Abstract
Antibodies empower numerous important scientific, clinical, diagnostic, and industrial applications. Ideally, the epitope(s) targeted by an antibody should be identified and characterized, thereby establishing antibody reactivity, highlighting possible cross-reactivities, and perhaps even warning against unwanted (e.g. autoimmune) reactivities. Antibodies target proteins as either conformational or linear epitopes. The latter are typically probed with peptides, but the cost of peptide screening programs tends to prohibit comprehensive specificity analysis. To perform high-throughput, high-resolution mapping of linear antibody epitopes, we have used ultrahigh-density peptide microarrays generating several hundred thousand different peptides per array. Using exhaustive length and substitution analysis, we have successfully examined the specificity of a panel of polyclonal antibodies raised against linear epitopes of the human proteome and obtained very detailed descriptions of the involved specificities. The epitopes identified ranged from 4 to 12 amino acids in size. In general, the antibodies were of exquisite specificity, frequently disallowing even single conservative substitutions. In several cases, multiple distinct epitopes could be identified for the same target protein, suggesting an efficient approach to the generation of paired antibodies. Two alternative epitope mapping approaches identified similar, although not necessarily identical, epitopes. These results show that ultrahigh-density peptide microarrays can be used for linear epitope mapping. With an upper theoretical limit of 2,000,000 individual peptides per array, these peptide microarrays may even be used for a systematic validation of antibodies at the proteomic level.
The immune system is endowed with a highly diverse repertoire of antibodies capable of targeting virtually any molecular structure. As specific affinity reagents, antibodies have become indispensable tools with a wide range of scientific and diagnostic applications (1, 2). Thus, antibodies are the main priority of several recent initiatives such as the Human Protein Atlas (3) and the ProteomeBinders consortium (4, 5) and of efforts to generate antibodies against cancer-related targets (6, 7), all of which aim to systematically generate affinity reagents, thereby facilitating the study of proteins and their role in biology and disease. As therapeutic agents, monoclonal antibodies have emerged as essential drugs with a wide range of clinical applications, making monoclonal antibodies one of the highest priorities of the pharmaceutical industry (8–11). The efficiency, accuracy, and safety of these antibody-mediated applications depend crucially on the selected antibodies being directed against the intended, and not against any unintended, target structure(s) (12). Specificity, the quintessential characteristic of an antibody, is therefore not only of scientific interest, but also of considerable practical importance.
For any antibody-based application, the establishment of specificity constitutes an important aspect of the validation process. Traditionally, the specificity of an antibody is examined in one or more in vitro assays (ELISA, Western blot, immunohistochemistry, flow cytometry, surface plasmon resonance, and many more (12–14)). Ideally, the entire epitope space should be examined; however, it is rarely possible to test more than a minor and ostensibly relevant part of the epitope space. What is relevant depends on the intended use; thus, the same antibody might exhibit sufficient and relevant specificity in one, but not in another, application (15). An important aspect of validating the specificity of an antibody is to determine the structure of the epitope that the antibody interacts with (12). Ideally, one would like to determine the three-dimensional structure of the binding complex using x-ray crystallography (16–18) or NMR1; however, such efforts are laborious and tend to have a low success rate and throughput. Many other epitope mapping approaches, such as fragmentation (19) or deuterium exchange in the presence or absence of antibody (20), directed mutagenesis, recombinant expression (including arrayed in situ cell-free translation approaches (20, 21)) of protein and peptide arrays, etc., have been suggested (12). Despite this plethora of methods, exact epitope information is lacking for the vast majority of antibodies used in life science research, and there is a significant need for simple and rapid methods to map epitopes. The availability of such methods would also support the selection of paired antibodies that each bind to separate parts of an antigen, thereby allowing one antibody to validate the results of another (12, 22).
Proteins constitute important immune targets, and many of the methods used to address antibody specificity are tailored for protein antigens. Traditionally, protein epitopes have been divided into discontinuous/conformational epitopes, which require that the native protein structure be intact, or continuous/linear epitopes, which may be represented by consecutive overlapping synthetic peptides encompassing the complete primary structure of the target antigen (15). The mapping resolution of linear epitopes depends on the peptide length, the overlap chosen for the initial epitope location, and the scale of the subsequent fine specificity analysis (e.g. N- and C-terminal truncations; amino acid scans; random single, double, or triple substitutions; etc.). The number of peptides required can be substantial, making the cost of peptides and the logistics of handling large panels of peptides a serious impediment of the in-depth characterization of linear epitopes. Most standard peptide synthesis equipment can synthesize only up to a few hundred single peptides simultaneously, although lately up to 8000 peptides have been synthesized in parallel on a cellulose membrane (23–25) using the SPOTTM technique. In addition to performing assays directly on the membrane (26), such peptides can be released and transferred onto glass slides using additional robotics and printing techniques (25). As alternatives to synthetic peptides, phages (27), bacteria (28), and yeast cells (29) have been used to express libraries of fragmented antigens (27) or of combinatorial peptides (30). These methods can potentially generate millions of peptides covering entire protein antigens, and they may, at least in some cases, mimic conformational epitopes (15, 31–33). Major drawbacks of these methods include the lack of control of the exact peptide sequences expressed and the need for separate sequencing of positive clones. None of these drawbacks are encountered with peptide microarrays.
Here, we present the first report on the feasibility of using ultrahigh-density peptide microarrays to address antibody specificities in casu mapping the fine specificity of polyclonal antibodies raised against linear protein epitopes. This allowed a fast and exhaustive analysis of the length requirements and a detailed analysis of the fine specificity of these antibodies. We suggest that specificity analysis of linear epitopes using ultrahigh-density peptide microarrays addressing the entire human proteome is within reach.
EXPERIMENTAL PROCEDURES
Derivatization of Synthesis Slides
Microscope slides (Nexterion E; Schott AG, Jena, Germany) for synthesis of the arrays were derivatized via incubation with 1 g/l bovine serum albumin in 0.5 m N-methylmorpholine (NMM)/acetate pH 8.5 for 3 h at room temperature. The slides were washed in water, N-methylpyrrolidone (NMP), and dichloromethane (DCM) and stored dry until use. Synthesis of the microarrays was performed directly on the BSA-coated slides using the epsilon amino groups of sterically exposed lysines as the starting point.
