Abstract
Advances in high resolution tandem mass spectrometry and peptide enrichment technologies have transformed the field of protein biochemistry by enabling analysis of end points that have traditionally been inaccessible to molecular and biochemical techniques. One field benefitting from this research has been the study of ubiquitin, a 76-amino acid protein that functions as a covalent modifier of other proteins. Seminal work performed decades ago revealed that trypsin digestion of a branched protein structure known as A24 yielded an enigmatic diglycine signature bound to a lysine residue in histone 2A. With the onset of mass spectrometry proteomics, identification of K-GG-modified peptides has emerged as an effective way to map the position of ubiquitin modifications on a protein of interest and to quantify the extent of substrate ubiquitination. The initial identification of K-GG peptides by mass spectrometry initiated a flurry of work aimed at enriching these post-translationally modified peptides for identification and quantification en masse. Recently, immunoaffinity reagents have been reported that are capable of capturing K-GG peptides from ubiquitin and its thousands of cellular substrates. Here we focus on the history of K-GG peptides, their identification by mass spectrometry, and the utility of immunoaffinity reagents for studying the mechanisms of cellular regulation by ubiquitin.
Post-translational modification by ubiquitin and ubiquitin-like proteins represents a major regulatory system in eukaryotic organisms (1, 2) and certain pathogenic bacteria (3). A conserved enzymatic cascade couples the C terminus of ubiquitin to the epsilon amino group of lysine residues on substrate proteins (4). Evidence has also emerged implicating ubiquitination via cysteine, serine, and threonine, as well as N-terminal residues (5–8). Depending on signaling context and the enzymes involved, protein substrates can be monoubiquitinated, multiubiquitinated, or polyubiquitinated. Ubiquitin-dependent processes are commonly modulated via formation of polyubiquitin chains, whereby the C terminus of a chain extending ubiquitin becomes linked to the N terminus or one of seven lysine residues (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, and Lys-63) within a substrate-bound ubiquitin molecule (9, 10). The functions of these structurally diverse modifications have been extensively studied through in vitro biochemistry (11–13), ubiquitin replacement in cellular models (14, 15), linkage specific antibodies (16–19), and mass spectrometry (20–26). A growing body of evidence indicates that polyubiquitin chains regulate biological processes not only by eliciting proteasomal degradation but also by altering subcellular localization, modulating enzymatic activity, and facilitating protein-protein interactions of ubiquitinated substrates (9, 24). Although in depth characterization of model substrates has led to significant advances in our understanding of biochemical mechanisms and cellular processes, much remains to be elucidated with relation to the dynamics of ubiquitination on individual substrates. The need for improved methodologies has driven innovation in the field of mass spectrometry proteomics, particularly in the characterization of protein ubiquitination sites. Here we focus on recent advances in ubiquitination site mapping and the questions that are driving this area of discovery biology.
Initial Characterization of Ubiquitination Sites
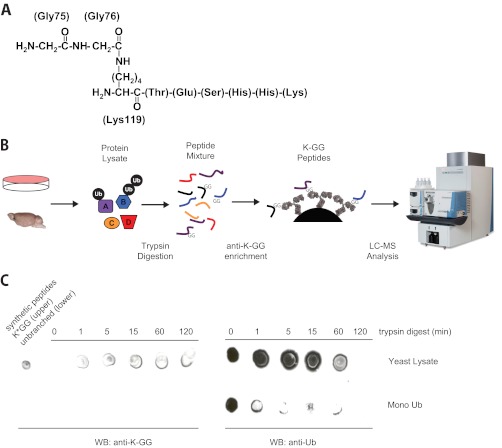
In 1977, while characterizing peptides from the unique chromosomal protein then known as A24, Ira Goldknopf and Harris Busch (27) described an isopeptide bond between lysine 119 of histone 2A and the tryptic C-terminal diglycine remnant of a “non-histone-like” sequence. The well chronicled flurry of work that followed established the non-histone-like molecule to be ubiquitin and defined its central role in cell regulation, leaving A24 as a remnant of history (28–32). However, from this groundbreaking work, two ideas emerged that define our current understanding of and experimental approaches toward protein ubiquitination. The fundamental characteristic of ubiquitin is indeed conjugation to protein substrates through an isopeptide bond. Moreover, the position of these substrate-ubiquitin bonds may be studied by generating and identifying tryptic K-GG signature peptides where the C-terminal diglycine of ubiquitin remains covalently attached to the substrate (Fig. 1A).
Fig. 1.
Identification of K-GG peptides. A, the first K-GG peptide originally identified by Goldknopf and Busch. The diglycine signature of trypsin-digested ubiquitin extends from Lys-119 of histone 2A. B, general scheme for immunoaffinity enrichment of K-GG peptides from either cultured cells or tissue lysates. Protein lysates are prepared under denaturing conditions, digested to peptides, and desalted, and K-GG peptides were captured using antibodies recognizing the isopeptide-linked diglycine signature. C, dot blot analysis performed showing specificity of anti-K-GG for tryptic peptides with isopeptide-linked diglycine signature. A mixture of either synthetic K-GG peptides (upper left) or unbranched ubiquitin peptides (lower left) was spotted adjacent to yeast protein lysate or purified monoubiquitin, respectively. A digestion time course was performed in parallel on both yeast lysate and monoubiquitin. Aliquots were collected at the indicated time points, spotted onto PVDF membrane, and immunoblotted using either anti-KGG or anti-ubiquitin (P4D1 epitope). Immunoreactivity confirms the presence of K-GG peptides in digested lysate. WB, Western blot; Ub, ubiquitin.
The earliest attempts at mapping ubiquitination sites on protein substrates focused on uncoupling ubiquitin-dependent degradation and stabilizing otherwise labile proteasome substrates. Work on the model substrate β-galactosidase (13), cell cycle regulator cyclin B1 (33), and the NF-κB inhibitor IκBα (34) established several themes. The ligase-binding site on a substrate often defines the region in which available lysines may be modified by ubiquitin. Although ubiquitination occurs within defined regions of a protein, individual lysines are typically dispensable. In this model, stabilization of a proteasome substrate through mutagenesis or truncation can be accomplished either by disrupting the ligase-substrate docking site or by eliminating all lysines capable of being targeted by the E31 ligase. In ubiquitin signaling, the concept of functional redundancy of adjacent lysines extends beyond proteasomal degradation to ligand mediated internalization and trafficking, as extensive combinatorial mutagenesis of the epidermal growth factor receptor kinase domain was required to yield loss of function (35, 36). More recently, monoubiquitination of SEC31 was shown to drive the assembly of large COPII coats on vesicles through a mechanism independent of any single lysine (37). Proliferating cell nuclear antigen, the clamp protein involved in post-replicative DNA repair, and Met4, a transcription factor involved with nutrient sensing, represent exceptions to functional redundancy, as single sites appear responsible for coordinating specific ubiquitin-dependent functions (38, 39).
