Abstract
Pristionchus pacificus is a nematode that is increasingly used as a model organism in evolutionary biology. The genome of P. pacificus differs markedly from that of C. elegans, with a high number of orphan genes that are restricted to P. pacificus and have no homologs in other species. To gain insight into the architecture of signal transduction networks in model nematodes, we performed a large-scale qualitative phosphoproteome analysis of P. pacificus. Using two-stage enrichment of phosphopeptides from a digest of P. pacificus proteins and their subsequent analysis via high accuracy MS, we detected and localized 6,809 phosphorylation events on 2,508 proteins. We compared the detected P. pacificus phosphoproteome to the recently published phosphoproteome of C. elegans. The overall numbers and functional classes of phosphoproteins were similar between the two organisms. Interestingly, the products of orphan genes were significantly underrepresented among the detected P. pacificus phosphoproteins. We defined the theoretical kinome of P. pacificus and compared it to that of C. elegans. While tyrosine kinases were slightly underrepresented in the kinome of P. pacificus, all major classes of kinases were present in both organisms. Application of our kinome annotation to a recent transcriptomic study of dauer and mixed stage populations showed that Ser/Thr and Tyr kinases show similar expression levels in P. pacificus but not in C. elegans. This study presents the first systematic comparison of phosphoproteomes and kinomes of two model nematodes and, as such, will be a useful resource for comparative studies of their signal transduction networks.
Pristionchus pacificus is a nematode that is established as a model in evolutionary developmental biology (2). Like Caenorhabditis elegans, which was the first multicellular organism to have its genome completely sequenced (3), it has several advantageous features: it is easy to cultivate in the laboratory, it feeds on E. coli, it has a short generation time of 4 days (at 20 °C), and, because it is a self-fertilizing hermaphrodite, it is amenable to forward and reverse genetic techniques. Its genome has recently been sequenced, revealing a high number of predicted genes that share no sequence similarity to genes from any other organisms (“orphan” or “pioneer” genes) (4). Like many other nematodes, P. pacificus exhibits phenotypic plasticity of its life cycle and is able to quickly adapt to different environmental conditions. Under favorable conditions, P. pacificus undergoes direct development, but it can arrest development to form a stress resistant dauer stage when the environmental conditions turn unfavorable. These examples of phenotypic plasticity have allowed nematodes to invade many different habitats (5). P. pacificus occupies a completely different ecological niche than C. elegans. It has a necromenic lifestyle in which the developmentally arrested dauer larva infests a scarab beetle and resumes development upon the beetle's death, feeding on the microorganisms that decompose the beetle's carcass (6). The estimated evolutionary distance between C. elegans and P. pacificus is 250 to 420 million years, which makes them very attractive models in evolutionary developmental biology (4).
We recently performed a comprehensive analysis of the proteome and transcriptome of P. pacificus, with the aim of refining its genome annotation. Retraining the gene prediction algorithm with gene expression data estimated the number of predicted open reading frames to 24,000. Comparison of our data to the predicted proteome of C. elegans revealed differences in the proteome structures of the two nematodes. Whereas the predicted proteome of P. pacificus showed a unimodal distribution of protein sizes, the proteome of C. elegans followed a clearly bimodal distribution. Interestingly, this bimodal distribution seemed to be connected to functions related to protein phosphorylation, suggesting a potential difference in protein phosphorylation between the two organisms (7).
To gain further insights into the proteome of P. pacificus, we performed a large-scale analysis of P. pacificus phosphoproteome using phosphopeptide enrichment and high accuracy mass spectrometry. Here we report the first comprehensive phosphoproteome map of P. pacificus, measured to a depth of almost 7,000 localized phosphorylation sites, and compare it to the recently reported phosphoproteome of C. elegans (8). We show that the two phosphoproteomes are of similar sizes but differ significantly in the frequencies of phosphorylated serine, threonine, and tyrosine residues. We define direct orthologs between the two organisms and show that this discrepancy is also pronounced at the ortholog level. We show that the products of orphan genes are significantly underrepresented among the detected P. pacificus phosphoproteins. Finally, we define the predicted kinome of P. pacificus and show that it is slightly smaller than that of C. elegans but contains all major classes of kinases.
MATERIALS AND METHODS
Culturing of Worms and Preparation of Protein Extracts
P. pacificus strain PS312 was grown on 10 cm NGM agar plates spotted with 2 ml E. coli OP50 solution. Plates were inoculated with between 50 and 100 worms and incubated at 25 °C. The mixed stage population was harvested shortly after the bacterial lawn was consumed, avoiding the starvation of the animals. After thorough washing with distilled water and 0.9% sodium chloride, worms were pelleted and prepared for proteomics measurements.1
For protein isolation, 100 μl of animals were solubilized in 300 μl denaturation buffer (6 m urea, 2 m thiourea, 10 mm Tris pH 8.0). After three cycles of freezing (liquid nitrogen) and thawing (37 °C), 100 μl of glass beads were added, and the solution was vortexed for 20 min. After centrifugation (20 min, 20.800 × g, 4 °C), the protein concentration of the supernatant was determined using the Bradford assay and further processed using the filter-aided sample preparation (FASP)2 method (9) (see below).