Synthesis
Peptide arrays were synthesized by Schafer-N (Copenhagen, Denmark) using a maskless photolithographic technique (34) in which 365 nm light with an energy density of ca. 20 mW/cm2 was projected onto 3′-nitrophenylpropyloxycarbonyl (NPPOC)-photoprotected (35, 36) amino groups on a glass surface in patterns corresponding to the synthesis fields. Details of the technique will be published elsewhere, but briefly, the patterns were generated using digital micromirrors and projected onto the synthesis surface using UV-imaging optics (supplemental Fig. S1A). In each layer of amino acids, the relevant amino acids were coupled successively to predefined fields after UV-induced removal (in 1 m diisopropylethylamine (DIEA) in NMP) of the photoprotection groups in those fields. The couplings were made using standard Fmoc-amino acids activated with O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate/DIEA in NMP. After coupling of the last Fmoc-amino acid in each layer, all Fmoc-groups were removed in 20% piperidine in NMP and replaced by NPPOC groups (37) coupled as the chloroformate in DCM with 0.1 m DIEA. The procedure was then repeated until all amino acids had been added to the growing peptide chains (supplemental Fig. S1B). Final cleavage of side protection groups was performed in TFA:1,2-ethanedithiol:water 94:2:4 v/v/v for 2 h at room temperature.
Epitope Mapping Using Peptide Arrays
Primary rabbit polyclonal antibodies were diluted to a concentration of around 100 ng/ml in PBS-Tween. Deprotected slides were blocked and hydrated overnight in a mixture of 1 g/l bovine serum albumin and 0.1% v/v detergent (Tween 20) in PBS and incubated for 1 h at room temperature with relevant polyclonal anti–protein epitope signature tag (PrEST) rabbit antibodies as primary reagents. After washing, the slides were incubated for 1 h at room temperature with Alexa Fluor 488-labeled goat-anti-rabbit IgG (Invitrogen, Carlsbad, CA) as a secondary reagent. Images of the stained arrays were recorded using an MVX10 fluorescence microscope equipped with an XM10 cooled digital camera (both from Olympus, Ballerup, Denmark) and analyzed using the analysis program PepArray (Schafer-N, Copenhagen, Denmark). See the supplementary information for a brief description of the PepArray program.
Epitope Mapping Using Cell-surface Display
Mapping using cell-surface display was performed as described elsewhere (28). Briefly, gene fragments encoding the different antigens were amplified separately via PCR (4.8 ml pooled), and the products were sonicated to generate random fragments. These were blunt-ended and phosphorylated before ligation into the cell-surface expression vector pSCEM2 and transformed into Staphylococcus carnosus. Cell aliquots of about 10-fold coverage of the library were incubated with about 1 ng antibody in reaction volumes of 70 μl PBS-P. Cells were washed and fluorescently labeled with Alexa 488 secondary goat-anti-rabbit antibodies (Invitrogen, Carlsbad, CA) and Alexa 647 labeled albumin for expression normalization and then washed again ahead of analysis via FACS. Single cells expressing antibody-binding peptides were sorted, sequenced, and aligned back to the target protein sequence.
RESULTS
Ultrahigh-density Peptide Microarrays
Peptide arrays were generated by a combined maskless photolithographic (34) and solid phase peptide synthesis strategy using a digital mirror device (1080P DMD (Digital Light Projections, Digital Light Innovations, Austin, TX) with 1920 × 1080 = 2,073,600 individually addressable micromirrors) to project 365 nm light onto NPPOC-photoprotected (35, 36) amino groups on a glass surface in patterns corresponding to the fields where the next amino acid extension should occur (supplemental Fig. S1A). Successively removing photoprotection groups extending the growing peptide chain with standard Fmoc-protected amino acids and exchanging the Fmoc-groups with NPPOC-groups after all extensions in a given layer (37) allowed individually predefined peptides to be built in each synthesis field (supplemental Fig. S1B). After synthesis of the peptide backbones, all side chain protection groups were removed via TFA treatment, leaving the peptides attached to the matrix through their C-terminals. Typically, each synthesis field was defined by a square measuring 2 × 2 (as in Fig. 1) or 3 × 3 mirrors. However, because synthesis fields defined by as few as one mirror could be discerned (Fig. 1), the maximum number of different synthetic peptides that can be realized with the current DMD device appears to be around 2,000,000 on a surface area of ∼2 cm2.
Fig. 1.
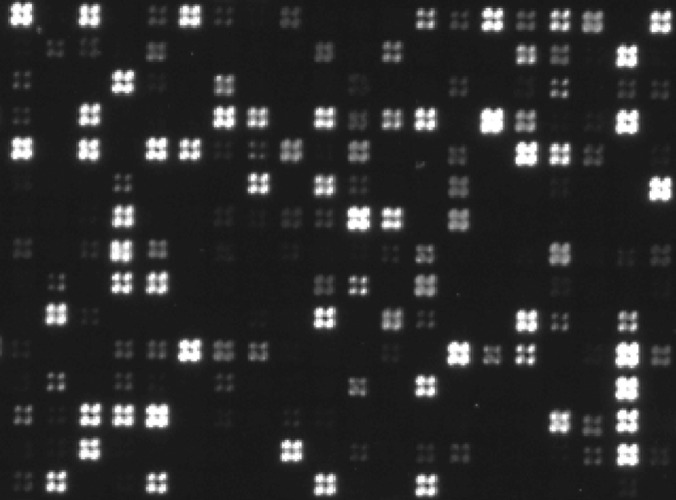
Image of a peptide microarray. Small section from a peptide array used for identification of different peptide epitopes, including the ones in Fig. 2A. The peptides were synthesized in quadratic fields defined by 2 × 2 mirrors (each mirror measuring 10 μm × 10 μm). One-mirror-wide empty regions separated the peptide fields. The fields were visualized via incubation with relevant rabbit antibodies followed by Alexa488-conjugated goat anti-rabbit IgG. The section shown corresponds to ca. 0.15% of the area of the entire array. Note that peptide synthesis at single mirror resolution can be observed.
Polyclonal Antibodies Specific for Linear PrEST Epitopes on Human Proteins
PrESTs are short (50 to 150 amino acids long) fragments of proteins that have been selected to be as sequence-dissimilar as possible to all other proteins in the corresponding proteome; that is, they aim to be unique and specific representatives of the proteins in question (38). As part of the Human Protein Atlas initiative (38), polyclonal rabbit antibodies were raised against PrESTs, which were expressed in E. coli and purified under denaturing conditions. Subsequently, the antibodies were affinity-purified using the same PrESTs as used for capture reagents. The specificities of the antibodies were validated with protein microarrays using immobilized PrESTs (see supplemental Fig. S2) and by Western blotting of lysates of human cell and tissues (data not shown). This immunization and purification strategy favors the generation of antibodies specific for linear epitopes. Theoretically, polyclonal antibodies could target multiple consecutive epitopes along the sequence of an extended protein in casu encompassing an entire PrEST; however, we have recently mapped a PrEST-specific polyclonal antibody to a few separate and distinct regions of its target protein, suggesting that large parts of a target sequence may be “epitope silent” (28, 39).
