Abstract
The application of mass spectrometry (MS) to the analysis of proteomes has enabled the high-throughput identification and abundance measurement of hundreds to thousands of proteins per experiment. However, the formidable informatics challenge associated with analyzing MS data has required a wide variety of data file formats to encode the complex data types associated with MS workflows. These formats encompass the encoding of input instruction for instruments, output products of the instruments, and several levels of information and results used by and produced by the informatics analysis tools. A brief overview of the most common file formats in use today is presented here, along with a discussion of related topics.
Mass spectrometry (MS) has accelerated the field of proteomics by enabling the high-throughput identification and abundance measurement of hundreds to thousands of proteins per experiment (1). The most common workflow for tandem mass spectrometry (MS/MS) (2) generally begins with the isolation of proteins from an original sample, digestion of the proteins into peptides with an enzyme such as trypsin, and separation of the peptides into multiple fractions to reduce the complexity in each fraction. Each fraction is then subjected to liquid chromatography (LC), ionized, and injected into the mass spectrometer. Individual species of peptide ions are isolated and fragmented to generate a fragment ion spectrum which may then be identified via software.
In nearly all of these high-throughput workflows, extensive analysis with software is required in order to translate the mass spectra into peptide identifications and perform abundance measurements (3). There are a wide variety of software tools available to assist with this analysis, including open-source software as well as proprietary and commercial products. As a result of efforts to enable the movement of complex data types among analysis tools and the sharing of data and results with others in the community (4), a wide variety of data formats have emerged. These formats may be broadly separated into open formats and proprietary formats. Open formats enable improved data sharing by allowing the data to be read by a variety of software tools without licensing restrictions. Open formats can be further separated into three categories: official standards, de facto standards, and other formats. Official standards are approved by a standards body, typically after a formal process of review and refinement, whereas de facto standards lack any official approval but are widely used by a large number of software tools and generally accepted as being a preferred mechanism of data exchange. Formats not falling into either of these two categories are simply referred to as other formats in this review.
Each of the major instrument vendors uses its own proprietary formats, continually updating the formats to support new features of their instruments. Open formats are generally created by the developers of analysis software and databases in order to enable the exchange of data between tools. Some formats have been developed by a single lab and are oriented around that lab's software, whereas other formats have emerged after a long process of collaborative development by a diverse group of contributors, often under the organization of a standards development group. The largest and most active standards development group in MS proteomics is the Human Proteome Organization (HUPO)1 Proteomics Standards Initiative (PSI) (5). The PSI aims to bring together representatives from commercial instrument manufacturers, software vendors, journal editors, and academic software developers and users to create common exchange formats and minimum information specifications that are then rigorously reviewed and approved as PSI standards.
Here we present an overview of the formats in common use in MS proteomics by popular software tools. The many formats cannot be described in great detail, but they are described very briefly, and relevant references or URLs are provided. Of course, many less common formats, especially simple tab-separated-value (TSV) formats of endless variety, cannot be covered here. Following the overview of all the formats covered here, a brief discussion of topics related to the application and evolution of these formats is presented.
Overview of Formats
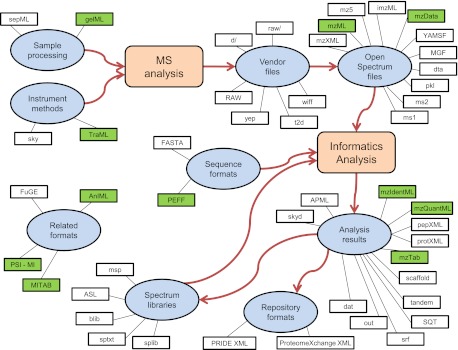
There are a large number of commonly used formats in MS proteomics at all stages of data analysis. Fig. 1 depicts an overview of all the formats discussed here, crudely organized by their niche in typical proteomics workflows. From the upper left and proceeding clockwise are formats in use to describe information relevant to sample preparation, mass spectrometer input, mass spectrometer output, software analysis results, and finally formats for repository submission and spectral libraries. Each of these formats is introduced briefly in this order later in the paper. A listing of the major software tools and libraries that implement many of these formats is provided in Table I.
Fig. 1.
Overview graph of the mass spectrometry proteomics formats discussed here. The overall workflow of MS proteomics is depicted by the large shapes and the arrows connecting them. Ovals represent the major data types within the workflow. The small rectangles represent the individual file formats associated by an edge to their general data type. Shaded formats are officially approved or soon-to-be-approved standards. Different formats associated with the same data type are not necessarily redundant or equivalent.
Table I. List of some of the most prominent software tools and libraries and the formats they support.
Most search engines, which all support a variety of formats, are not included.
| Tool | Formats | Reference |
|---|---|---|
| ProteoWizard | mzML, TraML, mzIdentML, mzXML, vendor formats | (53) |
| OpenMS | mzML, TraML, mzIdentML, mzData, mzQuantML, et al. | (14) |
| Trans-Proteomic Pipeline (TPP) | mzML, mzXML, pepXML, protXML (ProteoWizard) | (30, 55) |
| compomics-utilities | MSF, tandem, mzML, omx, dat, FASTA | (56) |
| jmzReader | mzML, mzXML, mzData, PRIDE XML, dta, MGF, ms2, pkl | (57) |
| jTraML | TraML | (13) |
| multiplierz | Vendor formats | (58) |
| PEFF Viewer | PEFF | |
| PRIDE Converter 2 | mzTab, PRIDE XML (jmzReader) | (47) |
| Mascot & Distiller | MGF, mzML, mzXML, mzIdentML, vendor formats | |
| SpectraST | msp, splib, blib, ASF, mzML, mzXML, pepXML, etc. | (42) |
| ProHits | PSI-MI (TPP formats) | (50) |
| Anubis | TraML, mzML, mzXML | (11) |
| Proteios | TraML, mzML, mzXML | (32) |
| Skyline | .sky, .skyd, mzML, mzXML, vendor formats | (16) |
| ATAQS | TraML, mzML, mzXML | (12) |
| Corra | APML, mzXML | (37) |
| Java MIAPE API | PRIDE XML, mzML, mzIdentML, GelML | (20) |
Pre-Mass Spectrometry Formats
There are relatively few common formats (i.e. those not specific to one vendor) for information specifically focused on information prior to MS. There are broadly two categories of such formats: one that describes sample handling, and one that contains formats for mass spectrometer target input.