Although these and many other studies involving mutagenesis have shaped our fundamental understanding of protein ubiquitination, a cautionary note remains: correlating loss of function with elimination of ubiquitin acceptor sites provides only indirect evidence for ubiquitination at the mutated residues. One alternative mechanism envisions lysine to arginine substitutions preventing ubiquitination by interfering with binding of the ligase to the substrate. The two essential pieces of information needed to discriminate between these closely related mechanisms are direct evidence of the linkage between ubiquitin and the modified lysine and confirmation that the abundance of the modified peptide responds to biological perturbation. Demonstrating ubiquitination in vivo is complicated further by the multiplicity of ubiquitin ligases that often act toward individual substrates. The specific lysine residues modified on a particular substrate may vary depending on which enzymes are involved and how each interacts with the substrate. Additional complexity is conferred by deubiquitinating enzymes, which have the potential to selectively edit ubiquitin signals. Focused quantitation of individual ubiquitination sites promises to further our understanding of ligase-substrate dynamics.
Nearly three decades passed between identification of the first diglycine-modified peptide by Edman sequencing and the emergence of LC-MS/MS as the primary tool for mapping ubiquitination sites. Initial proof of concept came through identification of the K-GG peptide of Lys-48-linked polyubiquitin (22, 40) and Lys-165 of the yeast G-protein coupled receptor Gpa1 (41). These K-GG peptides require cleavage between Arg-74 and Gly-75 of ubiquitin, typically by trypsin or ArgC proteases (Fig. 1A). An amino group extends from the attached diglycine remnant, giving these peptides an additional N terminus that plays a prominent role in current enrichment strategies.
Mass spectrometry has proven effective at mapping protein ubiquitination sites on individual substrates of interest. The two main strategies have been identification of K-GG peptides following in vitro ubiquitination of a purified substrate or immunoprecipitation of ubiquitinated substrates directly from cells. Analyses from cells have the advantage that modified proteins occur in biological context and can be induced by relevant stimuli, as in the cases of epidermal growth factor receptor, inositol 1,4,5-trisphosphate receptor, and dopamine transporter ubiquitination following treatment with epidermal growth factor, gonadotropin-releasing hormone, or protein kinase C activation, respectively (36, 42, 43). In the case of caspase-8, ubiquitination was shown to occur on Lys-461 in response to activation of death receptors by an extrinsic stimulus (44). Analogous studies have been carried out in yeast to identify ubiquitination sites on proteins including Gpa1, Met4, and Ypt7 (39, 41, 45), as well as in tissue from Alzheimer's patients to identify ubiquitination sites on the pathological form of Tau (46). Alternatively, physiologically relevant ubiquitination sites can be identified from in vitro ubiquitination reactions. In the context of neurodegeneration, ubiquitination sites have been mapped on α-synuclein and the E3 ligase CHIP (47, 48). For CHIP, studies demonstrated that ubiquitination on Lys-2 near the N terminus mediates interaction with the polyglutamine expanded ataxin-3 and may contribute to spinocerebellar ataxia (48). Mass spectrometry likewise identified lysine residues within the proteasome-associated ubiquitin receptor Rpn10 that when modified by ubiquitin affect its ability to interact with ubiquitinated proteasome substrates (49).
Efforts to successfully purify ubiquitinated substrates from lysate for identification by mass spectrometry initially employed epitope-tagged ubiquitin and immobilized metal affinity resins (22, 50). Subsequent efforts taking advantage of improvements in epitope tagging approaches (51–53) or tandem ubiquitin-binding domain affinity resins (54, 55) in combination with more sensitive mass spectrometers advanced these studies to the point that hundreds of ubiquitination sites could be identified from a single sample. Following isolation of an enriched population of ubiquitinated substrates, SDS-PAGE can be used to separate proteins based on size. For substrates modified by different numbers of ubiquitin molecules, a technical limitation is that the corresponding K-GG peptides are distributed across multiple gel bands (56). For single substrate ubiquitination site mapping, digestion and MS analysis can be performed on large gel sections spanning a range of 50–100 kDa, effectively pooling ubiquitinated substrate (36). Such an approach is less useful in global ubiquitin enrichment studies because of the added sample complexity. Even for single protein analyses, analysis of large gel sections can compromise sensitivity because of gel-based losses and abundant trypsin autolysis products. These limitations can be partially overcome by instead performing in solution digestion of the complex mixture followed by peptide level fractionation. Digesting proteins together that would otherwise have distributed across a wide mass range effectively pools K-GG peptides, improving sensitivity (56). This must be balanced against losses encountered in fractionation and the instrument time required to analyze fractionated samples. Given the technical challenges and the low stoichiometry of K-GG peptides relative to unmodified peptides from substrates and ubiquitin itself, methods that rely on protein level ubiquitin enrichment remain suboptimal for global ubiquitination site mapping. Maximal sensitivity requires focused and efficient enrichment of modified peptides from complex mixtures of unmodified peptides.
Enrichment of Diglycine-modified Peptides
Several recent papers have described immunoaffinity enrichment of K-GG peptides from complex protein lysates (Fig. 1B) (57–63). The method is based on a similar strategy originally described for enriching low abundance tyrosine phosphorylated peptides (64) and initially shown to be successful using polyclonal antibodies generated against the -GG signature of ubiquitin (65). In contrast to protein level enrichment of ubiquitin, which has yielded up to ∼750 unique ubiquitination sites in a single study (53), direct enrichment of K-GG peptides from cellular lysates has yielded over >10,000 unique sites on >4,000 proteins (57, 58). This focused approach has enabled discovery of ubiquitination sites to a similar depth as global phosphorylation and acetylation studies (66–69) and added support to the idea that many proteins are ubiquitinated at multiple lysine residues within their sequences. Antibodies broadly recognizing K-GG peptides have been generated using two distinct approaches. In one case, a peptide library immunization strategy was employed to generate a rabbit monoclonal antibody against peptides with the sequence CXXXXXXKGGXXXXXX (where X indicates any amino acid except Cys and Trp) (57). In the other case, an antigenic protein was created by chemical modification of purified histones using t-BOC-Gly-Gly-N-hydroxysuccinimide, followed by release of the BOC with TFA. This -GG-modified protein was injected into mice and used to generate a hybridoma for monoclonal production (59). Generation of diglycine-modified proteins such as histones, lysozyme, lactoglobulin, or whole cell lysates provided an additional benefit in visualizing antibody-antigen interactions during validation studies. Similarly, dot blots of undigested and digested lysates demonstrate the high level of specificity for these reagents (Fig. 1C).
A comparison of the K-GG peptides captured with these two antibodies suggests a notable difference in their preference toward or tolerance for certain amino acids adjacent to GG-modified lysines. Of the 23,439 ubiquitination sites reported by Kim et al. (57) and Wagner et al. (58), ∼4,300 are held in common. K-GG peptides enriched using the peptide library derived rabbit monoclonal reagent display increased frequency of small (Ala and Gly) and acidic residues (Asp and Glu) in positions adjacent to the diglycine-modified lysine. In contrast, no such preference toward small residues was observed for the mouse monoclonal, whereas Asp and particularly Glu appear to be disfavored (58). Instead, peptides captured by the GG-histone derived mouse monoclonal antibody more frequently displayed the hydrophobic residues Leu, Ile, Phe, Tyr, and Trp, particularly in the −3 to +3 region surrounding the modified lysine. Given the notable differences between existing studies in terms of lysis, digest conditions, ratios of input material to antibody, resin incubation, and wash times, a direct, quantitative comparison of two antibodies will be valuable to the community. Detailed analytical comparisons of binding kinetics and effects of the biological matrix on binding between immunoaffinity reagents and modified peptides will be particularly informative. Based on the existing data, an optimized mixture of the two currently available antibodies may be an even more effective tool for profiling ubiquitinated substrates and may further advance efforts aiming to match E3 ligases and deubiquitinating enzymes with the substrates they regulate.
