Abstract
Protein turnover through the ubiquitin-proteasome pathway controls numerous developmental decisions and biochemical processes in eukaryotes. Central to protein ubiquitylation are ubiquitin ligases, which provide specificity in targeted ubiquitylation. With more than 600 ubiquitin ligases encoded by the human genome, many of which remain to be studied, considerable effort is being placed on the development of methods for identifying substrates of specific ubiquitin ligases. In this review, we describe proteomic technologies for the identification of ubiquitin ligase targets, with a particular focus on members of the cullin-RING E3 class of ubiquitin ligases, which use F-box proteins as substrate specific adaptor proteins. Various proteomic methods are described and are compared with genetic approaches that are available. The continued development of such methods is likely to have a substantial impact on the ubiquitin-proteasome field.
The ubiquitin-proteasome system (UPS)1 is responsible for selective and timely protein turnover and is essential for proper cellular function (1, 2). The ubiquitin system also controls the activity of numerous proteins through nondegradative mechanisms. Perturbations in the UPS are linked to many diseases, such as cancers and neurodegeneration (3–5). Ubiquitylation occurs when a covalent isopeptide bond is formed between the C-terminal carboxyl group of 76 amino acid ubiquitin protein (referred to herein as Ub) and the ε-amino-group of a Lys residue of the target protein. This process is catalyzed through an E1-E2-E3 cascade, wherein Ub is first activated via an E1 Ub activating enzyme in an ATP-dependent manner to form a Ub-adenylate, which is then transferred to a conserved cysteine residue within the E1 to generate a high-energy thioester bond (6). The activated Ub is subsequently transferred to one of about two dozen E2 conjugating enzymes, again forming a thioester. E2 enzymes play a major role in determining both the number of Ub molecules added to a protein and the chain linkage types that are generated on poly-ubiquitylated proteins (6).
Charged E2s transfer Ub to substrates via an E3 Ub ligase. Two major classes of Ub-E3s are found in eukaryotes: the Homologous to the E6AP Carboxyl Terminus (HECT) E3 ligases and the Really Interesting New Gene (RING) E3 ligases (7–9). HECT domain E3s receive Ub from the E2 as a thioester and subsequently transfer Ub to an associated substrate. Thus, HECT domains have both a conserved motif that associates with the E2 enzyme and a catalytic cysteine residue. In contrast, RING domain proteins associate with charged E2s via their RING domain and then transfer directly to bound substrates. The RING E3 ligases can be divided into two types: the simple RING E3 ligases and the cullin-RING E3 ligases (CRLs) (9). CRLs are modular protein complexes comprising a substrate adaptor arm, a cullin protein that acts a scaffold, and the RING protein (9, 10). In contrast, simple RING E3s are typically single polypeptides that also contain substrate binding domains.
E3s add Ub to targets in three major ways—monoubiquitylation, multi-monoubiquitylation, and poly-ubiquitylation—and the exact type of modification is dictated by the combined activities and specificities of the E2 and the E3 (11, 12). In polyubiquitylation, a Ub molecule already linked to the substrate is the target for further ubiquitin conjugation. All seven lysine residues in ubiquitin, as well as the N-terminal amino group, can receive ubiquitin during the formation of a poly-ubiquitin chain. With the exception of lysine-63 (K63) linkages, all other lysine-based linkages increase in abundance upon inhibition of the proteasome, suggesting that each of these linkages may be acted upon by the proteasome (13, 14). However, K48-linked, and possibly K11-linked, poly-ubiquitin chains are the best understood as proteasomally targeted devices (11, 12).
Historically, the identification of substrates for specific ubiquitin ligases has been a challenge. This reflects, in part, the rapid turnover of most degradative ubiquitylation substrates, as well as the typically weak interactions between E3s and their substrates. Early landmark studies defining major classes of E3s and their targets often focused on genetic relationships for substrate identification, and in many cases a particular protein of interest was known to be unstable and candidate or genetic approaches were used to identify the E3 of interest (15, 27, 28, 59). In addition, for a small number of well-studied E3s, conserved targeting sequences in substrates were identified, which made it possible to search for proteins containing such sequences as candidate substrates (16–19). Through the definition of functional domains in E3s, it is now appreciated that perhaps 600 or more individual E3s exist in the human genome, many of which are either completely unstudied or poorly understood (9). Thus, a major challenge for the field concerns the question of how to identify substrates for poorly understood E3s. The majority of E3s bind only transiently to their targets, and the inherent instability of many ubiquitylation targets increases the difficulty of their detection. However, in recent years, new proteomic approaches have come to the forefront, not only with respect to the identification of substrates for individual E3s, but also for global characterization of ubiquitylation sites on proteins, as well as for the analysis of regulatory networks involving proteins in the ubiquitin system.
In this review, we discuss the major proteomic and genomic approaches that have been developed to identify substrates of the CRLs, a family of Ub-E3s with more than 200 members, as well as general approaches for identifying ubiquitylation sites in the proteome, and we describe the impact this has had on our understanding of how protein stability is regulated globally (20). The SKP1-CUL1-F-box (SCF) complex is the archetypal member of the CRL super-family and has a substrate adaptor module consisting of two components: SKP1 and the F-box protein (FBP) (21–23). SKP1 acts as a bridge between CUL1 and the FBP, and the FBP is responsible for substrate binding and is referred to as the specificity factor. CUL1 also associates with a key RING finger protein (RBX1 or RBX2) that recruits ubiquitin-charged E2s (9, 20). Of the ∼70 FBPs found in humans, fewer than half of them are characterized (24). However, from existing studies on FBPs, we know that they regulate numerous cellular processes, including cell cycle progression, transcription, and cell survival (25, 26). The SCF complex serves as a paradigm for several other CRLs, and similar methods for substrate identification are applicable for all CRLs (20). A key feature of substrate binding adaptor proteins is that they often contain conserved surfaces that selectively interact with particular sequence motifs in substrates, which are typically referred to as degrons (16, 17, 19, 29, 60, 61). In some cases, the identification of degrons for a particular adaptor can provide a means by which to search for substrates either in whole genomes or in candidate substrates identified by proteomics (16, 19, 30).