The PSI has developed a pair of formats for pre-MS sample handling, gelML (6) and sepML (7). The gelML standard format is specifically designed to encode information related to one-dimensional and two-dimensional gel electrophoresis prior to MS. It can encode basic sample origin information, how the gel is prepared, and details about how the gel bands or spots are excised from the gels. The two-dimensional gel spots are annotated with coordinates, shapes, density, and identifier information that can be referenced later in the MS. As with most PSI formats, gelML is designed to work with an accompanying controlled vocabulary (CV) called PSI-SEP. The CV ensures that a concept is referred to by its accession number (e.g. SEP:0021) for its term in the vocabulary, which is accompanied by a clear definition and synonyms, rather than by any number of variously spelled names (e.g. reversed-phase chromatography, reverse phase liquid chromatography, RPC, RP-HPLC, etc.).
The sepML format was developed to describe other kinds of separation methods prior to MS, such as strong cation exchange, free-flow electrophoresis, and reversed-phase LC. There are many attributes associated with such components of a proteomics workflow, and they can be neatly encoded with the help of CV terms, so that the meaning of such metadata is clear and will be encoded uniformly. The sepML format is coupled with the PSI-SEP CV. However, unlike most of the formats described herein, virtually no tools using sepML have emerged.
A few other very general formats are not specific to pre-MS sample processing metadata but do include components to encode at least some sample information. One example is the Functional Genomics Experiment (FuGE) (8) format, which attempts to provide components for information common to most experiments, as well as a framework for building components specific to a particular technology. FuGE is described in more detail below.
The other common type of pre-MS file format is for encoding acquisition information for the mass spectrometer. Most such formats are commonly referred to as “method files” and contain information describing exactly how the mass spectrometer will acquire data in a run. This can include both data-dependent acquisition modes and targeting instructions for selected reaction monitoring (SRM) or instructions for data-independent acquisition methods. Each vendor has defined its own format, or even multiple formats, for its method files, but these are coupled to the vendor's acquisition software, are not generally applicable to another vendor's instrument, and are thus not described further here.
However, among the mass spectrometer input formats, an open format for encoding SRM transitions has been gaining significant use. SRM transitions are the signatures needed to target specific peptide ions during an SRM experiment. It is a significant informatics challenge to select optimum transitions, and it is therefore of great value to share transitions once they have been optimized and successfully used in an experiment (9). The PSI has recently released the Transitions Markup Language (TraML) format (10). TraML can encode both simple transition lists and complex annotations of the optimization attributes associated with each transition. TraML has been designed to be applicable not just to proteomics, but also to metabolomics or other fields that aim to target nonpeptidic compounds. TraML can also encode inclusion lists, a simpler form of targeting, because they are conceptually quite similar to SRM target lists. TraML is not yet supported by instrument vendor software, although most vendors have affirmed a plan to support it. Several open-source packages such as Anubis (11), ATAQS (12), jTraML (13), OpenMS (14), PeptideAtlas (9), and Proteios (15) support TraML. The Skyline software program (16) has become a popular desktop application for the design and analysis of SRM experiments, and its open, XML-based format for transitions, the .sky format, although not an official standard like TraML, has become a common way to share transitions with other Skyline users.
MS Output Files
The most diverse group of file types is for encoding the mass spectra that are the result of MS runs. This group of formats may be broadly separated into three subgroups: proprietary vendor formats, complex open formats, and simple text formats.
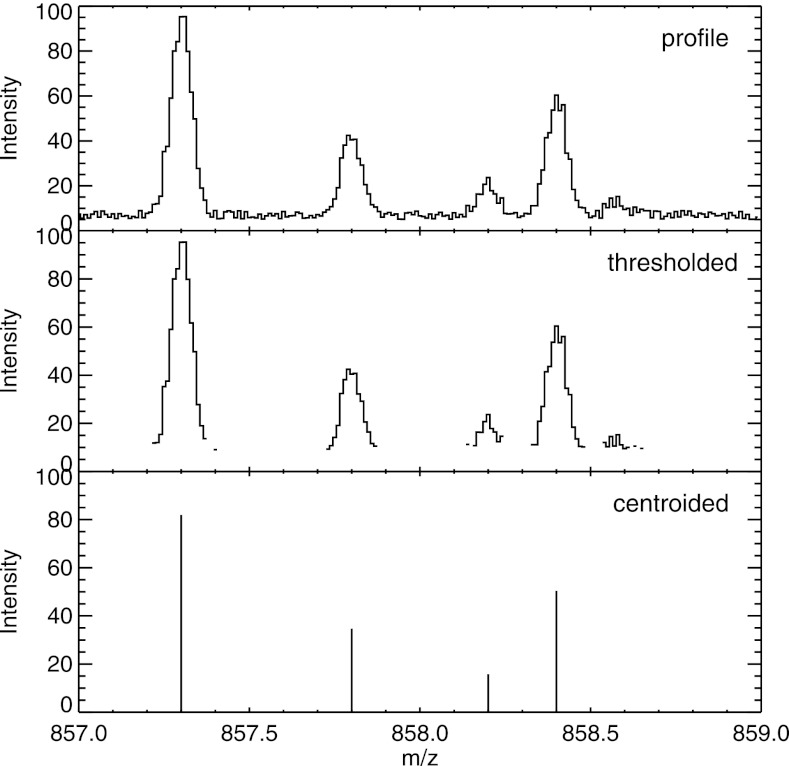
The mass spectra themselves can commonly be represented in two major ways: as “profile-mode” (also called “continuous”) spectra and as “peak lists” (also called “centroided” or “peak-picked”). Profile-mode spectra are encoded as regularly spaced data points such that individual ion features are sampled at a frequency higher than the instrument resolution so that each peak has a measureable shape. Note that the bin size may change as a function of m/z, as the original evenly spaced sampling may be in the time or frequency domain. A space-saving variant of this is sometimes called “thresholded,” in which all data points below the threshold are simply omitted. This allows full resolution in regions of significant signal and reduces the space required to encode below-threshold noise. Once an algorithm has been applied to a profile-mode spectrum to extract only the detectable peaks, a spectrum represented as the list of m/z and intensity pairs of the peaks is called “centroided.” All instruments natively collect profile-mode data, but vendor raw files written out after each run may contain one or more of these types of spectra, as selected by the user. An example of these different types is displayed in Fig. 2.