The Location and Length of Linear Epitopes
The ultrahigh-density peptide microarray technology was used to map the specificities of 22 polyclonal anti-PrEST antibodies (38). Initially, we addressed the location and length of the recognized epitopes by systematically scanning through the entire sequence of each PrEST using an overlapping peptide strategy with an offset of one amino acid (this is the smallest offset possible and thus allowed us to achieve the maximum resolution) and included all lengths from 2-mers to 20-mers. This experiment entailed the synthesis of more than 74,000 different peptides. To counter the possible influences of artificially introduced N-terminals or artificially tethered C-terminals, all peptides were extended with “padding sequences” (N-terminally with GAG and C-terminally with GAGADDD).2 Peptide microarrays were stained and recorded as detailed in Materials and Methods. Briefly, the slide was blocked, incubated for 1 h at room temperature with relevant polyclonal anti-PrEST antibodies as primary reagents, and stained for 1 h at room temperature with an Alexa Fluor 488-conjugated goat-anti-rabbit serum as a secondary reagent. Images of the stained arrays were captured with a fluorescence microscope and analyzed using a microarray analysis program.
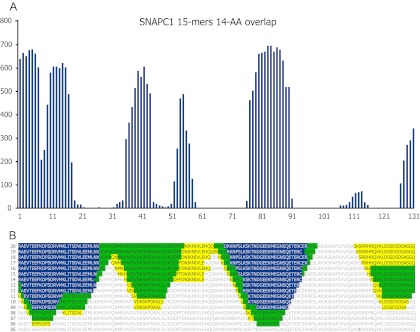
All possible 15-mers from each of the 22 PrESTs were assayed with the corresponding polyclonal anti-PrEST antibodies, and the locations of one or more epitope regions were suggested for each anti-PrEST antibody. As a representative example, an epitope location and length scan of a polyclonal antiserum generated against a 145 aa long PrEST representing the 43 kDa human polypeptide 1 of the small nuclear RNA activating protein complex SNAPC1 is shown (Fig. 2). A bar graph illustrates the background-subtracted signals obtained from overlapping 15-mers with an offset of one amino acid. Several distinct peaks of reactivity were located (Fig. 2A). To give an overview of the relationship between peptide length and reactivity, data obtained with different peptide lengths were converted to color-coded strings of the PrEST sequence indicating strong, intermediary, and weak reactivity of the corresponding peptides (Fig. 2B). This readily revealed the shortest recognizable sequences of the most dominant reactivities (e.g. strongly interacting 6-mer EFKDPS and 7-mer KTNDGEE peptides; intermediary interacting 8-mer KLITSDVL, 10-mer VDKSKPDKAL, and 9-mer LDSSDSDSA peptides). The minimum length requirement thus varied considerably from epitope to epitope. For this particular polyclonal antibody preparation, no signals were obtained for peptides shorter than six amino acid residues (excluding paddings).
Fig. 2.
Analyzing the length and fine specificity of polyclonal antibody epitopes. A, Bar chart displaying fluorescence signal obtained from antibodies binding to array-bound peptides synthesized as 15-mers overlapping by 14 amino acid residues. Rabbit antibodies were raised against a 145 residue PrEST coded by the human SNAPC1 gene. Alexa488-conjugated goat anti-rabbit IgG was used as secondary antibody. y-axis: fluorescence (AU); x-axis: residue number (n-terminal to the left). Each bar represents the signal obtained from a 15-mer peptide whose sequence starts at the indicated residue number. B, Different lengths of the SNAPC1 PrEST sequence (varying from 5-mers to 20-mers) were synthesized as overlapping peptides with an offset of one amino acid; that is, for each line the peptides were synthesized as n-mers with (n − 1) residue overlap. The fields were visualized by means of incubation with rabbit anti-SNAPC1 antibodies followed by Alexa488-conjugated goat anti-rabbit IgG. Results obtained from bar charts (as illustrated in Fig. 2A) are rendered as the PrEST sequence and color-coded to illustrate antibody-binding regions (yellow = low signal strength, green = intermediate signal strength, and blue = strong signal; colors from stronger signals are superimposed on colors from weaker signals). The lines represent results obtained with 20-mer peptides (upper line) down to 5-mer peptides (lower line); the peptide length of each line is indicated to the left.
Mapping the Fine Specificity of Polyclonal Antibodies Reacting with Linear Epitopes
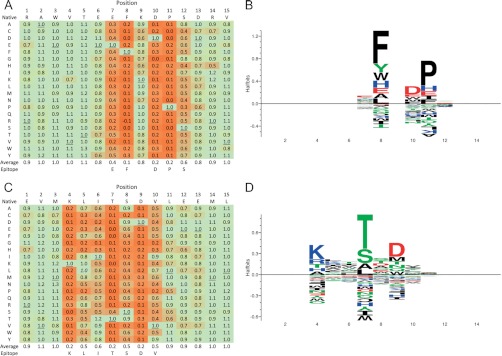
To study the fine specificity of polyclonal anti-PrEST antibodies, complete single amino acid substitution analyses were performed on the most prominent epitope regions suggested by the overlapping peptide scans described above. In an attempt to encompass the putative epitope in its entirety, each region was represented by a 15-mer peptide centered at the position of peak reactivity. All 20 naturally occurring amino acids were systematically tested as single substitutions in all 15 positions. The signal obtained from each singly substituted peptide was divided by the signal obtained with the native 15-mer peptide, and the resulting relative signal (RS) values were used to generate position-specific scoring matrices (PSSMs). As representative examples of such a single substitution analysis (SSA), the previously described EFKDPS and KLITSDVL epitopes are illustrated (Figs. 3A and 3C, respectively). For each position, the mean and standard deviation of the 20 RS values were calculated. Positions with maximum selectivity (i.e. where only one amino acid is acceptable) would be represented by an RS value of 1 for the essential amino acid and of 0 for all the other amino acids, leading to an average RS value of 0.05 for this position. In contrast, positions with minimum selectivity (i.e. where any amino acid is acceptable) would be represented by RS values of 1 for all amino acids, leading to an average RS value of 1. A one-way analysis of variance (ANOVA) (40) was done for each PSSM to determine whether two or more of the mean values differed significantly from each other. If so, then Tukey's least significant difference (LSD) was calculated to determine which of the mean values differed significantly (p < 0.01) from the null hypothesis of no selectivity (RS = 1). For the RAEVTEEFKDPSDRV region (Fig. 3A), the average RS values of the first six and last three positions did not deviate significantly from the null hypothesis (range: 0.83–1.06). In contrast, the null hypothesis was rejected for positions 7–8 and 9–12, where the average RS values were significantly less than 1 (range: 0.14–0.56). Note that position 9 featured a borderline selective position with an average RS value of 0.82, almost introducing a gap in the middle of this selectivity hot spot. Thus, the epitope contained within this 15-mer region could be defined as the 6-mer region containing the sequence EF-DPS (the most dominating residues are underlined, and any internal nonselective positions are indicated by a dash). Similarly, the epitope contained within the AVMKLITSDVLEEML region (Fig. 3C) could be defined as the 7-mer region containing the sequence KLITSDV (average RS values ranging from 0.15 to 0.60) surrounded by nonselective residues (average RS values ranging from 0.84 to 1.05). With a visual representation of the individual RS values employing a continuous color scale (green and red showing high and low binding, respectively), the fine specificities of these interactions were distinctly visible (see Figs. 3A and 3C). Note the strong discriminatory power of these polyclonal antibodies where most of the selective positions excluded even single conservative substitutions. An alternative visual representation, a sequence LOGO, illustrated the demarcated borders of the epitopes and the presence of particularly selective positions (Figs. 3B and 3D).