Approaches for Physical Detection of SCF Substrates
Immunoprecipitation followed by mass spectrometry (MS) (IP-LC/MS) has served as a primary route for the physical identification of targets for particular proteins of interest under largely physiological conditions. In this process, cell lines expressing most often an epitope-tagged version of the protein of interest are used for immunoprecipitation, and interacting proteins are identified via MS. However, there are several issues with this approach that limit its applicability in the identification of substrates of E3s. First, because most substrates bind transiently with the E3 or have a low binding energy, most substrates will not remain associated with the E3 during stringent washing of immune complexes. Second, because ubiquitylation itself most often promotes protein turnover, the levels of relevant in vivo substrates might be very low, limiting their detection. Third, many FBP–substrate interactions are signal dependent (for example, phosphorylation) (20). Because of these issues, the fraction of a substrate that is modified in the appropriate way for recognition by the FBP might be low, and this might limit one's ability to detect an interaction with an FBP. Thus, the low abundance of candidate substrates that remain associated with substrate specificity factors after immunoprecipitation is often very low relative to the dozens or hundreds of contaminating proteins that are typically detected in a standard immunoprecipitation MS experiment. As a result, specialized approaches are typically needed to allow the selective capture and identification of substrates in association with FBPs when contaminating proteins dominate proteomic datasets.
An early solution to the problem of substrate identification for SCF complexes developed by Michele Pagano and colleagues took advantage of a dual tagging strategy in which the FBP and associated substrates were immunoprecipitated and the immune complex was used to perform in vitro ubiquitylation reactions using an epitope-tagged version of ubiquitin (31). The idea was that any particular FBP-associated protein that was also a substrate for that particular SCF complex would become ubiquitinated. By recovering ubiquitylated proteins via the epitope tag on the ubiquitin molecule, it is possible to identify ubiquitylated proteins using proteomics. This approach was first applied to the FBP β-TrCP2, leading to the discovery of substrates such as Claspin, PCDC4, and REST (31–33). The identification of these proteins as substrates was aided by the fact that a consensus binding sequence for β-TrCP2 was known and could be searched for in any candidate substrate that emerged from the proteomics analysis (17, 60). However, this method has also been used in the identification of substrates for orphan FBPs—for example, identifying the circadian rhythm proteins CRY1 and CRY2 of FBXL3 (34).
While this approach, in principle, should provide a higher degree of selectivity, given that it is a two-step enrichment process, the often low-efficiency ubiquitylation reaction coupled with low recovery of ubiquitinated proteins renders this approach more difficult than single-step approaches. However, single-step approaches necessitate the successful filtration of contaminants away from potential candidate interacting proteins that are also ubiquitylation targets. A solution to this problem has emerged with the development of the Comparative Proteomics Analysis Software Suite (CompPASS), which uses a large database of unrelated but parallel IP-LC/MS experiments to filter out contaminants (35). The algorithm uses the abundance of interacting proteins based on spectral counts, the frequency of interaction across the database of parallel bait proteins, and the reproducibility of the interaction to provide empirical scores for each interacting protein. Proteins that pass a particular threshold for these scores are considered high confidence candidate interacting proteins (HCIPs). Typically, less than 3% of all proteins identified in association with a particular bait protein are regarded as HCIPs, pointing to the large number of contaminating proteins present in standard immune complexes (35).
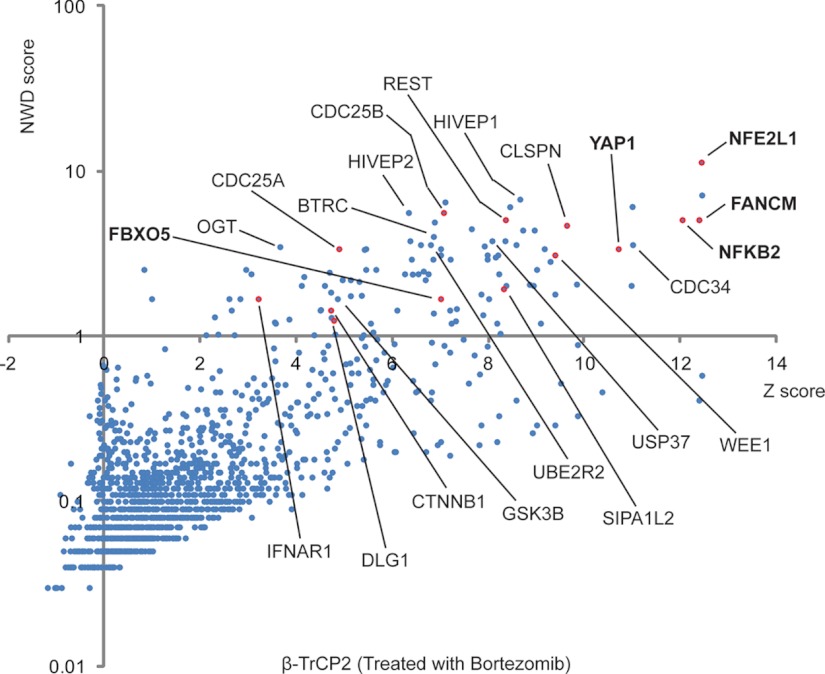