Fig. 2.
Example of a set of peaks depicted in “profile” mode as it is collected and commonly written by an instrument; “thresholded” mode, in which values below a certain threshold (or sometimes just zeros) are not written out to save space; and “centroided” mode, wherein only the detected peaks are written. Formats such as mzML can encode any one of these types per spectrum.
Each vendor has developed one or more formats of its own and continually extends them as new features are required by emerging instrumentation. The vendor formats come in three styles: single files per run, paired files, and folders containing several files per run. For Thermo Scientific instruments, all output is encoded in “raw files” with the .RAW extension. These may contain profile mode spectra or centroided spectra as selected by the user. They can even be mixed, with a popular configuration having MS1 scans saved as profile mode and MS2 scans saved as centroided spectra. Most AB SCIEX (Framingham, MA) instruments (with the exception of TOF-TOF instruments) are saved as files with the .wiff extension. These files might sometimes contain all information in a run or alternatively might contain only metadata and be paired with a file having a .wiff.scan extension that contains the spectra. An additional complexity is possible in that multiple runs may be saved to the same .wiff file, with each run given a unique name by the instrument operator. Data from Waters (Milford, MA) instruments and Agilent (Santa Clara, CA) instruments are stored in multiple files with a folder with the extension .raw or .d, respectively. These folders are typically treated as a unit, and individual files within these folders are hidden from the user. Other vendors operate on variations on these themes. These files can typically be read only by software from the instrument vendors themselves that is not freely available. Although the software is not freely available, works only on the format from one vendor, and is limited to the Microsoft Windows operating system, the software is typically very easy to use, is of high quality, and offers the fastest and most comprehensive way to explore the raw data interactively. As a result of repeated requests, most of the vendors do now provide freely available software application programming interfaces (APIs) in the form of Microsoft Windows dynamic link libraries. This has the advantage that software can be written to read these formats using relatively stable software libraries by the vendor. However, these libraries can be used only with software running on the Microsoft Windows operating systems, although there has been some success in getting them to work properly under Windows emulators (see, e.g., http://tools.proteomecenter.org/wiki/index.php?title=Msconvert_Wine). Further, although the software libraries are provided free of charge, they often come with extensive end-user license agreements that preclude unencumbered redistribution.
Because these formats are binary and difficult to read and parse, it becomes difficult to write software that operates on data from any vendor, although some search engines do use vendor files directly as input. Further, there is a concern that old proprietary binary data files might become unreadable as new software is adapted to read newer formats, older formats stop being supported, and older versions of software cease to function properly under newer operating environments, a concept sometimes termed “data rot” (17, 18). To address this problem, in 2003 (when vendor APIs were not available) there began an effort to develop open formats that could encode most of the important information from each run in a manner that could then be accessed by any tool with relative ease. The first such format was mzXML (19), produced at the Institute for Systems Biology (ISB). During a similar time frame, the HUPO PSI independently developed the mzData format (http://psidev.info/mzdata). The two formats were generally intended to encode the same information, but they employed different philosophies about how the data were to be encoded (see Ref. 21 for a discussion on this). The existence of two formats was widely regarded as an unwelcome distraction that caused extra work for software developers and confusion among users. To remedy this, the PSI and ISB came together to create a new format that would include the best features from both mzXML and mzData and eventually replace them. The new format, mzML (22), has been available in its present form since 2009, but it has been slow to catch on, mostly because mzXML and mzData are both quite capable. However, as newer workflows and features supported only by mzML become more prevalent (e.g. chromatograms from SRM experiments), the switch will eventually take place.
A common complaint about these XML-based formats is that the overhead in both file size and access time is significantly worse than with binary formats (23). This is thought to be generally offset by the performance gain in speed of development and ease of troubleshooting and handling. The recently introduced mz5 format (24) addresses file size concerns by translating mzML files into HDF5, a compact and well-supported binary storage mechanism, while still preserving all the structure of the mzML. In principle, a generic mechanism to port all PSI formats from XML to HDF5 might be possible.
With the advent of sequence search engines in the early days of MS/MS proteomics—well before the appearance of XML formats—it became common to convert the binary mass spectrometer output files into simple text files containing only the MS/MS spectra, a practice that is still common today. One of the first such formats was the simple dta format wherein each spectrum was written to a separate file containing one header line for the known or assumed charge and the mass of the precursor peptide ion, calculated from the measured m/z and the charge. This one line was then followed by all the m/z, intensity pairs that represent the spectrum. Other, highly similar formats are the pkl format and ms2 format, which differ only in subtleties of the header line format and content and support the added feature of being able to concatenate many spectra into one file. A more advanced solution was to place all precursor ion scans in one file and all product ion scans in a separate file, termed the MS1 and MS2 formats, respectively (25).