Protein Digestion
The soluble protein fraction was digested as described previously (7). Briefly, 5 mg of protein was reduced with a final concentration of 1 mm DTT and alkylated with a final concentration of 5.5 mm iodoacetamide. After the pH was adjusted to 8.0, 1 μg of trypsin was added per 100 μg of protein, and the mixture was incubated overnight at 37 °C.
The insoluble protein fraction was processed with a modified FASP protocol (9). The protein pellet was solubilized in 4% SDS, 100 mm DTT, and 100 mm Tris pH 7.6. An aliquot of the pellet was precipitated with chloroform/methanol and solubilized in denaturation buffer for Bradford analysis. Based on the Bradford measurement, a protein-SDS solution containing 5 mg of protein was diluted with urea buffer A (8 m urea in 100 mm Tris, pH 8.5) to a final volume of 6 ml and pipetted into the 15-ml Centriprep column YM-30 (Millipore, Billerica, MA). After the sample had been spun for 15 min at 6,000 g, 600 μl of iodoacetamide solution (550 mm) was added, and the sample was incubated for 1 h in the dark and then centrifuged for 15 min at 3,000 g. The protein was washed with UA three times, and the last centrifugation step was increased to 20 min. Six ml of ammonium bicarbonate was added, and the sample was centrifuged for another 15 min at 3,000 g. After that, Trypsin was added at a final concentration of 1 μg per 100 μg total protein and incubated overnight at 37 °C. After the next centrifugation step (15 min, 3,000 g), the peptides were collected in the flowthrough. Centrifugation was repeated with 3 ml water, and the flowthrough was collected for strong cation exchange (SCX).
Phosphopeptide Enrichment
After 5 mg of digested total protein lysate had been acidified to pH 2.7 with trifluoroacetic acid, the sample was loaded onto an ÄKTApurifier (GE Healthcare, Little Chalfont, UK) HPLC for SCX. The 16 resulting fractions were pooled according to the elution profile to 10 fractions for titanium dioxide enrichment. Five mg of TiO2 beads were resuspended in 50 μl of a 30 mg/ml 2.5 dihydrobenzoic acid, 80% acetonitrile in water solution. After 10 min of incubation at room temperature, the TiO2 loading solution was added to the sample and mixed for 30 min at room temperature using an orbital shaker. The beads were precipitated with centrifugation at 13,000 rpm for 2 min and washed with 1 ml Wash Solution I (30% acetonitrile (ACN), 3% TFA) for 10 min in a shaker and Wash Solution II (10) (80% ACN, 0.1% TFA) for 10 min in a shaker. The beads were then resuspended in 50 μl Wash Solution II and transferred to a 200 μl pipette tip plugged with one layer of Empore C8 tip. After the beads had been washed three times with 100 μl 40% ammonia solution (25% in water) in ACN pH 10.5, the eluate was reduced to 5 μl in a SpeedVac.
NanoLC-MS/MS Analysis
Enriched phosphopeptide mixtures were separated via Easy-LC nano-HPLC (Proxeon Biosystems, Odense, DK) coupled to an LTQ-Orbitrap-XL (Thermo Fisher Scientific) through a nano-LC-MS interface (Proxeon Biosystems). Chromatographic separation of the peptides was performed on a 15 cm fused silica emitter with a 75 μm inner diameter (Proxeon Biosystems), in-house packed with reversed-phase ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, DE). The peptide mixtures were injected onto the column in HPLC solvent A (0.5% acetic acid) at a flow rate of 500 nl/min and subsequently eluted with a 107 min segmented gradient of 2% to 80% of HPLC solvent B (80% acetonitrile in 0.5% acetic acid) at a flow rate of 200 nl/min.
The MS was operated in the data-dependent mode so as to automatically switch between MS and MS/MS acquisition. Survey full scan MS spectra were acquired in the mass range from m/z 300 to 2,000 in the orbitrap mass analyzer at a resolution of 60,000. An accumulation target value of 106 charges was set, and the lock mass option was used for internal calibration (11). The five most intense ions were sequentially isolated and fragmented in the linear ion trap using collision-induced dissociation (CID) at an ion accumulation target value of 5,000 and default CID settings. Multistage activation (at −98, −49, and −32.66 Th relative to the precursor ion) was used to optimize fragmentation of Ser/Thr phosphopeptides. The ions already selected for MS/MS were dynamically excluded for 90 s. The resulting peptide fragment ions were recorded in the linear ion trap. In total, 41 LC-MS measurements were performed, corresponding to 4 days of measurement time.
Data Processing and Analysis
MS data were processed with MaxQuant (12), version 1.0.14.3. Peak lists were generated and subsequently submitted to the Mascot search engine (Matrix Science, London, UK) to query a database consisting of the latest annotation of P. pacificus (dataset “HYBRID1 proteomics gene models”; 24,231 protein entries) (7), 4,256 E. coli proteins, 262 commonly observed protein contaminants, and 28,749 reversed sequences. The initial precursor mass tolerance was set to 7 ppm for Orbitrap data (full scans); fragment ion mass tolerance was set to 0.5 Da for ion trap data (MS/MS scans). Full trypsin specificity was required, and up to two missed cleavages were allowed. Carbamidomethylation on cysteine was defined as fixed modification; methionine oxidation, protein N-terminal acetylation, and phosphorylation on serine, threonine, and tyrosine were defined as variable modifications. The database search results were parsed by MaxQuant to assemble protein groups, peptides, and phosphorylation sites at a false discovery rate of 1%. All phosphorylation events having a reported localization probability of at least 0.75 were considered as localized (assigned to a specific amino acid). Subsequent downstream analysis of the result tables was done in R v2.11.1 (13).