Previous reports have suggested the potential for cross-talk between lysine acetylation and ubiquitination in the context of proteins such as p53 and the transcriptional coactivator p300 (70, 71). Comparison of the newest ubiquitination mapping studies to similar global acetylation mapping efforts reveals that ∼30% of sites previously identified as being acetylated were also found to be ubiquitinated (57, 58, 66, 69). Bioinformatic analysis suggests that peptides from this overlapping group displayed less significant increases in ubiquitination upon proteasome inhibition (58). Of these ∼1000 sites, a subset represent proteins involved in translation and mitochondrial metabolism, systems where these two modifications are known to exert significant regulation.
Despite the current data, further research remains to be done to ascertain the extent of cross-talk between lysine ubiquitination and acetylation. Defining the absolute and relative abundances of ubiquitination and acetylation at residues known to accommodate both modifications would shed light on a number of questions. A competitive mechanism whereby acetylation prevents ubiquitination through blocking available modification sites is mechanistically appealing, but in equilibrium would be expected to shift the stoichiometry of acetylation to very high levels for stabilized proteins. Although such a mechanism may operate for specific substrates or in certain subcellular locations, it seems unlikely to be broadly applicable. Because ubiquitinated and acetylated lysines often reside on surface-exposed patches, future work should consider whether individual acetylation events induce or inhibit ubiquitination through modulating interactions between ligases and substrates. More orchestrated mechanisms involving sequential addition and removal of modifications can also be envisioned but may only be defined through detailed time course and dose-response experiments with concurrent readouts of both pathways. A first step will be to understand the abundance of each modification in the basal state and the changes that occur following perturbation of the ubiquitination and acetylation systems with common proteasome and histone deacetylase inhibitors.
One limitation of current enrichment strategies is that tryptic digestion of substrate-bound ubiquitin-like modifiers NEDD8 and ISG15 generates K-GG signatures indistinguishable from those of ubiquitin. Although a detailed analysis comparing ubiquitin, NEDD8, and ISG15 levels has not been carried out in mammalian cells, work from yeast suggests that ubiquitin is expressed at >30× higher levels (not accounting for UBI3/UBI4 expression) than the yeast NEDD8 homolog Rub1 (72). Likewise, global proteomic profiling in nine mouse tissues revealed that peptides from ubiquitin were identified with markedly higher frequency than those from either NEDD8 or ISG15 (68). By extension, it has been argued that ubiquitin (as opposed to other ubiquitin-like modifiers) accounts for the vast majority of K-GG peptides identified during immunoaffinity enrichment. Spectral counting data from immunoaffinity enriched K-GG peptides lends some support to this conclusion, showing that K-GG peptides of ubiquitin far outnumber those from ISG15 or NEDD8. The utility of this data is limited, however, given that substrates modified by a single ISG15 or NEDD8 cannot be accounted for. To more directly assess the fraction of K-GG signature peptides resulting from neddylation, Kim et al. (57) took a two-pronged approach. In one arm of the study, SILAC was used to compare the abundance of individual K-GG peptides between untreated lysates and lysates incubated with the deubiquitinase USP2. USP2 has the ability to strip ubiquitin modifications from substrates, but not NEDD8 modifications, making it possible to assess the fraction of a given K-GG peptide derived from neddylation. In a complementary approach, a SILAC experiment was performed using MLN4924 to inhibit the NEDD8-activating enzyme NAE1. Based on these data, it was estimated that a maximum of 6% of K-GG peptides resulted from neddylation, even under conditions where the proteasome was inhibited and free ubiquitin pools were depleted (57).
Interestingly, because the ubiquitin E1 enzyme UBA1 is capable of charging NEDD8 (73), it is possible NEDD8 may substitute for ubiquitin as a covalent modifier of otherwise ubiquitinated substrates under conditions where the ubiquitin pool is depleted. For this reason, the finding that the K-GG peptide representing Lys-27-linked polyubiquitin was refractory to USP2 suggests two intriguing possibilities. Lys-27 is a fully conserved residue that plays a key role in maintaining structure of the ubiquitin fold. Future studies will hopefully reveal whether this form of polyubiquitin is indeed “invisible” to the deubiquitinating enzyme active site or rather that the Lys-27 polyubiquitin signature originates from a mixed chain of ubiquitin with either NEDD8 or ISG15.
Recent work has shown that a fraction of ubiquitinated peptides decrease in abundance after inhibition of the proteasome. These sites likely represent ubiquitination events that are not associated with proteasomal degradation. A subset of K-GG peptides might be expected to decrease in abundance because of degradation of their target substrates via the lysosomal pathway in response proteasome inhibition. Another critical subset of ubiquitinated substrates expected to display this pattern would be proteins that function as storage depots for cellular ubiquitin. Ubiquitinated histones have historically been considered as such and have been shown to release ubiquitin in response to stress (74). Recent proteomic studies in yeast indicate that approximately half of all conjugated ubiquitin is bound to substrates in the form of monoubiquitin or multiple monoubiquitin (75). In conjunction with current data, one hypothesis is that reserves of conjugated ubiquitin may be more widely distributed in cells than originally appreciated. In principle, distribution of the cellular ubiquitin pool would offer the cell flexibility in maintaining equilibrium under a wider array of conditions. Emphasizing the importance of dynamics between the free and conjugated ubiquitin pools, depletion of cellular ubiquitin is a key mediator of toxicity during inhibition of protein synthesis (76).
K-GG peptide enrichment will be critical to future work in defining the substrates of ubiquitin ligase and deubiquitinating enzymes. Two recent studies have taken advantage of immunoaffinity-based K-GG enrichment in this capacity, focusing on the quality control ligase HRD1 (61) and more broadly at cullin-RING ligases (60). In each case, K-GG enrichment studies were used in parallel with other methods to characterize substrates of each. HRD1 is a RING E3 ligase that functions in the endoplasmic reticulum-associated degradation pathway, and mutations in HRD1 have been associated with autoimmune disorders. Lee et al. (61) performed a SILAC comparison of control and HRD1 small interfering RNA-treated cells to compare ubiquitination events. HRD1 substrates were identified as those proteins whose peptides were decreased in abundance following ligase knockdown, either in K-GG-enriched samples or following multistep protein level enrichment of ubiquitinated substrates. In their K-GG immunoaffinity purification analyses, ∼1800 ubiquitination sites were identified on ∼900 proteins. Although the majority did not change upon HRD1 small interfering RNA, a series of putative substrates including several previously characterized proteins such as MHC, CD44, and integrins showed consistent decreases of ∼2-fold across three biological replicates. The two-pronged approach focusing on K-GG peptides and isolated ubiquitin conjugates in parallel is particularly appealing. As revealed in the study of HRD1, immunoaffinity enrichment of K-GG peptides provides a greater depth of sensitivity than protein level enrichment followed by in-gel digest. In contrast, analysis of isolated ubiquitin conjugates ensures that abundant, modified proteins are not overlooked because of ubiquitination in analytically challenging sequences that might not be well suited to either trypsin digestion or detection by the mass spectrometer. In ideal cases, complementary data were acquired through both methods as cross-validation.