Likely the most common text format is the Mascot Generic Format (MGF) file. This file is similar to the format described above in that it encodes multiple MS/MS spectra in a single file via m/z, intensity pairs separated by headers; in the case of MGF files, the headers can contain a bit more information, including search engine instructions (see http://www.matrixscience.com/help/data_file_help.html for additional description). The MGF file was developed by Matrix Science (London, UK), the maker of Mascot, the most widely used commercial search engine, but it is widely supported by many proteomics search engines. These simple text formats were created with the emergence of search engines and MS proteomics to enable the simple and reliable transmission of spectra to search engine software, a task for which most metadata are not necessary. However, more recently they have hindered the development of more advanced proteomics tools because so many valuable metadata are lost during the conversion to these very simple formats. The desire to preserve these important metadata and preserve the data from the MS1 scans to support isotopic labeling workflows led to the development of the complex open formats such as mzXML and mzData as described above.
MS/MS Shotgun Proteomics Postsearch Output Files
Shotgun MS/MS data are typically processed (3) through a sequence search engine that attempts to identify MS/MS spectra based on a list of protein sequences, a spectral library search engine that attempts to identify spectra based on a library of previously observed MS/MS spectra, or a de novo algorithm that attempts to assemble the peptide sequence based purely on the measured distances between peaks. Some software programs can combine these in a hybrid approach. Although there is common ground in the input formats to these programs, nearly every one employs a different output format, including simple text, tab-delimited text, HTML, binary, and XML formats. Often the output formats are highly coupled with the software used to view the search results.
SEQUEST originally employed the .out file format and has since advanced to SRF (SEQUEST results file) and MSF (Magellan storage file) formats. Mascot continues to use its own .dat format as output. The SQT format was developed as a more efficient alternative to the .out file (25). X!Tandem (26) employs a custom XML-based .tandem output format. OMSSA (27) uses its own custom XML-based .omx format, although it can also write out an ASN.1 format or a comma-delimited format. Most other search engines also emit their own custom comma- or tab-separated columnar output format, and they are not all listed here.
Because the formats were tightly linked to their respective search engines, it was difficult to write a single viewer that could support multiple search engines, and even more difficult to compare or combine the results of multiple search engines. Furthermore, capturing information about subsequent processing or filtering of the results was problematic. To address these shortcomings, the creators of the PeptideProphet (28) and ProteinProphet (29) tools (and mzXML as well) created the pepXML and protXML formats (30).
The pepXML format is intended to capture nearly all relevant output information from a search engine and support capture of the metadata associated with the modeling and filtering of search results. Much as mzXML was intended to enable the processing of data from multiple instrument vendors through a common set of software tools, the initial use case for pepXML was to support the analysis of different search engines through a common set of tools, namely, the Trans-Proteomic Pipeline (TPP) (30). Special TPP utilities were developed to transform the output of all supported search engines into pepXML. Then subsequent processing of the pepXML by PeptideProphet and other TPP tools would write back richer information, such as modeled probabilities of being correct and quantification information, into an updated pepXML file. The pepXML format was never officially approved as a standard, but it came to be a de facto standard that was supported by a variety of different tools beyond TPP on account of its openness, reasonable simplicity, and lack of capable alternative.
Although the pepXML format's primary currency is a peptide-spectrum match (PSM), a complementary format named protXML also was released (30), and its primary currency is a protein as inferred from the individual PSMs by the TPP tool ProteinProphet. Each protein could be a member of a complex group of proteins that shared peptides. The proteins were assigned probabilities of being identified in the sample and were accompanied by information about the individual peptide sequences and peptide ions that supported the identification of each protein. The protXML was also never approved as a standard, but it gained some popularity as a common format among tools beyond the TPP. As with pepXML, the modeling results that ProteinProphet provides can be encoded in protXML, and quantification results rolled up to the peptide and protein level can also be encoded in protXML.
Although the pepXML and protXML formats were becoming quite widely used, their primary purpose was to serve as a communication format among TPP tools. The formats' simplicity and inflexibility precluded a number of use cases and desired features, so the PSI set out to develop and standardize a next-generation format that would support all of the features of pepXML and protXML and many additional use cases. The format was initially called dataXML but was finally named mzIdentML (31). It supported many of the desired use cases originally designed, but it did not include the ability to encode quantification information. This was expressly omitted because of the desire to complete a 1.0 release in a timely manner with the limited human effort available, and the modeling of the very complex quantitative information was left to a later release. This newer format is currently still under development by the PSI Proteomics Informatics Working Group under the name mzQuantML.2 When complete, it will be able to encode far richer information about a wider variety of quantification strategies than pepXML and protXML can, at the expense of a far more complex format.
Because most of these native search engine formats, as well as standardized formats, are complex and can be difficult to work with, it has become commonplace for the output results of informatics processing to be transformed into simple tab-delimited formats for subsequent interpretation or analysis via non-proteomics-specific tools or integration with transcriptomic data. These simple output files do not contain all the information from their source file, but most of the salient information for further analysis is encoded. In an attempt to standardize this inevitable part of a common workflow, the PSI is developing the mzTab format (http://www.psidev.info/mztab). It is a relatively simple, tab-delimited format that can capture the most important information about peptide and protein identifications and quantification results, and it can be easily read into Excel, R-based applications, or custom analysis scripts. It is expressly not intended to replace the richer formats; rather, it provides a common format for a simplified, tabular representation of the final result. The mzTab format is currently undergoing review within the PSI document process (33).