Determination of Orthologous and Homologous Relationships
Pairwise orthologs and homologs between P. pacificus and C. elegans were inferred using bidirectional and unidirectional BLASTP, respectively (14, 15). We used Wormbase WS200 for C. elegans and the latest genome annotation for P. pacificus (7) as input. Global alignments between orthologous proteins were derived using Needle (16, 17).
Determination of Orphan/Pioneer Proteins
Orphan proteins were defined by two BLAST analyses. First we regarded every P. pacificus protein having no homologue in the NCBInr database (BLASTP E-value < 1 × 10−3) as a potential orphan. Second, we used the information derived from the pairwise BLAST analysis of the theoretical proteomes of P. pacificus and C. elegans as described above. Orphan proteins were required to have no homologues in the NCBInr database or in the Wormbase WS200.
Functional Annotation of the P. pacificus Proteome
Blast2GO software was used to derive Gene Ontology (GO) (18) terms via a BLAST search of the theoretical proteome of P. pacificus against the nonredundant NCBI protein database (downloaded on April 29, 2010) using default parameters. Information on specific pathways on the basis of Kyoto Encyclopedia of Genes and Genomes (KEGG) terms (19) was obtained from the KEGG Automatic Annotation Server (20) using default parameters. The classification of proteins into protein families was performed using Pfam (21). The significance E-value threshold was gathered by the software automatically. All types of annotation were merged and exported to an Excel sheet using R.
Functional Enrichment Analysis of the Detected Phosphoproteome
The frequencies of functional annotation terms assigned to the detected phosphoproteome were tested against the corresponding frequencies in the entire proteome using Fisher's exact test (one-sided). A minimum of five occurrences of each term was required in order for the term to be taken into account for analysis. Derived p values were further adjusted for multiple hypothesis testing using the method proposed by Benjaminii and Hochberg (22).
Draft Kinome Annotation
We considered all proteins having predicted Pfam domains “Pkinase,” “Pkinase_C,” or “Pkinase_Tyr” as potential kinases. In order to classify these kinases into kinase groups, families, and subfamilies, we performed a BLAST search of predicted kinase domains against all nematode-specific kinase domains contained in Kinbase. BLAST hits were considered significant if the reported E-value was below 1 × 10−20, resulting in a minimal bit score of 90.9. For further validation, we did a second BLAST search by querying the kinase domains contained in Kinbase against the predicted Pfam domains in the P. pacificus proteome and checking whether the results were consistent. All predicted Pfam domains that met these criteria were classified according to Kinbase annotation.
Phylogenetic distances between the domains were estimated by ClustalW and exported to Nexus format. Distances were logarithmized and imported into the Interactive Tree of Life online tool (23) to produce the phylogenetic trees. The trees were annotated with kinase groups using the classification obtained by the BLAST analysis described above.
Secondary Protein Structure Prediction
The secondary structures of all phosphorylated proteins detected in P. pacificus and C. elegans were calculated using PsiPred v3.3 (24) and PSIBLAST v2.2.23. Initial PSIBLAST searches were done against the Unriref90 database. Prior to the search, low complexity regions were removed from that database as described in the README file of the Psipred software.
Comparison with Transcriptome Data
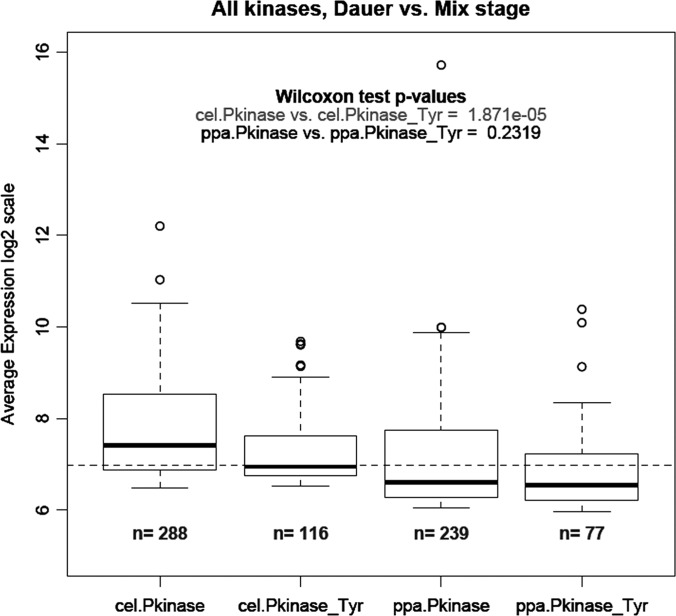
The gene expression data from a dauer versus mix-stage comparison in both C. elegans and P. pacificus were obtained from Sinha et al. (25). For P. pacificus, the gene predictions and, hence, the gene identifiers used in Ref. 25 are different from those used in Ref. 7, although the underlying genome assembly is the same. Thus, the mapping from a microarray probe to a gene prediction corresponding to Ref. 7 was calculated using stringent BLAST criteria (E-value < 1 × 10−10, 100% identity between the 60 bp microarray probe and the target gene). Probes that matched multiple genes were removed from the analysis, and fold-changes were calculated using the same parameters and methods as in Ref. 25. Pfam domain annotations of C. elegans were based on wormpep-210. We used kinase domain annotation for P. pacificus from supplemental Table 2. The average expression values of all the kinase genes (expression ratio “Dauer/Mix-stage”) for all genes annotated with a particular kinase domain (“Pkinase” or “Pkinase_Tyr”) were compared within species, and the significance of the difference was assessed based on two-sample Wilcoxon tests. The number n in Fig. 5 is the total number of genes belonging to a particular gene family. The “average expression” is defined as log2(RedSignal * GreenSignal) on an arbitrary scale. Hence the values can be compared only within a nematode species and should not be compared across nematodes. The P. pacificus and C. elegans fold-change and average expression data on kinases are included in supplemental Table 5.