In a study by Elledge and co-workers (60), K-GG enrichment was used to complement global protein stability profiling to examine substrates of the cullin-RING ligase family. As described by Kim et al. (57), these authors used the NAE1 inhibitor MLN4924 in a SILAC experiment to impair neddylation of and inactivate cullin E3 ligases. The results revealed that ∼10% of the ∼10,000 unique K-GG peptides decreased in abundance by at least 2-fold when neddylation was inhibited. Between biological replicates, >75% of these K-GG peptides were shown to decrease more than 2-fold in both. Global protein stability profiling provided an orthogonal validation strategy, reading out at the total protein level rather than at the level of ubiquitination. Particularly for rapidly turned over proteins, global protein stability profiling offers the opportunity to detect changes that would otherwise be overlooked if considering only ubiquitin-modified proteins or peptides.
Although proteasome inhibition stabilizes labile substrates and can improve detection, proteasome inhibitors themselves elicit biological changes that are worth considering when designing ubiquitination site mapping experiments. The cascade of events downstream of proteasome inhibition and consequent changes in transcription and translation may alter the substrates available to be modified, depending on the extent and duration of inhibition. Depletion of the free ubiquitin pool and the corresponding stress responses could likewise alter substrate prioritization by the ubiquitination and degradation machineries (75, 77). Because proteasome inhibition can trigger cell death programs, an additional consideration is the activation of ubiquitin ligases that regulate apoptosis. Although proteasome inhibitors increase the overall abundance of many K-GG peptides, it remains to be seen whether such increases affect sensitivity toward pathway-specific ubiquitination events of interest. The recent paper from Udeshi et al. (62) extends this one step further, assessing the effects of inhibiting deubiquination on the profile of ubiquitinated substrates. In that study, ∼4900 ubiquitin-modified peptides were quantified including a subset for which rapid deubiquitination appeared to be a key regulatory process. More work remains to be done to see whether these cellular treatments improve the ability to determine enzyme-substrate relationships using K-GG immunoaffinity enrichment.
Given the large number of uncharacterized ubiquitin ligases, it is conceivable that many ubiquitination events only occur following a specific biological stimulus. Such stimuli have the potential to alter the activity, specificity, and localization of enzymes within the ubiquitin system. An example of allosteric modulation occurs in plant growth and development with the SCFTIR1 ligase. The dual binding of the plant hormone auxin and inositol hexakisphosphate activates SCFTIR1, stimulating ligase activity by enhancing substrate binding to the TIR1 F-box protein (78). Similar observations have been made with regards to SCFFBXL5, a ligase in which the F-box component directly senses iron levels through its hemerythrin-like domain as a means of modulating its activity (79, 80). In the case of mammalian DNA damage signaling, recruitment of ubiquitin ligases such as RNF8, RNF168, and BRCA1 to sites of DNA damage is initiated through phosphorylation of H2AX (81). The activity of ubiquitin ligases toward their substrates may also be controlled via post-translational modification of the substrate, as in the case of the cellular oxygen sensor HIF1α by the VHL-Elongin BC-CUL2 ligase complex (82) or the prosurvival protein Mcl1 by the SCFFbw7 ligase (83, 84). Although K-GG immunoaffinity enrichment studies thus far have revealed important insights into the function of ubiquitin ligases in the basal state, connecting biological stimuli to E3 ligase activity and substrate targeting remains an area of significant interest.
Alkylating Reagents, C-terminal Lysines, and Pseudo K*GG Peptides
Iodoacetamide is commonly used in mass spectrometry proteomics to alkylate cysteine residues and create uniform, nonreactive populations of cysteine containing peptides for analysis. Previous work has also shown the potential for iodoacetamide to generate undesired modifications on amino acid side chains, as well as the N and C termini of peptides (85). The elemental composition of these modifications is C2H3N1O1, the same as the amino acid glycine. Because artifactual alkylation can occur twice on free amines such as the N terminus of a peptide or the side chain of lysine, it was recently reported that these modifications could be mistaken for diglycine remnants of ubiquitin (86). Previous work has shown that modification of free amines can be significant when peptides are incubated with iodoacetamide at high temperatures for extended periods of time (85). The use of chloroacetamide in place of iodoacetamide can limit, but does not eliminate, these artifactual 114.0429-Da lysine modifications. In consideration of these findings, detailed studies have been carried out using peptides from ubiquitin (24). This work has shown that for 1-h reactions at 37 °C in 50 mm iodoacetamide, ∼0.004% of ubiquitin peptides are adducted, although lower concentrations (10–30 mm) or lower temperatures (21 °C) decreased adducts to undetectable levels.
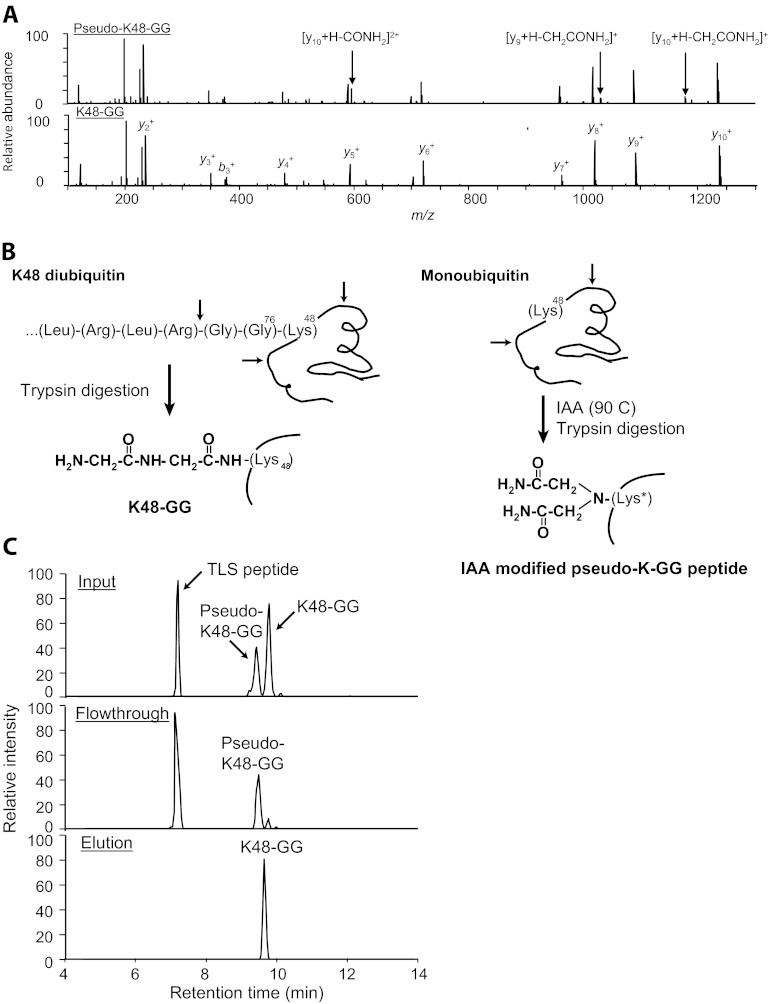
For existing data sets, a number of features can be used to identify the presence and determine the extent of acetamide adducts. As originally reported, bona fide K-GG peptides and doubly acetamide adducted “pseudo-K-GG” peptides are typically resolved by standard reversed phase HPLC gradients (86). For samples where enrichment of ubiquitin or the ubiquitin-modified substrate has been performed at the protein level, unmodified peptides from ubiquitin represent among the most abundant peptides in the sample. The absence of peak doublets for the predicted K-GG peptides of ubiquitin provides evidence against the presence of pseudo-K-GG peptides in the larger data set. Another analytical diagnostic is the presence of confidently scoring peptide spectral matches displaying pseudo-K-GG signatures on C-terminal lysines (87). Because trypsin cannot cleave at lysines modified by ubiquitin (88–90), a fully cleaved C-terminal K-GG signature may stem from post-digestion alkylation by iodoacetamide. We have found that ubiquitin peak doublets and C-terminal pseudo-K-GG peptides effectively diagnose samples that contain prevalent internal pseudo K-GG peptides. Interestingly, these more deceptive pseudo-K-GG peptides with the adduct on an internal lysine are themselves distinct from K-GG spectra, because they display a high frequency of −57 and −114 Da neutral losses, as seen the HCD MS/MS shown in Fig. 2A. This feature is uncommon among bona fide ubiquitinated peptides. Recent data have further shown that K-GG peptides are enriched >1000-fold relative to adducted peptides during immunoaffinity purification (Fig. 2). While in depth inspection of chromatographic and spectral data is paramount, careful application of reduction and alkylation procedures and the post-analysis diagnostic checks make the continued use of iodoacetamide a viable option for mapping ubiquitination sites.