MS1 Profiling Analysis Output Files
Another popular MS proteomics workflow involves only acquiring MS1 scans to build a map of detected features within an MS run. The maps are typically aligned among multiple conditions, and the resulting features, representing peptide ions, are cataloged and measured. Features that exhibit differential expression among the various runs in a way that provides insight into the nature of the samples are often targeted for identification.
This workflow begins with the same files as previously described for encoding the output of the mass spectrometer runs, including vendor proprietary formats and mzXML or mzML. These mass spectrometer files are then processed by any number of possible software packages such as SpecArray (34), msInspect (35), Superhirn (36), Corra (37), MaxQuant (38), PEPPeR (39), and others. The output of most of these programs is some sort of tab-delimited text file, but several programs support an open file format developed at ISB in collaboration with others called Annotated Peptide Markup Language (APML) (37). This format can encode information about all of the detected features and their attributes, including intensity information across any number of runs, as well as the results of statistical analysis. The concepts in APML are being incorporated into the development versions of mzQuantML and mzTab, but at present APML remains the only complex open format for MS1 profiling results.
Targeted Proteomics (SRM) Workflow Files
In a targeted proteomics, or SRM, workflow, the mass spectrometer is directed to target predetermined peptide ion signatures, rather than allowed to trigger on the most intense ions. The TraML format, the .sky format, and vendor-specific input files are described above in the pre-MS subsection. However, these files are only for input. For the results of analysis of SRM data, the landscape of formats is still evolving rapidly. The Skyline program (16) uses its own open XML-based .skyd file format to encode output. Other programs such as mProphet (40) and AuDIT (41) use their own TSV-based formats. Vendor analysis programs such as MultiQuant (AB SCIEX) use proprietary formats. The PSI is currently developing the mzQuantML and mzTab formats mentioned above with the aim of their supporting SRM results in addition to the results of many other kinds of quantitative proteomics analyses.
Spectral Library Formats
A new data type that has emerged in the past few years is one that can contain a set of previously identified spectra. In a typical workflow, the identification results from a shotgun analysis are combined into consensus spectra and those spectra stored with their peak annotations and other related metadata about the spectra used to create the combined spectrum. These spectral libraries may then be used to search new data with a spectral library search engine such as SpectraST (42), X!Hunter (43), or Bibliospec (44) in place of or in addition to a search with a sequence search engine like Mascot.
The National Institute for Standards and Technology provides the largest set of consensus libraries of peptide ion spectra, and they distribute their libraries free of charge in their own msp format. The SpectraST tool, part of the TPP, can create libraries and search data with libraries in its splib format. SpectraST also produces an sptxt format, a pure text format that is easy to read and parse and which is nearly the same as the msp format. X!Hunter and Bibliospec programs can also create and read their own formats, ASL and blib, respectively. SpectraST is able to convert any of these formats to splib/sptxt. There is currently no effort underway to create a standard spectral library format.
Sequence File Formats
Most searches of shotgun data are still performed with sequence search engines such as Mascot or X!Tandem. These tools require a list of potential protein sequences against which they analyze the input spectra. For most tools, this is a simple FASTA format, which consists of a series of entries of a single header line followed by one or more lines of sequence. The format of the header line is loosely defined and highly variable among different sources. Some search engines, such as OMSSA and InsPecT (45), transform the FASTA file into a custom format prior to searching to increase processing speed. Some search engines index the FASTA file according to taxonomy or protein molecular weight to facilitate searching.
In order to address the problem of wildly variable header line formats, the PSI has recently defined a new format, the PSI Extended FASTA Format (PEFF; http://www.psidev.info/peff). It follows the conventional FASTA format with the small addition of hash-mark initiated header lines (and therefore requires only a minor modification to older parsers for backward compatibility), but it imposes a very strict syntax in the header line in which quite rich information about the sequence entry, including sequence variant information, can be stored and uniformly parsed.
Related File Formats
A few related file formats are worth noting. One of the oldest proteomics data repositories is the PRIDE database (46). Submissions to PRIDE are performed in the PRIDE XML format. This format borrows heavily from the mzData format. It is expected that PRIDE XML will be replaced by mzIdentML eventually. Currently the easiest and most common way of creating PRIDE XML is with the PRIDE Converter tool (47), although other, less common methods exist from within proteomics analysis databases. Further, the ProteomeXchange consortium is developing a ProteomeXchange XML format to pass the metadata about an experiment from one proteomics data repository to another.
Several related formats are specific to MS proteomics but are related such that they are used in conjuction with MS formats or have contributed to their development. The PSI has developed a format for the exchange of molecular interactions, the PSI-MI format (48). It is used to exchange molecular interaction information among the interactions databases, usually after curation of a journal article by one of the databases, although some submissions by the original authors are made in PSI-MI. Many interactions lists are derived from MS proteomics techniques. However, because of the complexity of the PSI-MI format and the general desire of many users to have a simple list of interactions, the PSI has also developed the MI-TAB format (49), which is a relatively simple tabular format for encoding a list of interactions with minimal attributes about each interaction. Much of the rich information that can be encoded in the PSI-MI format cannot be encoded in MI-TAB, but often a simple summary of the interaction information will suffice. The MI-TAB is intended not to replace PSI-MI but rather to serve as a standardized format for cases in which the complexity of XML is undesirable and a simple tabular format will be used anyway. Molecular interactions may, of course, be determined using technologies besides MS, but some of the most common workflows involve MS, and some tools, such as ProHits (50), process MS data specifically in order to generate molecular interaction information.