Fig. 5.
Average expression of different protein kinase classes in dauer versus mixed stages of C. elegans (cel) and P. pacificus (ppa). Kinome annotation from supplemental Table 2 was applied to quantitative transcriptomics data derived from Sinha et al. (25).
RESULTS
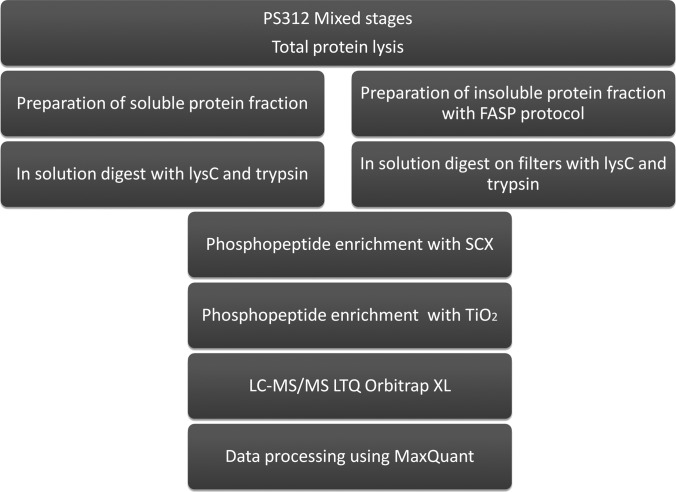
In this study we aimed to provide the reference phosphoproteome of the nematode model P. pacificus and compare it to the recently published phosphoproteome of C. elegans (8). To minimize experimental bias and enable direct comparison between the datasets, we employed similar sample preparation, measurement, and data processing workflows as in the phosphoproteomic study of C. elegans. Briefly, we lysed a well-fed mixed stage P. pacificus culture by rupturing the cuticle with freeze-thaw cycles and glass bead treatment. We extracted the proteins from the insoluble fraction in 4% SDS and processed them via the FASP protocol as described by Wisniewski et al. (9). We digested the soluble protein fraction in solution with trypsin and separately subjected both fractions to two stages of phosphopeptide enrichment, consisting of strong cation exchange and TiO2 chromatographies (26, 27). We performed LC-MS analysis on an Easy-LC (Proxeon Biosystems) coupled to an LTQ-Orbitrap XL MS (Thermo Fisher Scientific) and processed the data using the MaxQuant software suite (12). The workflow employed in this study is depicted in Fig. 1.
Fig. 1.
Biochemical workflow used in this study. A mixed population of P. pacificus worms was harvested, and the protein extract was split into soluble and insoluble fractions, which were processed further using the FASP protocol. After LysC/trypsin digestion, phosphopeptides were enriched by SCX and TiO2 chromatographies and measured on an LTQ Orbitrap XL mass spectrometer.
Detected Phosphoproteome of P. pacificus
The analysis of the P. pacificus phosphoproteome resulted in 60,358 identified MS/MS spectra that detected 9,872 nonredundant peptide sequences with a median absolute mass deviation of 255 ppb (supplemental Fig. 1). We detected 3,158 P. pacificus protein groups at a false discovery rate of 1%; of these, 2,508 were phosphorylated (Table I) and 1,518 were not detected in our previous large-scale proteomics study (7). This resulted in extension of the catalogue of P. pacificus proteins detected by MS to 5,547 (supplemental Fig. 2). In total, we localized 6,809 phosphorylation events to a specific amino acid residue with a median confidence level of 99.8%. The frequencies of phosphorylated serines, threonines, and tyrosines were found to be 87.8% (5,981 events), 11.1% (756 events), and 1.06% (72 events), respectively. All detected phosphorylation sites are presented in supplemental Table 1.
Table I. Number of (phospho)proteins detected in this study (at 1% false discovery rate (FDR)).