Fig. 3.
Fine specificity described by exhaustive single substitution scans. Single substituted analog peptides, scanned through all 15 positions of the native peptide sequence and including all 20 naturally occurring amino acids, were synthesized and tested for binding to the appropriate anti-PrEST antibody. The relative signals of the analog peptide and of the native peptide are shown and color shaded so that reddish hues are assigned to substitutions resulting in reduced binding of the antibody. A, Position specific scoring matrix (PSSM) representing RS values of a single substitution scan of the 15-mer peptide RAEVTEEFKDPSDRV. B, The EF-DPS epitope identified from the RAEVTEEFKDPSDRV peptide. C, PSSM representing RS values of single substitution scans of the 15-mer peptide AVMKLITSDVLEEML. The PSSM matrices were also visualized as sequence logos using the Sequence2Logo server (http://www.cbs.dtu.dk/biotools/Seq2Logo-1.0/). D, The KLITSDV epitope identified from the AVMKLITSDVLEEML peptide.
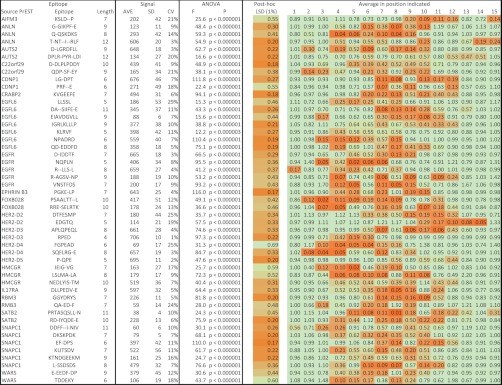
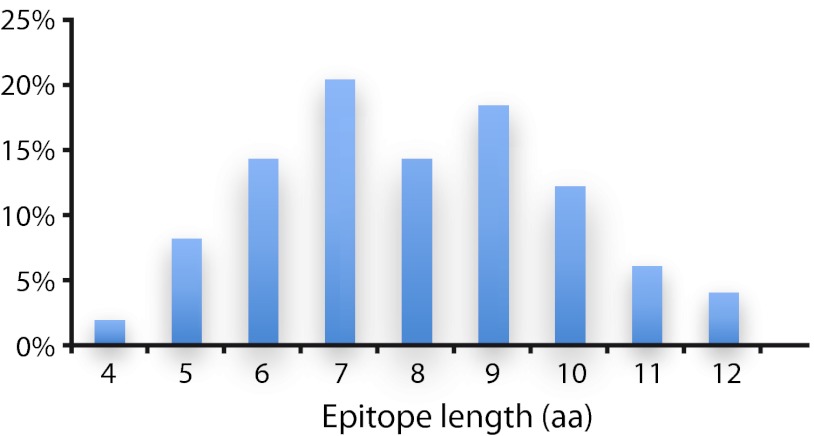
Complete fine specificity analyses were extended to 22 anti-PrEST antibodies involving 79 putative epitope regions. The majority of these analyses, 49 (62%; overview in Fig. 4), resulted in PSSMs showing a highly significant pattern of selective positions indicative of the presence of antibody epitopes (p < 0.00001, ANOVA); 95% of these epitopes appeared fully contained within the 15-mer stretch selected for the fine specificity analysis, whereas the remaining 5% extended to the N- or C-terminus of the selected 15-mer stretch and could potentially extend even further. Particularly noteworthy, the epitopes identified by the substitution analysis were in general well defined with sharply demarcated borders (Figs. 3 and 4). In general, the epitopes were from 5 to 10 amino acids long (range from 4 to 12 amino acids; Fig. 5). The epitopes contained highly dominant positions where only a few amino acid substitutions were acceptable, less dominant positions where several amino acid substitutions were acceptable, and nonselective positions where all amino acid substitutions were acceptable (as illustrated in Fig. 3). The SSA failed to identify epitopes in 30 of the 79 (38%) 15-mer regions selected for further analysis by the length scan. On a protein antigen basis, this analysis confirmed the presence of epitopes for 20 of the 22 examined antibodies (91%).
Fig. 4.
Overview of the main experimental SSA data leading to the identification of 49 epitopes in 20 of 22 source PrEST proteins. The “epitope” box contains the source PrEST protein, the identified epitope, and the length of the epitope. The “signal” box contains the statistical analysis of the signal strength of the 15 repeats of the native peptide sequence with average (AVE), standard deviation (SD), and calculated variation coefficient (CV). The “ANOVA” box contains the ANOVA analysis of the exhaustive SSA with the F value and the associated probability. The final box shows the epitope identification with the 1% LSD value (shaded from most discriminatory in yellow to least discriminatory in red) obtained by Tukeys post-hoc analysis and used to identify the positions with average values (as in Fig. 3) that deviate significantly from 1.00 (nonsignificant values are shaded green; shading starts at 1 LSD and is maximal (red) at 0). Note that even weak signals can result in highly significant epitope calling (see, e.g., the SNAPC1 epitope DKSKPDK).
Fig. 5.
Length distribution (in amino acid residues) of 49 different SSA-determined epitopes. The y-axis is the number of epitopes found of the length indicated. The x-axis is the epitope length (i.e. number of residues covered by the epitope region).