Fig. 2.
Selectivity of K-ε-GG antibody against pseudo-K-GG peptides (specificity of anti-K-GG antibody with Lys-48-GG peptides). A, HCD MS/MS spectra of pseudo-Lys-48-GG peptide are highly similar to that of bona fide Lys-48-GG peptide, except that the pseudo-Lys-48-GG peptide has several neutral loss peaks as indicated. B, preparation of Lys-48-GG and pseudo-K-GG peptides for characterization in anti-K-GG immunoaffinity enrichment. Lys-48-linked diubiquitin was digested with trypsin to produce the Lys-48-GG peptide. Pseudo-K-GG peptide at Lys-48 (and other lysines of ubiquitin) was generated by incubating monoubiquitin with iodoacetamide at high temperature (90 °C), followed by trypsin digestion. C, digested Lys-48 diubiquitin was mixed with digested monoubiquitin containing pseudo-K-GG peptides and incubated with K-ε-GG antibody affinity beads. After washing, associated peptides were eluted and characterized by LC-MS. Extracted ion chromatograms of three Ub peptides are shown (±0.01 m/z) in input (top panel), flow through (middle panel), and elution following antibody affinity purification (bottom panel). The monoisotopic m/z of these ions are 541.2798 (S57 peptide, TLSDYNIQK, +2) and 730.8964 for both pseudo-Lys-48-GG and Lys-48-GG (LIFAGK(GG)QLEDGR, +2).
Although current database search algorithms efficiently match post-translationally modified peptides, work on phosphorylation from many groups has shown that they fall short when it comes to determining the precise position of the modification within the peptide (91). One benefit of mapping ubiquitination sites in trypsin-digested samples, over modifications such as phosphorylation, is that the frequency of internal lysine residues is relatively low. Nonetheless, the mislocalization of diglycine signatures to C-terminal lysine residues still occurs when using standard search algorithms. Along with the recently published K-GG data sets, the field has seen the extension of tools, originally designed to localize phosphorylation modifications based on site-determining ions, for use with K-GG-modified peptides (57, 58). Even in the absence of a corrective tool, we routinely notice that peptide spectral matches proposed to carry C-terminal K-GG signatures commonly have an unmodified lysine residue within several positions of the C terminus, permitting efficient manual relocalization. To minimize false positive identifications where the reported peptide displays a K-GG signature on the C-terminal lysine, these peptide spectral matches have customarily been omitted from final lists of modified peptides. Because the presence of peptides with C-terminal K-GG signatures within a data set can be diagnostic and informative, we would instead argue in favor of marking and retaining these putative false positives within systematically filtered data sets to permit post hoc data assessment. An additional consideration is that in the rare cases when a protein sequence ends in a C-terminal lysine that is modified by ubiquitin, discarding these K-GG peptides would be inappropriate.
Along with the emergence of large scale K-GG data sets has come the need for robust and efficient methods of validating substrate-specific ubiquitination events. Although the reliance on automated tools for peptide spectral matching and site localization will remain for some time, interaction with the MS and chromatographic results remain essential to identifying systematic issues at the raw data level. Moving into the future, spectral libraries of validated K-GG-modified peptides, along with their relative retention times, would be a valuable resource to biology focused groups that will study individual substrates in detail. Likewise, isotopically labeled internal standard peptides that can be used to assay critical ubiquitination events would be valuable, as they have been in characterizing polyubiquitin linkages (21, 23). Methods that combine site-specific elimination of ubiquitin-modified lysines by mutagenesis, with focused, multiplexed mass spectrometry assays that read out the modified and unmodified forms of a protein should be expected to emerge as a means of assessing the temporal dimension of substrate-specific ubiquitination. Experimental systems capable of high throughput expression and enrichment method will likewise be valuable, as they begin to provide both Western blot and mass spectrometry validation of fleeting ubiquitination events demonstrated in other systems. Ultimately, however, orthogonal validation methods and functional readouts in physiologically relevant systems remain the gold standard. The multipronged strategy utilized to define HRD1 (61) substrates and the orthogonal functional validation of SCF substrates (60) provide a template that can be replicated moving into the future, whereas the growing availability of genetically engineered mouse models promises to open the door to true in vivo exploration.
Current Challenges in Ubiquitination Site Mapping
While ubiquitination site mapping by LC-MS/MS and database searching has revealed thousands of modification sites, bottom-up approaches have inherent limitations. By their nature, the proteolysis event that generates the branched K-GG signature peptide effectively destroys information about which polyubiquitin linkages were attached at the substrate modification site. Similarly, it is difficult to ascertain whether ubiquitination events at nonadjacent lysines co-exist or represent modifications on separate members of a population. Methods that permit analysis of concurrent modifications on a single protein molecule will open the door to connecting functions with combinatorial modification states (93). Middle-down approaches with partial proteolysis have shown potential with regards to characterizing intact polyubiquitin chains (25), although throughput has currently limited widespread application. Extension of these and top-down methods capable of distinguishing isoforms will be essential to determining the extent to which a “ubiquitin code” directs cellular processes.