MS data were searched against a decoy database containing P. pacificus and E. coli protein entries.
| Phosphoproteins | All proteins | |
|---|---|---|
| P. pacificus | 2,508 | 3,158 |
| E. coli | 11 | 23 |
Functional Classes and Kinase Motifs of Detected P. pacificus Phosphoproteins
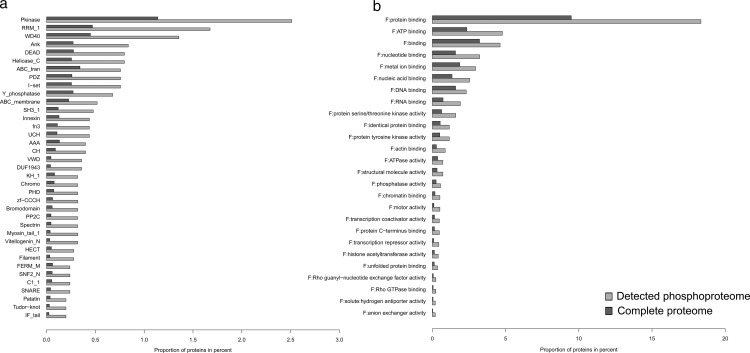
To gain insight into the functional distribution of proteins phosphorylated in P. pacificus, we first retrieved the latest functional annotation according to GO terms, KEGG pathways, enzyme commission numbers (ECs), and protein families and domains (Pfam) (supplemental Table 2). We then performed enrichment analyses of the GO, KEGG, Pfam, and EC terms of proteins detected as phosphorylated (supplemental Table 3). The GO term analysis showed an enrichment of functions related to protein and nucleoside binding, transcription repressor activities, and kinase regulator activities, terms commonly enriched in large phosphoproteome datasets. The Enzyme Class analysis showed significant enrichment of only two classes, protein tyrosine kinases (EC 2.7.10.0; 23 detected phosphoproteins) and protein serine kinases (EC 2.7.11.0; 35 detected phosphoproteins). This was expected because kinases and phosphatases themselves are commonly regulated by phosphorylation, and many kinases show autophosphorylation activity. In agreement with this, the Pfam analysis showed an enrichment of protein kinase domains, as well as phosphatase domains. Proteins with domains involved in protein–protein interactions and signaling were also overrepresented in comparison with the total gene predictions. Among the detected domains, WD40, VWD, Ankyrin, and PDZ domains were highly represented. Moreover, RNA binding domains such as rrm-1, helicase, and DEAD were also overrepresented. The results of the functional enrichment analysis are summarized in Fig. 2.
Fig. 2.
Functional enrichment analysis of the detected P. pacificus phosphoproteome. a, enrichment of Pfam terms; b, enrichment of GO terms (molecular function).
We next tested the representation of P. pacificus orphan gene products in the phosphoproteome. In total, we detected phosphorylation on 234 products of orphan genes (9.3% of the detected phosphoproteome). Compared with all orphan genes in the P. pacificus genome (9,957; 41.09% of the genome), this presented a significant underrepresentation (p < 3.64 × 10−303). However, it has to be noted that this class of gene products showed a similar underrepresentation at the proteome level (7), pointing to the fact that their underrepresentation in the phosphoproteome results from the lack of expression, not phosphorylation.
Next, we tested the enrichment of specific kinase target motifs on P. pacificus phosphoproteins detected in our dataset, as described by Zielinska et al. (8). On proteins phosphorylated on serine, three motifs were overrepresented—CAMK2 (RXX[pS]), CK2 ([pS]XXE), and PKA (RX[pS])—whereas on proteins phosphorylated on threonine, only the CAMK2 motif was overrepresented. For both phosphorylated residues there was also significant overrepresentation of proline adjacent to the phosphorylation site ([pS]P and [pT]P) (supplemental Fig. 3). No significant motifs were detected on proteins phosphorylated on tyrosine residues, most likely because of the small size of the dataset.
Comparison of P. pacificus and C. elegans Phosphoproteomes
We next compared the phosphoproteome of P. pacificus to the recently published phosphoproteome of C. elegans (8), in which 6,699 phosphorylation sites were localized on 2,365 proteins (Table II). The sizes of the two phosphoproteomes were very similar, and the enriched functional classes of detected phosphoproteins were almost identical, demonstrating that both nematodes likely use protein phosphorylation in similar biological processes. Interestingly, the two phosphoproteomes differed in frequencies of S/T/Y phosphorylation events. In C. elegans, the reported pSer, pThr, and pTyr frequencies were 80.2%, 18%, and 1.8%, whereas in P. pacificus they were 87.8%, 11.1%, and 1.1%, respectively. The frequencies of all phosphorylated amino acids were significantly different despite very similar overall frequencies of these amino acids in the proteomes of P. pacificus and C. elegans (Table II).
Table II. Numbers and frequencies of phosphorylation sites localized on serine, threonine, and tyrosine in P. pacificus and C. elegans.