The application of the CompPASS approach to the well-studied β-TRCP2 resulted in the identification of several known substrates that passed the metrics for being an HCIP, including β-catenin, CDC25A, CDC25B, Claspin, REST, ATF4, and YAP1, although in several cases, between two and five peptides for a particular substrate were identified (Fig. 1). Based on these results, the questions that emerge are whether this approach can be used to identify candidate substrates for new E3s for which no candidate substrates are known, and whether this approach can be used to identify proteins with very low total peptide scans (for example, one or two spectral counts). The answers to these questions came with the application of a comparative analysis of two IP-LC/MS experiments that are performed in parallel, wherein the specificity factor of interest is immunoprecipitated from cells that are untreated or from cells that are treated for 4 h with proteasome inhibitors such as MG132 or Bortezomib. Proteins that are normally being degraded via the particular FBP complex are stabilized, and under such conditions the steady-state abundance of substrates associated with the FBP increases proportionally. Thus, by comparing spectral counts for proteins that pass the CompPASS metrics in the presence and absence of proteasome inhibition, it is possible to identify those proteins that both selectively associate with the FBP of interest and increase in abundance in response to proteasome inhibition. Such proteins have the characteristic of being bona fide substrates. Thus far, this approach has been used to identify a new substrate for β-TRCP2, as well as the first substrate for the orphan FBP FBXO22. In the case of β-TRCP2, two spectral counts were identified for a novel candidate substrate, the mTOR inhibitor DEPTOR, in the presence of proteasome inhibitor, whereas no DEPTOR peptides were found in the absence of proteasome inhibitor despite comparable spectral counts for β-TRCP2 itself (36–38). Functional studies and reciprocal proteomics revealed that DEPTOR is a bona fide substrate and that the phosphodegron for recognition of DEPTOR by β-TRCP2 is generated by the dual action of both mTOR itself and casein kinase I (36, 38). Similarly, FBXO22 immune complexes contained four spectral counts for the lysine demethylase KDM4A specifically in the presence of proteasome inhibitor, and follow-up studies validated this demethylase as the first substrate for this FBP (36, 37). Interestingly, most CRL substrates are present in protein complexes with other regulatory proteins, and in many cases these regulatory proteins also increase in abundance in the immune complex when the proteasome is inhibited, revealing that the complex containing the target is recognized. The CompPASS approach also allows proteins that function as regulators of a particular CRL, which stably bind to the adaptor protein in the presence or absence of proteasome inhibition. For example, analysis of FBW8 led to the identification of Obscurin-like 1 (OSBL1) and CCDC8 as subunits of the CUL7-SKP1-FBW8 complex. Previous studies had indicated that CUL7 is mutated in 3-M Syndrome, a disease characterized by short stature and other developmental disorders (39). Interestingly, both OSBL1 and CCDC8 are also mutated in 3-M disease, but their physical link with CUL7 had been unknown (40). Functional studies demonstrated that OSBL1 is required to localize the CUL7-SKP1-FBW8 to Golgi in neurons, where it promotes turnover of GRASP65 (40).
Fig. 1.
Application of CompPASS-based proteomics for identification of substrates and binding partners of SCFβ-TRCP2. Dot plot depicting known β-TrCP2 interactors, including substrates, identified after IP-LC/MS and CompPASS analysis. Red dots indicate proteins with increased spectral counts in proteasome inhibitor (Bortezomib) treated sample relative to the untreated sample. Proteins that were not detected in the absence of Bortezomib are labeled in bold type.
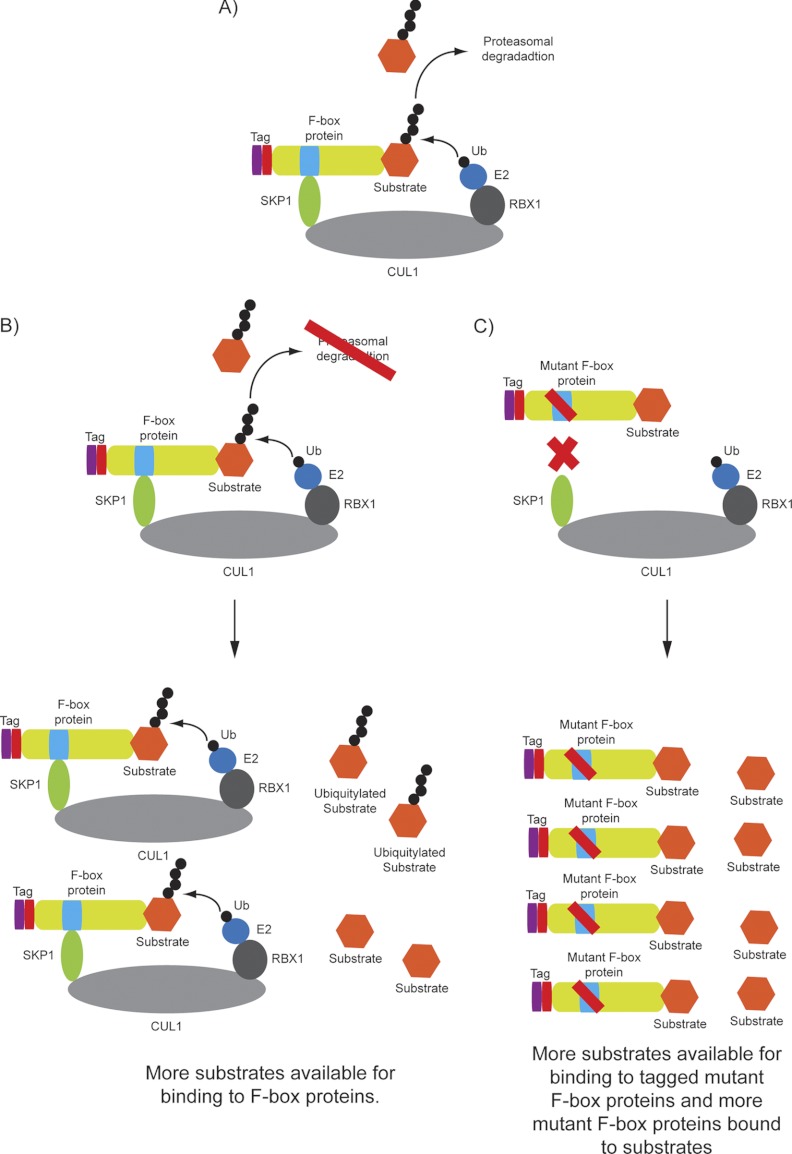
A further variation on this approach, sometimes referred to as “substrate trapping,” involves parallel analysis of IP-LC/MS results from cells expressing either the full-length wild-type FBP or the FBP in which the F-box has been mutated. FBPs lacking the F-box do not assemble with SKP1 and therefore cannot engage in an active CUL1 complex. When such FBP mutants are overexpressed, substrates are sequestered away from the endogenous FBP and are thereby stabilized. As with the proteasome inhibitor method described above, this approach yields higher levels of candidate substrates associated with the F-box mutant protein. As with other interaction-based approaches, overexpression of the F-box mutant protein could lead to the identification of spurious false positives, and therefore downstream experiments are necessary to validate any particular interaction.