Comparisons with a Bacterial Expression Epitope Mapping Strategy
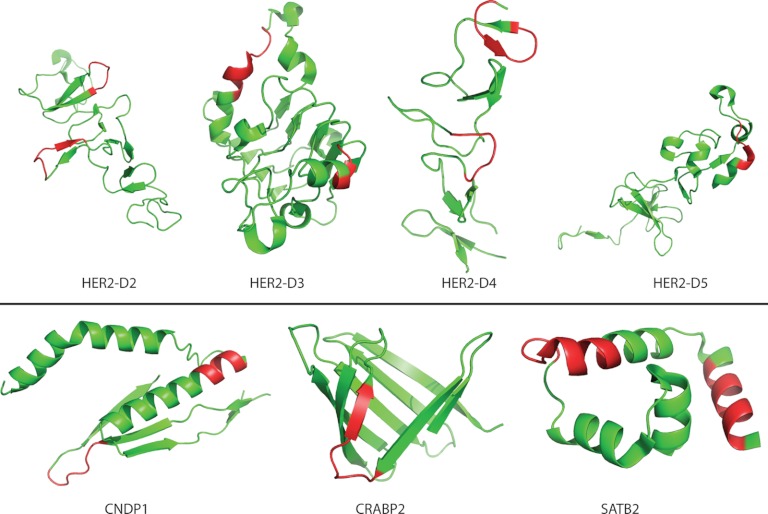
We have recently generated two alternative antibody-mapping approaches. In the first approach, Staphylococcus carnosus cells are used to display peptide libraries generated by random fragmentation of genes encoding protein antigens of interest (28). Briefly, libraries are labeled with the antibody of interest and sorted by means of flow cytometry, and the antigen fragments expressed by the sorted cells are determined via DNA sequencing. In the second approach, biotinylated synthetic peptides (15-mers with 10-amino-acid overlap; Sigma-Aldrich, St Louis, MO) spanning the PrEST in question were coupled to Luminex streptavidin coated beads with unique reporter dyes, stained with the polyclonal anti-PrEST antibodies, labeled with phycoerythrin-conjugated secondary reagent (Moss Inc., Pasadena, MD), and analyzed using LX200 instrumentation with Luminex IS 2.3 software (39). For 17 of the PrEST-specific polyclonal antibodies, data obtained with these two established methods could be compared with those obtained with the peptide microarray-driven approach presented here. More than 80% of fragments found through the bacterial surface or Luminex bead display approaches overlapped the reactive stretches identified by the 15-mer length scanning approach (illustrated in Fig. 2B) with at least four amino acids, which was the smallest epitope length observed for these antibodies (illustrated in Fig. 5). Thus, the different approaches could have identified the same epitopes. Using the SSA approach, some of the putative shared epitopes could be demonstrated. An example is given in Fig. 6, in which the middle line shows a 15-mer peptide microarray scanning (with an offset of 1) for the SNAPC1 PrEST target protein (the target protein sequence is color-coded for reactivity as described in Fig. 2B). The locations of relevant fragments identified by bacterial surface display and the locations of the epitopes identified via the SSA approach are shown above and below the sequence, respectively. The 15 PrESTs available for this comparison encompassed 74 reactive stretches (ranging from 4 to 45 amino acids long) identified by the bacterial surface or Luminex bead display approaches and 49 reactive stretches (ranging from 4 to 12 amino acids long) identified by the SSA approach; of these, 29 were shared between the display approaches and the peptide microarray SSA approach. Some measure of disagreement between these approaches is to be expected because the methods are very different in nature (e.g. the difference in expression of peptide versus protein (or protein fragment) or in continuous versus discontinuous epitopes, such that the bacterial surface display system could miss some epitopes because of the random nature of the fragmentation or could potentially display discontinuous epitopes). Nonetheless, a highly significant correlation between the display approaches and the peptide microarray SSA approach was observed when assigning each amino acid of the 15 PrESTs available for comparison of whether it had been identified by both, one or the other, or none of the approaches (p < 0.001, Chi-square test with Yates correction; a visual representation of this comparison is given in supplemental Fig. S3). Finally, the epitopes identified by the peptide microarray were projected onto the known structures of the underlying proteins (Fig. 7). Epitopes located on several different secondary structural elements including parallel beta-sheets, loops, and helical regions could be identified. This suggests that it might be possible to identify paired antibodies specific for five of seven of these PrESTs.
Fig. 6.
The specificity of anti-PrEST polyclonal antibodies directed against SNAPC1 analyzed using different approaches. The top line indicates the peptide sequences identified via bacterial surface display. The center line contains the entire PrEST sequence representing the SNAPC1 protein, and the color encoding indicates the strength of the reactivity of the antibodies as detected in the peptide arrays by a 15-mer length scan with an offset of 1 (from Fig. 2B; blue, green, and yellow indicate strong, intermediate, and weak reactivity, respectively). The bottom line indicates the peptide sequences identified by the heat map approach (underlined residues indicates highly selective positions; dashes indicates nonselective interaction within an epitope).
Fig. 7.
Epitope context and structure. Epitopes identified by the peptide microarray approach and reported in Fig. 4 were mapped onto the known structure of the underlying proteins. Epitopes located on several different secondary structural elements, including parallel beta-sheets, loops, and helical regions, could be identified. In five of the seven cases shown here, several distinctly separated epitopes were identified.
DISCUSSION
Using a photolithographic approach, one of us has recently developed an ultrahigh-density peptide microarray technology theoretically capable of expressing up to 2,000,000 individual peptides on a ca. 2 cm2 area (41, 42). This has been achieved through a combination of previously reported advances in peptide microarray technology and chemistry. In a seminal 1991 study, Fodor et al. (43) used photo-masks and activated amino acids, which had been synthesized individually with a photolabile protection group, to generate a rather costly photolithographic principle for the synthesis of pre-addressable peptide microarrays. However, this technology was outcompeted by the cheaper and simpler SPOT principle of peptide array synthesis, which was introduced around the same time (44); for a historical overview, see Ref 45. Resurrecting photolithography as a principle of peptide microarray synthesis, Gao and co-workers (46, 47) used a DMD and photo-generated acids to effect light-directed peptide synthesis; however, this technology is limited by the need for physical barriers to confine the acid and prevent diffusion to unwanted areas, which also limits the peptide density that can be achieved. Recently, Li et al. (37) reported a chemical strategy for the in situ addition of photo-cleavable protection groups to a growing peptide chain, thus reestablishing a nondiffusable elongation principle. We have combined these advances, allowing a high-resolution DMD-driven photolithographic strategy without the need for photo-masks, physical barriers, or unique amino acid reagents (see supplemental Fig. S1), and basically allowing the use of standard solid-phase peptide synthesis reagents (40, 41).
Fig. 1 shows that this ultrahigh-density peptide microarray approach can achieve single mirror resolution and thus theoretically generate up to 2,000,000 peptides per microarray. By the same token, each of the 10 μm × 10 μm synthesis fields expresses very little peptide (estimated to be in the attomolar range). At this stage, it is not technically possible to address the quantity or quality of the peptides synthesized in each field (however, using all >2,000,000 mirrors to synthesize one and the same peptide, we have isolated sufficient material from a slide to ascertain that the intended peptide was indeed synthesized (data not shown)). A future hope would be that a high-sensitivity and label-free technology such as mass spectrometry could be adapted to validate the identity and purity of the peptides synthesized in each field, and potentially even be able to identify any reactant(s) offered to the peptide microarray. In the absence of such a technology, we have here resorted to a less direct, functional validation approach.