To test whether the frequencies of pS, pT, and pY were significantly different between the two nematodes, we calculated p values using a two-sided binomial test.
| Total | pS | pT | pY | |
|---|---|---|---|---|
| Wormbase200 | 23,973 proteins | 7.81% | 5.85% | 2.75% |
| Ppa database | 24,231 proteins | 8.12% | 5.89% | 3.13% |
| C. elegans (Zielinska et al. (8)) | 6,699 (2,365 proteins) | 5,372 (80.19%) | 1,207 (18.02%) | 120 (1.79%) |
| P. pacificus (this study) | 6,809 (2,401 proteins) | 5,981 (87.84%) | 756 (11.1%) | 72 (1.06%) |
| Binomial p value | p < 2.2 × 10−16 | p < 2.2 × 10−16 | p < 1.18 × 10−6 |
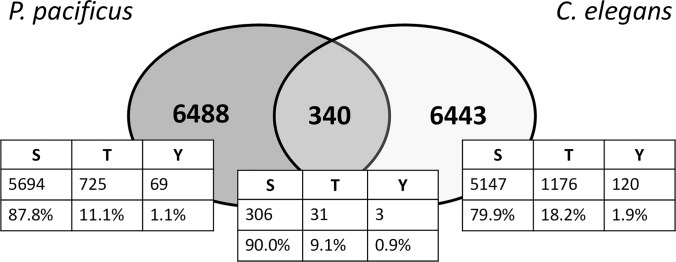
To gain insight into the potential origin of this discrepancy, we investigated the frequencies of pSer, pThr, and pTyr in orthologs shared between P. pacificus and C. elegans and therefore likely present in their common ancestor. Based on the bidirectional BLASTP approach, 619 phosphoproteins from our dataset were defined as orthologs between P. pacificus and C. elegans and phosphorylated in both species. On these orthologs, 340 phosphorylation sites were determined as conserved (Fig. 3; supplemental Table 4). Interestingly, the frequencies of pSer, pThr, and pTyr at the ortholog level (90%, 9.1%, and 0.9%, respectively) resembled more closely the frequencies measured in the phosphoproteome of P. pacificus than those in the phosphoproteome of C. elegans. This means that the basal phosphoproteome of P. pacificus might resemble the phosphoproteome of the common ancestor of P. pacificus and C. elegans.
Fig. 3.
Evolutionary conserved phosphorylated residues between P. pacificus and C. elegans. Venn diagram depicting the overlap of conserved phosphorylation sites on direct orthologs found to be phosphorylated in the phosphoproteome datasets.
Whereas different frequencies of tyrosine phosphorylation may be explained by different usages of this modification in signal transduction (see Discussion), different frequencies of detected serine and threonine phosphorylation are more difficult to explain, mostly because of the dual specificity of Ser/Thr kinases. A potential reason could be different representation of these amino acids in the unstructured protein regions that are more accessible to protein kinases. To test this hypothesis, we calculated the frequencies of all serine, threonine, and tyrosine residues in coiled coils and in helical and strand regions of proteins from detected phosphoproteomes of P. pacificus and C. elegans and compared them to detected phosphorylation sites (supplemental Fig. 4). As expected, this analysis did not reveal any significant differences in the frequencies of serine, threonine, and tyrosine in the two organisms, demonstrating that different accessibility is not the reason for the observed differences in frequencies of phosphorylated amino acids.
Predicted Kinome of P. pacificus and Its Comparison with C. elegans
We next compared the predicted kinomes of several sequenced model nematodes. To define the predicted kinomes of P. pacificus, we used Pfam annotation and considered all proteins containing a “P-kinase” domain as potential kinases (see Methods). After collapsing all C. elegans kinase isoforms, we compared the predicted kinome to that of P. pacificus. The kinome of P. pacificus contained 368 kinases (supplemental Table 2) and was 11% smaller than that of C. elegans, which contained 413 kinases (Table III); interestingly, the number of predicted tyrosine kinases was 20% lower in P. pacificus (94 kinases) and therefore was underrepresented relative to C. elegans (117 kinases). Of the 368 predicted kinases in P. pacificus, 77 were detected as phosphorylated in our study. Of those, 61 had direct orthologs and 30 were detected as phosphorylated in C. elegans (8). Interestingly, two of the three (66.6%) conserved pTyr residues were located on kinases (cdk-1, mbk-1), one of the 31 (3.2%) conserved pThr residues was located on a kinase (sek-1), and 12 of 306 (3.9%) conserved pSer residues were located on kinases (unc-82, unc-22, pkc-1, grk-1, ZK524.4, gcy-28, ZC581.9, B0495.2).
Table III. Number of predicted protein kinases in different nematodes according to Pfam annotations of protein kinase domains.
| Domains | Proteins | |
|---|---|---|
| C. elegans | 441 (119 pTyr) | 413 (117 pTyr) |
| P. Pacificus | 408 (102 pTyr) | 368 (94 pTyr) |
| B. malayi | 406 (89 pTyr) | 378 (83 pTyr) |
| M. incognita | 392 (57 pTyr) | 361 (53 pTyr) |
To classify P. pacificus kinases into groups, families, and subfamilies, we performed a bidirectional BLAST analysis of predicted kinase domains against C. elegans kinase domains contained in Kinbase. The BLAST analysis resulted in 282 highly confident hits, indicating that the catalytic domains of predicted kinases appeared to be conserved between the two nematodes. All eight major protein kinase groups present in C. elegans were also present in P. pacificus (Fig. 4; supplemental Fig. 5).