The substrate trapping approach can be performed either using spectral counting or, alternatively, in a stable isotope labeling by amino acids in cell culture (SILAC)-based manner wherein cells expressing wild-type FBP are grown in light media and cells expressing the F-box mutant protein are grown in heavy media. The first example of the use of this approach came with an analysis of the FBXL5 FBP, which led to the identification of two iron-regulatory proteins, IRP1 and IRP2, as substrates for FBXL5 (41, 42). A recent study compared SILAC to peptide counting using the substrate trapping approach for 3 FBPs (FBW7α, SKP2, and FBXL5) (43). This version of the substrate trapping approach is referred to as differential proteomics-based identification of ubiquitylation substrates (DiPIUS) in the SILAC mode and nonlabeled DiPIUS in the peptide counting mode. There are several interesting features of this analysis. First, with the exception of a small number of known substrates for these FBPs (c-MYC for FBW7α, p27 for SKP2, and IRP1 and -2 for FBXL5), little overlap was seen when the SILAC approach was compared with the peptide counting approach in HeLa cells. This might reflect rapid exchange between heavy and light substrate proteins present in the mixed lysate during the immunoprecipitation, which would tend to push potential substrate interacting proteins toward a heavy:light ratio approaching unity. In this case, actual substrates would be missed. Second, the sensitivity of detection might have been an issue, as the total number of known substrates identified for FBW7α and SKP2 was low. Third, the method of subtraction used to remove false positives or nonspecific interacting proteins relied on only two unrelated bait proteins, and if a particular protein was found with two of the three different FBPs under examination, the protein was considered a false positive. However, based on previous studies, this approach would be expected to substantially underestimate the actual number of nonspecific interacting proteins, because of stochastic sampling in the MS experiment (35). Previous studies suggest that a significantly larger number of unrelated bait proteins are required in order for one to survey the entire array of false positive proteins that may appear stochastically in association with any particular bait protein.
FBPs typically employ a conserved domain to interact with degrons on their targets, and mutations in key residues can result in a loss of substrate binding. Thus, mutants of this type provide an alternative strategy for comparative proteomics, wherein proteins associated with wild-type (WT), but not mutant, FBPs are candidate substrates. For example, the FBP Cyclin F contains a cyclin box fold, which has been shown to be important for substrate recognition (24, 44). By comparing the peptide profiles of WT and cyclin-box deleted Cyclin F mutant, Pagano and colleagues identified two novel SCFCyclinF substrates, RRM2 and CP110, for Cyclin F (45, 46).
Finally, in instances in which a particular protein is known to be degraded—for example, in a signal-dependent process—one can screen for candidate E3s using siRNA libraries targeting families of adaptor proteins or, alternatively, one can perform directed association studies using libraries of clone adaptor proteins. Such approaches have been used to identify REST and SPAR as substrates of β-TRCP, respectively (47, 48).
Quantitative Ubiquitin Remnant Profiling
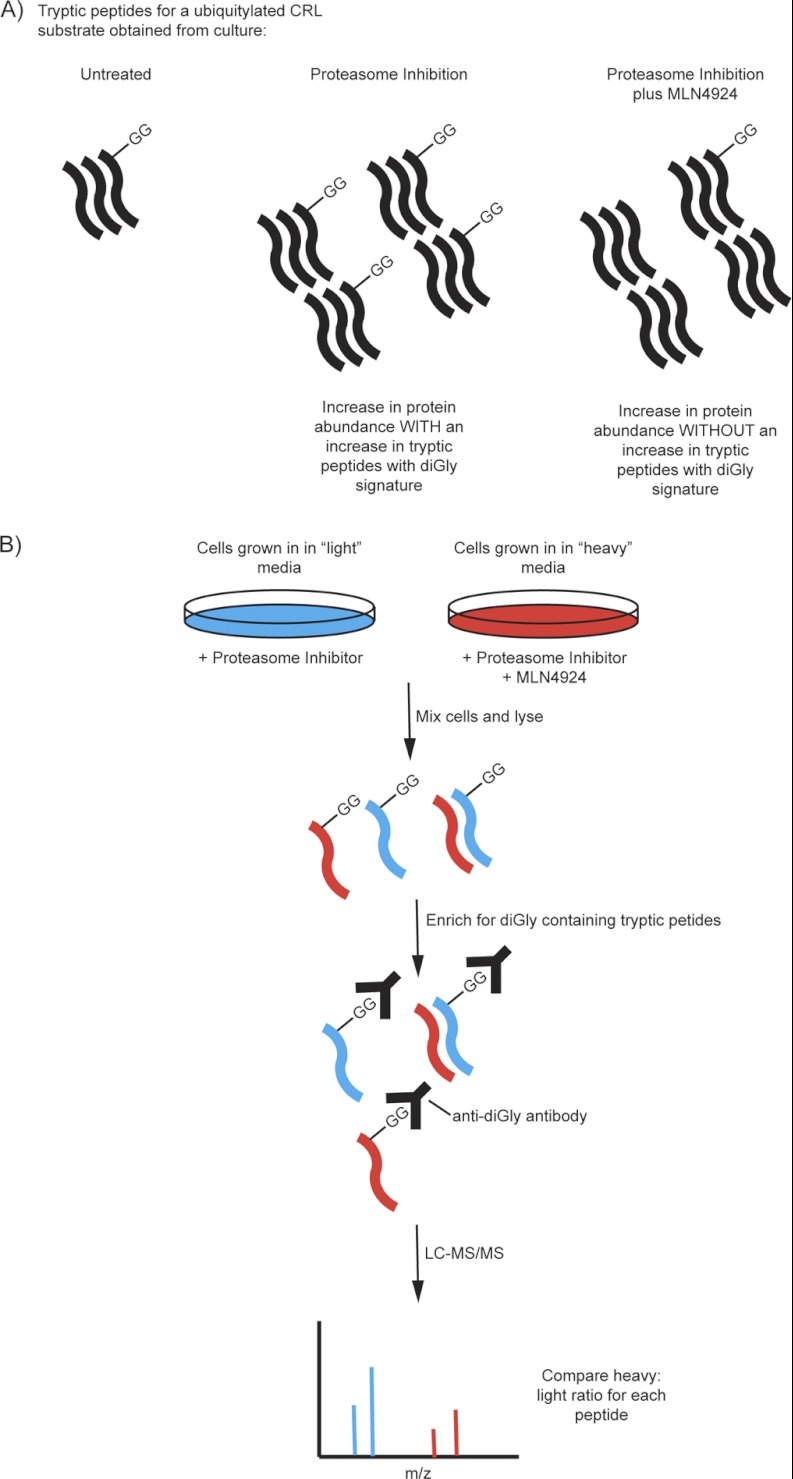
Ubiquitin is conjugated to lysine residues via an isopeptide bond with its C-terminal glycine residue, which is preceded by an Arg-Gly sequence. Thus, trypsin digestion of ubiquitylated substrates leaves a Gly-Gly peptide conjugated to the lysine of interest, and this modification can be identified based on its mass and fragmentation pattern. Early studies using this method for the detection of ubiquitinated peptides identified relatively small numbers of ubiquitylation sites, and in these experiments the low stoichiometry of ubiquitylation required that there be an enrichment step, for example, using ubiquitin binding proteins to select ubiquitinated proteins or expressing epitope-tagged ubiquitin (13). In principle, such approaches can present a bias in the number and types of proteins that are identified as being ubiquitylated, and any particular event may or may not represent an event present in the unperturbed cell. Moreover, although numerous proteins could be identified as being associated with ubiquitin, the actual number of diGly-modified sites that could be identified was small. In order to overcome these limitations, antibodies that can be used to immunoprecipitate diGly-modified peptides from trypsinized extracts have been developed (49). Importantly, this approach can be used in combination with SILAC to identify diGly-modified sites that are altered in response to a particular cellular perturbation (Fig. 2B). To date, more than 20,000 sites have been identified, and in many cases the increase in abundance of these peptides in response to proteasome inhibition has been determined (14, 49–51).