A complementary approach involves serial application of linkage specific enrichment at the protein level, followed by K-GG peptide immunoaffinity enrichment at the peptide level. Such studies have thus far been limited by the amounts of protein captured during high stringency linkage-specific immunoprecipitation and by the observation that an unexpectedly large fraction of substrates may be modified by more than a single polyubiquitin linkage (17, 21). Nonetheless, further improvements in enrichment methods and mass spectrometry will better enable researchers to probe these complex ubiquitin signals.
Another challenge associated with bottom-up mapping of ubiquitination sites is that many occur within analytically inaccessible sequences. Dual digestion methods where ubiquitinated proteins are serially digested, first with trypsin or ArgC to generate the K-GG signature and subsequently by an alternative enzyme like GluC, chymotrypsin, or pepsin, may be useful for mapping ubiquitination sites in regions of sequence where lysines and arginines are sparse. Because generation of the K-GG signature requires cleavage C-terminal to an arginine residue, modifications residing in highly basic regions are also inherently insensitive to trypsin-based mapping. Even when generated, the chromatographic profiles and irregular MS/MS spectra for highly basic peptides can pose additional challenges. Chemical modification methods that block lysines and convert trypsin into an arginine-specific protease hold potential for revealing modifications in these situations, as do second generation antibodies with the ability to recognize alternative ubiquitin remnants longer than the K-GG signature. Such antibodies, along with alternative digestion methods, would be useful in dispelling ambiguity between ubiquitin, NEDD8, and ISG15, while making it possible to systematically investigate other ubiquitin-like modifiers such as SUMO1/2/3.
Unexpected post-translational modifications pose a particular challenge in the context of matching K-GG peptides by traditional database search approaches. It remains unclear how frequently post-translational modifications within a protein sequence trigger ubiquitination of nearby lysine residues such that tryptic peptides contain both modifications. The existence of phosphodegrons (94) and the role of the ubiquitin-proteasome system in degrading oxidatively damaged proteins (95) suggest that certain ubiquitination events may only be identified if the correct concurrent modification is considered. Error tolerant or wildcard style searches have thus far provided only a limited solution. Application of automated, iterative search algorithms and advanced tools for discerning true positives from false positive matches in such searches should provide improvements in this area.
An area of acute interest within the ubiquitin field lies in the investigation of non-lysine ubiquitination. Multiple reports have described ubiquitination on residues such as cysteine, serine, and threonine (6, 7, 96, 97), although to date, there remains a dearth of mass spectral data to support the existence of ester- and thioester-linked ubiquitination. In the case of cysteine-linked thioester ubiquitin, the basic biochemistry of the ubiquitin system provides a strong mechanistic precedent. HECT and IBR/RBR family E3s are modified on cysteine residues as part of their biological activities, mandating that E2 enzymes have the capacity to transfer ubiquitin to certain cysteine residues. The inherent instability of cysteine-linked thioesters has been a limitation in structural studies (98) and likely explains the difficulty in detecting them by mass spectrometry.
Consensus is less clear for ester-linked ubiquitination through serine or threonine, because focused efforts toward mapping ester-linked -GG peptides have been undertaken by several groups without definitive resolution (97). It remains possible that ester-linked diglycine is unstable under standard LC-MS conditions or that the exact nature of this bond remains to be elucidated. Arguing against analytical instability, we have observed Ser-GG peptides from a ubiquitin charged E2 variant (C → S) under standard LC-MS conditions.2 Database searches for Ser/Thr-GG peptides in single protein ubiquitination analyses and among immunoaffinity enriched K-GG peptides have not yielded evidence to support the existence of ester-linked ubiquitin remnants. Definitive mass spectrometry evidence showing the biological nature of interaction between ubiquitin and Ser/Thr will be essential to establishing the significance of this modification in cellular regulation.
Quantitative analysis of specific ubiquitination sites has been performed using both metabolic labeling and isotopically labeled internal standards. With growing interest in multiplexing approaches, chemical labeling of K-GG peptides also has potential for dose-response and temporal profiling studies (99). One current challenge is that labeling must be performed following K-GG enrichment. The amount of starting material required to identify ubiquitination sites by K-GG enrichment is beyond the practical scale of iTRAQ or TMT labeling methods. Optimization studies have shown that for mouse brain lysate, the number of unique K-GG peptides identified correlates with input material up to ∼40 mg.2 Even if peptides were labeled prior to enrichment, modification of the N terminus of the K-GG signature itself by an amine reactive multiplexing reagent would interfere with immunoaffinity enrichment. Preliminary studies involving post-enrichment labeling have been successful by coupling carefully controlled sample handling and internal standards that report on IP efficiency across samples. It is unclear yet whether recent improvements in mass spectrometry methods for performing multiplexed chemical labeling (100, 101) will be sensitive enough for analysis of enriched K-GG peptides. Future advances in K-GG peptide enrichment and multiplexed quantitation hold great promise for characterization of ubiquitination in vivo where the need for replicate measurements is a paramount consideration.
While the published work using K-GG immunoaffinity enrichment has focused on identifying ubiquitination events globally from whole cell lysates, it seems likely that these methods may have additional utility in the analysis of a single proteins purified from eukaryotic cells. Identification of ubiquitination sites on individual substrates remains a challenge, particularly for proteins that are large in size or are substoichiometrically modified on many lysines. For these studies, two main strategies seem to hold promise. Performing substrate-specific enrichment by immunoprecipitation prior to enriching K-GG peptides is a viable strategy, particularly when the substrate is of relatively low abundance compared with other cellular targets of ubiquitin. For cells overexpressing a putative substrate, another approach might involve using targeted MS methods such as multiple reaction monitoring-initiated detection and sequencing workflow (MIDAS) (92) or inclusion list approaches to focus specifically on ions representative of K-GG peptides from the substrate. In this approach, pre-enrichment of the target would not necessarily be a prerequisite to K-GG immunoaffinity enrichment. Replacing in gel separation with K-GG immunoaffinity enrichment might be expected to increase sensitivity toward individual K-GG peptides by minimizing the losses stemming from post-digest extraction, avoiding dilution of individual peptides across multiple gel regions, and eliminating competition between modified and unmodified peptides in data-dependent MS analysis.
The extent to which K-GG immunoaffinity reagents can be used to identify low level substrate peptides from limited sample amounts remains uncharacterized. The presence of abundant polyubiquitin species represents a challenge, as these species can suppress ionization of lower abundance K-GG peptides. Although preclearing abundant K-GG peptides of polyubiquitin and histones might be possible, it seems more likely that the number of possible species may preclude establishment of a sufficiently comprehensive depletion reagent. Using linkage-specific antibodies for protein level depletion would seem particularly suboptimal, as this would altogether remove the ubiquitinated substrates covalently attached to polyubiquitin chains. Instead, fractionation of K-GG peptides before or after immunoaffinity enrichment seems the likely path forward for characterization of low abundance species (58, 62). The existing data suggest that it is now possible to detect ubiquitination events on important regulatory proteins, albeit from large amounts of starting material. Future work will focus on pushing the limits of detection and utilizing these new methods to understand the ever expanding roles of ubiquitin.