In this study, we have used an ultrahigh-density peptide microarray technology to map the location and fine specificity of a panel of polyclonal antibodies raised against short linear protein fragments uniquely representing human proteins (PrESTs). These antibodies were derived as part of the Human Protein Atlas initiative, which aims at generating specific antibodies against every protein of the human proteome (38). This initiative, together with other proteome-wide analysis initiatives, illustrates the need for new high-throughput technologies. Conventional solid phase peptide synthesis is obviously not able to provide the numbers of peptides needed to identify and validate proteome-wide reagents. Even array technologies based on pin synthesis or spotting approaches would be seriously challenged by these demands.
The ability to synthesize several hundred thousand peptides allowed us to address the specificity of 22 polyclonal antibodies using exhaustive and high-resolution length scans and SSA. This led to the identification of one or more epitopes for 20 of these 22 polyclonal antibody preparations. Five of the antibodies recognized only one epitope on their respective PrEST targets, whereas 15 of the antibodies recognized multiple discontinuous epitopes (up to seven epitopes per target). Our data confirm our previous report that antibodies within a polyclonal mixture can simultaneously be tested and used to identify linear peptide epitopes and that polyclonal antibodies, despite theoretically being able to target epitopes along the entire PrEST sequence, map to a few separate and distinct regions, suggesting that the majority of the target sequence is “epitope silent” (28, 39). Why some regions of these PrESTs remain epitope silent is not known. From a technical point of view, these epitope silent regions can be considered built-in negative experimental controls.
The length scans show the value of an exhaustive approach. As illustrated in Fig. 2B, some reactivity started being detectable at the level of 6-mer peptides, whereas others did not appear until at the level of 7-mer, 9-mer, or even longer peptides. This illustrates a fundamental problem in defining epitopes solely using overlapping peptides. Although short regions of strong reactivity probably represent dominant epitopes, this interpretation is confounded by the risk of the detected reaction being caused by two or more overlapping epitopes that might not have been resolved into individual epitopes. The closely positioned minimal epitopes EF-DPS and KLITSDVL illustrate this point. In this case, the minimal epitopes are sufficiently separated that each of them can be isolated and identified with short peptides; however, they would have been difficult to resolve if they had been more closely positioned.
One would be naturally inclined to compare the signal strengths of different peptide–antibody interactions and interpret them in terms of affinity. In this context, a word of caution is appropriate, because signal strength is determined by many factors. The relative contributions of these factors are not sufficiently controlled and/or known at this point. Thus, one should be careful when comparing different epitopes: a weak signal could theoretically be due to peptide synthesis failure, variations in peptide solvation, and/or the absence of high-affinity antibodies in reasonable concentrations. In this context, complete substitution analyses gave a very detailed, yet simple, description of the fine specificity of the epitope–antibody interactions and in many cases yielded highly significant results despite the weakness of the underlying signals. Thus, exhaustive SSAs followed by ANOVA and post hoc tests like Tukey's LSD proved to be an efficient way to perform epitope calls and identify positions of selectivity. Fig. 4 illustrates how this statistical analysis in terms of epitope calling is superior to the mere recording of signal strength, which would have led to several otherwise clearly selective epitopes being discarded.
To our knowledge, this is one of the largest collections of fully substituted antibody epitope mappings reported. Some information on the biology of antibody recognition of linear epitopes can be extracted. The lengths of the epitopes were mostly 7 to 9 amino acids long (range: 4–12). In general, the epitopes contained a few very selective positions where the original amino acid was almost exclusively preferred and the signal dropped dramatically if the original amino acid was substituted with any other amino acid. In a few positions, the signal dropped less dramatically when conservative substitutions were made. In yet other positions, no significant contributions to the specificity could be detected. Thus, highly stringent, more relaxed, and nonselective positions could be intermingled as shown for the EF-DPS epitope. Our data show that polyclonal antibodies can be extremely selective peptide binders. We have previously examined the peptide binding specificity of MHC molecules, which have evolved specifically to present oligopeptides to T lymphocytes. In line with the requirement of MHC molecules to sample many different peptides, the specificity requirements of MHC molecules are much more relaxed. Structurally, MHC molecules achieve this broad specificity through extensive interactions with the peptide backbone. By inference, one could speculate that the highly selective peptide–antibody interactions are dominated by peptide side-chain interactions.
As alluded to previously, there are some important limitations to the ability of the current peptide microarray technology to address protein-specific antibody epitopes, as peptides do not readily represent more complex structures such as discontinuous and/or post-translationally modified epitopes (15); obviously, some epitopes will be too large and/or complex to be included in current peptide microarrays. In terms of discontinuous epitopes, however, it remains to be seen whether a high-density peptide microarray technology will be able to assist in identifying components of discontinuous epitopes. In this context, it is encouraging that others have shown that two low-affinity peptide ligands, when joined, can form a complex high-affinity antibody target (31). In terms of post-translational modifications, whether a particular modified epitope can be generated by our peptide microarray technology depends on whether it is possible to generate the modification in question either during the peptide microarray synthesis or enzymatically after synthesis. A priori, it should be possible to include many modifications (e.g. phosphorylation, glycosylation, etc).
We envision that the location, length, and specificity of linear peptide epitopes conveniently can be identified through a two-step strategy. In the first step, all or most n-mer peptides from the target antigens are synthesized, after which the antibody-binding peptides are selected for synthesis with single-residue substitutions in the second step. A suitable choice of n in the first step seems to be 15, and the offset could be one or a few amino acids. Amino acid scans can then be made in the second step, for which an exhaustive analysis using each of the 20 common amino acids would require at least 1 + 15 × 19 = 286 syntheses for each 15-mer epitope candidate. Our data would suggest that the identification of important residues in a linear epitope can often be obtained from single residue scans made with only one or two amino acids. Thus, an even easier alternative would be to combine a 15-mer length scan with a single amino acid substitution scan. This might enable a simplified “single size fits all” approach. In this case, analyzing a target antigen with a length of 1000 amino acids (about 100 kDa) using 15-mer peptide scans with an offset of one including a single amino acid (say, alanine) scan of each peptide would require the synthesis of some 15,000 peptides. About 75,000 peptides would be needed to generate five copies of each peptide, and such a peptide microarray would still be able to hold all the peptides needed for parallel scans of another 10 similar-sized proteins.