Fig. 2.
Methods for enrichment of substrates in association with F-box proteins for identification by IP-LC/MS. A, diagram showing the ubiquitylation and subsequent degradation of SCF substrates. B, diagram showing that inhibiting the proteasome by treating cells with proteasomal inhibitors such as MG132 or Bortezomib results in the accumulation of SCF substrates and increases the fraction of substrates bound to the cognate FBP. C, diagram showing how the introduction of an FBP lacking the F-box domain can increase the cellular pool of its substrate, allowing more substrates to bind this mutant FBP.
There are several noteworthy aspects of these studies. First, although a large number of sites have been identified, it appears that this approach still does not identify all ubiquitinated proteins in a given extract, and numerous known ubiquitylation targets are not present in the published datasets. This likely reflects the very low abundance of particular ubiquitylated proteins in vivo. Second, peptide identification is largely stochastic. In biological duplicate IP-LC/MS experiments, a substantial fraction of the peptides are identified in only one of the two biological duplicates (14). This likely reflects the inability of the currently available antibodies to fully deplete diGLY-modified peptides from extracts. Third, although the number of sites that can be identified increases several-fold upon prolonged proteasome inhibition (14, 51), the vast majority of these sites appear in a manner that depends upon ongoing protein synthesis (14, 51). Thus, it is possible that a significant fraction of the ubiquitin modified proteome reported thus far represents ubiquitylation events that occur in the context of ubiquitin-proteasome stress, and might not represent a physiological control mechanism. Importantly, it is possible to classify candidate ubiquitylation targets as either those that are likely to be physiologically regulated or those that are likely to be protein quality control substrates by examining the kinetics of ubiquitylation. Fourth, there does not seem to be a strong bias for particular peptide sequences in cases in which it has been examined in detail (14). Thus, no specific ubiquitylation consensus sequence emerged from these studies, unlike the situation with sumoylation. Fifth, in addition to ubiquitin, two other ubiquitin-like proteins (NEDD8 and ISG15) are also conjugated to lysine residues in proteins and, when cleaved with trypsin, generate a diGly remnant peptide. One study (14) examined the contribution of peptides from these modifications using several approaches and found that a maximum of 6% of diGly-containing peptides are derived from ISG15 or NEDD8 in HCT116 cells. This number is likely to be much lower in actuality, given that the methods used could not completely differentiate between neddylation and ubiquitylation, but this also suggests that a large fraction of the ubiquitin-modified proteome that has been identified thus far is derived uniquely from ubiquitin. Interestingly, this study also demonstrated that ubiquitin depletion resulting from proteasome inhibition leads to mischarging of ubiquitin E1 by NEDD8 (14). NEDD8 is then transferred to what would normally be ubiquitin substrates, apparently through the action of ubiquitin E2s. This indicates that conditions that alter the endogenous balance of ubiquitin and NEDD8 can lead to a loss in fidelity of the ubiquitin conjugation and transfer system. Similar results were found by Hjerpe et al. (52).
Nevertheless, it is possible to employ quantitative diGly profiling to identify bona fide substrates of CRLs (Fig. 3). Physiologically regulated substrates accumulate diGly modified peptides rapidly in a time course in response to proteasome inhibition. If the activity of the E3 responsible for physiological ubiquitylation is inhibited, then the accumulation of specific diGly modified peptides will not increase even when cells are incubated with proteasome inhibitor. Thus, this provides a means by which to search for substrates of a particular ubiquitin ligase by looking for peptides that increase in abundance in the presence of proteasome inhibitor but not when the E3 is inhibited using either chemical inhibitors or RNAi. This approach not only allows the identification of candidate substrates of an E3, but also provides the identity of lysine residues in a substrate that are ubiquitylated. One potential complication is that the treatment of cells with proteosomal inhibitor might increase the amount of defective ribosomal products and misfolded proteins represented in the diGly modified protein collection. However, in practice this issue can be minimized by treating both the control sample and the pathway perturbed sample with proteasome inhibitors.
Fig. 3.
Quantitative MS approach using SILAC. A, diagram showing how different treatments can alter the abundance and number of diGly remnants found on tryptic peptides for a ubiquitylated CRL substrate. B, outline of the quantitative MS approach using SILAC together with the anti-diGly antibody.