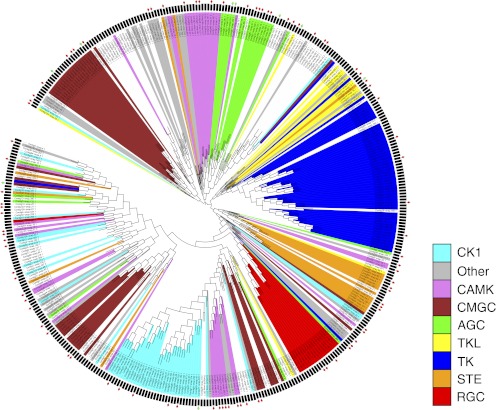
Fig. 4.
Phylogentic tree of the predicted P. pacificus kinome. The tree shows the phylogentic relationships of predicted kinase domains and their classification into kinase groups according to the C. elegans kinome. Phylogenetic distances are based on multiple sequence alignments of predicted kinase domains. The classification of domains into kinase groups is shown by the different colors of the branches. Red rectangles at the outer edge of the circle indicate kinases that are detected as phosphorylated; green rectangles indicate kinases that are detected as nonphosphorylated in our study.
Expression of Different Kinase Classes in C. elegans and P. pacificus
To assess the expression of different kinase classes in C. elegans and P. pacificus, we analyzed a recently published transcriptome dataset that addresses global changes in gene expression in the dauer and mixed populations of these two nematodes (25). Applying our Pfam-based kinome annotation, we extracted expression data for 404 kinases in C. elegans (288 Ser/Thr and 116 Tyr kinases) and 316 kinases in P. pacificus (239 Ser/Thr and 77 Tyr kinases). As expected, the transcriptome analysis showed good coverage of the kinome in both organisms, albeit slightly higher in C. elegans (404/413, 98%) than in P. pacificus (316/368, 86%). Interestingly, in C. elegans, the average expression of “Pkinase_Tyr” genes was significantly higher than the average expression of “Pkinase” genes in the dauer population. However, in P. pacificus, all kinase genes were expressed at a significantly higher level in the dauer population, and there was no difference in average expression between the two kinase categories, pointing to the fact that tyrosine kinases are expressed at levels similar to those of Ser/Thr kinases (Fig. 5). These data reveal that both nematodes express all classes of kinases and point to their potentially different usage in the dauer stage of the life cycle.
DISCUSSION
In this study, we have reported the first global phosphoproteomic dataset of the mixed stage population of the P. pacificus nematode. In order to increase the number of identified phosphorylation sites, we performed three biological replicates, two with the soluble and one with the insoluble protein fraction. In this way, we made all cellular compartments accessible to protein analysis. By using mixed stages, we aimed to get an in-depth catalog of phosphorylation sites of P. pacificus and compare it to the previously reported phosphoproteome of C. elegans, analyzed under similar conditions.
Although the two phosphoproteomes were very similar in terms of size, classes of phosphorylated proteins, and overrepresented kinase motifs, they were different in the extent of serine, threonine, and tyrosine phosphorylation. Interestingly, this difference might reflect the observed alterations in signal transduction during postembryonic development of these two species. Work over the past decade has compared signaling networks during vulva development and dauer formation between P. pacificus and C. elegans and identified substantial differences (28). In C. elegans, three vulva precursor cells (VPCs) are induced to form vulval tissue by a signal from the gonadal anchor cell. This signal is a secreted epidermal-growth factor (EGF)-type factor that is transmitted within the VPCs by EGFR-RAS-MAP kinase signaling and finally results in the initiation of cell division. A series of phosphorylation events by LIN-45/RAF, MEK-2/MAP kinase, and MPK-1/MAP kinase is at the center of C. elegans vulva induction (29). In P. pacificus, in contrast, vulva formation is regulated by a completely different regulatory mechanism (for a review, see (30)). While the same VPCs form vulval tissue, their induction requires regulatory input from Wnt signaling rather than EGF-MAP kinase signaling (31). This involves an unusual regulatory linkage of Wnt-type ligands and Frizzled-type receptors, as well as novel protein-interaction domains in LIN-18/Ryk/Derailed-type co-receptor (28). Thus, vulva induction in C. elegans is regulated by a kinase pathway involving a high extent of tyrosine phosphorylation, whereas the same process in P. pacificus depends much less on tyrosine phosphorylation. It has to be noted, however, that P. pacificus contains 1:1 orthologs for all of the EGF/Ras pathway genes/proteins known from C. elegans. Interestingly, Ppa-MPK-1 was the only kinase of the EGF/RAS pathway shown to be phosphorylated in our dataset. The functional significance of this finding, if any, has yet to be identified.
Similarly, work on dauer formation revealed potential differences in signaling activity during development. In C. elegans, the formation of dauer larvae, an arrested alternative life stage that facilitates the survival of harsh environmental conditions, involves insulin and TGF-β signaling activity that is coupled to transcriptional activity of the nuclear hormone receptor DAF-12 and the FOXO-transcription factor DAF-16 (5). In P. pacificus, both transcription factors have similar roles during dauer regulation, as indicated by the phenotype of mutations in the corresponding genes, whereas there is no report that would suggest similar roles of insulin and TGF-β signaling (6). However, as indicated above for vulva development, these differences in signaling activity in these two nematodes are not reflected in the copy number of genes encoding signaling components in the respective genomes. Thus, differences in phosphorylation patterns as revealed in our study can occur in the absence of major changes in the signaling pathways that act during development.