Following the development of a standard format for transcriptomics microarray data, MAGE-ML (51), there began an effort to develop a basic infrastructure to provide a set of reusable components that could serve as the basis for any format to encode the results from the analysis from any kind of functional genomics experiment. The result, called FuGE (8), has not been widely implemented, but several ideas and components originally developed for FuGE have been reused in other PSI formats. Finally, there is a related format infrastructure called AnIML (for “analytical information markup language”; http://animl.sourceforge.net/) that plans to support many different data types. Support for MS data is planned but not yet implemented.
DISCUSSION
The many formats presented here, except for the vendor mass spectrometer data formats, are used only within the proteomics community. Even the open XML formats that could be applied to other fields are slow to be adopted elsewhere. The mzML format can be readily used by any field using mass spectrometers. Yet only the metabolomics community is beginning to consider widespread adoption of the format. The use of the FASTA format, of course, extends far beyond proteomics, as it originated before proteomics. However, it remains to be seen whether other communities will adopt PEFF as well. Stated support for PEFF by the ubiquitous knowledge bases such as UniProt (52) makes this likely.
An alternate approach to standardized open formats is to create an API that can be used to access any of the formats directly without the need for an intermediate format. This enables software to be written that can work equally well on any of the native formats. This has been accomplished by the ProteoWizard project (53) for most of the mass spectrometer vendors, and by mzAPI (23), which provides an API for Thermo RAW and AB SCIEX wiff formats. The advantage is relief from the need to duplicate the data in two formats, which saves time, disk space, and workflow complexity. The primary disadvantage is that such schemes rely on the vendor API software to provide the access layer to the vendor files, and thus the scheme can work only on an operating system for which the vendor software libraries are available, which currently is true only of Microsoft Windows. This is unlikely to change. Further, the vendor software libraries are all written in C, C++, or C#, and interfacing with these from other languages is difficult, whereas writing parsers for the open formats in a variety of languages is relatively easy. Further, the raw data are often filtered or processed in some way prior to analysis, and the intermediate results need to be written out anyway prior to analysis. In theory, such filtering could be handled by search engines directly as they read the spectra from the original files, but it is unlikely that most search engines will be persuaded to implement this, and many searches are performed on Linux-based clusters, for which vendor support is not available. Therefore, such approaches can work well for Windows-only interactive applications. However, they seem unlikely to gain widespread usage for most applications. A few search engines, such as Spectrum Mill (Agilent Technologies Inc., Santa Clara, CA) and PEAKS (54), can read some vendor files directly.
Most of these open formats use the XML notation for encoding the data. This choice has been largely successful because XML is effective for encoding complex, structured data, has a large variety of industry-standard implementations of readers and writers and validators, and is easily read by developers, which makes troubleshooting parsing problems relatively straightforward. If a binary file cannot be read because of some problem, that is usually the end of the story, whereas if an XML file cannot be parsed or does not validate, a software developer can inspect the location in the file where the error occurs and might be able to manually repair the file, adapt a parser to handle the exception, or alert the writer of the file to the exact nature of the problem. This is a general feature of all text-based formats. Other, similar alternatives exist, such as JSON, but there seems to be little incentive to switch primarily on account of the XML infrastructure built by the PSI thus far.
One notable exception is the possibility of using resource definition framework (RDF) technology. This framework can use RDF XML for its encoding of information (serialization), although less popular RDF serialization alternatives exist. In its essence, RDF encodes a series of statements about resources, typically in subject-predicate-object expressions, such that these entities are either defined in common CVs or defined within the RDF document (see, e.g., http://www.w3.org/TR/rdf-primer/). In many ways, the extensive use of CVs in PSI document formats is a partial solution to what the inventors of RDF were themselves were trying to solve. In fact, during the early development of mzML (called dataXML at the time), RDF was strongly promoted as the framework for the new format. However, the majority of contributors to mzML were generally unfamiliar with RDF, and this was likely the major reason that a traditional XML format was selected. It might also be that RDF is better suited to the encoding of knowledge than to the encoding of pure data. Given that many of the PSI XML formats are heavily based on CVs and validation of the use of CVs, it might take only another half-step for future PSI formats to make the leap to RDF.
When the PSI formats discussed here are all complete and these formats have been widely implemented in most common software applications, it will be encouraged that analyses be performed and reported using all the PSI formats: TraML for transition lists or inclusion lists, mzML for mass spectrometer output results, mzIdentML for encoding the results of database searching and validation, and mzQuantML for encoding the quantitative results of the analysis. The use of such formats will ensure that the data and results can be readily accessed by everyone irrespective of what software they prefer to use.
CONCLUSION
There are a remarkably high number of different file formats commonly used in MS proteomics. These range from binary vendor-controlled formats to commonly used text representations of the data to community-driven, complex, XML-based formats. The variety of formats is indicative of the rapid advancement of the field. New formats are developed as new workflows and capabilities are developed. Yet, there is a willingness of many in the community to work together under the banner of the HUPO PSI to develop complex formats that can encode rich metadata that can serve everyone in the community and facilitate the reusability of well-annotated datasets and enable a common set of software tools to work with data from a variety of sources. These rich, open standards accelerate the pace of proteomics research in a way that the vendor formats and custom, private formats alone cannot do.
Acknowledgments
I thank Ruedi Aebersold and Terry Farrah for their comments during the preparation of the manuscript.
Footnotes
* This work has been funded in part by federal funds from the National Institute of General Medical Sciences under Grant No. R01 GM087221, P50 Grant No. GM076547 from the Center for Systems Biology, the EU FP7 grant “ProteomeXchange” (Grant No. 260558), the Luxembourg Centre for Systems Biomedicine, and the University of Luxembourg.
2 Walzer, M., Qi, D., Mayer, G., Uszkoreit, J., Eisenacher, M., Sachsenberg, T., Deutsch, E. W., Reisinger, F., Vizcaíno, J. A., Medina-Aunon, J. A., Albar, J. P., Kohlbacher, O., and Jones, A. R., “The mzQuantML data standard for quantitative studies in proteomics,” submitted for publication.