This approach has been used to identify candidate substrates of CRLs using the small molecule inhibitor of the NEDD8-activation cascade (53). NEDDylation is a critical post-translational modification that occurs on cullin molecules and is required for CRL-dependent ubiquitylation (54, 55). The addition of MLN4924, a specific inhibitor of the E1 enzyme, to cells renders this pathway inactive and blocks CRL-dependent ubiquitylation, as well as substrate turnover. Experiments profiling diGly sites on proteins in the presence of either proteasome inhibitor or proteasome inhibitor plus MLN4924 led to the identification of between 300 and 500 diGly modified peptides whose accumulation was blocked by MLN4924 (14, 50). Among these were numerous known sites for individual CRL family members, and there was a ∼30% overlap in the substrates identified in the two published studies, with a major difference being the cell line used, which could have contributed to the low overlap. Importantly, biological duplicates in the same cell line demonstrated a 70% overlap in proteins that scored as candidate CRL targets, indicating that the method is reasonably reproducible (14). However, it should be noted that the stochasticity issue noted above means that many low abundance peptides might not be identified in two biological duplicates. An important advantage of the diGly-capture approach is that it allows the immediate identification of ubiquitylated lysine residues in substrates. This can be useful in many ways, first and foremost in the design of non-ubiquitylatable mutant proteins that can be used to test the hypothesis that the ubiquitylation of a particular protein is important for a specific biological process. In addition, the identification of ubiquitylation sites can also aid in the identification of motifs that are recognized by the E3 or the E2, and which therefore might be important for specificity, as well as exposed regions of the target protein.
Quantitative Global Proteomics Method to Identify CRL Substrates
Inhibition of a ubiquitin ligase or a family of ubiquitin ligases would be expected to result in an increase in the abundance of the substrates that are normally degraded by the proteasome. Thus, in principle, SILAC-based approaches, wherein light cells have an intact E3 pathway whereas heavy cells have an inhibited E3 pathway, would be expected to identify proteins whose abundance is increased, assuming that sufficient depth can be reached in order to identify and quantify low abundance proteins that preferentially make up E3 targets. To date, a single study of this type has been reported for the identification of substrates of CRLs (56). In this experiment, A375 melanoma cells were used for SILAC and cells were either untreated or treated with the general CRL inhibitor MLN4924 for various times. Cells were mixed prior to lysis and fractionation by SDS-PAGE/in-gel digests. In total, 584 LC/MS/MS runs were collected, and ∼6000 proteins were identified with at least two peptides for each time point, with a false discovery rate of 1% (7689 total proteins over eight samples). This experiment also included a labeling switch for one of the time points. From this analysis, 120 proteins were found to increase in abundance by greater than 2-fold, suggesting that these might be CRL targets. Parallel transcriptional profiling revealed, however, that the mRNA abundance for 20 of these was also increased upon MLN4924 treatment, indicating an indirect affect on protein levels. Of the remaining 100 proteins, 7 were known CRL substrates. Thus, this experiment potentially identified numerous new CRL substrates, although further studies are required in order to validate these results and identify the particular CRL involved. Importantly, given the small number of known CRL substrates that were identified in this analysis, it would appear that sufficient depth of analysis was not achieved to globally identify CRL substrates using this approach. This likely reflects the fact that most substrates of CRLs are of very low abundance and therefore are difficult to detect even in an experiment of this scale.
Genetic Approaches for CRL Substrate Identification in Mammalian Cells
As an alternative approach, the Elledge laboratory has developed a novel methodology referred to as Global Protein Stability (GPS) profiling, which, in principle, allows the relative stability of proteins to be determined in a nearly genome-wide manner. GPS profiling is a fluorescence-based reporter system that combines fluorescence-activated cell sorting (FACS) with DNA microarray deconvolution to systematically examine changes in protein stability in live cells, for example, in response to inhibition of the CRL pathway. GPS vectors express a single transcript encoding both the DsRed open reading frame (ORF) and a GFP-ORF (representing a particular human gene) separated by an internal ribosome entry site (IRES), and because these proteins are produced from a single transcript, the GFP/red fluorescent protein (RFP) ratio serves as an indirect readout of the stability of the GFP-ORF protein in individual cells. By making ORF libraries representing a large fraction of the human genome, it is possible to screen a substantial fraction of the genome for proteins whose abundance is altered by various stimuli. Screening is performed by FACS sorting cells into eight bins that reflect the apparent stability of the particular GFP-ORF clone (50, 57). Upon perturbation (for example, the addition of the CRL neddylation inhibitor MLN4924 or expression of a dominant negative cullin), cells expressing GFP-ORF proteins that are stabilized will have a higher GFP/RFP ratio and will be shifted to a distinct bin. The distribution of ORF clones in individual bins before and after perturbation is determined by PCRing out the ORFs from each bin and individually hybridizing probes to custom microarrays that detect all the ORFs in the library. ORFs whose distribution changes toward a higher GFP/RFP ratio are candidate substrates, and this can be validated using conventional approaches. The initial GPS system reported (v1.0) contained 8000 human proteins, whereas the v2.0 library contains ∼12,500 proteins. These systems have been used to screen for substrates of CUL1-, CUL3-, and CUL4-based E3s, as well as for CRL targets using MLN4924 (50, 57).
With the v2.0 system, inhibition of CRLs globally with MLN4924 led to the identification of 244 proteins whose stability is significantly increased in the GPS system (50). Of the 190 cullin-interacting proteins present in the v2.0 library, 90 displayed an increase in stability upon MLN4924 addition, confirming that the GPS method can identify CRL substrates. In addition to chemical perturbation, this approach can also be used in combination with shRNA or dominant negative mediated pathway inhibition. Overexpression of dominant negative CUL4 and CUL3 vectors resulted in the identification of 279 and 188 candidate substrates, of which ∼75% in each screen were also found in the MLN4924 screen (50). Thus there is significant overlap in the identification of candidate CRL substrates using the GSP approach and different means of perturbing the pathway. Even more powerful in identifying candidate CRL substrates is the merging of quantitative diGly proteomics with the GPS system. For example, of the 295 genes in the v2.0 GPS library that correspond to proteins identified in parallel using the diGly capture approach in the presence of MLN4924, 108 (37%) were also identified in the GPS screen (50). Further studies are required in order to develop more inclusive libraries and to validate the very large number of substrates that have emerged using this approach together with diGly profiling.