The comparative analysis of the predicted kinomes of P. pacificus and C. elegans indicates that all major protein kinase groups are conserved between these two nematodes (supplemental Table 2), and recent transcriptome analysis suggests that all kinase classes are expressed (and presumably active) in both nematodes during the dauer stage of the life cycle (25). When compared with other protein classes, the kinome shows a relatively high level of conservation and low copy number variations. For example, many of the detoxification enzymes, such as cytochrome P450 proteins, show a more than 3-fold difference between the P. pacificus and C. elegans proteomes with 197 and 67 protein predictions, respectively (4). We speculate that the difference in cytochrome P450 enzymes reflects the adaptation to the different environments in which these nematodes are found. In contrast, the overall similarity of the two kinomes represents the conserved molecular and cellular processes, which evolved largely independent of ecological alterations. This evolutionary pattern becomes even stronger when data available for additional nematodes are considered: the numbers of predicted protein kinases of P. pacificus, C. elegans, the human parasite B. malayi, and the plant parasite M. incognita are surprisingly similar (Table III). Thus, the kinome represents a stable part of the nematode proteome, and most likely the analysis of a small number of selected model organisms will provide comprehensive insight into processes of phosphorylation.
Acknowledgments
We thank Matthias Mann for access to the Phosida database. We also thank Jonathan Goldberg for fruitful discussions on the kinome prediction.
Footnotes
* This work was supported by funding from Baden-Württemberg Stiftung (Juniorprofessoren-Programm), the German Research Foundation, and PRIME-XS (to B.M.).
 This article contains supplemental Figs. 1 to 5 and Tables 1 to 5.
This article contains supplemental Figs. 1 to 5 and Tables 1 to 5.
1 The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash: pxGey/Jh9q186pz5hyUKK13Idzf8sjFVLW+ZZbNgv0IkOAH71q31oIfK2vNyvp8wb7ItfBczkQ8O5W/llVxLtpPhpEoAAAAAAAA1Ow==.
The hash may be used to prove exactly what files were published as part of this manuscript's data set, and the hash may also be used to check that the data have not changed since publication. The data can also be viewed through the PHOSIDA database www.phosida.com (1).
2 The abbreviations used are:
- CID
- collision-induced dissociation
- EC
- enzyme commission number
- EGF
- epidermal-growth factor
- FASP
- filter-aided sample preparation
- GO
- Gene Ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- Pfam
- protein families and domains
- SCX
- strong cation exchange
- VPC
- vulva precursor cell.
REFERENCES
- 1. Gnad F., Ren S. B., Cox J., Olsen J. V., Macek B., Oroshi M., Mann M. (2007) PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 8, R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hong R. L., Sommer R. J. (2006) Pristionchus pacificus: a well-rounded nematode. Bioessays 28, 651–659 [DOI] [PubMed] [Google Scholar]
- 3.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 [DOI] [PubMed] [Google Scholar]
- 4. Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., Dinkelacker I., Fulton L., Fulton R., Godfrey J., Minx P., Mitreva M., Roeseler W., Tian H., Witte H., Yang S. P., Wilson R. K., Sommer R. J. (2008) The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coghlan A. (2005) Nematode genome evolution. WormBook 2005, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sommer R. J., Ogawa A. (2011) Hormone signaling and phenotypic plasticity in nematode development and evolution. Curr. Biol. 21, R758–R766 [DOI] [PubMed] [Google Scholar]
- 7. Borchert N., Dieterich C., Krug K., Schutz W., Jung S., Nordheim A., Sommer R. J., Macek B. (2010) Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zielinska D. F., Gnad F., Jedrusik-Bode M., Wisniewski J. R., Mann M. (2009) Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J. Proteome Res. 8, 4039–4049 [DOI] [PubMed] [Google Scholar]
- 9. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 10. Makarov A., Denisov E., Kholomeev A., Balschun W., Lange O., Strupat K., Horning S. (2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 11. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 12. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 13. R Development Core Team (2010) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 14. Remm M., Storm C. E., Sonnhammer E. L. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 15. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 16. Needleman S. B., Wunsch C. D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–453 [DOI] [PubMed] [Google Scholar]
- 17. Rice P., Longden I., Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 18. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K. F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34, D354–D357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M. (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 23. Letunic I., Bork P. (2007) Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 [DOI] [PubMed] [Google Scholar]
- 24. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 25. Sinha A., Sommer R. J., Dieterich C. (2012) Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans. BMC Genomics 13, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macek B., Mann M., Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 27. Olsen J. V., Macek B. (2009) High accuracy mass spectrometry in large-scale analysis of protein phosphorylation. Methods Mol. Biol. 492, 131–142 [DOI] [PubMed] [Google Scholar]
- 28. Wang X., Sommer R. J. (2011) Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLoS Biol. 9, e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sternberg P. W. (2005) Vulval development. WormBook 2005, 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sommer R. J. (2008) Homology and the hierarchy of biological systems. Bioessays 30, 653–658 [DOI] [PubMed] [Google Scholar]
- 31. Tian H., Schlager B., Xiao H., Sommer R. J. (2008) Wnt signaling induces vulva development in the nematode Pristionchus pacificus. Curr. Biol. 18, 142–146 [DOI] [PubMed] [Google Scholar]