Acknowledgments
We thank L. Phu and members of the Microchemistry and Proteomics Lab, as well as S. Gygi, W. Harper, W. Kim, E. Bennett, and C. Choudhary for thoughtful discussions.
Footnotes
* This work was supported in part by National Institutes of Health Grant RR025822 and funds from the American Lebanese Syrian Associated Charities (to J. P.).
2 D. Bustos and D. Kirkpatrick, unpublished data.
1 The abbreviations used are:
- E3
- ubiquitin-protein ligase
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- SILAC
- stable isotope labeling of amino acids in cell culture.
REFERENCES
- 1. Finley D., Ciechanover A., Varshavsky A. (2004) Ubiquitin as a central cellular regulator. Cell 116, S29–S32 [DOI] [PubMed] [Google Scholar]
- 2. Herrmann J., Lerman L. O., Lerman A. (2007) Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 100, 1276–1291 [DOI] [PubMed] [Google Scholar]
- 3. Pearce M. J., Mintseris J., Ferreyra J., Gygi S. P., Darwin K. H. (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 5. Ciechanover A., Ben-Saadon R. (2004) N-terminal ubiquitination: More protein substrates join in. Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 6. Cadwell K., Coscoy L. (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 [DOI] [PubMed] [Google Scholar]
- 7. Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., Hansen T. H. (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Herr R. A., Hansen T. H. (2012) Ubiquitination of substrates by esterification. Traffic 13, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behrends C., Harper J. W. (2011) Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18, 520–528 [DOI] [PubMed] [Google Scholar]
- 10. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: New molecular signals. “Protein Modifications: Beyond the Usual Suspects” review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofmann R. M., Pickart C. M. (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276, 27936–27943 [DOI] [PubMed] [Google Scholar]
- 12. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 14. Finley D., Sadis S., Monia B. P., Boucher P., Ecker D. J., Crooke S. T., Chau V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14, 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu M., Skaug B., Zeng W., Chen Z. J. (2009) A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol. Cell 36, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsumoto M. L., Wickliffe K. E., Dong K. C., Yu C., Bosanac I., Bustos D., Phu L., Kirkpatrick D. S., Hymowitz S. G., Rape M., Kelley R. F., Dixit V. M. (2010) K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 39, 477–484 [DOI] [PubMed] [Google Scholar]
- 17. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 18. Wang H., Matsuzawa A., Brown S. A., Zhou J., Guy C. S., Tseng P. H., Forbes K., Nicholson T. P., Sheppard P. W., Häcker H., Karin M., Vignali D. A. (2008) Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. Proc. Natl. Acad. Sci. U.S.A. 105, 20197–20202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsumoto M. L., Dong K. C., Yu C., Phu L., Gao X., Hannoush R. N., Hymowitz S. G., Kirkpatrick D. S., Dixit V. M., Kelley R. F. (2012) Engineering and structural characterization of a linear polyubiquitin-specific antibody. J. Mol. Biol. 418, 134–144 [DOI] [PubMed] [Google Scholar]
- 20. Kaiser S. E., Riley B. E., Shaler T. A., Trevino R. S., Becker C. H., Schulman H., Kopito R. R. (2011) Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 8, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Phu L., Izrael-Tomasevic A., Matsumoto M. L., Bustos D., Dynek J. N., Fedorova A. V., Bakalarski C. E., Arnott D., Deshayes K., Dixit V. M., Kelley R. F., Vucic D., Kirkpatrick D. S. (2011) Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell. Proteomics 10.1074/mcp.M110.003756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 23. Kirkpatrick D. S., Hathaway N. A., Hanna J., Elsasser S., Rush J., Finley D., King R. W., Gygi S. P. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 [DOI] [PubMed] [Google Scholar]
- 24. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu P., Peng J. (2008) Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal. Chem. 80, 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu P., Cheng D., Duong D. M., Rush J., Roelofs J., Finley D., Peng J. (2006) A proteomic strategy for quantifying polyubiquitin chain topologies. Isr. J. Chem. 46, 171–182 [Google Scholar]
- 27. Goldknopf I. L., Busch H. (1977) Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U.S.A. 74, 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hershko A., Ciechanover A., Heller H., Haas A. L., Rose I. A. (1980) Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. U.S.A. 77, 1783–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ciechanover A., Finley D., Varshavsky A. (1984) The ubiquitin-mediated proteolytic pathway and mechanisms of energy-dependent intracellular protein degradation. J. Cell. Biochem. 24, 27–53 [DOI] [PubMed] [Google Scholar]
- 30. Finley D., Ciechanover A., Varshavsky A. (1984) Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell 37, 43–55 [DOI] [PubMed] [Google Scholar]
- 31. Wilkinson K. D., Urban M. K., Haas A. L. (1980) Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 255, 7529–7532 [PubMed] [Google Scholar]
- 32. Ciechanover A., Finley D., Varshavsky A. (1984) Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 37, 57–66 [DOI] [PubMed] [Google Scholar]
- 33. King R. W., Glotzer M., Kirschner M. W. (1996) Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell 7, 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scherer D. C., Brockman J. A., Chen Z., Maniatis T., Ballard D. W. (1995) Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 92, 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang F., Goh L. K., Sorkin A. (2007) EGF receptor ubiquitination is not necessary for its internalization. Proc. Natl. Acad. Sci. U.S.A. 104, 16904–16909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 37. Jin L., Pahuja K. B., Wickliffe K. E., Gorur A., Baumgärtel C., Schekman R., Rape M. (2012) Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 39. Flick K., Ouni I., Wohlschlegel J. A., Capati C., McDonald W. H., Yates J. R., Kaiser P. (2004) Proteolysis-independent regulation of the transcription factor Met4 by a single Lys-48-linked ubiquitin chain. Nat. Cell Biol. 6, 634–641 [DOI] [PubMed] [Google Scholar]
- 40. Peng J., Gygi S. P. (2001) Proteomics: The move to mixtures. J. Mass Spectrom. 36, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 41. Marotti L. A., Jr., Newitt R., Wang Y., Aebersold R., Dohlman H. G. (2002) Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry 41, 5067–5074 [DOI] [PubMed] [Google Scholar]
- 42. Sliter D. A., Kubota K., Kirkpatrick D. S., Alzayady K. J., Gygi S. P., Wojcikiewicz R. J. (2008) Mass spectrometric analysis of type 1 inositol 1,4,5-trisphosphate receptor ubiquitination. J. Biol. Chem. 283, 35319–35328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miranda M., Wu C. C., Sorkina T., Korstjens D. R., Sorkin A. (2005) Enhanced ubiquitylation and accelerated degradation of the dopamine transporter mediated by protein kinase C. J. Biol. Chem. 280, 35617–35624 [DOI] [PubMed] [Google Scholar]
- 44. Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 45. Kleijnen M. F., Kirkpatrick D. S., Gygi S. P. (2007) The ubiquitin-proteasome system regulates membrane fusion of yeast vacuoles. EMBO J. 26, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cripps D., Thomas S. N., Jeng Y., Yang F., Davies P., Yang A. J. (2006) Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 281, 10825–10838 [DOI] [PubMed] [Google Scholar]
- 47. Rott R., Szargel R., Haskin J., Shani V., Shainskaya A., Manov I., Liani E., Avraham E., Engelender S. (2008) Monoubiquitylation of α-synuclein by seven in absentia homolog (SIAH) promotes its aggregation in dopaminergic cells. J. Biol. Chem. 283, 3316–3328 [DOI] [PubMed] [Google Scholar]