Strengths and Weaknesses of Proteomic and Genetic Approaches for CRL Substrate Identification
Each of the approaches described in this review has specific advantages, and it is likely that no single technique will routinely identify all CRL substrates in a given cell type. First, diGly; interaction proteomics, including the CompPASS approach; and global proteomics can miss proteins that are of particularly low abundance or that are insoluble under standard lysis conditions, whereas GPS profiling might be able to detect these, assuming the relevant genes are represented in the library. Alternatively, the GPS system might fail to identify proteins that cannot be epitope tagged and maintain regulation or which are not present in the library, whereas the proteomic approaches detect endogenous proteins. A further complementary aspect of these approaches is that diGly capture directly identifies ubiquitylation sites, which can then be immediately employed in the further design of functional studies. Moreover, proteomic approaches, unlike the GPS approach, do not directly filter out transcriptional effects of perturbations, an action that would require significant further validation. In addition, whereas the GPS system is useful for finding ubiquitylation targets that are degraded by the proteasome, the diGly and interaction proteomic approaches have the potential to identify ubiquitylation substrates that are not acted on by the proteasome. This is important in the context of CRL biology, as it was recently reported that the CUL3-KLHL12 complex mono-ubiquitinates its substrate SEC31 in order to alter its activity, rather than promoting its poly-ubiquitylation (58). Finally, it is evident that some cullin complexes have stably associated binding partners that play important regulatory roles. Interaction proteomics, unlike the other approaches, is uniquely suited for the identification of stable regulatory factors, which can be very important for placing CRLs and substrates into biological pathways, as evidenced by our identification of OBSL1 as a Golgi targeting subunit for the CUL7-SKP1-FBW8 complex (40). Thus, a complete understanding of CRL targets and biology will likely require a combination of approaches.
CONCLUSION
Ubiquitin proteomics has emerged as a major area of research in the ubiquitin field. Nevertheless, there remain several challenges with respect to the identification of not only substrates of CRLs but ubiquitin ligases in general. A central challenge concerns the identification of requisite signals that initiate the encounter of a particular substrate with an E3, including phosphorylation, hydroxylation, and glycosylation. Conserved motifs in both the substrate and the E3 will no doubt be critical for elucidating such signals and in extrapolating such motifs to build interaction networks and identify additional targets. Depending upon the cell type being employed, only a fraction of the potential targets of a particular CRL might actually be identifiable because of the absence of the requisite signal. In addition, the amplitude of any particular response might have ramifications for the amount of any particular substrate that can be captured via a proteomics approach. A major limitation in the current toolbox of techniques is the absence of methods for the identification of substrates in tissues. The approaches described above exclusively employ tissue culture cells, but lineage- and context-specific targets likely mean that many substrates of particular E3s will not be identified using current procedures. Thus, the use of quantitative deep proteome analysis might need to be developed to identify candidate substrates from tissues, for example, where a particular E3 has been inactivated by deletion. Alternatively, a more complete understanding of CRL targets, and of E3s generally, may be achieved through the development of larger collections of cell line systems that mimic a wide cross-section of tissue functionality. An additional area that is ripe for future development is that of protein arrays. Although this approach has not been used for CRL, previous work has shown that protein arrays can be used to identify substrates of HECT-domain E3s (62, 63). One potential limitation that will need to be overcome is that the majority of CRLs are thought to require substrate modification for efficient interaction. As purified, the proteins on arrays might not be properly modified, or, alternatively, the stoichiometry of modification might be too low to allow detection. In cases in which the proper signal is known, it might be possible to pre-treat proteins on the array with the relevant modification enzymes. Clearly, new and improved proteomic techniques and instrumentation will continue to have a major impact on our understanding of the CRL system and its role in health and disease.
Footnotes
* Work on the ubiquitin system in the Harper lab is supported by the National Institutes of Health (AG011085, GM054137, and GM070565) and by Millennium Pharmaceuticals. M.-K.M.T. is supported by a predoctoral fellowship from the Agency of Science, Technology and Research (A*STAR), Singapore.
Conflict of interest statement: J.W.H. is a consultant for Millennium Pharmaceuticals.
1 The abbreviations used are:
- CompPass
- Comparative Proteomics Analysis Software Suite
- CRL
- cullin-RING E3 ligase
- DiPIUS
- differential proteomics-based identification of ubiquitylation substrates
- FACS
- fluorescence-activated cell sorting
- FBP
- F-box protein
- GPS
- Global Protein Stability
- HCIP
- high confidence candidate interacting protein
- HECT
- Homologous to the E6AP Carboxyl Terminus
- IP-LC/MS
- Immunoprecipitation followed by mass spectrometry
- ORF
- open reading frame
- OSBL1
- Obscurin-like 1
- RING
- Really Interesting New Gene
- SCF
- SKP1-CUL1-F-box
- Ub
- ubiquitin protein
- UPS
- ubiquitin-proteasome system
- WT
- wild-type.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Shabek N., Ciechanover A. (2010) Degradation of ubiquitin: the fate of the cellular reaper. Cell Cycle 9, 523–530 [DOI] [PubMed] [Google Scholar]
- 3. Welcker M., Clurman B. E. (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 [DOI] [PubMed] [Google Scholar]
- 4. Frescas D., Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin S. M., Youle R. J. (2012) PINK- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 125, 795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ye Y., Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 8. Lipkowitz S., Weissman A. M. (2011) RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 11, 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 10. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wickliffe K., Williamson A., Jin L., Rape M. (2009) The multiple layers of ubiquitin-dependent cell cycle control. Chem. Rev. 109, 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Behrends C., Harper J. W. (2011) Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18, 520–528 [DOI] [PubMed] [Google Scholar]
- 13. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 14. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koepp D. M., Harper J. W., Elledge S. J. (1999) How the cyclin became a cyclin: Regulated proteolysis in the cell cycle. Cell 97, 431–434 [DOI] [PubMed] [Google Scholar]