In conclusion, ultrahigh-density peptide microarrays give rise to several advantages over existing methods, including comprehensive coverage of antigens using varying peptide length, short assay time, fast quantifiable fluorescent readout, and streamlined image analysis using tailored software to automatically identify binding regions. Once a polyclonal antiserum has been resolved into distinct peptide epitopes, it should even be possible to use these peptides to affinity purify multiple paired antibody species binding to separate parts of an antigen, thereby allowing one purified antibody preparation to validate the results of another (12). It also paves the way for whole proteome peptide microarrays. Ignoring post-translation modifications, all unique 13-mer peptide sequences in the entire humane proteome can be represented by ∼2,000,000 peptides scanning through the proteome using a peptide length of 18 and overlapping by 12 amino acids. We suggest that a peptide microarray representing the entire humane proteome is within reach.
Supplementary Material
Acknowledgments
Claus Schafer-Nielsen is the owner and CEO of Schafer-N. Soren Buus, Matthias Uhlén, Johan Rockberg, Björn Forsström, and Peter Nilsson declare no financial interests.
Footnotes
* The research leading to these results has received funding from the European Community's Seventh Framework Programme ([FP7/2007–2013]) under Grant No. 222773, PepChipOmics.
 This article contains supplemental Figs. S1 to S3.
This article contains supplemental Figs. S1 to S3.
2 Several different padding sequences with amino acids of different natures (negative, positive, hydrophobic, etc.) were tested. In general, the nature of the padding sequence did not affect the results qualitatively (data not shown).
1 The abbreviations used are:
- DCM
- dichloromethane
- DMD
- digital mirror device
- DIEA
- diisopropylethylamine
- Fmoc
- fluorenylmethyloxycarbonyl chloride
- NMM
- N-methylmorpholine
- NMP
- N-methylpyrrolidone
- NPPOC
- 3′-nitrophenylpropyloxycarbonyl
- ANOVA
- one-way analysis of variance
- PrEST
- protein epitope signature tag
- PSSM
- position-specific scoring matrix
- RS
- relative signal
- SSA
- single substitution analysis
- TFA
- trifluoroacetic acid
- LSD
- least significant difference.
REFERENCES
- 1. Gloriam D. E., Orchard S., Bertinetti D., Bjorling E., Bongcam-Rudloff E., Borrebaeck C. A., Bourbeillon J., Bradbury A. R., de Daruvar A., Dubel S., Frank R., Gibson T. J., Gold L., Haslam N., Herberg F. W., Hiltke T., Hoheisel J. D., Kerrien S., Koegl M., Konthur Z., Korn B., Landegren U., Montecchi-Palazzi L., Palcy S., Rodriguez H., Schweinsberg S., Sievert V., Stoevesandt O., Taussig M. J., Ueffing M., Uhlen M., van der Maarel S., Wingren C., Woollard P., Sherman D. J., Hermjakob H. (2010) A community standard format for the representation of protein affinity reagents. Mol. Cell. Proteomics 9, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubel S., Stoevesandt O., Taussig M. J., Hust M. (2010) Generating recombinant antibodies to the complete human proteome. Trends Biotechnol. 28, 333–339 [DOI] [PubMed] [Google Scholar]
- 3. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Bjorling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 4. Taussig M. J., Stoevesandt O., Borrebaeck C. A., Bradbury A. R., Cahill D., Cambillau C., de Daruvar A., Dubel S., Eichler J., Frank R., Gibson T. J., Gloriam D., Gold L., Herberg F. W., Hermjakob H., Hoheisel J. D., Joos T. O., Kallioniemi O., Koegl M., Konthur Z., Korn B., Kremmer E., Krobitsch S., Landegren U., van der Maarel S., McCafferty J., Muyldermans S., Nygren P. A., Palcy S., Pluckthun A., Polic B., Przybylski M., Saviranta P., Sawyer A., Sherman D. J., Skerra A., Templin M., Ueffing M., Uhlen M. (2007) ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. Nat. Methods 4, 13–17 [DOI] [PubMed] [Google Scholar]
- 5. Uhlen M., Graslund S., Sundstrom M. (2008) A pilot project to generate affinity reagents to human proteins. Nat. Methods 5, 854–855 [DOI] [PubMed] [Google Scholar]
- 6. Sahin U., Tureci O., Pfreundschuh M. (1997) Serological identification of human tumor antigens. Curr. Opin. Immunol. 9, 709–716 [DOI] [PubMed] [Google Scholar]
- 7. Haab B. B., Paulovich A. G., Anderson N. L., Clark A. M., Downing G. J., Hermjakob H., Labaer J., Uhlen M. (2006) A reagent resource to identify proteins and peptides of interest for the cancer community: a workshop report. Mol. Cell. Proteomics 5, 1996–2007 [DOI] [PubMed] [Google Scholar]
- 8. Reichert J. M., Valge-Archer V. E. (2007) Development trends for monoclonal antibody cancer therapeutics. Nat. Rev. Drug Discov. 6, 349–356 [DOI] [PubMed] [Google Scholar]
- 9. Piggee C. (2008) Therapeutic antibodies coming through the pipeline. Anal. Chem. 80, 2305–2310 [DOI] [PubMed] [Google Scholar]
- 10. Scolnik P. A. (2009) mAbs: a business perspective. MAbs 1, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beck A., Wurch T., Bailly C., Corvaia N. (2010) Strategies and challenges for the next generation of therapeutic antibodies. Nat. Rev. Immunol. 10, 345–352 [DOI] [PubMed] [Google Scholar]
- 12. Brennan D. J., O'Connor D. P., Rexhepaj E., Ponten F., Gallagher W. M. (2010) Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat. Rev. Cancer 10, 605–617 [DOI] [PubMed] [Google Scholar]
- 13. Kurien B. T., Dorri Y., Dillon S., Dsouza A., Scofield R. H. (2011) An overview of Western blotting for determining antibody specificities for immunohistochemistry. Methods Mol. Biol. 717, 55–67 [DOI] [PubMed] [Google Scholar]
- 14. Warford A., Flack G., Conquer J. S., Zola H., McCafferty J. (2007) Assessing the potential of immunohistochemistry for systematic gene expression profiling. J. Immunol. Methods 318, 125–137 [DOI] [PubMed] [Google Scholar]
- 15. Van Regenmortel M. H. (2009) What is a B-cell epitope? Methods Mol. Biol. 524, 3–20 [DOI] [PubMed] [Google Scholar]
- 16. Liu H. L., Hsu J. P. (2005) Recent developments in structural proteomics for protein structure determination. Proteomics 5, 2056–2068 [DOI] [PubMed] [Google Scholar]