- 48. Scaglione K. M., Zavodszky E., Todi S. V., Patury S., Xu P., Rodríguez-Lebrón E., Fischer S., Konen J., Djarmati A., Peng J., Gestwicki J. E., Paulson H. L. (2011) Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 43, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Isasa M., Katz E. J., Kim W., Yugo V., González S., Kirkpatrick D. S., Thomson T. M., Finley D., Gygi S. P., Crosas B. (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 38, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirkpatrick D. S., Weldon S. F., Tsaprailis G., Liebler D. C., Gandolfi A. J. (2005) Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics 5, 2104–2111 [DOI] [PubMed] [Google Scholar]
- 51. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: Application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 52. Meierhofer D., Wang X., Huang L., Kaiser P. (2008) Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J. Proteome Res. 7, 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10.1074/mcp.M110.003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M. S. (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi Y., Chan D. W., Jung S. Y., Malovannaya A., Wang Y., Qin J. (2011) A data set of human endogenous protein ubiquitination sites. Mol. Cell. Proteomics 10.1074/mcp.M110.002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peng J., Cheng D. (2005) Proteomic analysis of ubiquitin conjugates in yeast. Methods Enzymol. 399, 367–381 [DOI] [PubMed] [Google Scholar]
- 57. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10.1074/mcp.M111 013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu G., Paige J. S., Jaffrey S. R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee K. A., Hammerle L. P., Andrews P. S., Stokes M. P., Mustelin T., Silva J. C., Black R. A., Doedens J. R. (2011) Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J. Biol. Chem. 286, 41530–41538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Udeshi N. D., Mani D. R., Eisenhaure T., Mertins P., Jaffe J. D., Clauser K. R., Hacohen N., Carr S. A. (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Starita L. M., Lo R. S., Eng J. K., von Haller P. D., Fields S. (2012) Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics 12, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 65. Kirkpatrick D. S., Denison C., Gerber S. A., Rush J., Gygi S. P. (2004) A novel method for identification of protein ubiquitination sites in yeast by peptide immunoprecipitation and tandem mass spectrometry, 52nd ASMS Conference on Mass Spectrometry and Allied Topics, Peptides: Post Translational Modifications, Abstract 281 [Google Scholar]
- 66. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 67. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 68. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 70. Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., Yao T. P. (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jin Y. H., Jeon E. J., Li Q. L., Lee Y. H., Choi J. K., Kim W. J., Lee K. Y., Bae S. C. (2004) Transforming growth factor-β stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J. Biol. Chem. 279, 29409–29417 [DOI] [PubMed] [Google Scholar]
- 72. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 73. Whitby F. G., Xia G., Pickart C. M., Hill C. P. (1998) Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273, 34983–34991 [DOI] [PubMed] [Google Scholar]
- 74. Mimnaugh E. G., Chen H. Y., Davie J. R., Celis J. E., Neckers L. (1997) Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: Effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36, 14418–14429 [DOI] [PubMed] [Google Scholar]
- 75. Ziv I., Matiuhin Y., Kirkpatrick D. S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S. P., Haguenauer-Tsapis R., Reis N., Glickman M. H., Kleifeld O. (2011) A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell. Proteomics 10.1074/mcp.M111.009753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hanna J., Leggett D. S., Finley D. (2003) Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hanna J., Meides A., Zhang D. P., Finley D. (2007) A ubiquitin stress response induces altered proteasome composition. Cell 129, 747–759 [DOI] [PubMed] [Google Scholar]
- 78. Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 79. Salahudeen A. A., Thompson J. W., Ruiz J. C., Ma H. W., Kinch L. N., Li Q., Grishin N. V., Bruick R. K. (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vashisht A. A., Zumbrennen K. B., Huang X., Powers D. N., Durazo A., Sun D., Bhaskaran N., Persson A., Uhlen M., Sangfelt O., Spruck C., Leibold E. A., Wohlschlegel J. A. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huen M. S., Sy S. M., Chen J. (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 83. Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R. S., Zhai B., Wan L., Gutierrez A., Lau A. W., Xiao Y., Christie A. L., Aster J., Settleman J., Gygi S. P., Kung A. L., Look T., Nakayama K. I., DePinho R. A., Wei W. (2011) SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471, 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wertz I. E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D. J., Helgason E., Ernst J. A., Eby M., Liu J., Belmont L. D., Kaminker J. S., O'Rourke K. M., Pujara K., Kohli P. B., Johnson A. R., Chiu M. L., Lill J. R., Jackson P. K., Fairbrother W. J., Seshagiri S., Ludlam M. J., Leong K. G., Dueber E. C., Maecker H., Huang D. C., Dixit V. M. (2011) Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471, 110–114 [DOI] [PubMed] [Google Scholar]
- 85. Yang Z., Attygalle A. B. (2007) LC/MS characterization of undesired products formed during iodoacetamide derivatization of sulfhydryl groups of peptides. J. Mass Spectrom. 42, 233–243 [DOI] [PubMed] [Google Scholar]
- 86. Nielsen M. L., Vermeulen M., Bonaldi T., Cox J., Moroder L., Mann M. (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 [DOI] [PubMed] [Google Scholar]
- 87. Denis N. J., Vasilescu J., Lambert J. P., Smith J. C., Figeys D. (2007) Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics 7, 868–874 [DOI] [PubMed] [Google Scholar]
- 88. Kirkpatrick D., Haas W., Gygi S. (2008) Dissecting Ubiquitin Modifications for Analysis by Mass Spectrometry, Transworld Research Network, Kerala, India [Google Scholar]
- 89. Seyfried N. T., Xu P., Duong D. M., Cheng D., Hanfelt J., Peng J. (2008) Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 80, 4161–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shi Y., Xu P., Qin J. (2011) Ubiquitinated proteome: Ready for global? Mol. Cell. Proteomics 10.1074/mcp.R110.006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 92. Mollah S., Wertz I. E., Phung Q., Arnott D., Dixit V. M., Lill J. R. (2007) Targeted mass spectrometric strategy for global mapping of ubiquitination on proteins. Rapid Commun. Mass Spectrom. 21, 3357–3364 [DOI] [PubMed] [Google Scholar]
- 93. Hunter T. (2007) The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 94. Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. (2003) Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 [DOI] [PubMed] [Google Scholar]
- 95. Jung T., Grune T. (2008) The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 60, 743–752 [DOI] [PubMed] [Google Scholar]
- 96. Wang X., Herr R. A., Rabelink M., Hoeben R. C., Wiertz E. J., Hansen T. H. (2009) Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol. 187, 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shimizu Y., Okuda-Shimizu Y., Hendershot L. M. (2010) Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 40, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 99. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 100. Ting L., Rad R., Gygi S. P., Haas W. (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wenger C. D., Lee M. V., Hebert A. S., McAlister G. C., Phanstiel D. H., Westphall M. S., Coon J. J. (2011) Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 8, 933–935 [DOI] [PMC free article] [PubMed] [Google Scholar]