1 The abbreviations used are:
- API
- application programming interface
- APML
- Annotated Peptide Markup Language
- CV
- controlled vocabulary
- FuGE
- Functional Genomics Experiment
- HUPO
- Human Proteome Organization
- ISB
- Institute for Systems Biology
- LC
- liquid chromatography
- MGF
- Mascot Generic Format
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- PEFF
- PSI Extended FASTA Format
- PSI
- Proteomics Standards Initiative
- PSM
- peptide-spectrum match
- RDF
- resource definition framework
- SRM
- selected reaction monitoring
- TPP
- Trans-Proteomic Pipeline
- TraML
- Transitions Markup Language
- TSV
- tab-separated value.
REFERENCES
- 1. Beck M., Claassen M., Aebersold R. (2011) Comprehensive proteomics. Curr. Opin. Biotechnol. 22, 3–8 [DOI] [PubMed] [Google Scholar]
- 2. McCormack A. L., Schieltz D. M., Goode B., Yang S., Barnes G., Drubin D., Yates J. R., 3rd (1997) Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 69, 767–776 [DOI] [PubMed] [Google Scholar]
- 3. Deutsch E. W., Lam H., Aebersold R. (2008) Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol. Genomics 33, 18–25 [DOI] [PubMed] [Google Scholar]
- 4.Nature Methods (2008) Thou shalt share your data. Nat. Methods 5, 209 [Google Scholar]
- 5. Orchard S., Montechi-Palazzi L., Deutsch E. W., Binz P. A., Jones A. R., Paton N., Pizarro A., Creasy D. M., Wojcik J., Hermjakob H. (2007) Five years of progress in the Standardization of Proteomics Data 4(th) Annual Spring Workshop of the HUPO-Proteomics Standards Initiative, April 23–25, 2007, Ecole Nationale Superieure (ENS), Lyon, France. Proteomics 7, 3436–3440 [DOI] [PubMed] [Google Scholar]
- 6. Gibson F., Hoogland C., Martinez-Bartolome S., Medina-Aunon J. A., Albar J. P., Babnigg G., Wipat A., Hermjakob H., Almeida J. S., Stanislaus R., Paton N. W., Jones A. R. (2010) The gel electrophoresis markup language (GelML) from the Proteomics Standards Initiative. Proteomics 10, 3073–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orchard S., Hoogland C., Bairoch A., Eisenacher M., Kraus H. J., Binz P. A. (2009) Managing the data explosion. A report on the HUPO-PSI Workshop. August 2008, Amsterdam, The Netherlands. Proteomics 9, 499–501 [DOI] [PubMed] [Google Scholar]
- 8. Jones A. R., Miller M., Aebersold R., Apweiler R., Ball C. A., Brazma A., Degreef J., Hardy N., Hermjakob H., Hubbard S. J., Hussey P., Igra M., Jenkins H., Julian R. K., Jr., Laursen K., Oliver S. G., Paton N. W., Sansone S. A., Sarkans U., Stoeckert C. J., Jr., Taylor C. F., Whetzel P. L., White J. A., Spellman P., Pizarro A. (2007) The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat. Biotechnol. 25, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 9. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deutsch E. W., Chambers M., Neumann S., Levander F., Binz P. A., Shofstahl J., Campbell D. S., Mendoza L., Ovelleiro D., Helsens K., Martens L., Aebersold R., Moritz R. L., Brusniak M. Y. (2012) TraML—a standard format for exchange of selected reaction monitoring transition lists. Mol. Cell. Proteomics 11, R111.015040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teleman J., Karlsson C., Waldemarson S., Hansson K., James P., Malmstrom J., Levander F. (2012) Automated selected reaction monitoring software for accurate label-free protein quantification. J. Proteome Res. 11, 3766–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brusniak M. Y., Kwok S. T., Christiansen M., Campbell D., Reiter L., Picotti P., Kusebauch U., Ramos H., Deutsch E. W., Chen J., Moritz R. L., Aebersold R. (2011) ATAQS: a computational software tool for high throughput transition optimization and validation for selected reaction monitoring mass spectrometry. BMC Bioinformatics 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helsens K., Brusniak M. Y., Deutsch E., Moritz R. L., Martens L. (2011) jTraML: an open source java API for TraML, the PSI standard for sharing SRM transitions. J. Proteome Res. 10, 5260–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sturm M., Bertsch A., Gropl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS—an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hakkinen J., Vincic G., Mansson O., Warell K., Levander F. (2009) The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J. Proteome Res. 8, 3037–3043 [DOI] [PubMed] [Google Scholar]
- 16. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wiley H. S., Michaels G. S. (2004) Should software hold data hostage? Nat. Biotechnol. 22, 1037–1038 [DOI] [PubMed] [Google Scholar]
- 18. Martens L., Nesvizhskii A. I., Hermjakob H., Adamski M., Omenn G. S., Vandekerckhove J., Gevaert K. (2005) Do we want our data raw? Including binary mass spectrometry data in public proteomics data repositories. Proteomics 5, 3501–3505 [DOI] [PubMed] [Google Scholar]
- 19. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 20. Medina-Aunon J. A., Martinez-Bartolome S., Lopez-Garcia M. A., Salazar E., Navajas R., Jones A. R., Paradela A., Albar J. P. (2011) The ProteoRed MIAPE web toolkit: a user-friendly framework to connect and share proteomics standards. Mol Cell Proteomics 10, M111.008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deutsch E. (2008) mzML: a single, unifying data format for mass spectrometer output. Proteomics 8, 2776–2777 [DOI] [PubMed] [Google Scholar]