- 17. Obmolova G., Malia T. J., Teplyakov A., Sweet R., Gilliland G. L. (2010) Promoting crystallization of antibody-antigen complexes via microseed matrix screening. Acta Crystallogr. D Biol. Crystallogr. 66, 927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho H. S., Mason K., Ramyar K. X., Stanley A. M., Gabelli S. B., Denney D. W., Jr., Leahy D. J. (2003) Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 421, 756–760 [DOI] [PubMed] [Google Scholar]
- 19. Dhungana S., Williams J. G., Fessler M. B., Tomer K. B. (2009) Epitope mapping by proteolysis of antigen-antibody complexes. Methods Mol. Biol. 524, 87–101 [DOI] [PubMed] [Google Scholar]
- 20. Ramachandran N., Raphael J. V., Hainsworth E., Demirkan G., Fuentes M. G., Rolfs A., Hu Y., LaBaer J. (2008) Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He M., Stoevesandt O., Taussig M. J. (2008) In situ synthesis of protein arrays. Curr. Opin. Biotechnol. 19, 4–9 [DOI] [PubMed] [Google Scholar]
- 22. Hjelm B., Forsstrom B., Igel U., Johannesson H., Stadler C., Lundberg E., Ponten F., Sjoberg A., Rockberg J., Schwenk J. M., Nilsson P., Johansson C., Uhlen M. (2011) Generation of monospecific antibodies based on affinity capture of polyclonal antibodies. Protein Sci. 20, 1824–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winkler D. F., Andresen H., Hilpert K. (2011) SPOT synthesis as a tool to study protein-protein interactions. Methods Mol. Biol. 723, 105–127 [DOI] [PubMed] [Google Scholar]
- 24. Frank R. (2002) The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—principles and applications. J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 25. Halperin R. F., Stafford P., Johnston S. A. (2011) Exploring antibody recognition of sequence space through random-sequence peptide microarrays. Mol. Cell. Proteomics 10, M110.000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otvos L., Jr., Pease A. M., Bokonyi K., Giles-Davis W., Rogers M. E., Hintz P. A., Hoffmann R., Ertl H. C. (2000) In situ stimulation of a T helper cell hybridoma with a cellulose-bound peptide antigen. J. Immunol. Methods 233, 95–105 [DOI] [PubMed] [Google Scholar]
- 27. van Zonneveld A. J., van den Berg B. M., van Meijer M., Pannekoek H. (1995) Identification of functional interaction sites on proteins using bacteriophage-displayed random epitope libraries. Gene 167, 49–52 [DOI] [PubMed] [Google Scholar]
- 28. Rockberg J., Lofblom J., Hjelm B., Uhlen M., Stahl S. (2008) Epitope mapping of antibodies using bacterial surface display. Nat. Methods 5, 1039–1045 [DOI] [PubMed] [Google Scholar]
- 29. Chao G., Cochran J. R., Wittrup K. D. (2004) Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J. Mol. Biol. 342, 539–550 [DOI] [PubMed] [Google Scholar]
- 30. Pizzi E., Cortese R., Tramontano A. (1995) Mapping epitopes on protein surfaces. Biopolymers 36, 675–680 [DOI] [PubMed] [Google Scholar]
- 31. Williams B. A., Diehnelt C. W., Belcher P., Greving M., Woodbury N. W., Johnston S. A., Chaput J. C. (2009) Creating protein affinity reagents by combining peptide ligands on synthetic DNA scaffolds. J. Am. Chem. Soc. 131, 17233–17241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Timmerman P., Beld J., Puijk W. C., Meloen R. H. (2005) Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. Chembiochem. 6, 821–824 [DOI] [PubMed] [Google Scholar]
- 33. Heinis C., Rutherford T., Freund S., Winter G. (2009) Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 5, 502–507 [DOI] [PubMed] [Google Scholar]
- 34. Singh-Gasson S., Green R. D., Yue Y., Nelson C., Blattner F., Sussman M. R., Cerrina F. (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17, 974–978 [DOI] [PubMed] [Google Scholar]
- 35. Hasan A., Stengele K.-P., Giegrich H., Cornwell P., Isham R. K., Sachleben R. A., Pfleiderer W., Foote R. S. (1997) Photolabile protecting groups for nucleosides: synthesis and photodeprotection rates. Tetrahedron 53, 4247–4264 [Google Scholar]
- 36. Bhushan K. R., DeLisi C., Laursen R. A. (2003) Synthesis of photolabile 2-(2-nitrophenyl)propyloxycarbonyl protected amino acids. Tetrahedron Lett. 44, 8585–8588 [Google Scholar]
- 37. Li S., Marthandan N., Bowerman D., Garner H. R., Kodadek T. (2005) Photolithographic synthesis of cyclic peptide arrays using a differential deprotection strategy. Chem. Commun. (Camb). 5, 581–583 [DOI] [PubMed] [Google Scholar]
- 38. Persson A., Hober S., Uhlen M. (2006) A human protein atlas based on antibody proteomics. Curr. Opin. Mol. Ther. 8, 185–190 [PubMed] [Google Scholar]
- 39. Hjelm B., Fernandez C. D., Lofblom J., Stahl S., Johannesson H., Rockberg J., Uhlen M. (2010) Exploring epitopes of antibodies toward the human tryptophanyl-tRNA synthetase. N. Biotechnol. 27, 129–137 [DOI] [PubMed] [Google Scholar]
- 40. Bewick V., Cheek L., Ball J. (2004) Statistics review 9: one-way analysis of variance. Crit. Care 8, 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roder G., Geironson L., Darabi A., Harndahl M., Schafer-Nielsen C., Skjodt K., Buus S., Paulsson K. (2009) The outermost N-terminal region of tapasin facilitates folding of major histocompatibility complex class I. Eur. J. Immunol. 39, 2682–2694 [DOI] [PubMed] [Google Scholar]
- 42. Andreatta M., Schafer-Nielsen C., Lund O., Buus S., Nielsen M. (2011) NNAlign: a web-based prediction method allowing non-expert end-user discovery of sequence motifs in quantitative peptide data. PLoS One 6, e26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. (1991) Light-directed, spatially addressable parallel chemical synthesis. Science 251, 767–773 [DOI] [PubMed] [Google Scholar]
- 44. Frank R. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrtahedron 48, 9217–9232 [Google Scholar]
- 45. Volkmer R. (2009) Synthesis and application of peptide arrays: quo vadis SPOT technology. Chembiochem 10, 1431–1442 [DOI] [PubMed] [Google Scholar]
- 46. Pellois J. P., Zhou X., Srivannavit O., Zhou T., Gulari E., Gao X. (2002) Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol. 20, 922–926 [DOI] [PubMed] [Google Scholar]
- 47. Pellois J. P., Wang W., Gao X. (2000) Peptide synthesis based on t-Boc chemistry and solution photogenerated acids. J. Comb. Chem. 2, 355–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.