett. 1989;30:1927–1930. [Google Scholar]
- 26.Sherry AD, Brown RD, III, Geraldes CFGC, Koenig SH, Kuan KT, Spiller M. Inorg. Chem. 1989;28:620–622. [Google Scholar]
- 27.Aime S, Anelli PL, Botta M, Fedeli F, Grandi M, Paoli P, Uggeri F. Inorg. Chem. 1992;31:2422–3428. [Google Scholar]
- 28.Aime S, Barge A, Bruce JI, Botta M, Howard JAK, Moloney JM, Parker D, de Sousa AS, Woods M. J. Am. Chem. Soc. 1999;121:5762–5771. [Google Scholar]
- 29.Lloyd GO, Luckay RC. Acta Crystallogr., Sect. E: Struct. Rep. Online. 2005;E61:o3405–o3407. [Google Scholar]
- 30.Kubicek V, Havlickova J, Kotek J, Tircso G, Hermann P, Toth E, Lukes I. Inorg. Chem. 2010;49:10960–10969. doi: 10.1021/ic101378s. [DOI] [PubMed] [Google Scholar]
- 31.Martell AE, Smith RM, Motekaitis RJ. Critically Selected Stability Constants of Metal Complexes Database. 8 ed. College Station, TX: Texas A&M University; 2004. [Google Scholar]
- 32.Chaves S, Delgado R, Frausto da Silva JJR. Talanta. 1992;39:249–254. doi: 10.1016/0039-9140(92)80028-c. [DOI] [PubMed] [Google Scholar]
- 33.Burai L, Fabian I, Kiraly R, Szilagyi E, Brucher E. J. Chem. Soc., Dalton Trans. 1998:243–248. [Google Scholar]
- 34.Keire DA, Kobayashi M. Bioconjugate Chem. 1999;10:454–463. doi: 10.1021/bc980128u. [DOI] [PubMed] [Google Scholar]
- 35.Pasha A, Tircso G, Benyo ET, Brucher E, Sherry AD. Eur. J. Inorg. Chem. 2007:4340–4349. doi: 10.1002/ejic.200700354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar K, Chang CA, Francesconi LC, Dischino DD, Malley MF, Gougoutas JZ, Tweedle MF. Inorg. Chem. 1994;33:3567–3575. [Google Scholar]
- 37.Cacheris WP, Nickle SK, Sherry AD. Inorg. Chem. 1987;26:958–960. [Google Scholar]
- 38.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Chem. Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 39.Lauffer RB. Chem. Rev. 1987;87:901–927. [Google Scholar]
- 40.Bruce JI, Lowe MP, Parker D. In: The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. Merbach AE, Toth E, editors. Chichester: John Wiley & Sons; 2001. pp. 437–460. [Google Scholar]
- 41.GeninattiCrich SG, Cabella C, Barge A, Belfiore S, Ghirelli C, Lattuada L, Lanzardo S, Mortillaro A, Tei L, Visigalli M, Forni G, Aime S. J. Med. Chem. 2006;49:4926–4936. doi: 10.1021/jm0601093. [DOI] [PubMed] [Google Scholar]
- 42.Aime S, Botta M, Crich SG, Giovenzana G, Pagliarin R, Sisti M, Terreno E. Magn. Reson. Chem. 1998;36:S200–S208. [Google Scholar]
- 43.Aime S, Botta M, Fasano M, Paoletti S, Terreno E. Chem.-Eur. J. 1997;3:1499–1504. [Google Scholar]
- 44.Powell DH, Ni Dhubhghaill OM, Pubanz D, Helm L, Lebedev YS, Schlaepfer W, Merbach AE. J. Am. Chem. Soc. 1996;118:9333–9346. [Google Scholar]
- 45.Swift TJ, Connick RE. J. Chem. Phys. 1962;37:307–320. [Google Scholar]
- 46.Viswanathan S, Kovacs Z, Green KN, Ratnakar SJ, Sherry AD. Chem. Rev. 2010;110:2960–3018. doi: 10.1021/cr900284a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banci L, Bertini I, Luchinat C. Nuclear and Electronic Relaxation. Weinheim: VCH; 1991. [Google Scholar]
- 48.Korb JP, Ahadi M, Zientara GP, Freed JH. J. Chem. Phys. 1987;86:1125–1130. [Google Scholar]
- 49.Aime S, Barge A, Botta M, Parker D, De Sousa AS. J. Am. Chem. Soc. 1997;119:4767–4768. [Google Scholar]
- 50.Toth E, Connac F, Helm L, Adzamli K, Merbach AE. Eur. J. Inorg. Chem. 1998:2017–2021. [Google Scholar]
- 51.Bertini C, Luchinat I. Coord. Chem. Rev. 1996;150:1–292. [Google Scholar]
- 52.Jounaidi Y, Waxman DJ. Cancer. Res. 2000;60:3761–3769. [PubMed] [Google Scholar]
- 53.Kirkpatrick JP, Cardenas-Navia LI, Dewhirst MW. Int. J. Radiat. Oncol. Biol. Phys. 2004;59:822–833. doi: 10.1016/j.ijrobp.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 54.Moriyama EH, Jarvi M, Niedre M, Mocanu JD, Moriyama Y, Li B, Lilge L, Wilson BC. Proc. SPIE-Int. Soc. Opt. Eng. 2007;6449:64490F/1–64490F/8. [Google Scholar]
- 55.Kodibagkar VD, Gulaka PK, Rojas-Quijano F, Kovacs Z, Mason RP, Sherry AD. Proc. Internat. Soc. Magn. Reson. Med. Montreal, Canada: 2011. p. 1672. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.