- 22. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Rompp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Askenazi M., Parikh J. R., Marto J. A. (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat. Methods 6, 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilhelm M., Kirchner M., Steen J. A., Steen H. (2011) mz5: space- and time-efficient storage of mass spectrometry data sets. Mol. Cell. Proteomics. 10, O111.011379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., Yates J. R., 3rd (2004) MS1, MS2, and SQT—three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 26. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 27. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 28. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 29. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 30. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones A. R., Eisenacher M., Mayer G., Kohlbacher O., Siepen J., Hubbard S., Selley J., Searle B., Shofstahl J., Seymour S., Julian R., Binz P. A., Deutsch E. W., Hermjakob H., Reisinger F., Griss J., Vizcaino J. A., Chambers M., Pizarro A., Creasy D. (2012) The mzIdentML data standard for mass spectrometry-based proteomics results. Mol. Cell. Proteomics. 11, M111.014381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garden P., Alm R., Hakkinen J. (2005) PROTEIOS: an open source proteomics initiative. Bioinformatics 21, 2085–2087 [DOI] [PubMed] [Google Scholar]
- 33. Vizcaino J. A., Martens L., Hermjakob H., Julian R. K., Paton N. W. (2007) The PSI formal document process and its implementation on the PSI website. Proteomics 7, 2355–2357 [DOI] [PubMed] [Google Scholar]
- 34. Li X. J., Yi E. C., Kemp C. J., Zhang H., Aebersold R. (2005) A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol. Cell. Proteomics 4, 1328–1340 [DOI] [PubMed] [Google Scholar]
- 35. Bellew M., Coram M., Fitzgibbon M., Igra M., Randolph T., Wang P., May D., Eng J., Fang R., Lin C., Chen J., Goodlett D., Whiteaker J., Paulovich A., McIntosh M. (2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 36. Mueller L. N., Rinner O., Schmidt A., Letarte S., Bodenmiller B., Brusniak M. Y., Vitek O., Aebersold R., Muller M. (2007) SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7, 3470–3480 [DOI] [PubMed] [Google Scholar]
- 37. Brusniak M. Y., Bodenmiller B., Campbell D., Cooke K., Eddes J., Garbutt A., Lau H., Letarte S., Mueller L. N., Sharma V., Vitek O., Zhang N., Aebersold R., Watts J. D. (2008) Corra: computational framework and tools for LC-MS discovery and targeted mass spectrometry-based proteomics. BMC Bioinformatics 9, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 39. Jaffe J. D., Mani D. R., Leptos K. C., Church G. M., Gillette M. A., Carr S. A. (2006) PEPPeR, a platform for experimental proteomic pattern recognition. Mol. Cell. Proteomics 5, 1927–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reiter L., Rinner O., Picotti P., Huttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 41. Abbatiello S. E., Mani D. R., Keshishian H., Carr S. A. (2010) Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin. Chem. 56, 291–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 43. Craig R., Cortens J. C., Fenyo D., Beavis R. C. (2006) Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 5, 1843–1849 [DOI] [PubMed] [Google Scholar]
- 44. Frewen B. E., Merrihew G. E., Wu C. C., Noble W. S., MacCoss M. J. (2006) Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 45. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 46. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 47. Barsnes H., Vizcaino J. A., Eidhammer I., Martens L. (2009) PRIDE Converter: making proteomics data-sharing easy. Nat. Biotechnol. 27, 598–599 [DOI] [PubMed] [Google Scholar]
- 48. Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., Roechert B., Poux S., Jung E., Mersch H., Kersey P., Lappe M., Li Y., Zeng R., Rana D., Nikolski M., Husi H., Brun C., Shanker K., Grant S. G., Sander C., Bork P., Zhu W., Pandey A., Brazma A., Jacq B., Vidal M., Sherman D., Legrain P., Cesareni G., Xenarios I., Eisenberg D., Steipe B., Hogue C., Apweiler R. (2004) The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 22, 177–183 [DOI] [PubMed] [Google Scholar]
- 49. Orchard S., Jones A., Albar J. P., Cho S. Y., Kwon K. H., Lee C., Hermjakob H. (2010) Tackling quantitation: a report on the annual Spring Workshop of the HUPO-PSI 28–30 March 2010, Seoul, South Korea. Proteomics 10, 3062–3066 [DOI] [PubMed] [Google Scholar]
- 50. Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z. Y., Breitkreutz B. J., Ding Y., Colwill K., Pasculescu A., Pawson T., Wrana J. L., Nesvizhskii A. I., Raught B., Tyers M., Gingras A. C. (2010) ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28, 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spellman P. T., Miller M., Stewart J., Troup C., Sarkans U., Chervitz S., Bernhart D., Sherlock G., Ball C., Lepage M., Swiatek M., Marks W. L., Goncalves J., Markel S., Iordan D., Shojatalab M., Pizarro A., White J., Hubley R., Deutsch E., Senger M., Aronow B. J., Robinson A., Bassett D., Stoeckert C. J., Jr., Brazma A. (2002) Design and implementation of Microarray Gene Expression Markup Language (MAGE-ML). Genome Biol. 3, RESEARCH0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Apweiler R., Bairoch A., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O'Donovan C., Redaschi N., Yeh L. S. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., Ma B. (2012) PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11, M111.010587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barsnes H., Vaudel M., Colaert N., Helsens K., Sickmann A., Berven F. S., Martens L. (2011) compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Griss J., Reisinger F., Hermjakob H., Vizcaino J. A. (2012) jmzReader: A Java parser library to process and visualize multiple text and XML-based mass spectrometry data formats. Proteomics 12, 795–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parikh J. R., Askenazi M., Ficarro S. B., Cashorali T., Webber J. T., Blank N. C., Zhang Y., Marto J. A. (2009) multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]