- 16. Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr. (2005) The v-jun point mutation allows c-jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 17. Winston J. T., Strack P., Beer-Romero P., Chu C. Y., Elledge S. J., Harper J. W. (1999) The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Welcker M., Orian A., Jin J., Grim J. E., Harper J. W., Eisenman R. N., Clurman B. E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margottin-Goguet F., Hsu J. Y., Loktev A., Hsieh H. M., Reimann J. D., Jackson P. K. (2003) Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 4, 813–826 [DOI] [PubMed] [Google Scholar]
- 20. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 21. Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 22. Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J. (1997) A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221–230 [DOI] [PubMed] [Google Scholar]
- 23. Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274 [DOI] [PubMed] [Google Scholar]
- 24. Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 26. Silverman J. S., Skaar J. R., Pagano M. (2012) SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem. Sci. 37, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zachariae W., Nasmyth K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13, 2039–2058 [DOI] [PubMed] [Google Scholar]
- 28. Harper J. W., Burton J. L., Solomon M. J. (2002) The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16, 2179–2206 [DOI] [PubMed] [Google Scholar]
- 29. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 30. Mailand N., Bekker-Jensen S., Bartek J., Lukas J. (2006) Destruction of claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 [DOI] [PubMed] [Google Scholar]
- 31. Peschiaroli A., Dorrello N. V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N. E., Pagano M. (2006) SCFbetaTrCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23, 319–329 [DOI] [PubMed] [Google Scholar]
- 32. Guardavaccaro D., Frescas D., Dorrello N. V., Peschiaroli A., Multani A. S., Cardozo T., Lasorella A., Iavarone A., Chang S., Hernando E., Pagano M. (2008) Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature 452, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dorrello N. V., Peschiaroli A., Guardavaccaro D., Colburn N. H., Sherman N. E., Pagano M. (2006) S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314, 467–471 [DOI] [PubMed] [Google Scholar]
- 34. Busino L., Bassermann F., Maiolica A., Lee C., Nolan P. M., Godinho S. I., Draetta G. F., Pagano M. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 [DOI] [PubMed] [Google Scholar]
- 35. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao D., Inuzuka H., Tan M. K., Fukushima H., Locasale J. W., Liu P., Wan L., Zhai B., Chin Y. R., Shaik S., Lyssiotis C. A., Gygi S. P., Toker A., Cantley L. C., Asara J. M., Harper J. W., Wei W. (2011) mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 44, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan M. K., Lim H. J., Harper J. W. (2011) SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell Biol. 31, 3687–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duan S., Skaar J. R., Kuchay S., Toschi A., Kanarek N., Ben-Neriah Y., Pagano M. (2011) mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol. Cell 44, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huber C., Dias-Santagata D., Glaser A., O'Sullivan J., Brauner R., Wu K., Xu X., Pearce K., Wang R., Uzielli M. L., Dagoneau N., Chemaitilly W., Superti-Furga A., Dos Santos H., Megarbane A., Morin G., Gillessen-Kaesbach G., Hennekam R., Van der Burgt I., Black G. C., Clayton P. E., Read A., Le Merrer M., Scambler P. J., Munnich A., Pan Z. Q., Winter R., Cormier-Daire V. (2005) Identification of mutations in CUL7 in 3-M syndrome. Nat. Genet. 37, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 40. Litterman N., Ikeuchi Y., Gallardo G., O'Connell B. C., Sowa M. E., Gygi S. P., Harper J. W., Bonni A. (2011) An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates golgi morphology and dendrite patterning. PLoS Biol. 9, e1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salahudeen A. A., Thompson J. W., Ruiz J. C., Ma H. W., Kinch L. N., Li Q., Grishin N. V., Bruick R. K. (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vashisht A. A., Zumbrennen K. B., Huang X., Powers D. N., Durazo A., Sun D., Bhaskaran N., Persson A., Uhlen M., Sangfelt O., Spruck C., Leibold E. A., Wohlschlegel J. A. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yumimoto K., Matsumoto M., Oyamada K., Moroishi T., Nakayama K. I. (2012) Comprehensive identification of substrates for F-box proteins by differential proteomics analysis. J. Proteome Res. 11, 3175–3185 [DOI] [PubMed] [Google Scholar]
- 44. Noble M. E., Endicott J. A., Brown N. R., Johnson L. N. (1997) The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem. Sci. 22, 482–487 [DOI] [PubMed] [Google Scholar]
- 45. D'Angiolella V., Donato V., Forrester F. M., Jeong Y. T., Pellacani C., Kudo Y., Saraf A., Florens L., Washburn M. P., Pagano M. (2012) Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149, 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D'Angiolella V., Donato V., Vijayakumar S., Saraf A., Florens L., Washburn M. P., Dynlacht B., Pagano M. (2010) SCF(cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 466, 138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W., Elledge S. J. (2008) SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ang X. L., Seeburg D. P., Sheng M., Harper J. W. (2008) Regulation of postsynaptic RapGAP SPAR by polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J. Biol. Chem. 283, 29424–29432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu G., Paige J. S., Jaffrey S. R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., Dick L. R., Kurz T. (2012) Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 54. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liao H., Liu X. J., Blank J. L., Bouck D. C., Bernard H., Garcia K., Lightcap E. S. (2011) Quantitative proteomic analysis of cellular protein modulation upon inhibition of the NEDD8-activating enzyme by MLN4924. Mol. Cell. Proteomics 10, M111.009183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yen H. C., Elledge S. J. (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322, 923–929 [DOI] [PubMed] [Google Scholar]
- 58. Jin L., Pahuja K. B., Wickliffe K. E., Gorur A., Baumgartel C., Schekman R., Rape M. (2012) Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Willems A. R., Goh T., Taylor L., Chernushevich I., Shevchenko A., Tyers M. (1999) SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 354, 1533–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 61. Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007) Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–141 [DOI] [PubMed] [Google Scholar]
- 62. Persaud A., Alberts P., Amsen E. M., Xiong X., Wasmuth J., Saadon Z., Fladd C., Parkinson J., Rotin D. (2009) Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4–2 using proteome arrays. Mol. Syst. Biol. 5, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Persaud A., Rotin D. (2011) Use of proteome arrays to globally identify substrates for E3 ubiquitin ligases. Methods Mol. Biol. 759, 215–224 [DOI] [PubMed] [Google Scholar]